Abstract
The important role of lncRNAs and miRNAs in directing immune responses has become increasingly clear. Recent evidence conforms that miRNAs and lncRNAs are involved in NK cell biology and diseases through RNA–protein, RNA–RNA, or RNA–DNA interactions. In this view, we summarize the contribution of miRNAs and lncRNAs to NK cell lineage development, activation and function, highlight the biological significance of functional miRNAs or lncRNAs in NKTL and discuss the potential of these miRNAs and lncRNAs as innovative biomarkers/targets for NKTL early diagnosis, target treatment and prognostic evaluations.
Keywords: Activation, Development, Function, lncRNAs, miRNA, NK cell Biology, NK/T-cell lymphoma
Introduction
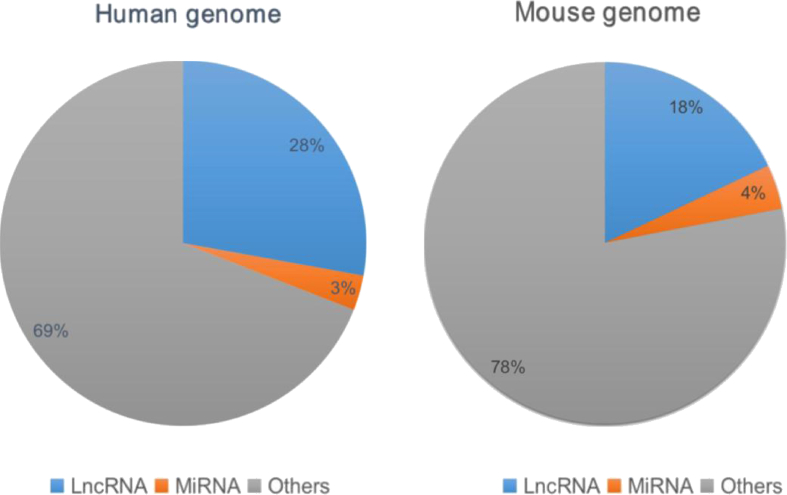
Non-coding RNAs are an important family in human genome and are broadly classified as lncRNAs, miRNAs, snoRNAs, snRNAs and short non-coding RNAs according to the length of the nucleotide. According to the latest GENCODE release (version 33) or (version M24) (http://www.gencodegenes.org), it indicates the abundance of lncRNA and miRNA in human or mouse. (Fig. 1). For miRNAs, they are 23 nucleotides shorter, are best characterized to introduce mRNA degradation or block mRNA translation through deadenylation and decapping methods. Vasudevan et al has confirmed that some miRNAs increase protein expression via binding to the 5′UTR, but it remains unclear whether this is widely applicable.1 The molecular mechanism of miRNAs are clearer than before and there are some classified reviews of miRNAs.2, 3, 4 For lncRNAs, they are longer than 200 nucleotides and another important non-coding member. Compared with mRNA, lncRNAs are usually shorter in length, contain fewer exons and are less conservative–only about 12% of lncRNAs can be found in other organisms, other than humans.5 Meanwhile, lncRNAs have precise tissue specificity and developmental stage-specific manner.5,6 According to their genomic localization with respect to the neighboring protein coding gene, lncRNAs are classified into five types: intronic lncRNA; antisense lncRNA; long intergenic non-coding RNA (lncRNA); sense lncRNA; bidirectional lncRNA. In fact, some lncRNAs do not belong to any of the above classifications. They often have two or more characteristics, which makes them more difficult for the classification and identification of lncRNAs. It has been found that lncRNAs mediate gene activation through multiple mechanisms: in the nucleus, lncRNAs directly regulate gene expression via cis-/trans-action or as protein bait for indirect regulation; in the cytoplasm, lncRNAs regulate gene expression by affecting mRNA stability and translation process or involvement in miRNA regulation as competitive endogenous RNA or binding with transcription factors to affect gene function.7, 8, 9 At present, we know more about how miRNAs and lncRNAs work and further confirm that various functional miRNAs and lncRNA play key roles in the innate or adaptive immune system.5,7 However, various secrets in lncRNAs are still unclear, such as the number and species of functional lncRNAs, the function of immune-related lncRNAs in vivo, the role of lncRNAs in different kinds of immune cells, like T, B, NK cells and so on.
Figure 1.
Abundance of lncRNA and miRNAs genes. The data respectively represents the latest GENCODE release (Human genome version 33 vs. Mouse genome version 24): the number of lncRNA gene:16,892(human) vs. 9959(mouse); the number of miRNA gene:1881 (human) vs. 2202 (mouse). (http://www.gencodegenes.org).
NK cells are an important kind of lymphocyte to defense against bacteria, viruses and tumors in human body. There are quite more activating and inhibitory receptors expression on NK cell surface. The balance of these two types of receptors regulates the activation of NK cells. Besides these, there are also some efficient molecules, such as granzyme B, perforin, IFN-γ, TNF-α in NK cells, which cause target cell lysis and death.10,11 For example, following activation by bacterial lipopolysaccharide (LPS), NK cells inhibit pathogen infection via the cytokines secretion to kill the cells infected with pathogen. Meanwhile, NK cells also secrete soluble factors such as IL-3, GM-CSF and M-CSF to recruit neutrophils, macrophages and dendritic cells to activate the adaptive immune response.12,13 Therefore, NK cells play an critical role in innate immunity or acquired immunity.
For NK cells, recent studies have focused on the function of receptors, cytotoxic molecules, transcriptional factors involved in immune response. With the study of miRNA or lncRNAs, there is accumulating evidence shows that miRNA or lncRNAs are involved in various innate or adaptive immune response.14, 15, 16, 17 For example, lnc-DC controls the development and differentiation of DCs by directly binding to STAT3 in the cytoplasm18; miRNA-17-92 cluster directly mediate the differentiation of the naïve CD4+ T cells into Th1, Th2, Th17, Treg and Tfh.19 In addition, increasing number of publications showed the critical effect of non-coding RNAs in NK cells, especially lncRNAs and miRNAs. Though the role of miRNAs in NK cells have been recently reviewed, new advance on the relationship between NK and miRNAs or lncRNA are rarely been summarized and reported. Here, we summarize NK cell related miRNAs and lncRNAs from the latest researches and discuss their critical roles in NK cell development, activation and function, with a focus on abnormal miRNAs and lncRNAs in NKTL. As our knowledge of the molecular programs, including miRNAs and lncRNAs, that regulate NK cells increases, this will lead to identification of novel molecules and pathways that may be manipulated to enhance or attenuate NK cell function and NKTL early diagnosis, target treatment and prognostic evaluations.
