Abstract
Insulin-resistance (IR) is one of the most important precursors of type 2 diabetes (T2D). Recent evidence suggests an association of depression with the onset of T2D. Accumulating evidence shows that depression and T2D share common biological origins, and DNA methylation examination might reveal the link between lifestyle, disease risk, and potential therapeutic targets for T2D. Here we hypothesize that integrative mining of IR and depression cohort data will facilitate predictive biomarkers identification for T2D. We utilized a newly proposed method to extract gene-level information from probe level data on genome-wide DNA methylation array. We identified a set of genes associated with IR and depression in clinical cohorts. By overlapping the IR-related nutraceutical-gene network with depression networks, we identified a common subnetwork centered with Vitamin D Receptor (VDR) gene. Preliminary clinical validation of gene methylation set in a small cohort of T2D patients and controls was established using the Sequenome matrix-assisted laser desorption ionization-time flight mass spectrometry. A set of sites in the promoter regions of VDR showed a significant difference between T2D patients and controls. Using a logistic regression model, the optimal prediction performance of these sites was AUC = 0.902,and an odds ratio = 19.76. Thus, monitoring the methylation status of specific VDR promoter region might help stratify the high-risk individuals who could potentially benefit from vitamin D dietary supplementation. Our results highlight the link between IR and depression, and the DNA methylation analysis might facilitate the search for their shared mechanisms in the etiology of T2D.
Keywords: Depression, DNA methylation, Insulin resistance, Nutraceuticals, Type 2 Diabetes, Vitamin D receptor
Introduction
Type 2 Diabetes (T2D) is a global health problem, and according to the World Health Organization, diabetes is a major cause of blindness, kidney failure, heart attacks, stroke, and lower-limb amputations. In 2016, an estimated 1.6 million deaths were attributed directly to diabetes.1 T2D is a multifactorial disease defined by the interaction of genetics and lifestyle factors, including malnutrition, obesity, physical activity, stress, and xenobiotics.2 Considering the urgent need for interventions to prevent T2D, natural alternatives, such as nutraceuticals that are safe, without risk, and with fewer regulatory hurdles than drugs are required for usage. Nutraceuticals have received considerable attention in recent years for their potential roles in preventing or treating several chronic diseases.3 A promising strategy to prevent T2D is to identify individuals with high risk and intervene with a nutraceutical that targets the corresponding dysregulated gene network. This strategy offers the potential to identify natural mimetics of metformin and rapamycin.4 The key is to identify the biomarkers, which (1) can be targeted by nutraceuticals and (2) epigenetically regulated in the early stage of disease onset.
Epigenetics was considered as a molecular link between environmental factors and T2D development.5,6 Therefore, an evaluation of DNA methylation might improve the understanding of the T2D pathogenesis, contribute to the development of novel treatments, and offer means to identify individuals at risk for developing the disease.7 Accordingly, the DNA methylation pattern associated with T2D in pancreatic islets and adipose tissue was also detected in leukocytes.8, 9, 10 A large, prospective, case–control study showed that differences in peripheral blood DNA methylation status might predict future T2D incidence.11 Insulin resistance (IR) is responsible for T2D development. T2D is a pathological condition wherein cells fail to respond to insulin, and earlier studies showed that IR is epigenetically regulated.12 A recent epigenome-wide association study of peripheral white blood cells identified epigenetic signatures associated with IR as measured by the Homeostatic Model Assessment of IR (HOMA-IR).13
Growing evidence shows that depression and T2D share common biological origins, particularly over activation of innate immunity leading to a cytokine-mediated inflammatory response and potential dysregulation of the hypothalamic–pituitary–adrenal axis.14 Proinflammatory cytokines might directly affect the brain, causing depressive symptoms. Our previous analysis of healthy individuals shows that long term isolation leads to changes in metabolism and mood state. Glucose metabolism and long-term depression are two significant dysregulated pathways in the Mars500 isolation,15 and 180 days isolation16 studies, thus suggesting a link between change in glucose metabolism and mood state in apparently healthy individuals.
