Abstract
PIWI-interacting RNA (piRNAs), once thought to be mainly functioning in germlines, are now known to play an essential role in somatic and cancerous tissues. Ping-pong cycle initiation and mitochondria-based phased production constitute the core of the piRNA biogenesis and these two processes are well conserved in mammals, including humans. By being involved in DNA methylation, histone marker deposition, mRNA degradation, and protein modification, piRNAs also contribute to carcinogenesis partly due to oncogenic stress-induced piRNA dysregulation. Also, piRNAs play important roles in cancer stemness, drug resistance, and tumor immunology. Results from liquid biopsy analysis of piRNA can be used in both cancer diagnoses and cancer prognoses. A combination of targeting piRNA with other therapeutic strategies could be groundbreaking cancer treatment.
Keywords: Carcinogenesis, Clinical utility, Mitochondria-based phased production, Ping-pong cycle, Piwi-interacting RNAs
Introduction
P-element-induced wimpy testis (PIWI-interacting RNAs (piRNAs) are the new members of the RNA interference family. They usually comprise of 21–35 nucleotides (nt) with a 2ʹ-O-methyl-modified 3ʹ termini and interact specifically with the PIWI subfamily of Argonaute proteins.1, 2, 3 Originally described as repeat-associated siRNAs (rasiRNAs), piRNAs mediate the silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline.4,5 It wasn't until 2006 when they were found to interact with PIWI proteins in many other organisms were they referred as piRNAs.1, 2, 3 Since then, there have been several impactful discoveries made in the piRNA-PIWI field (Fig. 1A). And their functions have been expanded from the well-known role of transposon silencing in germline cells to epigenetic modification or post-transcriptional gene silencing (PTGS) of protein-coding genes in somatic and cancerous tissues.6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20
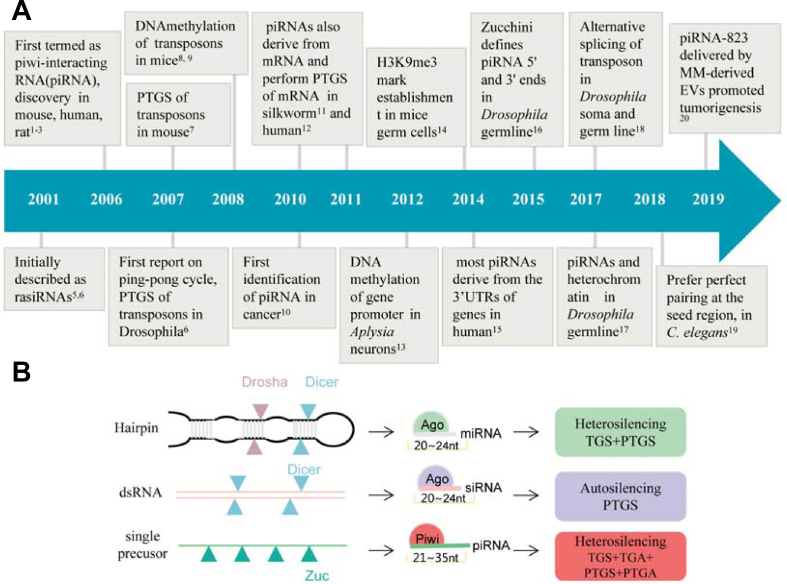
Figure 1.
Characteristics and progression of piRNAs. (A) The milestone events in the piRNA field. (B) piRNAs show distinct characteristics different from miRNAs and siRNAs, from genome source to RISC formulation and gene silencing style. Abbreviation: EV, exosome vesicle; MM, Multiple myeloma; PTGS, post-transcriptional gene silencing; PTGA, post-transcriptional gene activation; TGA, transcriptional gene activation; TGS, transcriptional gene silencing; H3K9me3, trimethylation of H3 lysine 9; UTR, untranslated region; RISC, RNA-induced silencing complex.
There are a few major differences that distinguish piRNAs from miRNAs and siRNAs. For example, as Argonaute proteins are divided into two subfamilies, the Argonaute (AGO) and PIWI families, piRNAs bind PIWI subfamily proteins to exert their effects while miRNAs and siRNAs bind the AGO subfamily proteins.21 In terms of gene regulation, siRNAs, miRNAs, and piRNAs are known to have the ability to silence or trigger the degradation of mRNA. Members of the piRNAs primarily function through epigenetic silencing in the nucleus although they can conduct miRNA-like transcript silencing in the cytoplasm.22,23 piRNAs, 21 to 35 nucleotides (nt) long, are derived from single strand precursors generated via a Dicer-independent mechanism. In contrast, miRNAs and siRNAs, 20 to 24 nt long, are derived from double-stranded RNA (dsRNA) or stem-loop precursors processed by Dicer RNase III.24 An additional distinguishing characteristic is that piRNAs possess 2′-O-methylation at the 3′-terminus in a variety of animals while 2′-O-methylation in siRNAs and miRNAs are mainly found in plants, despite a few reports of 2′-O-methylated siRNAs in Drosophila.25, 26, 27 Quantitatively speaking, piRNAs far outnumber miRNAs in the eukaryotic genome, for example, the human genome contains ∼ 8,400,000 annotated piRNAs while ∼ 2000 annotated hairpin miRNA precursors, and ∼2600 mature miRNA sequences.28, 29, 30 As a distinguishing characteristic, all piRNAs possess 2′-O-methylation at the 3′-terminus, a unique feature making them distinct from miRNAs and siRNAs. Moreover, both miRNAs and piRNAs predominantly target genes different from their origins by inhibiting translation or enhancing target mRNA degradation (heterosilencing). In contrast, siRNAs target and degrade mRNAs transcribed from the same genes they originated (autosilencing) (Fig. 1B).31
Since the discovery of piRNAs in HeLa cells in 2010, the role of piRNAs in malignant cells have also emerged.10 This review article aimed to discuss the activity of piRNAs at the transcriptional and post-transcriptional levels first followed by the emerging roles of piRNAs in cancer stemness, drug resistance, and tumor immunology. Besides, the applications of piRNAs in liquid biopsy and cancer therapies will also be highlighted.
PIRNA: mitochondria-based biogenesis
piRNA biogenesis
As the root of piRNA dysregulation in cancer is likely the result of disorders in piRNA biogenesis, it is necessary to get insight into the piRNA biogenesis mechanism first. Recent studies of flies and mice,32, 33, 34 and data from an evolutionarily broad range of non-model species,35 suggest that piRNA biogenesis is a highly conserved process, and in most animals the ping-pong and phased piRNA pathways collaborate to make complex populations of piRNAs (Fig. 2).
Figure 2.
piRNA biogenesis in Drosophila. Abbreviations: Ago3, Argonaute3; Aub, Aubergine; Armi, Armitage; CH3, depicts a 2-O-methyl group; Zuc, Zucchini.
Transcription in nuclear
piRNA clusters are a discrete number of genomic loci that stood out as hotspots responsible for producing the vast majority of piRNAs in most species.36 Based on the ability to generate piRNAs from one or both genomic strands, piRNA clusters are roughly divided into unistrand and dual-strand clusters. Both types of piRNA clusters are transcribed by RNA polymerase II (pol II) (Fig. 2). The products of unistrand clusters depend on canonical genic transcription units, especially the conserved transcriptional factor A-MYB, and will undergo canonical RNA processing in the nucleus, i.e., 5′-capping, splicing, and polyadenylation.37, 38, 39, 40, 41 On the other hand, dual-strand piRNA clusters do not possess their own promoters and are regulated by non-canonical transcriptional mechanisms driven by nuclear transcription factors including Rhino (Rhi), Deadlock (Del), and Cutoff (Cuff).38,42, 43, 44, 45 In flies, piRNA precursors transcribed from genomic clusters are capable of targeting transposons for repression.6 However, in humans, most genomic piRNAs are derived from human protein-coding genes or long non-coding RNA (lncRNA) genes rather than transposons, indicating that human piRNAs may have functions beyond transposon silencing.1,3,41,46,47
Ping-pong cycle
In most animals, piRNA precursors are exported from the nucleus and localize in the nuage where biogenesis via the ping-pong cycle takes place (Fig. 2).6,48 In the germ cells of Drosophila, Argonaute 3 (Ago3) and Aubergine (Aub) play instrumental roles in the ping-pong cycle. Ago3 is biased towards a sense strand and adenine at nucleotide 10, whereas Aub prefers an antisense strand and uridine at nucleotide 1.6,49 Via their slicer activity, Ago3 and Aub cleave target RNAs at positions 10 and 11 of the guide strand, respectively.50 More specifically, a trigger piRNA will combine with Ago3 and scan all of the transcripts that exit the nucleus. After identifying a target with a complementary sequence (antisense), the Ago3/piRNA complex binds to the precursor transcript and cleaves it, thereby generating antisense piRNA. The 5′-end of the cleaved transcript then becomes a responder piRNA (antisense) and binds to Aub. This complex can go on to cleave cluster transcripts (sense); the sense piRNA product of this cleavage event can reinitiate the cycle. In mice, ping-pong cycle is primarily driven by PIWIL2/MILI and PIWIL4/MIWI2 in pre-pachytene piRNAs of pre-natal male mice.8,51,52 The ping-pong signal is also found in bovine, macaque, and human ovarian or testicular piRNA biogenesis.53 And in fetal human ovaries, ping-pong cycle may be formed by PIWIL2/HILI and PIWIL4/HIWI2.53,54
Shuttling process
In Drosophila, pre-piRNAs produced in the nuage/Yb bodies are then shuttled by Armitage (Armi) to the outer membrane of mitochondria (OMM) for phased production.55,56 At the OMM, Armi is anchored to a binding platform by Gasz and Daedalus (Daed).57 Both Gasz and Daed ensure the proper localization of Armi, proximal to dimerized and active Zuc. In mice, the shuttling protein might be Moloney leukemia virus 10 like-1 (MOV10L1), an RNA helicase analogous to Armi.58, 59, 60, 61, 62, 63 Gasz has also been confirmed to be a component of intramitochondrial cement (IMC) that co-localizes with MILI, TDRD1, and mouse VASA homolog (MVH).64,65 In humans, MOV10L1 is a testis-specific isoform of MOV10. The latter has been reported to colocalize and interact with Ago1 and Ago2 proteins to mediate miRNA-guided mRNA cleavage in humans.49,58 There are hints that MOV10L1 gene polymorphisms are associated with male infertility in azoospermic men with complete maturation arrest.66 Given the critical role of piRNAs in germ cell maturation and infertility, there may be an interaction between MOV10L1 and piRNAs.67 The exact role of MOV10L1 and any cofactors that may shuttle PIWI-piRNA complex from perinuclear granules to IMC in human piRNA biogenesis is yet to be confirmed, but certainly warrants further investigation.