Regulation of lncRNAs and miRNAs in the development of NK cell
Murine & human NK cell development
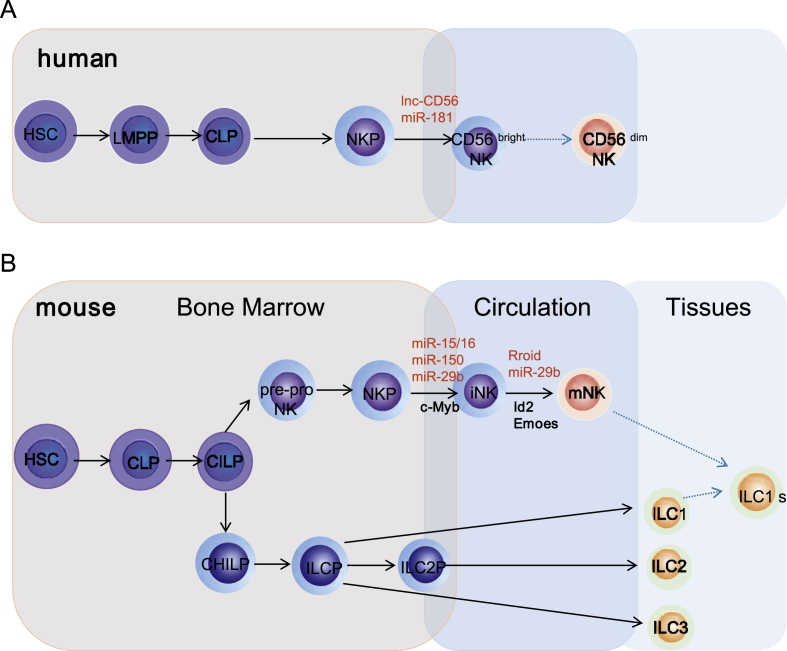
In the last decades, there has been two major advanced researches in NK cells, including the diversity of NK cell receptors and the diversity of NK cell function. Both the activity and function of NK cells are close to the development and differentiation of NK cells. The maturation of murine NK cells is a continuous process; according to their phenotypic and functional characteristics, NK cells into two different development stages, in first stage: hematopoietic stem cells (HSC) differentiated into common lymphoid precursors (CLP), and then differentiated into the earliest precursor of NK cells, namely pre-pro NK cells. During the process, some key transcription factor, such as PU.1,20,21 ETS-422 are involved in the development of NK cells by regulating the generation of pre-pro NK cells. And next, pre-pro NK cells up-regulate the expression of CD122 (IL-2Rβ) and further develop into NK cell precursors (NKP), which is mainly regulated by transcription factor E4BP4.23 In second stage: accepting IL-15 stimulus NKPs differentiate to immature NK cells (iNK), which relies on the expression of transcription factor TOX,24 T-bet.25 And then iNKs receive MHC-I domestication signals and finally develop into mature NK cells (mNK), this needs various transcription factor, such as Id2,26 Blimp-1,27,28 Emoes,29 Aiolos,30 Rfx7.31 Generally, the traditional development of NK cells is quite clear as introduced above. However, the emergence of innate lymphoid cells (ILC) presents a new challenge to the definition of traditional NK cells and their precursors. The new study suggests that CLPs will further differentiate into the common ILC precursors (CILP). And CILPs further develops into the common helper-like ILC precursors (CHILP) and NKPs.32 Since then, the relationship between pre-pro NK cells and CILPs becomes more complicated, and that between ILC and traditional NK cell development intermediates, which need to be confirmed and differentiated by further study. (Fig. 2A).
Figure 2.
The key lncRNA and miRNA involved in developmental origin of murine and human NK cells. A.In human, HSCs firstly transit into CD45RA+ LMPPs. By expressing CD38, CD7, CD10 and IL-7Rα, LMPPs next differentiate into CLPs with potential to make lineage commitments into NKPs. Expression of CD122 further marks NKPs transit into immature NK cells (iNK). The appearance of CD56 indicates a final transition of iNK cells into mNK cells.CD56bright NK cells are considered less mature and CD56dim NK cells probably differentiation from CD56bright NK cells (dotted arrow) that is yet to be validated. B.In murine, HSCs firstly transit into CLPs and then differentiate into the CILPs. CILPs further develop into the common helper-like ILC precursors (CHILP) and earliest precursor of NK cells, namely pre-pro NK cells. Pre-pro NK cells up-regulate the expression of CD122 and further develop into NKPs. Accepting IL-15 stimulus, NKPs differentiate to iNK. iNKs receive MHC-I domestication signals and finally develop into mature NK cells. CHILPs further differentiate into ILCPs and finally transit into ILC1, ILC2, ILC3.
However, the developmental path of murine NK cells does not fully explain the development of human NK cells. Human NK cells have been reported to mature in the BM and secondary lymphoid organs such as lymph nodes, spleen.33 HSCs firstly transit into CD45RA+ lymphoid-primed multipotential progenitor (LMPP).34 By expressing CD38,35 CD7,36 CD1037 and IL-7Rα,38 LMPPs next differentiate into CLPs with potential to make lineage commitments into Pro-B, Pre-T and NKPs. Expression of CD122 further marks the irreversible fate decision of NKPs into iNKs. The appearance of CD56 (NCAM) indicates a final transition of iNKs into mNK. According to the different expression densities of CD56, human NK cells are divided into two sub-populations: CD56bright NK cells and CD56dim NK cells. CD56bright NK cells are considered less mature and CD56dim NK cells probably differentiation from CD56bright NK cells.39 Distinct stages through which human NK cells develop are less understood compared to that of the murine counterparts. (Fig. 2B).
Regulation of lncRNAs in NK cells development
During hematopoietic stem cells (HSC) differentiated to ST-HSCs (Short Time Hematopoietic Stem Cells), Monnier P et al found that H19 lncRNA can regulate maternal imprinting during embryo formation by binding to MBD1 (methy1-CpG-binding-domain)1 protein and recruiting histone–lysine–methyltransferase complexes. The deficiency of H19 expression in male parent of hematopoietic system in vivo will decrease the number of LT-HSCs (Long Time Hematopoietic Stem Cells) and increase the number of ST-HSCs.40 (Fig. 4A).
Figure 4.
The molecular mechanism of InCRNA in NK cells or NKTL. (A) The H19 RNA is required for the recruitment of MBD1 to inhibit Igf2 gene. The H19 lncRNA–MBD1 complex through its interaction with histone lysine methyltransferases KMT, and then acts by bringing repressive H3K9me3 marks on the differentially methylated region of Igf2 DMR1. (B) With IL15 signaling, Rroid promotes the deposition of phosphorylated STAT5 at the Id2 promoter region through long–range interaction and finally increases the activity of Id2. (C) LncRNA GAS5 increase the cytotoxic effect of NK cell via binding to miR-544 and down-regulating the expression of miR-544.
NK cells are classified to Group 1 innate lymphoid and play a “cytotoxic” role. Mowel WK et al firstly found in mouse that a nuclear lncRNA-Rroid that highly and specially expressed in ILC1s, but not ILC2s or CD4+ILC3s by gene brower tracks of ATAC sequence and RNA sequence from mouse indicated cell populations–CLP, ILC1, ILC2, CD4+ILC3.41 As mentioned earlier, lncRNAs regulate target gene expression by direct or indirect molecular mechanism. Similarly, Rroid can repress the expression of E-protein target genes, promote ILC1 lineage identity, sustain ILC1 homeostasis and function by indirectly promoting the activity of Id2, a key transcription factor for mNKs.41 This indicates lncRNAs are distinctive among different kinds of immune cells and act as crucial roles in the development and identity homeostasis of immune cells. (Fig. 4B).
NK cells are a group of innate immune cell which specifically express adhesion molecule CD56. In order to identify NK cell-specific lncRNAs, Zhang et al first broadly analyzed lncRNA expression in three different human NK cell samples. They detected various novel lncRNAs related to NK cell development, differentiation and function by analyzing human primary lymphocyte lncRNA expression profiles.42 Among these lncRNAs, they found the lnc-CD56 is positively correlated with the expression of adhesion molecule CD56. Interestingly, catRAPID predicted the interactions between lnc-CD56 and critical transcription factors of NK cell–TBX21, IRF2, IKZF2, ELF4, EMOES.42 Thus, additional studies of lnc-CD56 in human NK cells will be required to confirm its roles in NK cell biology.
