Abstract
In the era of antibiotic resistance, in silico prediction of bacterial resistome profiles, likely to be associated with inactivation of new potential antibiotics is of utmost importance. Despite this, to the best of our knowledge, no tool exists for such prediction. Therefore, under the rationale that drugs with similar structures have similar resistome profiles, we developed two models, a deterministic model and a stochastic model, to predict the bacterial resistome likely to neutralize uncharacterized but potential chemical structures. The current version of the tool involves the prediction of a resistome for Escherichia coli and Pseudomonas aeruginosa. The deterministic model on omitting two diverse but relatively less characterized drug classes, polyketides and polypeptides showed an accuracy of 87%, a sensitivity of 85%, and a precision of 89%, whereas the stochastic model predicted antibiotic classes of the test set compounds with an accuracy of 72%, a sensitivity of 75%, and a precision of 83%. The models have been implemented in both a standalone package and an online server, uCAREChemSuiteCLI and uCARE Chem Suite, respectively. In addition to resistome prediction, the online version of the suite enables the user to visualize the chemical structure, classify compounds in 19 predefined drug classes, perform pairwise alignment, and cluster with database compounds using a graphical user interface.
Availability
uCARE Chem Suite can be browsed at: https://sauravsaha.shinyapps.io/ucarechemsuite2/, and uCAREChemSuiteCLI can be installed from:
1. CRAN (https://cran.r-project.org/package=uCAREChemSuiteCLI) and
2. GitHub (https://github.com/sauravbsaha/uCAREChemSuiteCLI).
Keywords: Drug resistance, Escherichia coli, Prediction, Pseudomonas aeruginosa, Resistome, uCARE chem suite, uCAREChemSuiteCLI
Introduction
The emergence and spread of antimicrobial resistant (AMR) bacterial strains pose a serious threat to the current health care system. To address this crisis, numerous national and international agencies, such as the Infectious Diseases Society of America (IDSA), the US Food and Drug Administration (FDA), the Centers for Disease Control and Prevention (CDC), the World Health Organization (WHO), and the European Medicines Agency (EMA), have taken initiatives to facilitate and incentivize novel antimicrobial discovery.1 Despite tangible progress in the development of novel antimicrobials, there is a pressing need to bring new antimicrobials into the clinical development process.1,2
The bacterial resistance phenotype can be attributed to one or a few resistance mechanisms. These strategies involve mechanisms such as inactivating the drug molecule through enzymatic degradation or modification of the drug scaffolds; altering or overexpressing of the drug target, reducing affinity of a drug towards the target; limiting entry of a drug inside the cell membrane through altering the permeability of the plasma membrane; and extrusion of a drug from the cell by efflux pumps.3 Mechanistic understanding of these resistance mechanisms for a specific drug or drug class is critical for accelerating the novel drug discovery process.4
Drugs with similarities in chemical structure, mechanism of action, and pharmacological effect are often used interchangeably (the drug class effect).5 Because cross-resistance to antibiotics within a class is a frequent phenomenon,6 we propose that the drug class effect is not limited to pharmacological properties but will also include the resistome profile, i.e., similar drug molecules will share a similar resistome profile.
With this rationale, an online suite and a command line R package, uCARE Chem Suite and uCAREChemSuiteCLI, respectively, were developed to predict the resistome profile of E. coli and P. aeruginosa likely to cause resistance against novel candidate drug(s). Here we introduce and discuss the architecture, usage, and utilities provided by the tool.