Mitochondria-dependent phased production
After being shuttled to the OMM, long piRNA precursors will be phased by Zucchini (Zuc)/MitoPLD, a highly conserved mitochondria-anchored endonuclease responsible for generating both the 5′- and 3′-termini of piRNAs.16,68, 69, 70, 71, 72 In Drosophila or mice, Zucchini (Zuc)/MitoPLD could cleave long piRNA precursors approximately every 25 or 40 nt, producing shorter trailer piRNAs following one another (Fig. 2).36,57 As this process occurs, Armi helps to unwind piRNA precursor and PIWI protein helps to position Zuc, thereby dictating the distinct “phasing” of cleavage.35 In humans, the mitochondria-based piRNA phased production process has been experimentally verified.73 HIWI2 is enriched in the IMC of the primordial follicles of human oocytes and spermatogonia. Interestingly, HILI is largely excluded from the IMC by HIWI2, suggesting the specific subcellular compartmentalization of PIWI proteins in humans. The exact details of piRNA phased production in humans is not confirmed, but it is thought that human MitoPLD is involved. As a member of the Phospholipase D (PLD) superfamily, human MitoPLD localizes on the OMM.74,75 Whether or not HIWI2 and MOV10L1 guide human MitoPLD is undiscovered.
Finally, the piRNAs are subjected to 3′ trimming by nibbler and are 2′-O-methylated by Hen1.76,77 These PIWI-piRNA products are then transferred to the nucleus or remain in the cytoplasm to exert their effects.
piRNA biogenesis and cancer
A variety of cancers involve ectopic piRNA expression (Fig. 3). In essence, the ectopic piRNA expressions are caused by the changes in piRNA biogenesis. Alterations in piRNA biogenesis promote tumorigenesis mainly through the following ways:
-
(1)
The ping-pong amplification loop is a post-transcriptional silencing (PTGS) that combines the silencing of target mRNAs with piRNA production.36 In Drosophila and mice, the ping-pong cycle despair often results in transposon burst.8,36 In humans, disruption of piRNA biogenesis might result in activation of oncogenes.78,79
-
(2)
Mitochondria are well known as the powerhouse of cells for their function of ATP production and suicidal weapon store for signal transduction.80 The importance of mitochondrial proteins in piRNA biogenesis, especially Zuc/MitoPLD, further supports the mitochondrial regulation of cellular parameters, and possibly carcinogenesis.36
-
(3)
Mature piRNAs control target genes via DNA methylation, histone marker deposition, mRNA decay and degradation, and protein modification. However, a disruption in piRNA biogenesis may lead to the disorders of piRNA-mediated transcriptional and post-transcriptional gene regulation; oncogenes can escape silencing, while tumor-suppressive genes are mistakenly silenced.
-
(4)
piRNA biogenesis may be related to cancer stemness. For example, PIWIL2, an important participant in piRNA biogenesis, cannot be detected in most normal adult organs in humans, except in testes, as well as in a wide variety of tumors.81 These facts suggest that piRNA biogenesis maybe active in germ cells, quiescent in normal tissues, but revive in cancer tissues, consistent with the changing trend of stemness in the corresponding states of organisms. In addition, piRNAs and PIWI proteins are reported to be related to cancer stem-cell maintenance.82,83 In this regard, piRNA biogenesis may be involved in cancer stemness.
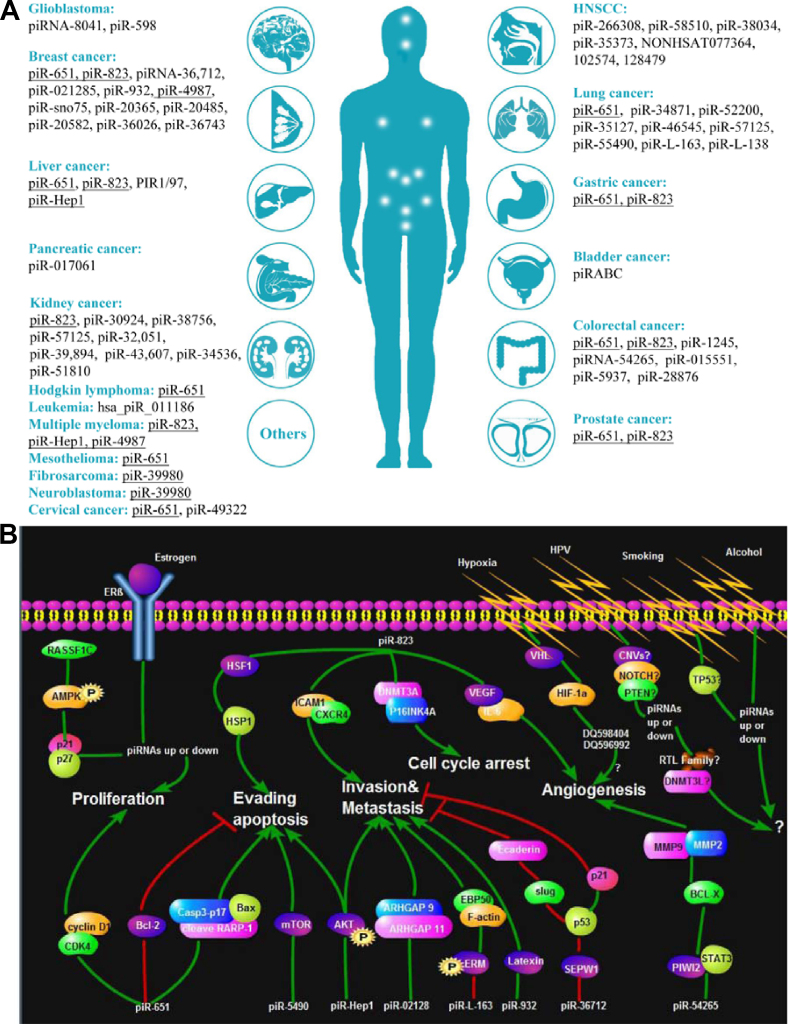
Figure 3.
Dysregulated piRNAs and their cellular signaling pathways in cancer. (A) Ectopically expressed piRNAs in cancers. The white dots refer to the site of corresponding organs. Among all the piRNAs, piR-651, piR-823, piR-4987, piR-39980, and piR-Hep1 exhibit pan-cancer expression patterns, and they are highlighted by underlines. (B) piRNA dysregulation results in the activation or repression of participating molecules in cellular signaling pathways. This leads to malignant cell phenotypes such as cell proliferation, evading apoptosis, metastasis, invasion, and cell cycle arrest. The positive effect is shown as green line, while the negative effect is shown as red line. Abbreviation: HNSCC, head and neck squamous cell carcinoma. ARHGAP, Rho GTPase activating proteins; ERβ, Estrogen receptor; CASP3, caspase 3, CDK4, Cyclin dependent kinase 4; DNMT, DNA methyltransferase; CNV, copy number variation; CXCR4, C-X-C motif chemokine receptor 4; HSP1, heat shock protein 1; HSF1, heat shock transcription factor 1; HPV, human papillomavirus; ICAM1, intercellular adhesion molecule 1; IL6, interleukin 6; mTOR, mechanistic target of rapamycin kinase; PTEN, phosphatase and tensin homolog; SEPW1, selenoprotein W 1; STAT3, signal transducer and activator of transcription 3; TDRD1, tudor domain containing 1; TP53, tumor protein p53; MMP, matrix metallopeptidases; VEGF, vascular endothelial growth factor; VHL, von Hippel-Lindau tumor suppressor.
In summary, piRNA biogenesis is a complex process and highly conserved in humans. Even modest changes in this process are expected to influence the piRNA expression level and cellular function profoundly. The following section discusses how the piRNA expression levels are crucial for cancer development.
PIRNAS and cancer: cause and effect
The first report of the role of piRNAs in cancer occurred in 2010 when piRNAs were discovered in HeLa cells.10 Since that initial discovery, many publications have attempted to elucidate the role piRNAs serve in cancer development and progression (Fig. 3A).20,55,78,79,82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127 Ectopically expressed piRNAs identified in numerous cancers are shown in Fig. 3A and Table 1. Many piRNAs, especially piR-823 and piR-651, have been reported to be dysregulated in various types of cancers, including but not limited to breast, stomach, lung, prostate, bladder, colorectal, kidney, liver, lymphomas and leukemias (Table 2).
Table 1.
Summary of altered piRNA expression in cancer.