In summary, lncRNAs are a key regulator of NK cell lineage. Presently, there are less functional lncRNAs which have been found in the development of NK cells. Mowel WK et al. provided a convenient and effective method for screening conditional lncRNAs nearby Id2 in NK cells.41 Besides, with the popularity of mass spectrometry flow cytometry and single-cell analysis techniques, single-cell tracking from HSC to mNK may simplify the study of NK cell development and help to discover more functional lncRNAs involved in NK development. Besides lncRNAs, miRNAs are crucial for NK cell development. There are more functional miRNAs in the development of NK cell which can be divided into positive or negative functional regulation.
Regulation of miRNAs in NK cells development
MiRNAs positively regulate the development of NK cells. The expression of miR-15/16 in NK cells is relatively high and promotes NK cell differentiation and development. Hence, there are a large number of immature NK cells in miR-15/16 knockout mice. When miR-15/16 and c-Myb gene are both knocked out, the number of immature NK cells gets less. It has seen that miR-15/16 promotes NK cell maturation by inhibiting c-Myb.43 Similarly, for miR-150, some animal experiments proved that miR-150 could promote NK cell development through the inhibition of c-Myb.44 MiR-181 can regulate CD34+ HSC to differentiate into mature NK cells. One paper shows that nemo-like kinase (NLK) mRNA is the miR-181 target gene. NLK blocks NK cell maturation by inhibiting Notch signaling pathway, while miR-181 can inhibit NLK gene expression. Thus miR-181 positively regulate NK cell development.45
MiRNAs negatively regulate the development of NK cells. During the process of NK cell maturation, miR-583 is a miRNA which changes the most. Over-expression of miR-583 can inhibit NK cell differentiation. If IL-2R-γ 3′-untranslated region (3′-UTR) basic group mutated, the inhibitory function of miR-583 will disappear. Therefore, miR-583, as a negative regulatory factor, acts on IL-2R-γ 3′-UTR to inhibit NK cell differentiation.46 TBET and EOMES are both key transcription factors taking part in the differentiation and development of NK cells. It has been found in mouse that TBET and EOMES can been meaningfully decreased by miRNA-29b to inhibit NK cell maturation.47 In addition, Over-expression of miRNA-29b induces NK cell depletion, indicating a potential mechanism of miRNA-mediated in immune escape.47 (Table 1).
Table 1.
Gene regulated by lncRNAs and miRNAs in NK cell development.
| LncRNA/MiRNA | Target gene | Model | Biological significance | Reference |
|---|---|---|---|---|
| lnc-CD56 | CD56 | human | enhances the expression of CD56 | 42 |
| miR-583 | IL-2Rγ | human | acts as a negative regulator of NK cell differentiation by silencing IL2Rγ | 46 |
| miR-181 | Nemo-like kinase | human | promotes NK cell development through the suppression of Nemolike-kinase | 45 |
| Rroid | Id2 | mouse | regulates the function and lineage identity of NK cells | 41 |
| miR-29b | TBET, EOMES | mouse | deletion of miR-29b in NK cells reverses the depletion of NK cell subset in leukemic mice | 47 |
| miR-150 | c-Myb | mouse | differentially controls the development of NK cell lineages by targeting c-Myb | 44 |
| miR-15/16 | c-Myb | mouse | miR-15/16 regulation of Myb controls the NK cell maturation program | 43 |
Regulation of lncRNAs and miRNAs in NK cell activation
NK cells express a series of activating and inhibitory receptors on their surface. Whether NK cells are activated relied on the balance between these activating and inhibitory signal of NK cells. Broadly speaking, the currently recognized activating receptors are NKP30,48 NKG2D,49 NKP44,50 NKP46,51 CD244/2B4,51 NKP46,51 CD16a,52 CD226,52 while inhibitory receptors are CD94/NKG2A,53 CD158b.54 Among these receptors, NKG2D is the most famous receptor. Its high expression promotes NK cell activation and increases their killing activity against target cells.49 However, many miRNAs have been found that they can positively or negatively regulate NKG2D signaling.55,56 A high level of miR-34a can up-regulate NKG2D ligand expression in hepatocytes of HCC patients.55 In addition, miR-182 also enhances NK cell cytotoxicity which leads to HCC (hepatocellular carcinoma cells) by regulating NKG2D expression. Furthermore, over-expression of miR-182 increases NK cell cytotoxicity against liver carcinoma via modulating NKG2D expression.56 Moreover, miR-30c augment the cytotoxicity of NKL cells (an human NK cell line) by up-regulating NKG2D expression. Further study found that only 40% of NK cells from freshly isolated PBMC (Peripheral Blood Mononuclear Cell) express a high level of NKG2D after miR-30c over-expression, suggesting that miRNA may show a perplexing and synergistic effect in NK cell-mediated immune response.57 In the latest research, miRNA-186 also can promote NKG2D-meidated cytotoxicity by directly inhibiting MYCN, AURKA, TGFRB1 and TGFRB2 expression.58 NKG2DLs are the ligand of NKG2D, including MHC class I-related chain A/B (MICA/B), and usually expressed on the surface of some tumor cells or pathogen-infected cells, such as breast cancer, cervical cancer and leukaemia.49,59,60 It has been reported in breast cancer that miR-17-92 can specifically bind to the 3′UTR of MICA/B.60 Thus, silencing miR-17-92 can enhance NK cell-mediated cytotoxicity by up-regulating NKG2DLs expression.
MiRNAs can not only positively regulate the NKG2D or NKG2DL expression, some miRNAs also can negatively mediate the activation of NKG2D signaling. TGFβ1 is a major negative regulator of NKG2D expression.61 MiRNA-1245 can down-regulate NKG2D expression and impair NKG2D-mediated immune responses in NK cells though up-regulating TGFβ1 expression from post-transcription level.62 Similarly, Tang P et al. found that increased levels of miR-20a in cancer cells may functionally inhibit NKG2D-mediated cytotoxicity of NK cell by down-regulating MICA/B expression of colorectal cancer cells.63 These studies also provide a potential evidence between cancer cells and immune escape from NK cells.
Except NKG2D, CD226 is also an important activating receptor. The activity of CD226 signal can increase the cytotoxicity of NK cells. Research showed that miRNA-30c-1 promoted NK cells cytotoxicity against human liver cancer cells by binding to the transcription factor hmbox1, a direct target gene of miR-30c-1.64 Further study found that miRNA-30c-1 acts on CD226 to promote the production of TNF-α and enhance the killing effect of NK cells.64 (Table 2). NK cells are different from other immune cells and not dominated by any single receptor. Presently, except for the NKG2D and CD226 receptors, the relationship between other receptors and lncRNA or miRNA have been rarely reported. Therefore, more researches are urgently needed to investigate the role of lncRNAs or miRNAs on NK cell activity.61 The regulatory network diagram of miRNA or lncRNA on the receptor-ligand signal axis of NK cells is shown in Fig. 3A. In summary, the function of NK cells depends on the balance of signals transmitted by the activating receptors and inhibitory receptors after recognizing the corresponding ligands on the target cell surface. Although NK cells express a variety of activating or inhibitory receptors, studies have found that miRNAs are mainly involved in the expression of CD226 and NKG2D. Therefore, the regulatory relationship between miRNAs and other receptors is still unclear, which is worth more researches to focus on.