Materials and methods
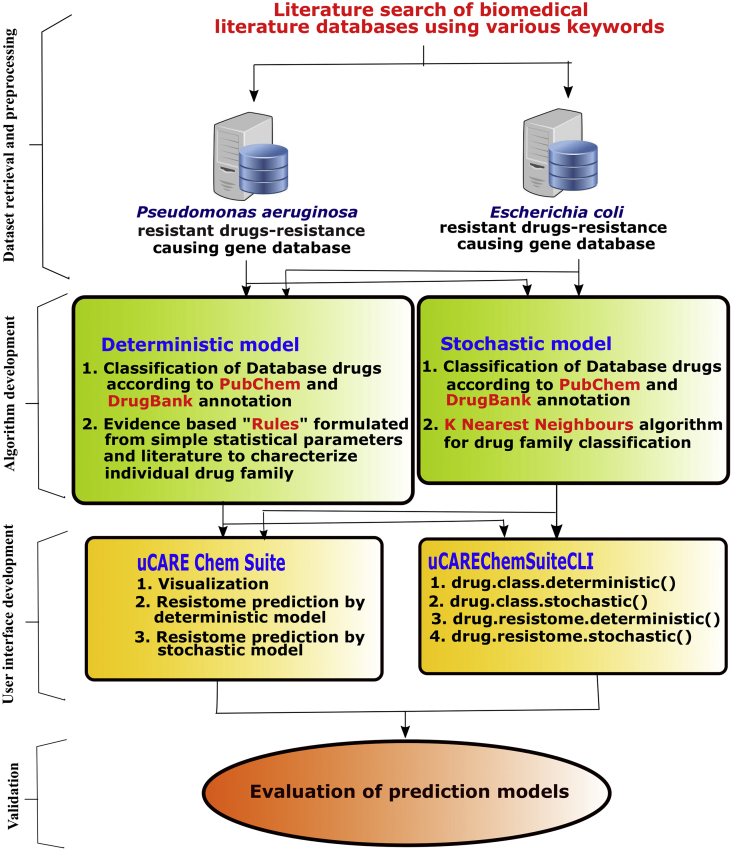
uCARE Chem Suite and uCAREChemSuiteCLI were entirely written in R. Several R packages—ChemmineR for drug(s) feature extraction and analysis;7 Shiny for uCARE Chem Suite's web implementation; devtools for uCAREChemSuiteCLI package building; and roxygen2 for documentation were also utilized. The workflow of the present work is presented in Fig. 1.
Figure 1.
The workflow of the present work.
Development of prediction models
Dataset retrieval and preprocessing
Escherichia coli and Pseudomonas aeruginosa were chosen because of the availability of data and their critical effects on human health.8 A keyword search on literature databases, including PubMed and PubMed Central and other meta search engines, such as Google, was carried out to create a database of drugs that have become ineffective due to antimicrobial resistance and of the genes involved in their resistance for both of the microorganisms. The keywords used were “antibiotic resistance in Escherichia coli/E. coli”, “multiple drug resistance in Escherichia coli/E. coli”, and “drug resistance genes in Escherichia coli/E. coli” for E. coli and “antibiotic resistance in Pseudomonas aeruginosa/P. aeruginosa”, “multiple drug resistance in Pseudomonas aeruginosa/P. aeruginosa”, and “drug resistance genes in Pseudomonas aeruginosa/P. aeruginosa” for P. aeruginosa. The articles were manually and exhaustively read to generate a database of drug specific resistome profiles for each of the two pathogens (Tables S1 and S2). This was followed by segregating the drugs according to their drug classes, as derived from PubChem and DrugBank annotations (Table 1). Drugs without well-defined class labels such as acriflavine, florfenicol were marked a separate class.
Table 1.
Classes of the database drugs derived from PubChem and DrugBank annotations.
| Drug class names | |
|---|---|
| Acriflavinea | Florfenicola |
| Aminocoumarinsb | Fluoroquinolonesb |
| Aminoglycosidesb | Nitrofuransb |
| Aminoquinonesb | Peptide drugsb |
| Anisolesb | Polyketidesb |
| Anthracyclinesb | Pyridopyrimidinesb |
| Benzalkoniuma | Quinolonesb |
| Beta lactamsb | Rhodaminea |
| Chloramphenicola | Sulfonamidesb |
| Drug cocktailc | Thiolactomycina |
Drug.
Drug class.
Drug group.
Based on the assumption that drugs with a similar structure (drug class) will have a similar resistome, the goal of the study was to predict the class of a candidate drug structure and to subsequently extract and assign resistance-associated genes from the database to the predicted drug class. Therefore, to determine the class of an unknown candidate drug, two algorithms, a deterministic model and a stochastic model, were developed.