| Cancer | piRNA | Expression | Related gene | Mechanical points | Clinical Utility | Sample Type |
Detection Technique | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TCGA | Tissue | cell | serum | mice | ||||||||
| Breast cancer | piRNA-36,712 | ↓ | SEPW1P, P53,P21 and E-cadherin | Inhibits SEPW1 expression, which may suppress P53, leading to the upregulated Slug but decreased P21 and E-cadherin levels. | Prognostic | yes | yes | yes | no | yes | TCGA | 84 |
| piR-021285 | ↑ | ARHGAP11A | Attenuated 5′-UTR/first exon methylation of proinvasive ARHGAP11A gene | Diagnostic | yes | no | yes | yes | no | Genome wide screen RT-PCR | 78 | |
| piR-932 | ↑ | Latexin | piR-932 and PIWIL2 may promote metastasis through Latexin methylation | Theraputic | no | yes | yes | no | yes | piRNA microarray RT-PCR | 82 | |
| piR-36026 | ↑ | SERPINA1, LRAT | Directly interacts with SERPINA1 or LRAT and induce caspase-3 and PI in the nucleus. | Diagnostic | no | no | yes | no | no | RT-PCR Molecular beacon |
85, 86, 87 | |
| piR-36743 | ↑ | – | – | Diagnostic | no | no | yes | no | no | RT-PCR Molecular beacon | 85,87 | |
| piR-sno75 | ↓ | TRAIL, MLL3, KDM6A | piR-sno 75/PIWIL1/4 complex upregulates the transcription of tumor necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL) by recruiting H3K4 methylase MLL3 and H3K27 demethylase KDM6A | Theraputic | no | yes | yes | yes | no | Small RNA sequencing RT-PCR | 137 | |
| piR-20365 | ↑ | – | – | Prognostic | no | yes | no | no | no | piRNA microarray RT-PCR | 88 | |
| piR-20485 | ||||||||||||
| piR-20582 | ||||||||||||
| 17 piRNAs | ↑ | – | – | Diagnostic | yes | yes | no | no | no | piRNA microarray RT-PCR | 89 | |
| 8 piRNAs | ↓ | |||||||||||
| Bladder cancer | piRABC | ↓ | TNFSF4 | Inhibit cell proliferation, colony formation, and promote cell apoptosis by increasing the expression of TNFSF4 protein | Diagnostic | no | yes | yes | no | no | piRNA microarray RT-PCR | 79 |
| Cervical cancer | piR-30074 | – | – | Regulating transformation of stem cells | Theraputic | no | no | yes | no | no | – | 125 |
| piR-49322 | ↑ | LINE1 | Suppress LINE1 retrotransposon along with HILI | – | no | no | yes | no | no | high throughput sequencing | 10 | |
| Colorectal cancer | piR-1245 | ↑ | – | An inverse correlation between piR-1245 and a panel of tumor suppressor genes (predicted) | Prognostic | yes | yes | yes | no | yes | TCGA piRNA microarray RT-PCR |
90 |
| piRNA-54265 | ↑ | STAT3 signaling pathway | Induce p-STAT3 and BCL-XL, MMP2 and MMP9, reduce cleaved-CASP9, 3 and 7 | Diagnostic Theraputic |
no | yes | yes | yes | yes | TCGA GEO RT-PCR |
91 | |
| piR-015551 | ↓ | – | piR-015551 may be generated from LNC00964-3, which may be involved in the development of CRC | Risk assesment | no | yes | no | no | no | piRNA microarray RT-PCR | 92 | |
| piR-5937 | ↓ | – | Differentiate between cancer patients and healthy donors, diagnostic performance in patients with stage I disease; higher sensitivity than currently used biomarkers CEA and CA19-9. | Diagnostic | no | no | no | yes | no | piRNA microarray RT-PCR |
93 | |
| piR-28876 | ||||||||||||
| piR-30074 | – | – | Regulating transformation of stem cells | Theraputic | no | no | yes | no | no | – | 94 | |
| Gastric cancer | 93 piRNAs | ↓ | – | – | – | yes | TCGA | 95 | ||||
| 7 piRNAs | ↑ | |||||||||||
| Liver cancer | PIR1/97 | ↑ | – | Involved in cell adhesion. | – | no | yes | yes | no | no | piRNA microarray RT-PCR |
96 |
| Lung cancer | piR-34871 | ↑ | RASSF1C | RASSF1C modulate piRNA through attenuation of the AMPK pathway, as over-expression of RASSF1C resulted in reduction of p-AMPK, p21, and p27 protein levels. | Theraputic | no | yes | yes | no | no | piRNA microarray RT-PCR |
98 |
| piR-52200 | ||||||||||||
| piR-35127 | ↓ | |||||||||||
| piR-46545 | ||||||||||||
| piR-57125 | ↓ | – | Most significantly associated with distant metastasis | Prognostic | 99 | |||||||
| piR-55490 | ↓ | mTOR | Directly binds to 3′-UTR of mTOR mRNA and induce its degradation | Theraputic | no | yes | yes | no | yes | RT-PCR | 100 | |
| piR-L-163 | ↓ | ERM proteins, F-actin, EBP50 | Binds directly to phosphorylated ERM proteins (p-ERM), which is critical for p-ERM's binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein 50 (EBP50). | Theraputic | no | no | yes | yes | no | RNA sequencing RT-PCR |
101 | |
| piR-L-138 | ↑ | p60-MDM2 | piR-L-138 is upregulated by CDDP-based agents and inhibits apoptosis through direct interaction with p60-MDM2. | Theraputic | no | no | yes | no | no | RT-PCR | 149 | |
| piR-30074 | – | – | Regulating transformation of stem cells | Theraputic | no | no | yes | no | no | – | 28 | |
| Leukemia | hsa_piR_011186 | ↑ | CDKN2B, DNMT1, Suv39H1, EZH2 | hsa_piR_011186 promoted cell-cycle progression, decreased apoptosis by forming complex with DNMT1, Suv39H1 and/or EZH2 proteins, which inhibit CDKN2B gene through modulation of the methylation of DNA and histone H3 in its promoter region. | Diagnostic | no | no | yes | no | no | RT-PCR | 126 |
| Kidney cancer | piR-34536 | ↓ | – | – | Prognostic | no | yes | no | yes | no | RT-PCR | 102 |
| piR-51810 | – | – | ||||||||||
| piR-30924 | ↑ | – | Associated with tumor recurrence and overall survival. | Prognostic | no | yes | no | no | no | piRNA microarray RT-PCR | 104 | |
| piR-38756 | ||||||||||||
| piR-57125 | ↓ | |||||||||||
| piR-32,051 | ↑ | – | Associated with ccRCC metastasis, late clinical stage and poor cancer-specific survival. | Prognostic | no | yes | no | no | no | piRNA microarray RT-PCR | 105 | |
| piR-39,894 | ||||||||||||
| piR-43,607 | ||||||||||||
| Glioblastoma | piRNA-8041 | ↓ | Cellular stress and survival pathway | Suppressed the heat shock protein and related DNAJ Protein chaperone families, MAPK/ERK signaling pathway proteins; (PREICTED) | Therapeutic | no | yes | yes | no | yes | piRNA microarray RT-PCR |
106 |
| piR-598 | ↑ | – | variant rs147061479 in piR-598 increases glioma risk by abolishing the tumor-suppressive function of piR-598 | Diagnostic | yes | no | yes | no | no | genome-wide profiling RT-PCR |
107 | |
| HNSCC | NONHSAT077364 | ↑ | HPV, PIWIL4, RTL, DNMT3L, PTEN, NOTCH, HNSCC-associated CNVs | Positively correlated with PIWIL4 and DNMT3L; negatively relative to RTL by direct binding interaction; upregulated by PTEN mutation or NOTCH mutation or downregulated by HNSCC associated CNVs | Diagnostic | yes | no | yes | no | no | TCGA RT-PCR |
109 |
| NONHSAT102574 | ||||||||||||
| NONHSAT128479 | ||||||||||||
| piR-266308 | ↑ | Alcohol | Low expression of piR-58510 and piR-35373 significantly correlated with improved patient survival. Alcohol consumption may cause dysregulation of piRNA expression and upregulation of human PIWIL 4. | Prognostic | yes | no | yes | no | no | RT-PCR | 110 | |
| piR-58510 | ||||||||||||
| piR-38034 | ||||||||||||
| piR-35373 | ||||||||||||
| 13 smoking-related piRNAs | ↑/↓ | Smoking, PIWIL1, TP53 | Associated with tumor stage, patient survival, and smoking-altered PIWIL1 protein expression. correlated with TP53 mutation, TP53–3p co-occurrence, and 3q26, 8q24, and 11q13 amplification. | Prognostic | yes | no | no | no | no | TCGA | 111 | |
| Hypoxic cancers | 40 hypoxia regulated piRNAs | Hypoxia, HIF-1α and VHL | Hypoxia-regulated piRNAs are regulated by VHL/HIF signaling in vitro, HIF-1α stabilization induced by VHL knockdown was responsible for the increase in piRNA expression. | Prognostic | yes | no | yes | no | no | TCGA RT-PCR |
112 | |
| 36 piRNAs | ↑ | |||||||||||
| 4 piRNAs | ↓ | |||||||||||
| Pancreatic cancer | piR-017061 | ↓ | – | – | Theraputic | no | yes | no | no | no | Next-generation sequencing | 113 |
Abbreviation: ARHGAP, Rho GTPase activating proteins; CNV, copy number variation; ccRCC, clear cell renal cell carcinoma; CRC, colorectal cancer; CDKN2B, cyclin dependent kinase inhibitor 2B; ERβ, estrogen receptor; CASP3, caspase 3; CNV, copy number variation; GEO, gene expression omnibus; HPV, human papillomavirus; mTOR, mechanistic target of rapamycin kinase; LRAT, lecithin retinol acyltransferase; PTEN, phosphatase and tensin homolog; PIWIL, piwi like RNA-mediated gene silencing 2; RT-PCR, quantificational real time-polymerase chain reaction; RRM2, ribonucleotide reductase regulatory subunit; RTL, Retrotransposon Gag like; SEPW1, selenoprotein W 1; SERPINA1, serpin family A member 1; STAT3, signal transducer and activator of transcription 3; TCGA, The Cancer Genome Atlas; TDRD1, tudor domain containing 1; TNFSF4, TNF superfamily member 4; TP53, tumor protein p53; MMP, matrix metallopeptidases; VHL, von Hippel-Lindau tumor suppressor; UTR, untranslated region.
Table 2.
Summary of piRNAs showing pan-cancer expression patterns.