Table 2.
Gene regulated by lncRNAs and miRNAs in NK cell activivation.
| Receptor | LncRNA/MiRNA | Target gene | Biological significance | Reference |
|---|---|---|---|---|
| NKG2D | miR-34a | / | increases their susceptibility to NK cell-mediated cytolysis in non-transformed liver cells | 55 |
| miR-17-92 | MICA/B | regulates NLG2L and NK cell-mediated cytotoxicity | 60 | |
| hcmv-miR-UL112 | MICB | down-regulates MICB expression during viral infection, leading to decreased binding of NKG2D and reduced killing by NK cells | 61 | |
| miR-182 | NKG2D/NKG2A | augments NK cell cytotoxicity against liver cancer via modulating NKG2D and NKG2A expressions | 56 | |
| miR-20a | MICA | regulates sensitivity of CRC cells to NK cells by targeting MICA | 63 | |
| miR-1245 | NKG2D | microRNA-1245 down-regulation significantly increases the expression of NKG2D expression in NK cells | 62 | |
| CD226 | miRNA-30c-1 | hm-box1 | promots NK cell cytotoxicity against hepatoma cells by targeting HMBOX1 | 64 |
Figure 3.
The key lncRNAs and miRNAs involved in activation and function of NK cells. (A) NKG2D, CD226 represented by gray triangle are activating receptors of NK cells; NKG2A represented by green triangle is inhibitory receptors of NK cells. MICA/MICB, HLA-E and CD112 are the ligands of NKG2D, NKG2A and CD225, respectively. Green arrow represents the promotion of receptors or ligands expression. Red arrow indicates the inhibition of receptors or ligands expression. (B) The key lncRNAs and miRNAs involved in function of NK cells. IFN-γ, TNF-α, Perforin and Granzyme B are molecules via which NK cells play a cytotoxic role. Red coloured words represent the promotion of corresponding molecule expression. Black coloured words indicate the inhibition of corresponding molecule expression.
Regulation of lncRNAs and miRNAs in NK cell function
Activation of NK cells is the foundation for them to be effective to anti-infection, anti-virus and anti-tumor. As mentioned earlier, the activation of inhibitory receptors on the surface of NK cells can inhibit the activation of NK cells by inhibiting intracellular calcium signaling through SHP-1, SHP-2 and inhibiting phosphorylation level of critical factors in NK cells. However, if the ligand binds to the activating receptors of NK cell surface, tyrosine activating motif of intracellular receptors activates the downstream PTKs, which will increase the secretion of IFN-γ, granzyme B and perforin to play NK cell cytotoxicity.65
lncRNAs are involved in the regulation of many cellular processes. Stein N et al. found that lncRNA IFNG-AS1 can sharply increase IFN-γ secretion and strongly inhibit pathogens infection in mice after IFNG-AS1 over-expression in primary NK cells or an NK cell line.66 In addition, Fang P et al. also found that knockdown of GAS5 in activated NK cells will reduce the clearance ratio of NK cells against liver cancer cells through decreasing IFN-γ secretion and the percentage of CD107+ NK cells. On the contrary, lncRNA GAS5 over-expression can increase the cytotoxic effect of NK cell on liver cancer cell by down-regulating the expression of miR-544, suggesting that lncRNAs have an important effect on the release of IFN-γ67 (Fig. 4).
Apart from IFN-γ, NK cells mainly secrete perforin and granzymes to kill target cell. At present, lncRNAs are mainly found to be involved in the regulation of IFN-γ secretion. MiRNAs are main regulator in the expression of perforin or granzymes B. It has been reported Ni F et al. found NK cells in human peripheral blood highly expressed miR-362-5p.68 The miRNA can up-regulate CD107a expression in NK cells, promote perforin and granzyme B secretion, and increase NK cell cytotoxicity by targeting the tumor suppressor factor –CYLD (cylindromatosis).68 After NK cells are activated by IL-15, miRNA-27a-5p also can positively regulated perforin and granzyme B mRNA expression by directly combining to 3′-UTR of them. In order to prevent NK cells from excessive activation when miR-27a-5p is over-expression, granzyme B and perforin expression are decreased.69
Being contrary with the effect of miRNA-362-5p, miRNA-223 and miRNA-27a both negatively regulate NK cell cytotoxicity.70,71 MiRNA-223 directly inhibits granzyme B mRNA translation in NK cells.70 MiRNA-27a also inhibit NK cell cytotoxic effect by simultaneously inhibiting granzyme B and perforin mRNA translation in NK cells.71 MiRNA-378 is a negative regulated factor of NK cell cytotoxicity. Research showed that miRNA-378 expresses less in NK cells of DENV (Dengue virus) infected patients. When adding miRNA-378 agomir to mice infected with DENV, the expression of granzyme B was inhibited and DENV replication increased.72 Moreover, besides miRNA-378, miRNA-30e is also involved in the negative regulation in the expression of granzyme B and perforin.73 If miRNA-378 or miRNA-30e was inhibited in NK-92 nucleus, the cytotoxicity and tumor killing ability of NK-92 cells sharply increase. Interestingly, in the NK cells of miRNA-233 knockout mice, the secretion of granzyme B and NK cell cytotoxicity are different from those of normal wild mice.74 It suggests that miRNA-233 does not work alone, and there may be other factors or miRNAs involved in this process. All-trans retinoic acid (ATRA) can inhibit NK cell killing ability by inhibiting target gene expression. It has been confirmed that miRNA-23a express less after NK cells are activated and is a negative regulator of cathepsin C (CTSC). CTSC is a downstream target gene of miRNA-23a.75 After CTSC was activated, granzyme B secretion can increase. However, ATRA can induce miRNA-23a up-regulation, thus inhibiting CTSC expression, and result in impaired NK cell killing function. (Table 3). In summary, miRNAs can be specifically paired with the 3′-UTR of granzyme B and perforin mRNA to cause degradation of the mRNA. It reduces the stability of target gene mRNA or inhibits protein translation, regulates the secretion of granzyme and perforin of NK cells, thereby regulate NK cytotoxicity. The regulatory effect of miRNA or lncRNA on the function of NK cells is shown in Fig. 3B. Though miRNAs largely participate in the regulation of NK cell cytotoxic molecules, especially IFN-γ, lncRNAs related to NK cell function are rarely reported, which is related to the difficulty in finding functional lncRNAs. However, lncRNAs have an absolute advantage in non-coding RNAs, and their modes of action and mechanisms are more complex compaired with miRNAs. Therefore, more researches are needed to reveal the relationship between the NK cell function and lncRNAs, which will provide a theoretical basis for the application of miNRAs or lncRNAs in NK cells.
Table 3.