For identifying an early intervention epigenetic biomarker for T2D, we started with the global nutraceutical target gene network to investigate the association between genes involved in IR and depression, from two large clinical datasets. We choose genes in the center of the overlapped subnetwork of IR, nutraceutical-gene network, and depression networks, to design a DNA methylation assay and analyzed using the Sequenome matrix-assisted laser desorption ionization-time flight mass spectrometry (MALDI-TOF MS). We tested the identified epigenetic biomarker in a small cohort of patients with T2D and controls. Although our results showed the validity of this biomarker, it requires further studies to establish an early risk monitor-nutraceutical intervention loop for the prevention of T2D.
Material and methods
Nutraceutical target gene network
The nutraceuticals and their target protein information were downloaded from the DrugBank database (version 5.1, https://www.drugbank.ca/drugs), with the filter “Nutraceutical” category.17
SimPo algorithm for DNA methylation chip data analysis
A recent study showed a difference between the gene body and promoter methylation (MeGDP) and gene expression, and the correlation coefficient is as high as 0.67.18 Therefore, MeGDP could be used as a predictor for gene expression levels. A higher MeGDP suggests a higher gene expression value, thus suggesting a higher degree of association with the relevant phenotype. Based on the above research results, we propose a SimPo (the Statistical Difference of DNA Methylation between Promoter and Other regions algorithm) method to calculate the DNA methylation status of different genes.27
Using the Illumina HM450K chip methylation data as the input, the SimPo algorithm calculates the DNA methylation value of cg-probes, which are located in the TSS200 promoter and outside the TSS200 regions. We used the statistical difference method t-test in SimPo algorithm, and the degree of difference (SimPo score) is used to characterize the DNA methylation of corresponding genes:
Where
: average DNA methylation value of all probes annotated in TSS200 region; : average DNA methylation value of all probes annotated outside TSS200 region; m: number of probes annotated in TSS200 region; n: number of probes annotated outside TSS200 region; : variance of DNA methylation values of probes annotated in TSS200 region; : variance of DNA methylation values of probes annotated outside TSS200 region.
Phenotype associated gene identification
IR-related DNA methylation data used in this study contains the methylation profiling of 474 peripheral white blood cells (GSE115278, Illumina 450 k bead chip).13,19 Each sample was annotated for the corresponding HOMA-IR value. Depression-related DNA methylation data (GSE125105, Illumina humanity 450 bead chip) analyzed in this study were obtained from the 699 samples recruited by Max Planck Institute of Psychiatry (MPIP), including 489 samples with phenotype of depression and 210 controls.26
We used SimPo method to transform probe level data set into gene level data set. Then we used t-test to identify phenotype related gene lists in this association study.
Type 2 diabetes patient data
We analyzed the blood samples from a total of 24 patients with T2D (male: 13, female: 11, age: 33–68) from the First Affiliated Hospital of PLA General Hospital, and 47 healthy controls (male: 16, female: 31, age: 40–73) from the FoShan New RongQi Hospital. All peripheral whole blood samples were treated with EDTA anticoagulant (Table 1).
Table 1.
Clinicopathological characteristics of the samples (n = 71).