| piRNA | Cancer | Expression | Related gene | Mechanical points | Clinical Utility | Sample Type |
Detection Technique | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TCGA | Tissue | cell | serum | mice | ||||||||
| piR-651 | Breast cancer | ↑ | Estrogen and Androgen | Modulated by estrogen and androgen hormone levels | Theraputic | no | no | yes | no | no | RT-PCR | 114 |
| ↑ | – | – | Diagnostic | no | yes | yes | no | no | piRNA microarrayRT-PCR | 115 | ||
| Cervical cancer | ↑ | – | – | Diagnostic | no | no | yes | no | no | piRNA microarrayRT-PCR | 115 | |
| Gastric cancer | ↓ | – | The piR-651 level in gastric adenocarcinoma was higher than that in gastric signet ring cell carcinoma. | Diagnostic Theraputic |
no | no | no | yes | no | RT-PCR | 116 | |
| ↑ | – | Promote cancer cell growth | Diagnostic | no | no | yes | no | no | piRNA microarrayRT-PCR | 115 | ||
| Hodgkin lymphoma | ↓ | – | – | Diagnostic | no | yes | yes | yes | no | RT-PCR | 117 | |
| Liver cancer | ↑ | – | – | Diagnostic | no | no | yes | no | no | piRNA microarrayRT-PCR | 115 | |
| Lung cancer | ↑ | Caspase-3-p17, cleaved-PARP-1, Bcl-2 | piR651 inhibitor induced cell apoptosis, with caspase-3-p17 and cleaved-PARP-1 increased, whereas Bcl-2 expression decreased | Diagnostic | no | no | yes | no | no | RT-PCR | 118 | |
| ↑ | – | – | Diagnostic | no | no | yes | no | no | piRNA microarrayRT-PCR | 115 | ||
| ↑ | Cyclin D1 and CDK4 | piR-651 induces NSCLC progression through the cyclin D1 and CDK4 pathway | Diagnostic Theraputic |
no | yes | yes | no | yes | RT-PCR | 119 | ||
| ↑ | – | promote cell proliferation, apoptosis, migration and invasion | Theraputic | no | no | yes | no | no | RT-PCR | 120 | ||
| Mesothelium | ↑ | – | – | Diagnostic | no | no | yes | no | no | piRNA microarrayRT-PCR | 115 | |
| Prostate cancer | ↑ | Estrogen and Androgen | Modulated by estrogen and androgen hormone levels | Theraputic | no | no | yes | no | no | RT-PCR | 114 | |
| piR-823 | Breast cancer | ↑ | Estrogen and Androgen | Modulated by estrogen and androgen hormone levels | Theraputic | no | no | yes | no | no | RT-PCR | 114 |
| Colorectal cancer | ↑ | HSF1 (heat shock protein (HSP) 27, 60, 70) | Upregulate phosphorylation and transcriptional activity of HSF1 | Theraputic | no | yes | yes | no | no | RT-PCR | 121 | |
| Gastric cancer | ↓ | – | The piR-823 level was positively associated with tumor-node-metastasis stage and distant metastasis. | Diagnostic Theraputic |
no | no | no | yes | no | RT-PCR | 116 | |
| ↓ | – | Inhibited cell growth in vitro and in vivo. | Theraputic | no | yes | yes | no | yes | piRNA microarray RT-PCR | 122 | ||
| Kidney cancer | ↑/↓ | – | piR-823 is down-regulated in tumor tissue, but positively correlated with worse outcome, indicating its complex role in RCC pathogenesis (↑blood serum and urine↓tumor tissue) | Diagnostic | no | yes | no | yes | no | RT-PCR | 123 | |
| Liver cancer | ↑ | Gradually increased from cirrhosis to low- and high-grade dysplastic nodules and to hepatocellular carcinoma (HCC) | Diagnostic | no | no | yes | no | no | RT-PCR | 55 | ||
| Multiple myeloma | ↑ | VEGF, IL-6, ICAM-1, CXCR4, Bax, Bcl2 | Promoted cell proliferation, tube formation, and invasion by enhancing VEGF, IL-6, ICAM-1, CXCR4 and attenuating apoptosis. (inhibition of Caspase-3 activation, downregulation of Bax, and upregulation of Bcl2) and production of ROS and NO | Prognostic Therapeutic | no | no | yes | yes | yes | RT-PCR | 20 | |
| ↑ | DNMT3B | Myeloid-derived suppressor cells confer stem-like qualities to multiple myeloma cells by upregulating piRNA-823 expression and activating DNMT3B activation | Theraputic | no | yes | yes | no | yes | RT-PCR | 83 | ||
| ↑ | DNMT3A and 3B, p16INK4A | induce de novo DNA methyltransferases, DNMT3A and 3B and inhibit methylation-silenced tumor suppressor, p16INK4A. |
Theraputic | no | yes | yes | no | no | RT-PCR | 124 | ||
| Prostate cancer | ↑ | Estrogen and Androgen | Modulated by estrogen and androgen hormone levels | Theraputic | no | no | yes | no | no | RT-PCR | 114 | |
| piR-4987 | Breast cancer | ↑ | – | Up-regulated piR-4987 expression was associated with lymph node positivity | Prognostic | no | yes | no | no | no | piRNA microarray RT-PCR | 88 |
| Multiple myeloma | ↑ | – | Upregulated in stage III MM patients compared with healthy individuals | Diagnostic | no | yes | no | no | no | RT-PCR | 124 | |
| piR-Hep1 | Liver cancer | ↑ | AKT | Promotes cell viability, motility, and invasiveness by activation of AKT phosphorylation | – | no | yes | yes | no | no | RT-PCR | 97 |
| Multiple myeloma | ↑ | – | Upregulated in stage III MM patients compared with healthy individuals | Diagnostic | no | yes | no | no | no | RT-PCR | 124 | |
| piR-39980 | Fibrosarcoma | ↓ | RRM2 | piR-39980 promotes apoptosis and inhibits proliferation by repressing RRM2 through direct targeting at its 3′-UTR through extensive sequence complementary binding | Therapeutic | no | no | yes | no | no | RT-PCR | 108 |
| Neuroblastoma | ↑ | JAK3 | Inhibition of piRNA induces NB cells senescence by modulating JAK3 through target binding. piR-39980 desensitizes the effect of doxorubicin and inhibit drug-induced apoptosis. | Theraputic | no | no | yes | no | no | piRNA profiling RT-PCR | 127 | |
Abbreviation: BCL2, BCL2 apoptosis regulator; CDK4, Cyclin dependent kinase 4; CXCR4, C-X-C motif chemokine receptor 4; DNMT, DNA methyltransferase; JAK3, Janus kinase 3; HSP, heat shock protein; HSF1, heat shock transcription factor 1; VEGF, vascular endothelial growth factor; IL6, interleukin 6; ICAM1, intercellular adhesion molecule 1; MM, Multiple myeloma; RRM2, ribonucleotide reductase regulatory subunit M2; RT-PCR, quantificational real time-polymerase chain reaction; TCGA, The Cancer Genome Atlas; UTR, untranslated region.
piRNA dysregulation promotes cancer cell malignant phenotypes
Tumor-suppressor piRNAs act as defenders against carcinogenesis by suppressing cell proliferation, increasing apoptosis, disrupting colony formation, decreasing migration and invasion abilities, and arresting cell cycles.79,84,100,101,106; Additionally, xenografts with tumor suppressor piRNA overexpression have been found to often show a slower growth rate and exhibit decreased metastasis.84,100,106 Such reports include cases of increased piR-8014 in human glioblastoma, piR-36712 in breast cancer (BrCa), piRABC in bladder cancer and piR-55490 or piR-L-163 in lung cancer.79,84,100,101,106
In contrast, oncogenic piRNAs promote cancer progression by increasing cell proliferation, inhibiting apoptosis, promoting colony formation and angiogenesis, and enhancing migration and invasion20,90,91,118,121; Xenografts with oncogenic piRNA overexpression often show a faster rate of growth and exhibit increased angiogenesis and metastasis.20,91 Such reports include cases of increased piRNA-823 in multiple myeloma, piR-54265 in colorectal adenocarcinoma, piRNA-651 in non-small cell lung cancer, and piR-823 or piR-1245 in colorectal cancer (Fig. 3B).20,83,90,91,118,121
Mechanism of piRNA dysregulation and cancer cell malignant phenotypes
To become functional, mature piRNAs usually have to be loaded into PIWI to form the RNA-induced silencing complex.128 piRNA–PIWI complexes are thought to be primarily involved in target regulation through four mechanisms: transcriptional gene silencing (TGS) or activation (TGA) and PTGS or activation (PTGA). PTGS mechanisms regulate target transcripts directly through cleavage and require the catalytic activity of PIWI proteins. The ping-pong cycle is a representative example of PTGS mechanisms.32,129 TGS or TGA, in contrast, does not require the cleavage activity of PIWI proteins, although it does require the activity of enzymes such as histone-modifying enzymes or DNA methyltransferases.129,130 With the contribution of these enzymes, piRNA–PIWI RISC can lead to a change in the mRNA output indirectly via chromatin remodeling. Thus, piRNAs take part in many aspects of tumorigenesis, from epigenetic regulation to mRNA degradation and protein modification (Fig. 4).
Figure 4.
Mechanism of piRNAs in cancer. (A) TGS or TGA by piRNAs via aberrant DNA methylation: In cancer, the piRNA–PIWI complex represses tumor-suppressor genes (TSGs) via recruiting DNA methyltransferases to the promoter and activates oncogenes by recruiting TET to the gene body. piRNA variants fail to bind to target regions and to form a piRNA–PIWI–DNMT complex, resulting in gene body hypomethylation and oncogene activation. (B) TGS or TGA by piRNAs via histone modification: the piRNA–PIWI complex induces the transformation between heterochromatin (condensed) and euchromatin (open) by recruiting methyltransferases or demethylases to histones and depositing active (H3K4me3) or repressive histone markers (H3K9me3 or H3K27me3). (C) PTGS by piRNAs via mRNA decay or deregulation: the piRNA–PIWI complex recruits the TRAMP complex and leads to mRNA decay. The piRNA–PIWI complex results in mRNA degradation via the slicer activity of PIWI. Whether or not other factors are needed is unknown. (D) PTGS or PTGA by piRNAs via protein modification: piRNA loading induces a conformational change in PIWI and enhances its interaction with proteasomes, resulting in protein phosphorylation or ubiquitination. Abbreviation: CDS, coding DNA sequence; DNMT, DNA methyltransferase; H3K9, H3 lysine 9; H3K27, H3 lysine 27; H3K4, H3 lysine 4; KDM, lysine demethylase 6A; PTGS, post-transcriptional gene silencing; PTGA, post-transcriptional gene activation; TF, transcription factor; SUV39H1, suppressor of variegation 3–9 homolog 1; TET, tet methylcytosine dioxygenase 1; TGA, transcriptional gene activation; TGS, transcriptional gene silencing; UTR, untranslated region.
TGS and TGA by piRNAs via aberrant DNA methylation
Active genes are normally identified by the absence of promoter DNA methylation with the presence of gene body CpG methylation. In cancer, the methylation status is reversed with promoter hypermethylation or genome hypomethylation (Fig. 4A).131,132 piRNAs are hypothesized to have the ability to both inactivate tumor suppressor genes via aberrant promoter hypermethylation and activate oncogenes via gene body hypomethylation (Fig. 4A).78,124,133 After direct sequence-specific binding to target CpG sites, piRNAs recruit methyltransferases such as DNMT1, DNMT3A, and DNMT3B to the respective region (Fig. 4A).124,126,133 Since binding of transcription factors and DNA methylation are mutually exclusive, CpG methylation in promoter regions by piRNAs has been found to result in transcriptional repression of tumor suppressor genes p15 and p16.124,126,134 In terms of hypomethylation the exact mechanism is still unclear. It is thought to involve a TET methylcytosine dioxygenase (TET)-dependent manner, as TET in collaboration with transcription factors may lead to DNA demethylation and thus gene activation (Fig. 4A).131 Interestingly, piRNA mutations may contribute to cancer if the variant fails to bind to target regions and fails to form the piRNA-PIWI-DNMT complex. The exact roles of piRNA in DNA methylation and hypomethylation are worth further investigation.