Gene regulated by lncRNAs and miRNAs in NK cell function.
| Cytotoxic molecules | LncRNA/MiRNA | Target gene | Biological significance | References |
|---|---|---|---|---|
| Granzyme B | miR-362–5p | CYLD | enhances the expression of granzyme-B via NK-κB pathway | 68 |
| miR-223 | GzmB | contributes to control of GzmB translation in resting NK cells | 74 | |
| miR-27a | GzmB | suppresses NK-cell cytotoxicity by silencing GzmB expression | 71 | |
| miR-378 | GzmB | suppresses GrzB expression in NK cells | 72 | |
| miR-30e | GzmB | suppresses GrzB expression in NK cells | 73 | |
| miR-23a | CTSC | decreases CTSC expression and granzyme B activity | 75 | |
| Perforin | miR-362–5p | CYLD | enhances the expression of perforin via NK-κB pathway | 68 |
| miR-27a | Prf1 | silences Prf1 expression in NK cells | 71 | |
| miR-30e | Prf1 | suppresses Prf1 expression in NK cells | 73 | |
| miR-150 | Prf1 | represses NK cell lytic activity by targeting perforin-1 | 44 | |
| miR-378 | miR-378 | inhibites GrzB expression in DENV-infected patients | 74 | |
| TNF-a | miR-146a | STAT1 | negatively regulates TNF-α expression via STAT1 signaling | 87 |
| miR-30c-1 | hmbox1 | enhances NKL cell cytotoxicity through up-regulation of TNF-α | 64 | |
| IFN-γ | lncRNA GAS5 | miR-544/RUNX3 | increases RUNX3 expression and IFN-γ secretion | 69 |
| lncRNA IFNG-AS1 | / | induces upon NK cell activation and increases IFNγ secretion | 66 | |
| miR-155 | SHIP-1/Noxa | increase IFN-γ production in HCV-infected patients | 95 | |
| miR-155 | Tim-3 | regulates IFN-γ production in NK cells via Tim-3 signalling | 96 | |
| miR-146a | STAT1 | negatively regulates IFN-γ expression via STAT1 signaling | 98 | |
| miR-362–5p | CYLD | enhances the expression of IFN-γ via NK-κB pathway | 68 | |
| miR-34a | / | enhances cytolysis and interferon-γ production by NK-92MI cells | 55 | |
| miRNA-29b | DNMTs | regulates INF-γ expression via decreasing methylation of IFN-γ | 99 | |
| miR-15/16 | c-Myb | enhances the expression of granzyme-B in NK cells | 43 |
The role of lncRNAs and miRNAs in NK/T cell lymphoma
Extranodal nasal-type natural killer/T-cell lymphoma (NKTL), an Epstein–Barr virus (EBV) associated lymphoma, is mostly derived from cytotoxic NK cells and a small part is derived from NK-like T cells. Study has reported that the EBV infection rate in NKTL patients is up to 90%.76 Normally, though cytotoxic NK cells will eradicate viral infections, it is interesting to investigate how the EBV evades and transforms NK cells in NKTL. IFN-γ is an important cytotoxic cytokine of NK cells and main transcribed by transcription factor T-bet. The EBV-encoded miR-BART20-5p inhibits both T-bet and IFNG in NKTL, which allows the survival of EBV inside NK cells.77,78 Apart from miR-BART20-5p, there are also some other EBV-encoded miRNA help NKTL tumorigenesis and progression as mentioned below: BART9 is involved in NKTL proliferation by regulating LMP-1 expression;79 miR-BART8 cause progression of NKTL through inhibition of the IFN-γ-STAT1 pathway.78 The more information of abnormal EBV-encoded miRNAs in NKTL is summarized in Table 4. Though miR-142-3p and miR-205 may not EBV-encoded miRNA, they are down-regulated in the EBV-positive vs EBV-negative lymphomas.80 Down-regulated miR-142-3p and miR-205 can contribute to lymphomas tumorigenesis through up-regulating the oncogenic BCL6 and the proinflammatory cytokine interleukin 1 alpha (IL1A) expression. Clearly, the EBV encodes at least 44 miRNAs, but the pathogenesis of most EBV-encoded miRNAs in NKTL remain to be explored.
Table 4.
The role of lncRNAs and miRNAs in NKTL.
| Noncoding RNA | Encoded by EBV (Yes/No) | Target gene | Underexpressed Overexpressed in NKTL | Biological significance | References |
|---|---|---|---|---|---|
| BART9 | Yes | LMP-1 | Overexpressed | BART9 is involved in NKTL proliferation by regulating LMP-1 levels | 79 |
| miR-BART20-5p | Yes | T-bet(TBX21) | – | promotes NKTL progress via the PTEN-AKT-mTOR/RICTOR pathway | 85 |
| miR-BART20-5p | Yes | – | – | promotes the development of NKTL through inhibition of the IFN-γ-STAT1 pathway | 78 |
| miR-BART8 | Yes | – | – | causes progression of NKTL through inhibition of the IFN-γ-STAT1 pathway | 78 |
| miR-142-3p | No | IL1-a | Underexpressed | regulates the expression of IL1A | 85 |
| lncRNA MALAT1 | No | ZH2, SUZ12 | Overexpressed | MALAT1 is related to poor prognosis | 91 |
| miR-21,miR155 | No | – | Overexpressed | leads to activation of the PI3K-AKT pathway | 82 |
| miR-150 | No | AKT2, DCK1 | Underexpressed | induces continuous activation of the PI3K-AKT pathway | 86 |
| miRNA-146a | No | TRAF6 | Underexpressed | downregulates NF-κB activity and is related to prognosis | 87 |
| miR-15a | No | Myb, cyclin D1 | Underexpressed | promotes cell proliferation and predicts poor prognosis in NKTL | 94 |
| miR-223 | No | PRDM1 | Overexpressed | The downregulation of the tumour suppressor PRDM1 is mediated by miR-223 and that PRDM1-positive staining might have prognostic value for NKT patients | 92 |
| miR-155 | No | BRG1 | Overexpressed | activates STAT3/VEGFC signaling, promotes lymphangiogenesis and controlled the viability of NKTCL cells | 84 |
| miR-155 | No | Foxo3a | – | Reduction in miRNA-155 expression can inhibit the proliferation of SNK-6 lymphoma cells and promote their apoptosis | 89 |
| miR-494-3p | No | PTEN | Overexpressed | promotes NKTL progress through the PTEN-AKT-mTOR/RICTOR pathway | 85 |
| miR-142-3p | No | RICTOR | Overexpressed | inhibites RICTOR, with secondary suppression of AKT in YT cells | 80 |
| circulating miR-221 | No | – | – | a reverse correlation with performance status and the overall survival after treatment | 101 |
| miR-16 | No | Bmi1 | Underexpressed | enhances p21 expression via downregulation of Bmi1, thereby inducing cellular senescence | 102 |
| miR-205 | No | BCL6 | Underexpressed | regulates the expression of the oncogenic BCL6 | 80 |
| miR-34a,miR-181c | No | PDGFRα | – | are involved in the oncogenic progression of NKTL through the regulation of PDGFRα, STAT3, and K-RAS | 88 |
| miR-143,miR-20b, miR34a | No | – | – | miR-20b, miR34a, miR-143 expression showed inverse correlations with STAT3 mRNA expression in NKTCL tissues | 99 |
MiRNA deregulation in NKTL
Mention to the pathogenesis of NKTL, besides EBV infection, like p53, C-kit, K-ras, C-MYC et al gene mutations and abnormal activation of JAK-STAT, NF-κB, Wnt/β-catenin are also important factors. These deregulated single genes and deregulated signaling pathways mainly cause the increasing of tumor cell proliferation or the inhibition of tumor cell apoptosis.81 To understand the pathogenetic role of miRNA deregulation in NKTL, Ng et al performed a comprehensive genome-wide miRNA expression profiling of NKTL tissues and NK cell lines compared with normal NK cells and found differentially expressed miRNAs in NKTL are predominantly down-regulated. Re-expression of down-regulated miRNAs, such as miR-101, miR-26a, miR26b, miR-28-5, and miR-363, may reduce the growth of the NK cell line and modulated the expression of their predicted target genes, suggesting the potential functional role of the deregulated miRNAs in the oncogenesis of NKTL.82 On the other hand, there are also some microRNAs, such as miR-21 and miR-155 have been shown to be over-expressed in ENKTL (Extranodal NK/T cell lymphoma), with pro-oncogenic consequences. All above these suggest the potential functional role of the deregulated miRNAs in the oncogenesis of NKTL.82
Mechanically, abnormal miRNAs usually promote or inhibit the expression of key genes and key signaling pathway to help NKTL tumorigenesis and progression. In NKTL, the activation of AKT signal, NF-κB, STAT3 can promote cancer cell proliferation, inhibit cell apoptosis and induce tumorigenesis. In NKTL, there are some abnormal miRNAs can activate these signaling passway by acting on the targets. For example, Aberrant over-expression of miR-21,83 miR-155,84 miR-494-3p85 and down-regulation of miR-15086 in NKTL lead to activation of the PI3K-AKT pathway; aberrant down-regulation of miRNA-146a in NKTL lead to activation of the NF-κB pathway.87 Over-expressed miR-155 also can activate STAT3/VEGFC signaling and promoted lymphangiogenesis.84 In addition, miR-34a and miR-181c may be involved in the oncogenic progression of NKTCL through the regulation of STAT3 pathway;88 miRNA-342-3p may contributes to the development of NKTL via the TIAM1 pathway.80 Other abnormal miRNAs participate in the pathogenesis of NKTL by inhibiting or promoting key genes that regulate growth, development, proliferation, and apoptosis of cells. For example, miR-181c and miR-34a may be involved in the oncogenic progression of NKTCL by targeting PDGFRα and K-RAS;88 abnormal upregulation of miR-155 may be associated with regulation of FOXO3a gene can promote the proliferation of SNK-6 lymphoma cells and inhibit their apoptosis.89 This table summarizes the mechanisms by which miRNA deregulation contributes to lymphomagenesis in NKTL (Table 4).