| Variable | T2D Patients (n = 24) | Control (n = 47) |
|---|---|---|
| Male/Female | 13/11 | 16/31 |
| Age | 50.8 ± 9.5 | 58.8 ± 10.4 |
| Fasting blood glucose (FBG) | 9.13 ± 2.24 | 5.28 ± 0.52 |
| 2hPBG | 14.53 ± 4.21 | 7.03 ± 1.35 |
| HbA1c | 8.19 ± 1.64 | 5.57 ± 0.32 |
MassARRAY analysis
We identified the sequence of 500 bp up- and down-stream from the CpG position in promoter region of VDR gene (probe cg02522757 and cg13556224 annotated position in HM450K chip) by UCSC genome browser (http://genome.ucsc.edu/), and designed two primer sets for the methylation analysis of the amplicon-cg02522757 and amplicon-cg13556224 region by EpiDesigner software (http://epidesigner.com; Table 2). Quantitative DNA methylation analysis of the above two amplicons was carried out by the MassARRAY platform (SEQUENOM) following the manufacturer's protocols. Briefly, DNA was treated with sodium bisulfite, PCR amplified and subjected to bisulfite reactions. The DNA methylation status of the samples was tested quantitatively by MALDI-TOF MS. Finally, 18 CpG units in the amplicon-cg02522757 region, and 4CpG units in amplicon-cg13556224 region (one to two CpG sites per unit) were generated by the EpiTyper v1.0.5 software (Supplementary Material).
Table 2.
Sequence of MassARRAY primer relative to amplicon-cg02522757 and amplicon-cg13556224.
| Amplicons | Primer | Sequence (5′ -> 3′) |
|---|---|---|
| amplicon-cg02522757 | 10F | aaatccaatcctctcttaccaaaa |
| T7R | ttttaatttgtgggattaggttgag | |
| amplicon-cg13556224 | 10F | tttcaccttatccctctaaaccata |
| T7R | tattttttgagatttggaattgtgg |
Results
Insulin-resistance and depression related DNA methylation remodeling genes
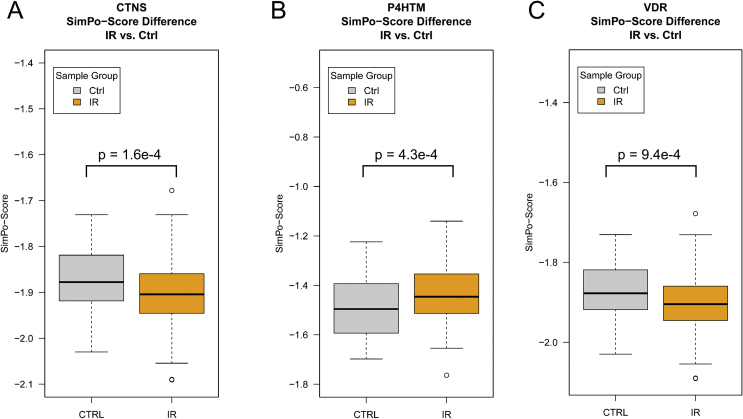
We used the t-test to measure the significant differences in DNA methylation beta value distribution between the promoter (TSS200) and other regions, and used SimPo score to summarize this difference. SimPo score reflects the specific DNA methylation remodeling in the promoter regions and might correlate with a select gene expression profile. The IR analysis identified 15 genes that demonstrated a significant SimPo score in insulin-resistant vs. healthy individuals (t-test p-value < 0.05) (Fig. 1, Table 3). Additionally, we tested the SimPo score difference of the above 15 genes in the depression cohort. The results showed that CTNS, VDR, RARA, NQO2, and SGPL1 also showed significant differences between individuals with depression and healthy controls (t-test p-value < 0.05) (Fig. 1, Table 3). Next, the potential nutraceuticals were queried for the related protein in the DrugBank database. The results identified all the above genes with nutraceuticals that could remodel the gene functions.
Figure 1.
List of genes with significant differences in SimPo-Score between insulin-resistant and healthy individuals.
Table 3.