TGS and TGA by piRNAs via histone modification
The N-terminal of histone proteins undergoes extensive modifications including but not limited to methylation, acetylation, phosphorylation, ubiquitination, and SUMOylation.135 Activating histone markers include histone H3 lysine 4 trimethylation (H3K4me3), H3 and H4 acetylation, and histone H3 lysine 36 (H3K36me2 and H3K36me3). These activating markers help to shape the three-dimensional structure of the nucleosome into a euchromatin conformation with open chromatin available for transcription. Repressive histone markers such as trimethylation of H3 lysine 9 and 27 (H3K9me3 and H3K27me3) create a heterochromatin state with condensed chromatin (Fig. 4B).136 piRNA can switch the chromatin conformation freely from heterochromatin to euchromatin by recruiting different histone-modifying enzymes. Such enzymes include mixed lineage leukaemia 3 (MLL3) for H3K4 methylation, suppressor of variegation 3–9 homolog 1 (SUV39H1) for H3K9 methylation, enhancer of zeste homolog 2 (EZH2) for H3K27 methylation, lysine demethylases 1A and 5B (KDM1A and 5B) for H3K4 demethylation, as well as KDM6A for H3K27 demethylation (Fig. 4B).126,137,138 It has been reported that the piR-011186/Ago3 complex represses the expression of CDKN2B by recruiting Suv39H1 and/or EZH2 to modify the methylation levels of H3K9 and H3K27 in its promoter region.126 On the other hand, by complexing with PIWIL1/4 and subsequently recruiting MLL3 and KDM6A, piR-sno75 is capable of simultaneously enhancing H3K4 methylation and H3K27 demethylation, and ultimately upregulates the expression of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL).137 These observations demonstrate the flexibility of different piRNA/PIWI complexes in the regulation of gene expression through specific chromatin modification.
PTGS by piRNAs via mRNA decay or degradation
piRNA-induced repression may be mediated at the PTGS level via direct binding followed by PIWI protein-mediated cleavage or deadenylation and degradation of the target mRNA (Fig. 4C).79,84,86,90,100,109 Typically, piRNAs target the 3′-untranslated region (3′-UTR) of an mRNA, which may be attributed to the fact that most human genomic piRNAs are derived from 3′-UTR (Fig. 4C).15,79,84,86,90,100 However, the coding sequence or 5′-UTR has been identified as other possible binding regions (Fig. 4C).86,90 In mice, a large increase in the expression of targeted RNAs in PIWIL1 slicer mutants indicates the piRNA-mediated degradation of mRNA depends on the slicer activity of PIWIL1. Furthermore, piRNAs can also trigger mRNA deadenylation and decay in human somatic cells; a similar report has been made in Drosophila and mice.139,140
In CD4 primary T-lymphocytes, piR-30840 binds to the pre-mRNA intron via sequence complementarity.141 After that, the complex acts together with PIWIL4 and Ago4 to recruit the Trf-Air2-Mtr4 polyadenylation (TRAMP) complex to this region. This induces the decay of targeted pre-mRNA through nuclear exosomes. In this way, piR-30840 significantly downregulates interleukin-4 (IL-4) and subsequently inhibits the development of Th2 CD4+ T-lymphocytes (Fig. 4C).141 This mechanism portrays the versatility of piRNA downregulating mechanisms, specifically in post-transcriptional gene silencing.
PTGS and PTGA by piRNAs via protein modification
Dysregulation of phosphorylation or ubiquitination of proteins is a defining feature of numerous cancers.142 piRNAs may promote tumor formation in this way by upregulating the phosphorylation and transcriptional activity of signaling proteins such as heat shock transcription factor 1 (HSF1), Akt, or signal transducer and activator of transcription 3 (STAT3).91,97,121,143 piR-Hep1 overexpression in hepatocellular carcinoma (HCC) promotes cell viability, motility, and invasiveness by inducing AKT phosphorylation and thus results in augmentation of PI3K/AKT signaling pathway.97 By binding to the PIWI domain of PIWIL2, piR-54265 facilitates the formation of the PIWIL2/STAT3/phosphorylated-SRC (p-SRC) complex, thereby inducing the phosphorylating activation of STAT3 by p-SRC and promoting malignancy of CRC cells.91 It has been proposed that the molecular mechanism may be that piRNA loading leads to a conformational change in PIWI which enhances its interaction with proteasomes (Fig. 4D).144
Although piRNAs can act as either tumor suppressors or tumor promoters, the general mechanisms used by piRNA-mediated inhibition or activation are the following four-step process. (1) Formation of the PIWI-piRNA complex because a specific PIWI protein is essential for a functional piRNA128; (2) Searching and interacting with their specific targets; (3) Recruiting of specific enzymes such as DNA methyltransferases and/or histone/protein-modifying enzymes129,145; (4) Silencing or activating their targets. However, there are some exceptions. For example, in piRNA-mediated mRNA degradation, the PIWI-piRNA complex themselves are fully functional without any additional enzymes.
Emerging role of piRNAs in cancer stemness and drug resistance
Cancer stem cells (CSCs) are regarded as small and unique sources of cells with self-renewal and differentiation capacity and can generate a variety of cell types that constitute a tumor.146 These cells, despite making up a small proportion of the total number of tumor cells, may serve as the primary contributors to progression, metastasis, recurrence, and drug resistance.147 In recent studies researchers have been investigating the role of piRNAs in cancer stem or stem-like cells and the various mechanisms they may be involved in. It has been reported that the expression of piR-932 and PIWIL2 is significantly higher in breast CSCs which have been induced to epithelial-to-mesenchymal transition (EMT) by transforming growth factor-β1 (TGF-β1).82 Further functional study shows that the piR-932-PIWIL2 complex depresses the Latexin tumor suppressor gene via the aberrant methylation of its promoter region CpG island, thus contributing to the progression of breast cancer.82 In another instance, granulocytic-myeloid-derived suppressor cells (G-MDSCs) have been found to confer stem-like properties to multiple myeloma cells by upregulating piR-823 expression and activating DNMT3B activation.83 Interestingly, the knockdown of piR-823 reverses the upregulation of CSC marker genes and proteins induced by G-MDSCs in multiple myeloma cells. This suggests that piR-823 is an essential factor in the maintenance of multiple myeloma stem cells (MMSC) stemness.
Drug resistance is an important feature of CSCs and one of the primary reasons for therapeutic failure.148 The biological functions of piRNAs have been found to play an important roles in chemoresistance.84,91,149 piR-L-138 inhibits apoptosis in lung squamous cell carcinoma cells and has been shown to provide chemoresistance toward cisplatin-based chemotherapy via interaction with p60-mouse double minute 2 homolog (MDM2).149 Additionally, increased levels of piR-54265 binds to PIWIL2 and activates STAT3 signaling, thereby conferring chemoresistance to 5-fluorouracil (5-FU) and oxaliplatin in colorectal cancer cells.91 A separate study conducted by the same authors suggests that a lower level of piR-36712 induces chemoresistance towards paclitaxel or doxorubicin in breast cancer.84 piR-39980 decreases the effects of doxorubicin and inhibits drug-induced apoptosis by modulating Janus Kinase 3 (JAK3) through target binding.127 Collectively, piRNAs may play a crucial role in maintaining the stem-cell properties of cancer cells, especially in drug resistance.
The uncertain meaning of piRNAs in tumor immunology
Studies on the involvement of piRNAs in the mammalian immune system are few, let alone tumor immunology. The limited number of reports indicate that piRNAs are abundant in human CD4 primary T-lymphocytes, monocytes or dendritic cells (DCs), and they play an important role in regulating gene expression in these highly differentiated somatic cells.141,150 piR-30840 significantly decreases interleukin-4 (IL-4) expression via sequence complementarity and binding to its pre-mRNA intron, subsequently restraining the development of Th2 T-lymphocytes. The piR-30840/PIWIL4/Ago4 complex further interacts with the TRAMP complex, and induces IL-4 decay in nuclear exosomes.141 Another study suggests that tRNA-Glu-derived piRNA [td-piR(Glu)] depresses CD1A transcription by inducing H3K9 methylation on monocytes/dendritic cells. The depression can be reversed by IL-4, inhibiting the biogenesis of td-piR(Glu) and upregulating CD1a molecules. As CD1a is a specific DC marker responsible for the presentation of lipid antigen to T-lymphocytes,151 it suggests that piRNAs can act as a mediator signal for lipid Ag presentation in monocytes/dendritic cells.150 These novel mechanisms provide evidence for the importance of piRNAs in immune cells. Given the impact of immune cells in tumor progression, piRNAs may contribute to the cancer immune system via IL-4 decay or CD1a dysregulation or other unidentified methods.
Potential clinical applications of piRNAs in cancer
Diagnostic and prognostic value of piRNAs
piRNAs have been proven to be effective diagnostic indicators in cancer, and appear to have higher sensitivity than currently used biomarkers carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA 19-9).93,116 Due to their distinct expression in postoperative and preoperative patients, peripheral blood piRNAs can be applied in postoperative long term monitoring and disease progression.93,116 Patients with increased levels of onco-piRNAs or low levels of tumor-suppressive piRNAs often show worse progression-free survival (PFS) or overall survival (OS) correlated with overall unfavorable prognosis.84,90,91,110,102 For example, PFS time and OS time are significantly shortened in patients with colorectal cancer (CRC) who have high piR-54265 levels compared with patients who have low piR-54265 levels.91 Additionally, piR-36,712 levels contain an independent prognostic value in breast cancer in that low levels are associated with worse clinical outcomes, shorter PFS, and higher rates of axillary lymph node metastasis.84 These clear pieces of evidence provide support for the idea that piRNAs are promising indicators for prognostic value in cancer.
Liquid biopsy of circulating piRNAs
Liquid biopsy is a commonly used technique that has been developed to detect cancer at an early stage and allows for tracking of tumor progression over time. In 2011, it was discovered that piRNAs could be a promising factor of analysis in liquid biopsy.122 Area under the receiver operating characteristic curves (AUCs) of piR-651, piR-823, and combinations of the two were 0.841, 0.822, 0.860 for gastric cancer detection in that 2011 study.122 Since then, a variety of circulating piRNAs have been reported as excellent candidates for liquid biopsy of cancer.91,93,102,117 Interestingly, exosomal piRNAs in the serum of cancer patients have also been quantified and are often more stable than miRNAs for the protection of PIWI proteins.20 Specifically, a higher level of piRNA-823 has been detected in extracellular vesicles secreted from multiple myeloma than from healthy controls and is positively associated with the stage of the disease. This indicates that the quantification of piRNAs in exosomes is also an attractive and promising inquiry.20
Therapeutic targeting of piRNAs
Molecular beacon (MB)-based bioimaging systems, which simultaneously combine the diagnosis and therapy of cancer, have attracted considerable attention in the cancer theragnosis field. Specific advantages include non-invasiveness, low cost, easy accessibility, and real-time quantitative imaging. MBs have also been applied in piRNA bioimaging.86,87 For example, endogenous piR-36026 in breast cancer cells has been successfully visualized by using a double stranded piR-36026 MB labeled with Cy5.5 fluorescent dye and black hole quencher 2 (BHQ2).86 In addition, the labeled piR-36026 displays a therapeutic response by binding to endogenous piR-36026 and blocking its hybridization with TSGs serpin peptidase inhibitor, clade A, member 1 (SERPINA1) and lecithin retinol acyltransferase (LRAT).86 This results in the upregulation of SERPINA1 and LRAT followed by caspase-3 activation and eventually cancer cell death.86
Moreover, labeling piRNA MBs with different Cyanine (Cy) fluorescent dyes allows the simultaneous visualization of multiple endogenous piRNAs in a single cell. For example, the dual transfection of Cy5.5 labeled piR-36026 MB and Cy3 labeled piR-36743 MB allows for the detection of both piRNAs in breast cancer cells.87 Furthermore, the quantitative measurement of different Cy5.5 and Cy3 signal intensities can successfully distinguish between the subtypes of breast cancer.87 In general, piRNA MBs may be a potential cancer theragnosis platform that allows for piRNA visualization, subtype identification, and TSG activation.