Clinical value of lncRNA and miRNA dysregulation in NKTL
Presently, the studies of lncRNA and miRNA mainly focused on cancer and an increasing number of researches showed that nc-RNAs can be defined as new biomarkers for disease diagnosis and new targets for drug treatment. For example, lncRNA-PCA3 in urine, has been identified as the most specific biomarker for proadenocarcinoma and it will be used in clinical soon.90 There are also some new miRNA or lncRNA biomarkers involved in NKTL. It has been found in NKTL that MALAT1 is most highly expressed in lncRNAs connected to the polycomb repressive complex (PRC) and is related to poor prognosis of NK cell lymphoma patients by directly binding to EZH2 an SUZ1285.91,92 Except lncRNAs, more miRNAs also are involved in NK cell lymphoma. For example, the down-regulation of the tumor suppressor--PRDM1 in NKTL samples is mediated by miR-223 and the PRDM1-positive staining might have prognostic value for evaluating the clinical outcome of NKTL patients.92 PRDM1 is an tumor suppressor gene and indirect target gene of miR-223. There are several publications showed PRDM1 might be a favorable predictor of overall survival and failure-free survival in EN-NK patients.87,93 Except contribution to prognosis, miRNAs also can be used as a target of disease treatment. Lin J et al found that miR-BART20-5p will inhibit the translation of T-bet, the master transcription factor for cytotoxic NK cells, and decrease p53 expression, a tumor suppressor gene. Therefore, an antagonist for miR-BART20-5p might be an effective therapeutic agent through inducting the expression of T-bet and p53. In addition, Komabayashi found over-expression of miR-15a can decrease MYB and cyclin D1 levels thereby blocking G1-S transition and cell proliferation.94 Further study also found in NK/T cell lymphoma tissues, reduced miR-15a expression, was associated with poor prognosis of NK/T cell lymphoma patients.94 Therefore, miR-15a may be a potential target for anti-tumor therapy and a prognostic predictor for NKTL.55,95, 96, 97, 98, 99, 100, 101(Table 4.)
Mechanisms of miRNA dysregulation in NKTL
Deregulated miRNA patterns are frequently linked to a variety of human cancers including lymphomas. The mechanisms of dysfunctional miRNAs in NKTL are beginning to be delineated. MYC, a key transcriptional regulator known to cause extensive repression of miRNA, has been reported to be over-expressed in NKTL and may cause the widespread downregulation of miRNAs in NKTL.82,102 Moreover, the oncogenic EBV is strongly associated with the pathogenesis of NKTL and expresses 44 mature miRNAs and two noncoding EBV-encoded RNAs (EBERs). It has been confirmed that EBV promotes the deregulation of miRNAs since down-regulation of miR-142–3p,85 MiR-20580 and up-regulation of miR-15584 in NKTL. Besides, epigenetic deregulation has been shown to result in the deregulation of miRNAs in NKTL, such as miR-146a.87 Paik JK et al. found that promoter methylation of miR-146a gene was observed in SNK6 and YT cells, as well as in NKTL tissues with low miR-146a expression, and miR-146a expression was induced by the conversion of methylation status with a demethylating agent in SNK6 and YT cells, which suggests promoter methylation of miRNA has contributed to the deregulation of miRNAs in NKTL.87
Summary and future outlook
Mounting evidence underscores the critical role of miRNAs and lncRNA as regulators of NK cell biology and carcinogenesis and progress of NKTL. MiRNAs or lncRNA finely tune the level of translatable mRNAs in response to specific developmental or stimulatory cues and change cellular requirements, which provides the possibility to control the cytotoxicity and cytokine secretion of NK cells. Meanwhile, lncRNA or miRNA has various target genes, which increases uncertainty and difficulty for directionally modifying the biological function of NK cells or curing diseases by regulating miRNAs or lncRNAs.
NKTL is a malignant tumor caused by the aberrant development NK cells. Abnormally expressed miRNAs and lncRNAs of NK cells contribute to the occurrence and progression of NKTL. Restoring key miRNAs and lncRNAs with abnormal expression may provide the possibility for the treatment of NKTL. Presently, lncRNA and miRNA related to NK cell biological activity and NKTL still have a certain distance in practical application. On one hand, compared with miRNAs, lncRNAs are much less studied in NK cell but they are the most component of non-coding RNA. To realize lncRNA application value in NK cell function and NKTL rapid diagnosis, treatment and prognosis evaluation, it is necessary to discover and study more functional lncRNAs in physiological or pathological NK cells. On the other hand, technically, transfection of NK cell is more difficult. Improving the transfection efficiency of miRNA mimics or miRNA antagomir in NK cells will help to verify the function of key functional miRNAs in NK cells and realize the application value of miRNAs in NK cell-related diseases, such as NKTL.
Although this review has focus on how miRNAs or lncRNAs participates in the development, activation, function of NK cells and NKTL occurrence from a relatively comprehensive and detailed perspective, the mechanism of how they works is not sufficiently elaborated. In summary, there has been discovered more functional miRNAs and lncRNAs with potential NKTL diagnosis and prognostic evaluation as introdued in this review. In order to realize the clinical application of miRNAs and lncRNAs in NKTL, more researches from different dimensions are needed to surpport the feasibility of miRNAs and lncRNAs as new NKTL diagnostic markers, indicators of patients’ prognostic and the targets of the therapeutic drugs.