Genes identified by SimPo score to reflect the DNA methylation remodeling in insulin resistance and depression.
| No. | Gene | p-value (IR vs. Ctrl) | p-value (Depression vs. Ctrl) | Nutraceutical targeting the protein |
|---|---|---|---|---|
| 1 | CTNS | 0.000155 | 0.002457 | Cystine |
| 2 | P4HTM | 0.000427 | 0.357627 | Ascorbic acid |
| 3 | VDR | 0.000935 | 0.010636 | Calcitriol, Calcifediol, Ergocalciferol, Cholecalciferol, Alfacalcidol, Vitamin D |
| 4 | AADAT | 0.000955 | 0.355463 | Pyridoxal Phosphate, Glutamic Acid |
| 5 | BCAT2 | 0.00112 | 0.87388 | Pyridoxal Phosphate, Glutamic Acid, l-Leucine, l-Isoleucine |
| 6 | PDCD6 | 0.001821 | 0.357192 | Calcium |
| 7 | RARA | 0.002744 | 0.012328 | Tretinoin |
| 8 | GRID1 | 0.003623 | 0.059968 | Glutamic Acid |
| 9 | NQO2 | 0.004228 | 4.65E-07 | NADH, Menadione, Melatonin |
| 10 | SGPL1 | 0.006147 | 0.005774 | Pyridoxal Phosphate |
| 11 | PPIH | 0.008855 | 0.331482 | Proline |
| 12 | GSTO2 | 0.012418 | 0.29847 | Glutathione |
| 13 | ADK | 0.012601 | 0.079088 | Adenosine phosphate |
| 14 | AHR | 0.027996 | 0.951787 | Ginseng |
| 15 | GRM8 | 0.039308 | 0.460064 | Glutamic Acid |
P1: SimPo-score-based t-test p-value for insulin resistance vs. healthy individual.
P2: SimPo-score-based t-test p-value for depression vs. healthy individual.
Overlapping nutraceutical-gene networks in insulin resistance and depression
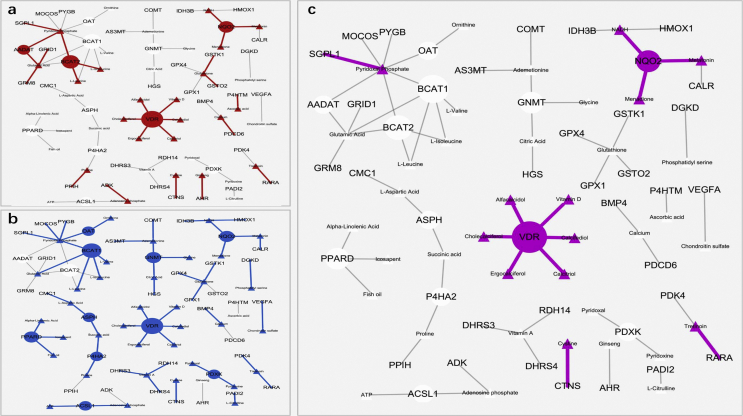
To identify the common DNA methylation remodeling modules in IR and depression, we generated the nutraceutical-gene graphs for IR and depression analyses and their overlaps (Fig. 2). The criteria for the identified genes included phenotype association (IR or depression) with a t-test p-value < 0.05, and links added in the DrugBank database for all the associated nutraceuticals. Edges in red demonstrated the nutraceutical-gene interactions for IR analysis (Fig. 2A), and blue for the T2D analysis (Fig. 2B). Edges in purple demonstrated the same nutraceutical-gene interactions appear in both analyses (Fig. 2C). A subnetwork centered with the VDR gene which links to six nutraceuticals was highlighted.
Figure 2.
The insulin-resistance and depression-related nutraceutical-gene network. All the genes with a SimPo score t-test P < 0.05 were plotted. The triangles represent nutraceuticals, and the circles represent genes. All edges (gray or colored) mean nutraceutical-gene interactions annotated in the DrugBank database. (A) The IR-related nutraceutical-gene networks (red edges). (B) The depression-related nutraceutical-gene networks (blue edges) (C) Purple edges represent overlapping in both IR and depression analysis.
Sequenome time-of-flight mass spectrometry
We further studied at the base-pair resolution the DNA methylation patterns in the genomic regions of VDR gene by MALDI-TOF MS test. Using annotation of two probes cg02522757 (TSS200 regions) and cg13556224 (TSS1500 regions) of VDR, we designed a MALDI-TOF MS method to measure methylation level in a small clinical cohort, which includes patients diagnosed with T2D and matched controls.