piRNA targeted therapy in combination with standard chemotherapeutic drugs has demonstrated promising anticancer effects in both in vitro and in vivo experiments. Both in vitro and in vivo trials have achieved greater efficacy in combined treatment of breast and colorectal cancer cells. Using a piR-54265 inhibitor (CRC) or a piR-36,712 analog (BrCa) in combination with chemotherapeutic agents such as 5-FU, paclitaxel, and doxorubicin were found to be more efficacious than treating with each agent alone.84,91 In addition, the combination of piRNA- and miRNA-targeting particles has been shown to achieve a satisfactory anticancer effect. After incubation with nano-sized complexes of miRNA-152 and piRNA-30074, CaCo2 colorectal adenocarcinoma cells is transformed into CD4+ cells. Increased levels of apoptotic markers caspase 9 and mechanistic target of rapamycin kinase (mTOR) and decreased levels of marker of proliferation ki-67 (MKI-67) has been detected in these cells.94 The same effect has been observed in A-549 lung adenocarcinoma cells and Girardi heart cells (cervical cancer combined with right atrium cancer) after incubation with the piRNA-30074 and antago-miR-155 complex103,125 These findings support the concept that the incorporation of piRNA-targeting particles with chemotherapy or miRNA-targeted therapy may lead to new pathways for cancer treatment research. In the future, additional combination therapeutic strategies, including the combination of piRNAs with immunotherapies such as immune checkpoint inhibitors [Programmed Death 1 (PD1)/cytotoxic T-lymphocyte-associated protein 4 (CTLA4)], can be designed to provide novel opportunities for addressing cancer treatment and drug resistance.
Conclusions and future perspectives
The last two decades have brought to light the true potential of piRNAs in liquid biopsy, prognosis, prediction of treatment response, and target therapy in cancer. piRNAs contribute to carcinogenesis via aberrant DNA methylation, histone marker deposition, mRNA degradation, and protein modification. However, most of the current researches focus on the downstream effects of piRNA dysregulation, and only a few studies investigated the upstream factors that cause piRNA dysregulation. The root of piRNA dysregulation in cancer is likely the result of disorders in piRNA biogenesis. Although the process of piRNA biogenesis in Drosophila and mice is well defined, the exact mechanism in humans remains unclear. Additionally, it is unknown whether or not piRNA biogenesis relevant proteins, such as MitoPLD or MOV10L1, is associated with piRNA dysregulation in cancer.
What's more, if mitochondria-mediated phased production is the final stage of piRNA biogenesis, then how do mature piRNAs return from mitochondria to nucleus to exert transcriptional regulation? In flies, most of the mature piRNAs produced on OMM are loaded into Piwi, one step essential for piRNAs nuclear translocation.16,33,72 piRNA loading triggers conformational change in Piwi and exposes a nuclear localization signal (NLS) at Piwi's N terminus which leads to importin-α-mediated transportation to the nucleus.152 Piwi could also interact with nucleoporin 358 (Nup358) to enter the nucleus.153 In mice, piRNAs are loaded with MIWI2 and subsequently translocated to the nucleus.6 In humans, however, the nucleus translocation mechanism remains elusive. Is PIWI loading necessary for piRNA nuclear entry in a similar way as in Drosophila and mice? If so, which protein is involved? It has been reported that HIWI/HIWI2-piR-sno75 complex could activate TRAIL gene by recruiting histone methytransferases MLL3 and KDM6A to its promoter. Since histone modification happens, it indicates that HIWI/HIWI2 may be relevant to nucleus import of piRNAs.137 However, the PIWI proteins which could be detected in nucleus are in the form of HIWI/HIWI2 and HILI.141,154,155 So, HILI may also be related in this process. Therefore, more systematic research on piRNA biogenesis and nuclear translocation machinery in humans is needed before we could better understand the piRNA dysregulation in cancer. Only then can we combat cancer by targeting piRNA imbalance-mediated carcinogenesis.
Despite the well-established PIWI-piRNA pathway during normal or cancer development, there are emerging hints that PIWI could function in a piRNA independent way. Li et al demonstrate that in the absence of piRNAs, human PIWIL1 plays an oncoprotein role in activating the anaphase promoting complex/cyclosome (APC/C) E3 complex, which then results in proteolysis of a critical cell adhesion-related protein, Pinin, and finally enhances the metastasis of pancreatic ductal adenocarcinomas (PDACs).156 Another study also shows that piRNA-unloaded PIWIL1 promotes gastric cancer through interaction with the UPF1- mediated nonsense-mediated mRNA decay (NMD) mechanism. Furthermore, mutating piRNA-binding residues of PIWIL1 does not compromise its oncogenic function. These piRNA-independent manner of PIWIL1 are consistant with an earlier report which failed to detected potentially functional piRISCs formed by PIWIL1 and piRNAs in a colon cancer cell line (COLO205).157 These lines of evidence support an piRNA-independent manner of oncogenic PIWIL1 in carcinogenesis. More importantly, all these studies uncover an unexpected fact, that is piRNAs are barely detectable in a series of cancer cells.156,157 It raises the question that whether the previous dysregulated piRNAs identified in various types of cancer cells are authentic piRNAs. A recent report indicates that ubiquitous background of small RNA-sequencing (RNA-seq) data may contaminate databases and abundant RNA fragments have sometimes been misinterpreted as piRNAs.158 So the authenticity of some of the reported somatic and cancerous piRNAs and their interaction between PIWI proteins deserve further investigation.
Authors contribution
Zhou Fuyou: Conceptualization, Supervision, Validation, Writing-review & editing. Zhang Hao: Conceptualization, Supervision, Validation, Writing-review & editing. Anthony Concilla: Investigation, Data curation, Writing-review & editing. Zhang Dianzheng: Investigation, Data curation, Writing-review & editing. Su Jingfen: Investigation, Data curation, Writing-original draft. Zhao Fang: Investigation, Data curation. Shen Fangfang: Investigation, Data curation.
Conflict of Interests
The authors declare that they have no competing interests.
Acknowledgements
The authors thank the members of the Fuyou Zhou laboratory for discussions and critical comments on the manuscript. This study was supported by National Natural Science Foundation, China [grant number U1504814] and the Major Projects of the Science and Technology Department, Henan Province [grant number 161100311200; 161100311300].
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Hao Zhang, Email: haozhang@jnu.edu.cn.
Fu-You Zhou, Email: ayzhoufuyou@163.com.
References
- 1.Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 2.Grivna S.T., Beyret E., Wang Z., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20(13):1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravin A., Gaidatzis D., Pfeffer S. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 4.Aravin A.A., Lagos-Quintana M., Yalcin A. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5(2):337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 5.Aravin A.A., Naumova N.M., Tulin A.V., Vagin V.V., Rozovsky Y.M., Gvozdev V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11(13):1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke J., Aravin A.A., Stark A. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 7.Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316(5825):744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 8.Aravin A.A., Sachidanandam R., Bourc'his D. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuramochi-Miyagawa S., Watanabe T., Gotoh K. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y., Li C., Zhang K. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Rep. 2010;43(9):635–641. doi: 10.5483/BMBRep.2010.43.9.635. [DOI] [PubMed] [Google Scholar]
- 11.Sana J., Faltejskova P., Svoboda M., Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med. 2012;10 doi: 10.1186/1479-5876-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito T., Magliocca S., Formicola D., Gianfrancesco F. piR_015520 belongs to Piwi-associated RNAs regulates expression of the human melatonin receptor 1A gene. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0022727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajasethupathy P., Antonov I., Sheridan R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149(3):693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pezic D., Manakov S.A., Sachidanandam R., Aravin A.A. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 2014;28(13):1410–1428. doi: 10.1101/gad.240895.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha H., Song J., Wang S. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics. 2014;15(1) doi: 10.1186/1471-2164-15-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han B.W., Wang W., Li C., Weng Z., Zamore P.D. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015;348(6236):817–821. doi: 10.1126/science.aaa1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen P.R., Tirian L., Vunjak M., Brennecke J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature. 2017;549(7670):54–59. doi: 10.1038/nature23482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teixeira F.K., Okuniewska M., Malone C.D., Coux R.X., Rio D.C., Lehmann R. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature. 2017;552(7684):268–272. doi: 10.1038/nature25018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D., Tu S., Stubna M. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science. 2018;359(6375):587–592. doi: 10.1126/science.aao2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B., Hong J., Hong M. piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene. 2019;38(26):5227–5238. doi: 10.1038/s41388-019-0788-4. [DOI] [PubMed] [Google Scholar]
- 21.Sheu-Gruttadauria J., MacRae I.J. Structural foundations of RNA silencing by argonaute. J Mol Biol. 2017;429(17):2619–2639. doi: 10.1016/j.jmb.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gou L.T., Dai P., Yang J.H. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2015;25(2) doi: 10.1038/cr.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X.A., Yin H., Sweeney S., Raha D., Snyder M., Lin H. A major epigenetic programming mechanism guided by piRNAs. Dev Cell. 2013;24(5):502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozata D.M., Gainetdinov I., Zoch A., O'Carroll D., Zamore P.D. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20(2):89–108. doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- 25.Yu B., Yang Z., Li J. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Yang Z., Yu B., Liu J., Chen X. Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr Biol. 2005;15(16):1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren G., Chen X., Yu B. Small RNAs meet their targets: when methylation defends miRNAs from uridylation. RNA Biol. 2014;11(9):1099–1104. doi: 10.4161/rna.36243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Zhang P., Lu Y. piRBase: a comprehensive database of piRNA sequences. Nucleic Acids Res. 2019;47(D1):D175–D180. doi: 10.1093/nar/gky1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnan P., Damaraju S. The challenges and opportunities in the clinical application of noncoding RNAs: the road map for miRNAs and piRNAs in cancer diagnostics and prognostics. Int J Genomics. 2018;2018 doi: 10.1155/2018/5848046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Han B.W., Tipping C. Slicing and binding by Ago3 or Aub trigger piwi-bound piRNA production by distinct mechanisms. Mol Cell. 2015;59(5):819–830. doi: 10.1016/j.molcel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senti K.A., Jurczak D., Sachidanandam R., Brennecke J. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 2015;29(16):1747–1762. doi: 10.1101/gad.267252.