Building the deterministic model
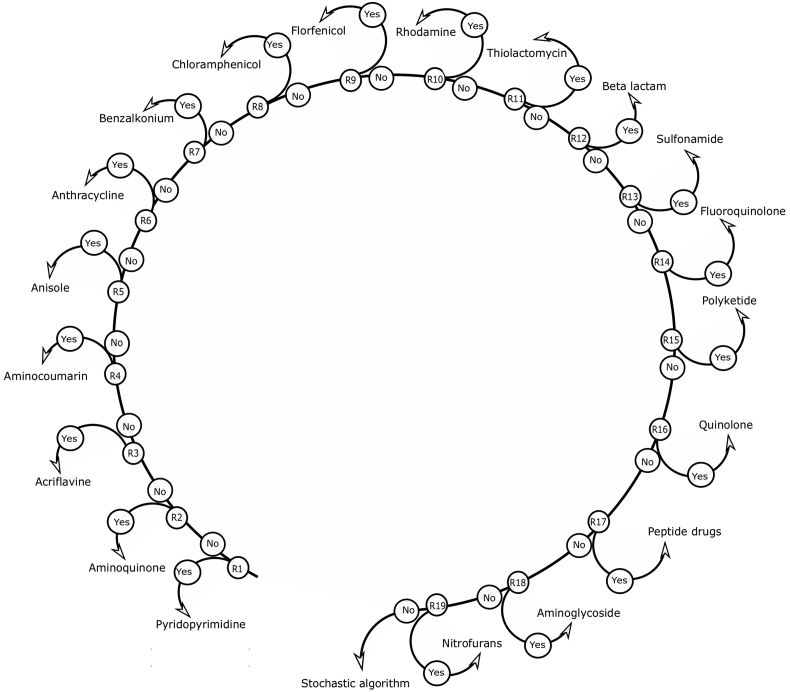
The goal of developing the deterministic model was to pin down structural and chemical features that are unique and specific to a drug family either through published biomedical literature or by using simple statistical parameters, such as the central tendency of different atoms. For example, any compound with a β-lactam ring, i.e., any four-membered aliphatic ring with 3 carbon atoms and 1 nitrogen atom, is likely to be a β-lactam drug.9 In this way, drug classes were manually and exhaustively studied, and statistical parameters were evaluated to formulate a set of evidence-based rules (Table S3) nested in various if-else conditions (Fig. 2) to characterize drug class of an unknown compound. The diversity in chemical structures in the class “drug cocktails” is huge, so finding class-specific features was not feasible for this present work. Therefore, the drug cocktail class was omitted when developing the deterministic model.
Figure 2.
Algorithm based on an evidence-based deterministic model to predict the drug class of the chemical compound.
Bias in the prediction model due to underrepresentation of drugs in few drug classes (Tables S1 and S2) led to the development of a stochastic model.
Building the stochastic model
The stochastic model we developed facilitated and provided a good fit for the prediction of drug classes that were either not well characterized or marginally represented in the database. To predict the drug class, we utilized the k-nearest neighbor algorithm, in which the distance between neighbors was determined by the Tanimoto similarity scores between the query compound and the database compounds. The Tanimoto index was chosen because of its simplicity and credibility as a coefficient of choice for computing molecular similarities.10 The Tanimoto coefficient as a similarity score for compound A and B can be defined as:
| Tanimoto coefficient, SA,B = c/[a + b +c], |
The variable c is the number of atom pairs common to both compounds, while a and b are the numbers of their unique atom pairs.
Evaluation of the models
To evaluate the models, a test set of data was generated by downloading the top 100 SDF (structure-data file) structures from PubChem with the keyword “antibiotic”. The classes of these drugs were then determined through PubChem and Drugbank annotations. Of these 100 drugs, 24 were omitted due to lack of their drug class information. Furthermore, accuracy, sensitivity, and precision were calculated to evaluate the drug classification quality of the models. These measures of assessment were defined with the following formulas:
| Accuracy = (TP + TN)/(TP + TN + FP + FN) |
| Recall (Sensitivity) = TP/(TP + FN) |
| Precision = TP/(TP + FP) |
where TP is true positive, TN is true negative, FP is false positive, and FN is false negative.11
User interface for the prediction algorithms
uCARE Chem Suite
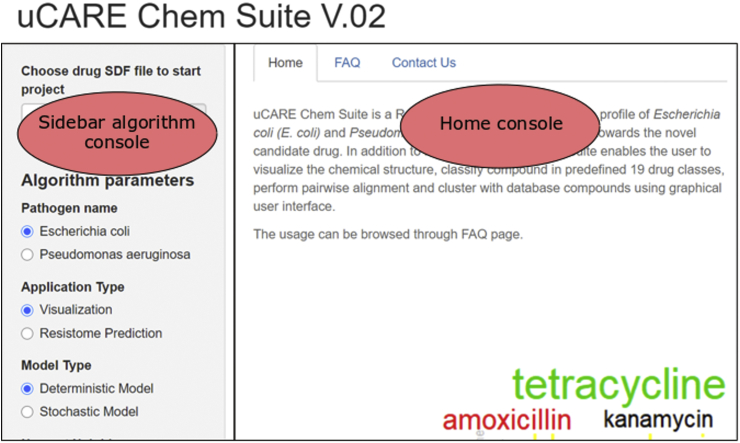
uCARE Chem Suite is a cross-platform online tool with a user-friendly graphical user interface for researchers without any previous programming experience. The dashboard of the tool has two console columns, a sidebar algorithm console and a visualization console (Fig. 3).