In the amplicon-cg02522757 regions, we identified six units with significant differences in DNA methylation values between patients with T2D vs. healthy controls (Fig. 3). In the amplicon-cg13556224 region, one site showed significant difference (P < 0.05, data not shown).
Figure 3.
Methylation level change of six units in amplicon-cg02522757 between patients with T2D and healthy controls.
Type 2 diabetes risk prediction model
We constructed a risk prediction model to classify patients with T2D vs. normal controls using the available methylation data from six units in the amplicon-cg02522757 and one unit in amplicon-cg13556224 region. A Logistic Regression model was constructed, and the performance was validated by Leave One Out Cross Validation (LOOCV).
First, a prediction model based on six units in amplicon-cg02522757 was constructed. The optimal prediction performance was AUC = 0.893 (odds ratio = 15.2). The sensitivity of the model was 0.884, when the specificity was 0.889 (Fig. 4A). Then, we updated the model with the additional input of one unit in the amplicon-cg13556224. The optimal predictive performance achieved was AUC = 0.902 (odds ratio = 19.76), and the sensitivity of the model was 0.837 when the specificity was 0.944 (Fig. 4B). The results showed the potential predictive ability of the VDR methylation level for T2D risk.
Figure 4.
T2D prediction model validation based on methylation level of amplicon-cg02522757 and amplicon-cg13556224 region in VDR. (A) ROC of T2D predictive model-1 based on amplicon-cg02522757 (AUC = 0.893, and OR = 15.2). (B) ROC of T2D predictive model-2 based on the amplicon-cg02522757 and amplicon-cg13556224 (AUC = 0.902, and OR = 19.76).
Discussion
A previous study suggested that longitudinal research is needed to identify risk factors and mechanisms for depression in patients with diabetes, particularly in early stages.14 Our preliminary results presented here demonstrated that IR and depression share a common nutraceutical-gene network module, and the center genes in this subnetwork could predict the risk of T2D. This observation highlights a potential approach to prevent the onset of T2D by identify high-risk individuals and intervene with nutraceutical, which targets the core genes. For example, if the VDR status could be monitored routinely, the individual with a high risk for T2D could test their vitamin D level regularly and take nutraceuticals to target the VDR protein as a potential preventative method. Considering that the single gene test based on MALDI-TOF MS is considerably cheaper than Illumina DNA methylation microarray, this interactive detection-prevention measure could be adopted in the clinic. Thus, larger prospective studies are needed to confirm these findings in future studies.
In this study, we focused on constructing a bioinformatical analysis pipeline and performed a preliminarily validation to prove the feasibility of our strategy. The criterion we chose to select candidate gene was as follows. First, we selected genes based on the nutraceutical-gene network analysis (Fig. 2C). We considered a gene as candidates not only for its phenotype association, but also for its nutraceutical preventing worth. Thus, we mapped the IR and depression association genes in the gene-nutraceutical connectivity network, and integrate the gene-phenotype and gene-nutraceutical information by overlapping the nutraceutical-gene network for IR and depression (Fig. 2C). Second, by searching the hub in the IR-depression overlapped network, we selected genes with the most connected nutraceuticals, which means the gene was more valuable for further clinical studies both in T2D risk monitoring and potential nutraceutical preventative applications. Thus, we focused on VDR gene for the latter clinical cohort validation.