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z., Chen K.M., Pandey R.R. PIWI slicing and EXD1 drive biogenesis of nuclear piRNAs from cytosolic targets of the mouse piRNA pathway. Mol Cell. 2016;61(1):138–152. doi: 10.1016/j.molcel.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gainetdinov I., Colpan C., Arif A., Cecchini K., Zamore P.D. A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol Cell. 2018;71(5):775–790. doi: 10.1016/j.molcel.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czech B., Munafo M., Ciabrelli F. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet. 2018;52:131–157. doi: 10.1146/annurev-genet-120417-031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goriaux C., Desset S., Renaud Y., Vaury C., Brasset E. Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep. 2014;15(4):411–418. doi: 10.1002/embr.201337898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohn F., Sienski G., Handler D., Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157(6):1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 39.Li X.Z., Roy C.K., Dong X. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell. 2013;50(1):67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolcun-Filas E., Bannister L.A., Barash A. A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development. 2011;138(15):3319–3330. doi: 10.1242/dev.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Özata D.M., Yu T., Mou H. Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat Ecol Evol. 2020;4(1):156–168. doi: 10.1038/s41559-019-1065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klattenhoff C., Xi H., Li C. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138(6):1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pane A., Jiang P., Zhao D.Y., Singh M., Schüpbach T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011;30(22):4601–4615. doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Thomas A., Stuwe E., Li S. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014;28(15):1667–1680. doi: 10.1101/gad.245514.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu B., Cassani M., Wang M. Structural insights into Rhino-mediated germline piRNA cluster formation. Cell Res. 2015;25(4):525–528. doi: 10.1038/cr.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ha H., Song J., Wang S. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics. 2014;15(1) doi: 10.1186/1471-2164-15-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams Z., Morozov P., Mihailovic A. Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep. 2015;13(4):854–863. doi: 10.1016/j.celrep.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 48.Gunawardane L.S., Saito K., Nishida K.M. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315(5818):1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 49.Meister G., Landthaler M., Peters L. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15(23):2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 50.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beyret E., Liu N., Lin H. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res. 2012;22(10):1429–1439. doi: 10.1038/cr.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabayama Y., Toh H., Katanaya A. Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes. Nucleic Acids Res. 2017;45(9):5387–5398. doi: 10.1093/nar/gkx027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roovers E.F., Rosenkranz D., Mahdipour M. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 2015;10(12):2069–2082. doi: 10.1016/j.celrep.2015.02.062. [DOI] [PubMed] [Google Scholar]
- 54.Sasaki T., Shiohama A., Minoshima S., Shimizu N. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003;82(3):323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 55.Rizzo F., Rinaldi A., Marchese G. Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma. Oncotarget. 2016;7(34):54650–54661. doi: 10.18632/oncotarget.10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ge D.T., Wang W., Tipping C., Gainetdinov I., Weng Z., Zamore P.D. The RNA-binding ATPase, armitage, couples piRNA amplification in nuage to phased piRNA production on mitochondria. Mol Cell. 2019;74(5):982–995. doi: 10.1016/j.molcel.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Munafo M., Manelli V., Falconio F.A. Daedalus and Gasz recruit Armitage to mitochondria, bringing piRNA precursors to the biogenesis machinery. Genes Dev. 2019;33(13–14):844–856. doi: 10.1101/gad.325662.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang P.J., McCarrey J.R., Yang F., Page D.C. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27(4):422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 59.Zheng K., Xiol J., Reuter M. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci U S A. 2010;107(26):11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frost R.J., Hamra F.K., Richardson J.A., Qi X., Bassel-Duby R., Olson E.N. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci U S A. 2010;107(26):11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu Q., Pandey R.R., Leu N.A., Pillai R.S., Wang P.J. Mutations in the MOV10L1 ATP hydrolysis motif cause piRNA biogenesis failure and male sterility in mice. Biol Reprod. 2016;95(5) doi: 10.1095/biolreprod.116.142430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vourekas A., Zheng K., Fu Q. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015;29(6):617–629. doi: 10.1101/gad.254631.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng K., Wang P.J. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet. 2012;8(11) doi: 10.1371/journal.pgen.1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eddy E.M. Fine structural observations on the form and distribution of nuage in germ cells of the rat. Anat Rec. 1974;178(4):731–757. doi: 10.1002/ar.1091780406. [DOI] [PubMed] [Google Scholar]
- 65.Ma L., Buchold G.M., Greenbaum M.P. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5(9) doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarkardeh H., Totonchi M., Asadpour O. Association of MOV10L1 gene polymorphisms and male infertility in azoospermic men with complete maturation arrest. J Assist Reprod Genet. 2014;31(7):865–871. doi: 10.1007/s10815-014-0240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hong Y., Wang C., Fu Z. Systematic characterization of seminal plasma piRNAs as molecular biomarkers for male infertility. Sci Rep. 2016;6 doi: 10.1038/srep24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishimasu H., Ishizu H., Saito K. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491(7423):284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 69.Pane A., Wehr K., Schupbach T. Zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12(6):851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voigt F., Reuter M., Kasaruho A., Schulz E.C., Pillai R.S., Barabas O. Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nuc. RNA. 2012;18(12):2128–2134. doi: 10.1261/rna.034967.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watanabe T., Chuma S., Yamamoto Y. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20(3):364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohn F., Handler D., Brennecke J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348(6236):812–817. doi: 10.1126/science.aaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomes Fernandes M., He N., Wang F. Human-specific subcellular compartmentalization of P-element induced wimpy testis-like (PIWIL) granules during germ cell development and spermatogenesis. Hum Reprod. 2018;33(2):258–269. doi: 10.1093/humrep/dex365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi S.Y., Huang P., Jenkins G.M., Chan D.C., Schiller J., Frohman M.A. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8(11):1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y., Liu X., Bai J. Mitoguardin regulates mitochondrial fusion through MitoPLD and is required for neuronal homeostasis. Mol Cell. 2016;61(1):111–124. doi: 10.1016/j.molcel.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 76.Hayashi R., Schnabl J., Handler D., Mohn F., Ameres S.L., Brennecke J. Genetic and mechanistic diversity of piRNA 3'-end formation. Nature. 2016;539(7630):588–592. doi: 10.1038/nature20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T. The 3' termini of mouse Piwi-interacting RNAs are 2'-O-methylated. Nat Struct Mol Biol. 2007;14(4):349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 78.Fu A., Jacobs D.I., Hoffman A.E., Zheng T., Zhu Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015;36(10):1094–1102. doi: 10.1093/carcin/bgv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chu H., Hui G., Yuan L. Identification of novel piRNAs in bladder cancer. Canc Lett. 2015;356(2 Pt B):561–567. doi: 10.1016/j.canlet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 80.Wallace D.C. Mitochondria and cancer. Nat Rev Canc. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee J.H., Schütte D., Wulf G. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15(2):201–211. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- 82.Zhang H., Ren Y., Xu H., Pang D., Duan C., Liu C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol. 2013;22(4):217–223. doi: 10.1016/j.suronc.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 83.Ai L., Mu S., Sun C. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Cancer. 2019;18(1) doi: 10.1186/s12943-019-1011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan L., Mai D., Zhang B. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-0940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hashim A., Rizzo F., Marchese G. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget. 2014;5(20):9901–9910. doi: 10.18632/oncotarget.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee Y.J., Moon S.U., Park M.G. Multiplex bioimaging of piRNA molecular pathway-regulated theragnostic effects in a single breast cancer cell using a piRNA molecular beacon. Biomaterials. 2016;101:143–155. doi: 10.1016/j.biomaterials.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 87.Park Y.K., Jung W.Y., Park M.G. Bioimaging of multiple piRNAs in a single breast cancer cell using molecular beacons. Medchemcomm. 2017;8(12):2228–2232. doi: 10.1039/c7md00515f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang G., Hu H., Xue X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15(7):563–568. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- 89.Krishnan P., Ghosh S., Graham K., Mackey J.R., Kovalchuk O., Damaraju S. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget. 2016;7(25):37944–37956. doi: 10.18632/oncotarget.9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weng W., Liu N., Toiyama Y. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer. 2018;17(1) doi: 10.1186/s12943-018-0767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mai D., Ding P., Tan L. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics. 2018;8(19):5213–5230. doi: 10.7150/thno.28001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chu H., Xia L., Qiu X. Genetic variants in noncoding PIWI-interacting RNA and colorectal cancer risk. Cancer. 2015;121(12):2044–2052. doi: 10.1002/cncr.29314. [DOI] [PubMed] [Google Scholar]
- 93.Vychytilova-Faltejskova P., Stitkovcova K., Radova L. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomark Prev. 2018;27(9):1019–1028. doi: 10.1158/1055-9965.EPI-18-0318. [DOI] [PubMed] [Google Scholar]
- 94.Klimenko O.V., Shtilman M. Reprogramming of CaCo2 colorectal cancer cells after using the complex of poly-(N-vinylpyrrolidone) with small non-coding RNAs. Toxicol Rep. 2019;6:186–192. doi: 10.1016/j.toxrep.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martinez V.D., Enfield K.S.S., Rowbotham D.A., Lam W.L. An atlas of gastric PIWI-interacting RNA transcriptomes and their utility for identifying signatures of gastric cancer recurrence. Gastric Cancer. 2016;19(2):660–665. doi: 10.1007/s10120-015-0487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miao P.Z., Yang Y., Chen E.B., Zhu G.Q., Wang B., Dai Z. [Differential expressions analysis of piwi-interacting RNAs in hepatocellular carcinoma] Zhonghua Gan Zang Bing Za Zhi. 2018;26(11):842–846. doi: 10.3760/cma.j.issn.1007-3418.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 97.Law P.T., Qin H., Ching A.K. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58(6):1165–1173. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 98.Reeves M.E., Firek M., Jliedi A., Amaar Y.G. Identification and characterization of RASSF1C piRNA target genes in lung cancer cells. Oncotarget. 2017;8(21):34268–34282. doi: 10.18632/oncotarget.15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Daugaard I., Veno M.T., Yan Y. Small RNA sequencing reveals metastasis-related microRNAs in lung adenocarcinoma. Oncotarget. 2017;8(16):27047–27061. doi: 10.18632/oncotarget.15968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peng L., Song L., Liu C. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol. 2016;37(2):2749–2756. doi: 10.1007/s13277-015-4056-0. [DOI] [PubMed] [Google Scholar]