Authors contribution
FengXia Gao performed the selection of literature, drafted the manuscript, and prepared the figures. SiRong He participated in the design and discussion. AiShun Jin carried out the design of this review and revised the manuscript. All authors contributed to this manuscript. All authors read and approved the final manuscript.
Conflict of Interests
The authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Funding
This research received no external funding.
References
- 1.Vasudevan S., Tong Y., Steitz J. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasko J., Wong J. Nuclear microRNAs in normal hemopoiesis and cancer. J Hematol Oncol. 2017;10(1) doi: 10.1186/s13045-016-0375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hausser J., Zavolan M. Identification and consequences of miRNA-target interactions--beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 5.Denaro N., Merlano M., Lo Nigro C. Long non-coding RNAs as regulators of cancer immunity. Mol Oncology. 2019;13(1):61–73. doi: 10.1002/1878-0261.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X., Sun M., Wu Y. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumour Biol. 2015;36:503–513. doi: 10.1007/s13277-015-3106-y. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J Exp Clin Canc Res. 2017;36(1) doi: 10.1186/s13046-017-0666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J., Tao H., Deng Z., Chao L., Li J. Non-coding RNA-mediated epigenetic regulation of liver fibrosis. Metab Clin Exp. 2015;64:1386–1394. doi: 10.1016/j.metabol.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y., Zheng Y., Jia L., Li W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/smad 3/HDAC signaling pathway by deriving miR-675. Stem Cells (Dayton) 2015;33:3481–3492. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 10.Jewett A., Kos J., Fong Y. NK cells shape pancreatic and oral tumor microenvironments; role in inhibition of tumor growth and metastasis. Semin Canc Biol. 2018;53:178–188. doi: 10.1016/j.semcancer.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Lin W., Man X., Li P. NK cells are negatively regulated by sCD83 in experimental autoimmune uveitis. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-13412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt L., Eskiocak B., Kohn R. Enhanced adaptive immune responses in lung adenocarcinoma through natural killer cell stimulation. Proc Natl Acad Sci USA. 2019;116:17460–17469. doi: 10.1073/pnas.1904253116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoyab M. Regulation of granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor expression by Oncostatin M. Blood. 1993;82:33–37. [PubMed] [Google Scholar]
- 14.Henao-Mejia Long noncoding RNAs and the regulation of innate immunity and host-virus interactions. J Leukoc Biol. 2019;106(1):83–93. doi: 10.1002/JLB.3MIR0918-354R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Boxberger N., Hecker M., Zettl U. Dysregulation of inflammasome priming and activation by MicroRNAs in human immune-mediated diseases. J Immunol. 2019;202:2177–2187. doi: 10.4049/jimmunol.1801416. [DOI] [PubMed] [Google Scholar]
- 17.Yang T., Ge B. miRNAs in immune responses to Mycobacterium tuberculosis infection. Canc Lett. 2018;431:22–30. doi: 10.1016/j.canlet.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 18.Wang P., Xue Y., Han Y. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science (New York, N.Y.). 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 19.Baumjohann D. Diverse functions of miR-17-92 cluster microRNAs in T helper cells. Canc Lett. 2018;423:147–152. doi: 10.1016/j.canlet.2018.02.035. [DOI] [PubMed] [Google Scholar]
- 20.Francesco C., Sandrine I., Rodney P., Olivier L., Harinder S., James D. Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97:2625–2632. doi: 10.1182/blood.v97.9.2625. [DOI] [PubMed] [Google Scholar]
- 21.Geogopoulos K. Lack of natural killer cell precursors in fetal liver of ikaros knockout mutant mice. Nat Immun. 1998;16:137–145. doi: 10.1159/000069438. [DOI] [PubMed] [Google Scholar]
- 22.Ramirez K., Chandler K., Spaulding C. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36:921–932. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gascoyne D., Long E., Veiga-Fernandes H. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 24.Brian D.L.T. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue–inducer cell and NK cell lineages. Nat Immunol. 2010;11:945–952. doi: 10.1038/ni.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Townsend M.J., Weinmann A.S., Matsuda J.L. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity. 2004;20(4):477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 26.Narni-Mancinelli E., Ugolini S., Vivier E. Tuning the threshold of natural killer cell responses. Curr Opin Immunol. 2013;25:53–58. doi: 10.1016/j.coi.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Kallies A., Carotta S., Huntington N. A role for Blimp 1 in the transcriptional network controlling natural killer cell maturation. Blood. 2011;117:1869–1879. doi: 10.1182/blood-2010-08-303123. [DOI] [PubMed] [Google Scholar]
- 28.Walker J.A., Clark P.A., Crisp A. Polychromic reporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity. 2019;51(1):104–118. doi: 10.1016/j.immuni.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steven L. The transcription factors T-bet and eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes M., Huntington N., Thong R. Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J. 2014;33:2721–2734. doi: 10.15252/embj.201487900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castro W., Chelbi S., Niogret C. The transcription factor Rfx7 limits metabolism of NK cells and promotes their maintenance and immunity. Nat Immunol. 2018;19:809–820. doi: 10.1038/s41590-018-0144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudensky A. Interactions between innate and adaptive lymphocytes. Nat Rev Immunol. 2014;14:631–639. doi: 10.1038/nri3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abel A., Yang C., Thakar M., Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Front Immunol. 2018;9:1869. doi: 10.3389/fimmu.2018.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weissman I. Lymphoid development from hematopoietic stem cells. Int J Hematol. 1999;69:217–226. [PubMed] [Google Scholar]
- 35.Ogawa M. CD38 expression by hematopoietic stem cells of newborn and juvenile mice. Leukemia. 2003;17:171–174. doi: 10.1038/sj.leu.2402785. [DOI] [PubMed] [Google Scholar]
- 36.Whiteside T. Expression and function of CD7 molecule on human natural killer cells. J Immunol. 1994;152:517–526. [PubMed] [Google Scholar]
- 37.Luetke-Eversloh M., Killig M., Romagnani C. Signatures of human NK cell development and terminal differentiation. Front Immunol. 2013;4 doi: 10.3389/fimmu.2013.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renoux V., Zriwil A., Peitzsch C. Identification of a human natural killer cell lineage-restricted progenitor in fetal and adult tissues. Immunity. 2015;43:394–407. doi: 10.1016/j.immuni.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Malmberg K. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116:3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 40.Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci USA. 2013;110:20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mowel W.K., McCright S.J., Kotzin J.J. Group 1 innate lymphoid cell lineage identity is determined by a cis -regulatory element marked by a long non-coding RNA. Immunity. 2017;47(3):435–449. doi: 10.1016/j.immuni.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R., Ni F., Fu B. A long noncoding RNA positively regulates CD56 in human natural killer cells. Oncotarget. 2016;7:72546–72558. doi: 10.18632/oncotarget.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan R.P., Leong J.W., Schneider S.E. MicroRNA-15/16 antagonizes Myb to control NK cell maturation. J Immunol. 2015;195(6):2806–2817. doi: 10.4049/jimmunol.1500949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanier L. miR-150 regulates the development of NK and iNKT cells. J Exp Med. 2011;208(13):2717–2731. doi: 10.1084/jem.20111386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cichocki F., Felices M., McCullar V. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171–6175. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yun S., Lee S., Kim J. Integrated mRNA-microRNA profiling of human NK cell differentiation identifies MiR-583 as a negative regulator of IL2Rγ expression. PloS One. 2014;9 doi: 10.1371/journal.pone.0108913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mundy-Bosse B., Scoville S., Chen L. MicroRNA-29b mediates altered innate immune development in acute leukemia. J Clin Invest. 2016;126:4404–4416. doi: 10.1172/JCI85413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mantovani S., Oliviero B., Lombardi A. Deficient natural killer cell NKp30-mediated function and altered NCR3 splice variants in hepatocellular carcinoma. Hepatology. 2019;69:1165–1179. doi: 10.1002/hep.30235. [DOI] [PubMed] [Google Scholar]