A recent study identified VDR as a critical modulator of inflammation and β-cell survival.20 β-cell dysfunction due to inflammatory stress and IR is the primary cause of disease progression in patients with T2D. Vitamin D via VDR exerts a direct effect on the expression of several hundred target genes, implying numerous effects on the epigenome.21 It was hypothesized that vitamin D modulates the epigenome of immune cells during antigen perturbation and other immunological challenges, thus suggesting the importance of optimal vitamin D levels for effective epigenetic programming, in particular in the innate immune system.22 Some studies showed that the β-cells contain high levels of the vitamin D receptor; however, its role in maintaining the β-cell maturity is unclear. Further research is required to establish a link between β-cells and VDRs.23 Vitamin D increases glucose-stimulated insulin secretion from insulin-producing beta cells (INS1E).24 Moderate swimming exercise improved these consequences through modulation of vitamin D status. Future studies should be designed to investigate the effect of the combination of vitamin D intake with exercise in diabetic patients.25
One of the limitations of our method is, the DNA methylation microarray includes a limited number of probes in the key genomic regions of promoters. We will extend this method to identify a higher number of epigenetically regulated genes by exploring other high-throughput methods. Initially, an 850 k chip upgrade will provide more information on the DNA methylation regulation of the promoter and body region of genes. Since the price of second-generation sequencing is decreasing gradually, the base-pair resolution of NGS data on DNA methylation will empower the method to reveal more informative genes in T2D. Secondly, for the statistical analysis, our method is dependent on the t-test statistic of two distributions, one is the methylation distribution of promoter region probes, another is the methylation distribution of other region probes. More sensitive bioinformatics could be developed to identify more epigenetic regulated genes in the etiology of T2D.
Ethics statement
The institutional ethics committee of the First Affiliated Hospital of PLA General Hospital approved this study. The study was conducted following the ethical guidelines of the Declaration of Helsinki. Written informed consent was obtained from all participating patients before enrollment in the study.
Authors contribution
Fengji Liang and Yuan Quan conducted the data mining and bioinformatics analyses; Ying Chen and Andong Wu helped in preparing the clinical samples; Ruifeng Xu helped in drafting the manuscript; Yuexing Zhu and Jianghui Xiong designed the bioinformatic analysis; Fengji Liang, Yuan Quan, and Jianghui Xiong wrote the manuscript.
Conflict of Interests
The authors declare no conflict of interest.
Funding
This research was funded by the grants from the Science, Technology and Innovation Commission of Shenzhen Municipality [grant numbers JCYJ20151029154245758, CKFW2016082915204709].
Availability of data and materials
The datasets supporting the results of this article are included within the article. For the relevant raw data of clinical samples, we declare that materials described in this manuscript will be freely available to any scientist wishing to use them for non-commercial purposes, without breach of participant confidentiality.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.(Who) .W.H.O. Diabetes. http://www.who.int/diabetes/en/2018http://www.who.int/diabetes/en/
- 2.Cornelis M.C., Hu F.B. Gene-environment interactions in the development of type 2 diabetes: recent progress and continuing challenges. Annu Rev Nutr. 2012;32:245–259. doi: 10.1146/annurev-nutr-071811-150648. [DOI] [PubMed] [Google Scholar]
- 3.Nasri H., Baradaran A., Shirzad H., Rafieian-Kopaei M. New concepts in nutraceuticals as alternative for pharmaceuticals. Int J Prev Med. 2014;5(12):1487–1499. [PMC free article] [PubMed] [Google Scholar]
- 4.Aliper A., Jellen L., Cortese F. Towards natural mimetics of metformin and rapamycin. Aging (N Y) 2017;9(11):2245–2268. doi: 10.18632/aging.101319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling C., Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58(12):2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gluckman P.D. Epigenetics and metabolism in 2011: epigenetics, the life-course and metabolic disease. Nat Rev Endocrinol. 2011;8(2):74–76. doi: 10.1038/nrendo.2011.226. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson E., Ling C. DNA methylation links genetics, fetal environment, and an unhealthy lifestyle to the development of type 2 diabetes. Clin Epigenetics. 2017;9 doi: 10.1186/s13148-017-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toperoff G., Aran D., Kark J.D. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21(2):371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacos K., Gillberg L., Volkov P. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun. 2016;7 doi: 10.1038/ncomms11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crujeiras A.B., Diaz-Lagares A., Sandoval J. DNA methylation map in circulating leukocytes mirrors subcutaneous adipose tissue methylation pattern: a genome-wide analysis from non-obese and obese patients. Sci Rep. 2017;7 doi: 10.1038/srep41903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers J.C., Loh M., Lehne B. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3(7):526–534. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J., Goldberg J., Bremner J.D., Vaccarino V. Global DNA methylation is associated with insulin resistance: a monozygotic twin study. Diabetes. 2012;61(2):542–546. doi: 10.2337/db11-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arpon A., Milagro F.I., Ramos-Lopez O. Epigenome-wide association study in peripheral white blood cells involving insulin resistance. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-38980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moulton C.D., Pickup J.C., Ismail K. The link between depression and diabetes: the search for shared mechanisms. Lancet Diabetes Endocrinol. 2015;3(6):461–471. doi: 10.1016/S2213-8587(15)00134-5. [DOI] [PubMed] [Google Scholar]
- 15.Liang F., Lv K., Wang Y. Personalized epigenome remodeling under biochemical and psychological changes during long-term isolation environment. Front Physiol. 2019;10 doi: 10.3389/fphys.2019.00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quan Y., Liang F., Zhu Y. Integrated analysis of DNA methylation and biochemical/metabolic parameter during the long-term isolation environment. Front Physiol. 2019;10 doi: 10.3389/fphys.2019.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart D.S., Feunang Y.D., Guo A.C. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Li Y., Li W. Guide Positioning Sequencing identifies aberrant DNA methylation patterns that alter cell identity and tumor-immune surveillance networks. Genome Res. 2019;29(2):270–280. doi: 10.1101/gr.240606.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salas-Perez F., Ramos-Lopez O., Mansego M.L. DNA methylation in genes of longevity-regulating pathways: association with obesity and metabolic complications. Aging (N Y) 2019;11(6):1874–1899. doi: 10.18632/aging.101882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Z., Yoshihara E., He N. Vitamin D switches BAF complexes to protect beta cells. Cell. 2018;173(5):1135–1149. doi: 10.1016/j.cell.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlberg C. Molecular endocrinology of vitamin D on the epigenome level. Mol Cell Endocrinol. 2017;453:14–21. doi: 10.1016/j.mce.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Carlberg C. Vitamin D signaling in the context of innate immunity: focus on human monocytes. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.02211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neelankal John A., Jiang F.X. An overview of type 2 diabetes and importance of vitamin D3-vitamin D receptor interaction in pancreatic beta-cells. J Diabet Complicat. 2018;32(4):429–443. doi: 10.1016/j.jdiacomp.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Bornstedt M.E., Gjerlaugsen N., Pepaj M., Bredahl M.K.L., Thorsby P.M. Vitamin D increases glucose stimulated insulin secretion from insulin producing beta cells (INS1E) Int J Endocrinol Metabol. 2019;17(1) doi: 10.5812/ijem.74255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aly Y.E., Abdou A.S., Rashad M.M., Nassef M.M. Effect of exercise on serum vitamin D and tissue vitamin D receptors in experimentally induced type 2 Diabetes Mellitus. J Adv Res. 2016;7(5):671–679. doi: 10.1016/j.jare.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zannas A.S., Jia M., Hafner K. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-κB–driven inflammation and cardiovascular risk. Proc Natl Acad Sci U S A. 2019;116(23):11370–11379. doi: 10.1073/pnas.1816847116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quan Y., Liang F., Deng S.M., Zhu Y., Chen Y., Xiong J. Mining the Selective Remodeling of DNA Methylation in Promoter Regions to Identify Robust Gene-Level Associations With Phenotype. Front Mol Biosci. 2020;8 doi: 10.3389/fmolb.2021.597513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the results of this article are included within the article. For the relevant raw data of clinical samples, we declare that materials described in this manuscript will be freely available to any scientist wishing to use them for non-commercial purposes, without breach of participant confidentiality.