- 101.Mei Y., Wang Y., Kumari P. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nat Commun. 2015;6 doi: 10.1038/ncomms8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao C., Tolkach Y., Schmidt D. Mitochondrial PIWI-interacting RNAs are novel biomarkers for clear cell renal cell carcinoma. World J Urol. 2019;37(8):1639–1647. doi: 10.1007/s00345-018-2575-1. [DOI] [PubMed] [Google Scholar]
- 103.Klimenko O.V. Joint Action of the Nano-Sized System of Small Non-coding RNAs with DDMC Vector and Recombinant IL-7 Reprograms A-549 Lung Adenocarcinoma Cells into CD4 + Cells. Immunotherapy. 2017 [Google Scholar]
- 104.Busch J., Ralla B., Jung M. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J Exp Clin Cancer Res. 2015;34(1) doi: 10.1186/s13046-015-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y., Wu X., Gao H. Piwi-interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol Med. 2015;21:381–388. doi: 10.2119/molmed.2014.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jacobs D.I., Qin Q., Fu A., Chen Z., Zhou J., Zhu Y. piRNA-8041 is downregulated in human glioblastoma and suppresses tumor growth in vitro and in vivo. Oncotarget. 2018;9(102):37616–37626. doi: 10.18632/oncotarget.26331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jacobs D.I., Qin Q., Lerro M.C. PIWI-interacting RNAs in gliomagenesis: evidence from post-GWAS and functional analyses. Cancer Epidemiol Biomark Prev. 2016;25(7):1073–1080. doi: 10.1158/1055-9965.EPI-16-0047. [DOI] [PubMed] [Google Scholar]
- 108.Das B., Roy J., Jain N., Mallick B. Tumor suppressive activity of PIWI-interacting RNA in human fibrosarcoma mediated through repression of RRM2. Mol Carcinog. 2019;58(3):344–357. doi: 10.1002/mc.22932. [DOI] [PubMed] [Google Scholar]
- 109.Krishnan A.R., Qu Y., Li P.X. Computational methods reveal novel functionalities of PIWI-interacting RNAs in human papillomavirus-induced head and neck squamous cell carcinoma. Oncotarget. 2018;9(4):4614–4624. doi: 10.18632/oncotarget.23464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saad M.A., Ku J., Kuo S.Z. Identification and characterization of dysregulated P-element induced wimpy testis-interacting RNAs in head and neck squamous cell carcinoma. Oncol Lett. 2019;17(3):2615–2622. doi: 10.3892/ol.2019.9913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Krishnan A.R., Korrapati A., Zou A.E. Smoking status regulates a novel panel of PIWI-interacting RNAs in head and neck squamous cell carcinoma. Oral Oncol. 2017;65:68–75. doi: 10.1016/j.oraloncology.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martinez V.D., Firmino N.S., Marshall E.A. Non-coding RNAs predict recurrence-free survival of patients with hypoxic tumours. Sci Rep. 2018;8(1) doi: 10.1038/s41598-017-18462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Muller S., Raulefs S., Bruns P. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Canc. 2015;14 doi: 10.1186/s12943-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oner C., Turgut Cosan D., Colak E. Estrogen and androgen hormone levels modulate the expression of PIWI interacting RNA in prostate and breast cancer. PloS One. 2016;11(7) doi: 10.1371/journal.pone.0159044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cheng J., Guo J.M., Xiao B.X. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412(17–18):1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 116.Cui L., Lou Y., Zhang X. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem. 2011;44(13):1050–1057. doi: 10.1016/j.clinbiochem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 117.Cordeiro A., Navarro A., Gaya A. PiwiRNA-651 as marker of treatment response and survival in classical Hodgkin lymphoma. Oncotarget. 2016;7(29):46002–46013. doi: 10.18632/oncotarget.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang S.J., Yao J., Shen B.Z. Role of piwi-interacting RNA-651 in the carcinogenesis of non-small cell lung cancer. Oncol Lett. 2018;15(1):940–946. doi: 10.3892/ol.2017.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li D., Luo Y., Gao Y. piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int J Mol Med. 2016;38(3):927–936. doi: 10.3892/ijmm.2016.2671. [DOI] [PubMed] [Google Scholar]
- 120.Yao J., Wang Y.W., Fang B.B., Zhang S.J., Cheng B.L. piR-651 and its function in 95-D lung cancer cells. Biomed Rep. 2016;4(5):546–550. doi: 10.3892/br.2016.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yin J., Jiang X.Y., Qi W. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Canc Sci. 2017;108(9):1746–1756. doi: 10.1111/cas.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheng J., Deng H., Xiao B. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Canc Lett. 2012;315(1):12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 123.Iliev R., Fedorko M., Machackova T. Expression levels of PIWI-interacting RNA, piR-823, are deregulated in tumor tissue, blood serum and urine of patients with renal cell carcinoma. Anticancer Res. 2016;36(12):6419–6423. doi: 10.21873/anticanres.11239. [DOI] [PubMed] [Google Scholar]
- 124.Yan H., Wu Q.L., Sun C.Y. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29(1):196–206. doi: 10.1038/leu.2014.135. [DOI] [PubMed] [Google Scholar]
- 125.Klimenko O.V. Complex of small non-coding RNAs piR-30074 and antago-miR-155 and miR-125b with DDMC carrier transforms Girardi heart cells into CD4+ cells. J Cancer and Tumor Int. 2016;4 [Google Scholar]
- 126.Wu D., Fu H., Zhou H., Su J., Zhang F., Shen J. Effects of novel ncRNA molecules, p15-piRNAs, on the methylation of DNA and histone H3 of the CDKN2B promoter region in U937 cells. J Cell Biochem. 2015;116(12):2744–2754. doi: 10.1002/jcb.25199. [DOI] [PubMed] [Google Scholar]
- 127.Roy J., Das B., Jain N., Mallick B. PIWI-interacting RNA 39980 promotes tumor progression and reduces drug sensitivity in neuroblastoma cells. J Cell Physiol. 2020;235(3):2286–2299. doi: 10.1002/jcp.29136. [DOI] [PubMed] [Google Scholar]
- 128.Mani S.R., Juliano C.E. Untangling the web: the diverse functions of the PIWI/piRNA pathway. Mol Reprod Dev. 2013;80(8):632–664. doi: 10.1002/mrd.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.De Fazio S., Bartonicek N., Di Giacomo M. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480(7376):259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 130.Sienski G., Dönertas D., Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151(5):964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Greenberg M.V.C., Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20(10):590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 132.Baylin S.B., Jones P.A. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8(9) doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fu A., Jacobs D.I., Zhu Y. Epigenome-wide analysis of piRNAs in gene-specific DNA methylation. RNA Biol. 2014;11(10):1301–1312. doi: 10.1080/15476286.2014.996091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stadler M.B., Murr R., Burger L. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 135.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 136.Baylin S.B., Jones P.A. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Canc. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.He X., Chen X., Zhang X. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015;43(7):3712–3725. doi: 10.1093/nar/gkv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nagamori I., Kobayashi H., Nishimura T. Relationship between PIWIL4-mediated H3K4me2 demethylation and piRNA-dependent DNA methylation. Cell Rep. 2018;25(2):350–356. doi: 10.1016/j.celrep.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 139.Rouget C., Papin C., Boureux A. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467(7319):1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Watanabe T., Cheng E.C., Zhong M., Lin H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015;25(3):368–380. doi: 10.1101/gr.180802.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhong F., Zhou N., Wu K. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015;43(21):10474–10491. doi: 10.1093/nar/gkv954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Needham E.J., Parker B.L., Burykin T., James D.E., Humphrey S.J. Illuminating the dark phosphoproteome. Sci Signal. 2019;12(565) doi: 10.1126/scisignal.aau8645. [DOI] [PubMed] [Google Scholar]
- 143.Lu Y., Zhang K., Li C. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhao S., Gou L.T., Zhang M. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell. 2013;24(1):13–25. doi: 10.1016/j.devcel.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 145.Sienski G., Donertas D., Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151(5):964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 147.Toh Tan Boon, Lim Jhin Jieh, Chow Edward Kai-Hua. Epigenetics in cancer stem cells. Mol Cancer. 2017;16(1) doi: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lacerda L., Pusztai L., Woodward W.A. The role of tumor initiating cells in drug resistance of breast cancer: implications for future therapeutic approaches. Drug Resist Updates. 2010;13(4–5):99–108. doi: 10.1016/j.drup.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 149.Wang Y., Gable T., Ma M.Z. A piRNA-like small RNA induces chemoresistance to cisplatin-based therapy by inhibiting apoptosis in lung squamous cell carcinoma. Mol Ther Nucleic Acids. 2017;6:269–278. doi: 10.1016/j.omtn.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang X., He X., Liu C. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J Immunol. 2016;196(4):1591–1603. doi: 10.4049/jimmunol.1500805. [DOI] [PubMed] [Google Scholar]
- 151.Moody D.B., Young D.C., Cheng T.Y. T cell activation by lipopeptide antigens. Science. 2004;303(5657):527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 152.Yashiro R., Murota Y., Nishida K.M. Piwi nuclear localization and its regulatory mechanism in Drosophila ovarian somatic cells. Cell Rep. 2018;23(12):3647–3657. doi: 10.1016/j.celrep.2018.05.051. [DOI] [PubMed] [Google Scholar]
- 153.Parikh R.Y., Lin H., Gangaraju V.K. A critical role for nucleoporin 358 (Nup358) in transposon silencing and piRNA biogenesis in Drosophila. J Biol Chem. 2018;293(24):9140–9147. doi: 10.1074/jbc.AC118.003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.He W., Wang Z., Wang Q. Expression of HIWI in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. BMC Cancer. 2009;9 doi: 10.1186/1471-2407-9-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee J.H., Jung C., Javadian-Elyaderani P. Pathways of proliferation and antiapoptosis driven in breast cancer stem cells by stem cell protein piwil2. Canc Res. 2010;70(11):4569–4579. doi: 10.1158/0008-5472.CAN-09-2670. [DOI] [PubMed] [Google Scholar]
- 156.Li F., Yuan P., Rao M. piRNA-independent function of PIWIL1 as a co-activator for anaphase promoting complex/cyclosome to drive pancreatic cancer metastasis. Nat Cell Biol. 2020;22(4):425–438. doi: 10.1038/s41556-020-0486-z. [DOI] [PubMed] [Google Scholar]
- 157.Genzor P., Cordts S.C., Bokil N.V., Haase A.D. Aberrant expression of select piRNA-pathway genes does not reactivate piRNA silencing in cancer cells. Proc Natl Acad Sci U S A. 2019;116(23):11111–11112. doi: 10.1073/pnas.1904498116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tosar J.P., Rovira C., Cayota A. Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun Biol. 2018;1 doi: 10.1038/s42003-017-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]