Figure 3.
Home page of uCARE Chem Suite.
The sidebar algorithm console comprises sections for uploading the input file, menus for selecting the study organism and prediction algorithm, and other clickable menus for optimizing the prediction models. The home console hosts the home page, which briefly describes the tool, links to frequently asked questions (FAQ), and links to contact information.
The input needed for the tool is a file with atoms, bonds, connectivity, and coordinates of the candidate drug molecule in the SDF format. uCARE Chem Suite's interface, once the input file is submitted, is divided into three sections: (a) visualization, (b) resistome prediction by deterministic model, and (c) resistome prediction by stochastic model.
-
a.
Visualization
The visualization section provides annotations about the drug structure via three tabs, “Chemical Properties”, “Atomic Properties”, and “Bond Attributes”.
The Chemical Properties tab links to information such as chemical formula, molecular weight, and structural formula; the Atomic Properties tab links to Atom block [Specifies the atomic symbol and any mass difference, charge, stereochemistry, and associated hydrogens for each atom] and Bond block [Specifies the two atoms connected by the bond, the bond type, and any bond stereochemistry and topology (chain or ring properties) for each bond] information12; ring attributes, such as the total number of rings, ring type, ring structure, and bond matrix can be accessed from the Bond Attributes tab.
-
b.
Resistome prediction by the deterministic model
When selecting resistome prediction using the deterministic model, the user is provided with the class of drug molecule predicted by the exhaustive rule-based deterministic model via the Drug Classification tab, a list of database drugs from the same family via the Nearest Drug/s tab, and the resistome profile via the Resistance Gene List tab. The user also has the ability to collate and download the results via the Export/Downloads tab.
-
c.
Resistome prediction by stochastic model
Resistome prediction by the stochastic model provides the following tabs: (i) Drug Classification, (ii) Database Query Search, (iii) Query-DB Clustering, and (iv) Database.
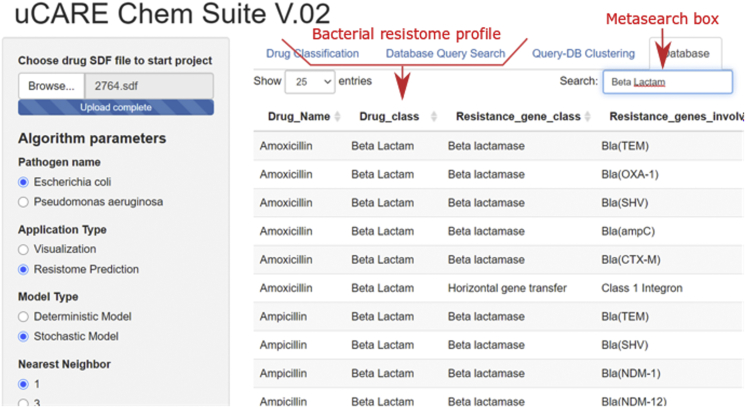
The Drug Classification tab provides the drug class predicted by the stochastic model. The Database Query Search tab provides a report of aligment between the query compound structure and all the database compound structures. The alignment report consist of database drug compounds ranked in descending order by their Tanimoto similarity scores. The Query-DB Clustering tab allows the clustering of a query compound with database compounds either by atom pair descriptor similarity or by fingerprint similarity scores; and allows plotting of clustering results in tree or circular plots. The Database tab provides access to the complete resistome database for E. coli and P. aeruginosa by selecting either Escherichia coli or Pseudomonas aeruginosa in the Pathogen name section of the sidebar console. Furthermore, the determination of a resistome in the stochastic model can be carried out by submitting an antibiotic-class name into the metasearch box provided in the Database tab (Fig. 4).
Figure 4.
Meta search box of Database tab.
uCAREChemSuiteCLI
uCAREChemSuiteCLI is a command line R package and can be easily installed from an R terminal. It has been developed and distributed with an MIT license so that the researchers can use, reuse, modify, and integrate the package into their own software. The package consists of four functions, viz., drug.class.deterministic() and drug.class.stochastic() for drug class prediction, whereas drug.resistome.deterministic() and drug.resistome.stochastic() can be utilized for resistome prediction.
The package can either be installed from the CRAN or the GitHub repositories. A package manual, README.md, has been provided in the GitHub repository to instruct the user in step-by-step package installation, usage descriptions, and examples of specific functions.
Results and discussion
Resistome databases
The aim of the current work was to establish the relationship between drug chemical structures and resistance-associated genes by analyzing previously reported drug-specific resistome profiles. A literature search of biomedical literature databases resulted in two distinct resistome datasets, a dataset for Escherichia coli with 152 data points from 37 articles (Table S1) and a dataset for Pseudomonas aeruginosa with 122 data points from 29 articles (Table S2). Further manual annotation of the resistome profile information from the literature in DrugBank and PubChem generated two distinct databases for individual organisms, each of which contained 6 fields: drug name, drug class, drug PubChem ID, resistance gene class, resistance genes involved, and bibliography (PMID).
The E. coli database consisted of 56 antibiotics, 18 characterized antibiotic classes, 12 resistance mechanism/gene classes, and 64 unique genes/mutations associated with E. coli antibiotic resistance [Fig. S1-S4]; the P. aeruginosa database contained 48 antibiotics, 11 characterized antibiotic classes, 8 resistance mechanism/gene classes, and 37 unique genes/mutations associated with P. aeruginosa antibiotic resistance [Fig. S5-S8].
Tetracycline and trimethoprim were the two most frequently reported antibiotics in the E. coli database, whereas imipenem and meropenem were the most frequent antibiotics in the P. aeruginosa database. Although they are a last resort for many other Gram-negative bacterial infections, antibiotics such as imipenem and meropenem in the carbapenem class of antibiotics are considered the first line of defense against P. aeruginosa infections.13 Multiple reports of carbapenems in the non-redundant resistome profile database are indicative of an alarming development of multiple mechanisms of resistance against carbapenems in P. aeruginosa.
In addition to these drugs, which were highly represented, the database also contained drugs with scarce data. As many as 22 drugs from E. coli and 26 drugs from P. aeruginosa had only one gene report in the database, which indicates substrate specificity of genes towards drugs.
Since Fleming's landmark discovery of penicillin, the most common antibiotic class prescribed against bacterial infections globally remains the β-lactams, and their global sales account for billions of US dollars.14, 15, 16 Despite the huge representation of tetracycline and trimethoprim in the E. coli database, the β-lactam class was found to be the most frequent class in both databases. The ubiquitous presence of β-lactams in the database is likely a reflection of the fact that they are the preferred prescription against bacterial infections.
Outer membrane proteins and efflux pumps in the database constituted more than 50% of the P. aeruginosa resistance-associated gene data, indicating the significance of these mechanisms in the antibiotic resistance of P. aeruginosa.17,18 New Delhi Metallo-β-lactamase (NDM) was the most prominent gene in the E. coli resistance gene database and was represented in 25% of the data in the entire database. The voluminous NDM data in the database most likely can be attributed to the rapid dissemination of this gene across the globe.19, 20, 21, 22, 23 It is worth noting that, since the emergence of NDM, the majority of NDM gene detection studies have been limited to bacterial strains in the Enterobacteriaceae family.
Despite the identification of NDM-containing P. aeruginosa strains in Serbia, France, India, Singapore, and North America, no direct NDM-associated carbapenem resistance has been reported in P. aeruginosa.24, 25, 26, 27, 28 In addition to the rarity of the NDM-containing P. aeruginosa reports,29 the study by Shanthi et al (2014) found that NDM-1 was not a major mechanism of carbapenem resistance in P. aeruginosa.26 However, recently Ding et al (2018) demonstrated an increase of meropenem minimum inhibitory concentration (MIC) in an NDM-acquired P. aeruginosa strain.13 The therapeutic options against P. aeruginosa are already limited. It will be no less than an “antibiotic apocalypse” if NDM genes further evolve in P. aeruginosa.
Assessment of the deterministic model
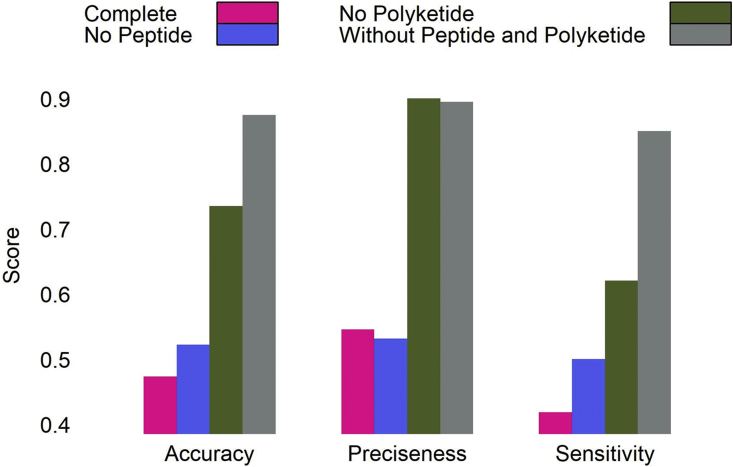
The goal behind developing a deterministic model was to determine the physiochemical properties unique to each class. However, it is evident from our results that generalization is not possible when classes as diverse as polyketides and polypeptides are underrepresented in the database.
Therefore, by omitting these classes, the assessment measures drastically improved from a complete set accuracy of 47%, a sensitivity of 42%, and a precision of 54% to an accuracy of 87%, a sensitivity of 85%, and a precision of 89% (Fig. 5).
Figure 5.
Accuracy, precision, and sensitivity of a deterministic model for a complete test set, a test set without peptide compounds, a test set without polyketide compounds, and a test set without polyketide and peptide compounds.
Assessment of the stochastic model
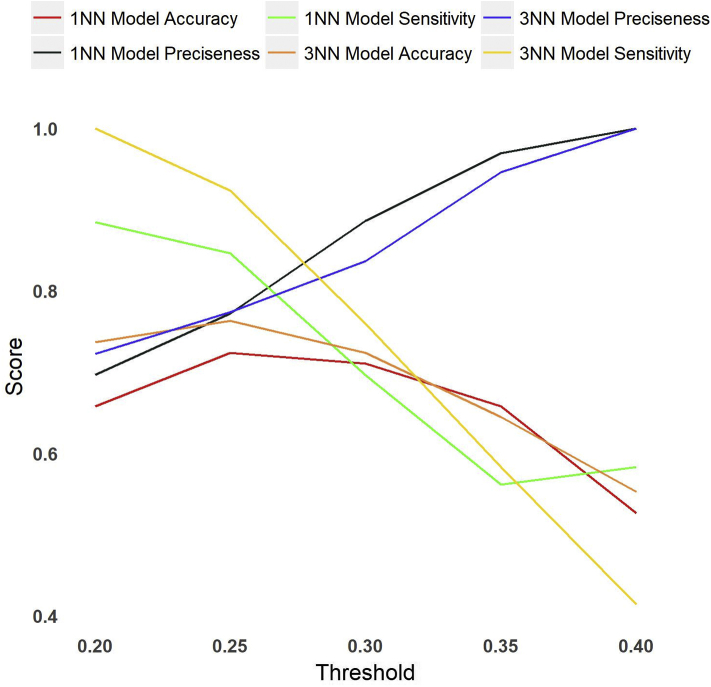
Iterative predictions were carried out with different parameters, i.e., nearest neighbor values of 1 and 3 and threshold similarity scores of 0.2, 0.25, 0.3, 0.35, and 0.4, to evaluate the quality of the stochastic model using different parameters. Assessment of the stochastic model showed a nearest neighbor value of 3 with a threshold similarity score of 0.3 to be our optimal parameter set for drug class prediction. The model predicted classes of test set drug molecules with an accuracy of 72%, a precision of 83%, and a sensitivity of 75% (Fig. 6).
Figure 6.
Accuracy, precision, and sensitivity of the stochastic model using different parameters, i.e., nearest neighbor values of 1 and 3 and threshold similarity scores of 0.2, 0.25, 0.3, 0.35, and 0.4.
Application, limitations, and future developments
Application
The motivation behind the present work was to develop a platform to facilitate collaborative research among researchers from various fields of science and to contribute a tool that could be used against the menace of antimicrobial drug resistance. Though the current version of the tool addresses the need for computational chemistry for experimental researchers working on microbiological aspects, it also opens avenues for bioinformaticians with expertise in structural modeling and pathway biology. Two of the research directions that the current tool will aid are described below.
Prediction of the resistome for candidate compounds
In a study, siderophore sulfactam BAL30072 was proposed to have antimicrobial properties against Gram-negative bacteria. Antimicrobial activity of this compound was compared with many other drugs, and its activity was checked against different resistance-associated genes, specifically β-lactamases and two component systems.30
Prediction of the resistome will broaden the understanding of the bacterial response to novel candidate compounds by providing a comprehensive list of genes that are likely to cause resistance. In addition, it will provide an opportunity for bioinformaticians with expertise in pathway biology to deduce the dynamics of bacterial response holistically, enabling them to develop and analyze pathway models of bacterial resistomes.
Antimicrobial resistance surveillance system
Reports of NDM in P. aeruginosa are scarce. However, Ding et al (2018) demonstrated that the alarming spread of NDM in P. aeruginosa is not in the distant future.13 As the back-end database is regularly updated, epidemiologists can utilize the database to carry out AMR-associated epidemiological studies.
Limitations and future development
The caveat for the current work was the low accuracy of the classifiers, causing a generalization error. This was mainly due to unbalanced data. Therefore, the immediate focus of tool development will be to address the generalization error. In addition, steps will be taken to increase the applicability of the tool by augmenting the back-end database, incorporating pathway information, embedding docking and virtual screening tools, and developing graphic user interface (GUI)-based standalone software.
Conclusion
We believe that implementing efficacy tests of compounds against their resistome space in the early phases of the traditional clinical trial pipeline will significantly curtail the cost and time of the drug discovery process. The present version of uCARE Chem Suite and uCAREChemSuiteCLI represents an attempt to utilize the relationship between drug chemistry and the resistome (chemical resistomics) to predict the E. coli and P. aeruginosa resistome. We expect that this tool will facilitate the pumping of potential novel drug compounds into clinical trial pipelines.
Conflict of Interests
None to declare.
Acknowledgements
The authors acknowledge the support of the Department of Computational Biology and Bioinformatics, Sam Higginbottom University of Agricultural, Technology and Sciences, Prayagraj, Uttar Pradesh, India. The authors would also like to thank Dr. Vipul Gupta for his technical help and Dr. Bruce Barron, Dr. Edmund F. and Dr. Rhoda E. Perozzi for English manuscript editing and proofreading.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.06.008.
Contributor Information
Saurav Bhaskar Saha, Email: saurav.saha@shiats.edu.in.
Vijai Kumar Gupta, Email: vijaifzd@gmail.com.
Pramod Wasudeo Ramteke, Email: pramod.ramteke@shiats.edu.in.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Talbot G.H., Jezek A., Murray B.E. The infectious Diseases society of America's 10 × '20 initiative (10 new systemic antibacterial agents US Food and drug administration approved by 2020): is 20 × '20 a possibility? Clin Infect Dis. 2019;69(1):1–11. doi: 10.1093/cid/ciz089. [DOI] [PubMed] [Google Scholar]
- 2.Simpkin V.L., Renwick M.J., Kelly R., Mossialos E. Incentivising innovation in antibiotic drug discovery and development: progress, challenges and next steps. J Antibiot (Tokyo) 2017;70(12):1087–1096. doi: 10.1038/ja.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crofts T.S., Gasparrini A.J., Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol. 2017;15(7):422–434. doi: 10.1038/nrmicro.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y., Ding S., Shen J., Zhu K. Nonribosomal antibacterial peptides that target multidrug-resistant bacteria. Nat Prod Rep. 2019;36(4):573–592. doi: 10.1039/c8np00031j. [DOI] [PubMed] [Google Scholar]
- 5.Soares I., Carneiro A.V. Drug class effects: definitions and practical applications. Rev Port Cardiol. 2002;21(9):1031–1042. [PubMed] [Google Scholar]
- 6.Fields F.R., Lee S.W., McConnell M.J. Using bacterial genomes and essential genes for the development of new antibiotics. Biochem Pharmacol. 2017;134:74–86. doi: 10.1016/j.bcp.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y., Charisi A., Cheng L., Jiang T., Girke T. ChemmineR: a compound mining framework for R. Bioinformatics. 2008;24(15):1733–1734. doi: 10.1093/bioinformatics/btn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2017. https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/
- 9.Neu H.C. Beta-Lactam antibiotics: structural relationships affecting in vitro activity and pharmacologic properties. Rev Infect Dis. 1986;8(Suppl 3):S237–S259. doi: 10.1093/clinids/8.supplement_3.s237. [DOI] [PubMed] [Google Scholar]
- 10.Bajusz D., Rácz A., Héberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J Cheminf. 2015;7 doi: 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Šimundić A.M. Measures of diagnostic accuracy: basic definitions. EJIFCC. 2009;19(4):203–211. [PMC free article] [PubMed] [Google Scholar]
- 12.Dalby A., Nourse J.G., Hounshell W.D. Description of several chemical structure file formats used by computer programs developed at Molecular design limited. J Chem Inf Comput Sci. 1992;32(3):244–255. [Google Scholar]
- 13.Ding Y., Teo J.W.P., Drautz-Moses D.I., Schuster S.C., Givskov M., Yang L. Acquisition of resistance to carbapenem and macrolide-mediated quorum sensing inhibition by Pseudomonas aeruginosa via ICETn43716385. Commun Biol. 2018;1 doi: 10.1038/s42003-018-0064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mölstad S., Lundborg C.S., Karlsson A., Cars O. Antibiotic prescription rates vary markedly between 13 European countries. Scand J Infect Dis. 2002;34(5):366–371. doi: 10.1080/00365540110080034. [DOI] [PubMed] [Google Scholar]
- 15.Crowder M.W., Spencer J., Vila A.J. Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc Chem Res. 2006;39(10):721–728. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- 16.Thakuria B., Lahon K. The beta lactam antibiotics as an empirical therapy in a developing country: an update on their current status and recommendations to counter the resistance against them. J Clin Diagn Res. 2013;7(6):1207–1214. doi: 10.7860/JCDR/2013/5239.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pechère J.C., Köhler T. Patterns and modes of beta-lactam resistance in Pseudomonas aeruginosa. Clin Microbiol Infect. 1999;5:S15–S18. doi: 10.1111/j.1469-0691.1999.tb00719.x. [DOI] [PubMed] [Google Scholar]
- 18.Aparna V., Dineshkumar K., Mohanalakshmi N., Velmurugan D., Hopper W. Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation. PloS One. 2014;9(7) doi: 10.1371/journal.pone.0101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yong D., Toleman M.A., Giske C.G. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrestha B., Tada T., Miyoshi-Akiyama T. Identification of a novel NDM variant, NDM-13, from a multidrug-resistant Escherichia coli clinical isolate in Nepal. Antimicrob Agents Chemother. 2015;59(9):5847–5850. doi: 10.1128/AAC.00332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soliman A.M., Khalifa H.O., Ahmed A.M., Shimamoto T., Shimamoto T. Emergence of an NDM-5-producing clinical Escherichia coli isolate in Egypt. Int J Infect Dis. 2016;48:46–48. doi: 10.1016/j.ijid.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Chen D., Gong L., Walsh T.R. Infection by and dissemination of NDM-5-producing Escherichia coli in China. J Antimicrob Chemother. 2016;71(2):563–565. doi: 10.1093/jac/dkv352. [DOI] [PubMed] [Google Scholar]
- 23.Li X., Fu Y., Shen M. Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob Resist Infect Contr. 2018;7 doi: 10.1186/s13756-018-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jovcic B., Lepsanovic Z., Suljagic V. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother. 2011;55(8):3929–3931. doi: 10.1128/AAC.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flateau C., Janvier F., Delacour H. Recurrent pyelonephritis due to NDM-1 metallo-beta-lactamase producing Pseudomonas aeruginosa in a patient returning from Serbia, France, 2012. Euro Surveill. 2012;17(45) [PubMed] [Google Scholar]
- 26.Shanthi M., Sekar U., Kamalanathan A., Sekar B. Detection of New Delhi metallo beta lactamase-1 (NDM-1) carbapenemase in Pseudomonas aeruginosa in a single centre in southern India. Indian J Med Res. 2014;140(4):546–550. [PMC free article] [PubMed] [Google Scholar]
- 27.Teo J.W., La M.V., Jureen R., Lin R.T. Emergence of a New Delhi metallo-β-lactamase-1-producing Pseudomonas aeruginosa in Singapore. Emerg Microb Infect. 2015;4(11) doi: 10.1038/emi.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mataseje L.F., Peirano G., Church D.L., Conly J., Mulvey M., Pitout J.D. Colistin-nonsusceptible Pseudomonas aeruginosa sequence type 654 with blaNDM-1 arrives in North America. Antimicrob Agents Chemother. 2016;60(3):1794–1800. doi: 10.1128/AAC.02591-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dortet L., Poirel L., Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed Res Int. 2014;2014 doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page M.G.P., Dantier C., Desarbre E. In vitro properties of BAL30072, a novel siderophore sulfactam with activity against multiresistant gram-negative bacilli. Antimicrob Agents Chemother. 2010;54(6):2291–2302. doi: 10.1128/AAC.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.