- 49.Paczulla A., Rothfelder K., Raffel S. Publisher Correction: absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature. 2019;572 doi: 10.1038/s41586-019-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parodi M., Favoreel H., Candiano G. NKp44-NKp44 ligand interactions in the regulation of natural killer cells and other innate lymphoid cells in humans. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costello R., Boehrer A., Sanchez C. Differential expression of natural killer cell activating receptors in blood versus bone marrow in patients with monoclonal gammopathy. Immunology. 2013;139:338–341. doi: 10.1111/imm.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guillamón C., Martínez-Sánchez M., Gimeno L. NK cell education in tumor immune surveillance: DNAM-1/KIR receptor ratios as predictive biomarkers for solid tumor outcome. Cancer Immunology Research. 2018;6:1537–1547. doi: 10.1158/2326-6066.CIR-18-0022. [DOI] [PubMed] [Google Scholar]
- 53.Sivori S., Vacca P., Del Zotto G., Munari E., Mingari M., Moretta L. Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell Mol Immunol. 2019;16(5):430–441. doi: 10.1038/s41423-019-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGrath E., Ryan E., Lynch L. Changes in endometrial natural killer cell expression of CD94, CD158a and CD158b are associated with infertility. AJRI (Am J Reprod Immunol) 2009;61:265–276. doi: 10.1111/j.1600-0897.2009.00688.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhou M., Zhao C., Chen X. MicroRNA-34a promotes MICB expression in hepatocytes. Carcinogenesis. 2018;39:1477–1487. doi: 10.1093/carcin/bgy128. [DOI] [PubMed] [Google Scholar]
- 56.Abdelrahman M., Fawzy I., Bassiouni A. Enhancing NK cell cytotoxicity by miR-182 in hepatocellular carcinoma. Hum Immunol. 2016;77:667–673. doi: 10.1016/j.humimm.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 57.Chen L. MicroRNA-30c promotes natural killer cell cytotoxicity via up-regulating the expression level of NKG2D. Life Sci. 2016;15(151):174–181. doi: 10.1016/j.lfs.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 58.Neviani P., Wise P., Murtadha M. Natural killer-derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms. Canc Res. 2019;79:1151–1164. doi: 10.1158/0008-5472.CAN-18-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho H., Chung J., Kim S. MICA/B and ULBP1 NKG2D ligands are independent predictors of good prognosis in cervical cancer. BMC Cancer. 2014;14 doi: 10.1186/1471-2407-14-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen J., Pan J., Du C. Silencing NKG2D ligand-targeting miRNAs enhances natural killer cell-mediated cytotoxicity in breast cancer. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanah M. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Espinoza J., Takami A., Yoshioka K. Human microRNA-1245 down-regulates the NKG2D receptor in natural killer cells and impairs NKG2D-mediated functions. Haematologica. 2012;97:1295–1303. doi: 10.3324/haematol.2011.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang S., Fu H., Xu Q., Zhou Y. miR-20a regulates sensitivity of colorectal cancer cells to NK cells by targeting MICA. Biosci Rep. 2019;39(7) doi: 10.1042/BSR20180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen L. miR-30c-1∗ promotes natural killer cell cytotoxicity against human hepatoma cells by targeting the transcription factor HMBOX1. Canc Sci. 2012;103:645–652. doi: 10.1111/j.1349-7006.2012.02207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malmberg K., Carlsten M., Björklund A., Sohlberg E., Bryceson Y., Ljunggren H. Natural killer cell-mediated immunosurveillance of human cancer. Semin Immunol. 2017;31:20–29. doi: 10.1016/j.smim.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Mandelboim O. IFNG-AS1 enhances interferon gamma production in human natural killer cells. iScience. 2019;25(11):466–473. doi: 10.1016/j.isci.2018.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang P., Xiang L., Chen W. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3. Innate Immun. 2019;25:99–109. doi: 10.1177/1753425919827632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ni F., Guo C., Sun R. MicroRNA transcriptomes of distinct human NK cell populations identify miR-362-5p as an essential regulator of NK cell function. Sci Rep. 2015;5 doi: 10.1038/srep09993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Regis S., Caliendo F., Dondero A. TGF-β1 downregulates the expression of CXCR1 by inducing miR-27a-5p in primary human NK cells. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fehniger T., Wylie T., Germino E. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010;20:1590–1604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim T., Lee S., Yun S. Human microRNA-27a∗ targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood. 2011;118:5476–5486. doi: 10.1182/blood-2011-04-347526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu S., Chen L., Zeng Y. Suppressed expression of miR-378 targeting gzmb in NK cells is required to control dengue virus infection. Cell Mol Immunol. 2016;13:700–708. doi: 10.1038/cmi.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang P., Gu Y., Zhang Q. Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J Immunol. 2012;189:211–221. doi: 10.4049/jimmunol.1200609. [DOI] [PubMed] [Google Scholar]
- 74.Johnnidis J., Harris M., Wheeler R. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 75.Sanchez-Martínez D., Krzywinska E., Rathore M. All-trans retinoic acid (ATRA) induces miR-23a expression, decreases CTSC expression and granzyme B activity leading to impaired NK cell cytotoxicity. Int J Biochem Cell Biol. 2014;49:42–52. doi: 10.1016/j.biocel.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 76.Bacchi C. Clinicopathologic and molecular features of 122 Brazilian cases of nodal and extranodal NK/T-Cell lymphoma, nasal type, with EBV subtyping analysis. Am J Surg Pathol. 2011;35:1195–1203. doi: 10.1097/PAS.0b013e31821ec4b5. [DOI] [PubMed] [Google Scholar]
- 77.Lewis L. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang W., Lin C. EBV-encoded miR-BART20-5p and miR-BART8 inhibit the IFN-γ-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am J Pathol. 2014;184:1185–1197. doi: 10.1016/j.ajpath.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 79.Ramakrishnan R., Donahue H., Garcia D. Epstein-Barr virus BART9 miRNA modulates LMP1 levels and affects growth rate of nasal NK T cell lymphomas. PloS One. 2011;6 doi: 10.1371/journal.pone.0027271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alles J., Menegatti J., Motsch N. miRNA expression profiling of Epstein-Barr virus-associated NKTL cell lines by Illumina deep sequencing. FEBS open bio. 2016;6:251–263. doi: 10.1002/2211-5463.12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Mel S., Soon G., Mok Y. The genomics and molecular biology of natural killer/T-cell lymphoma: opportunities for translation. Int J Mol Sci. 2018;19(7) doi: 10.3390/ijms19071931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ng S., Yan J., Huang G. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood. 2011;118:4919–4929. doi: 10.1182/blood-2011-07-364224. [DOI] [PubMed] [Google Scholar]
- 83.Yamanaka Y., Tagawa H., Takahashi N. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–3275. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 84.Chang Y., Cui M., Fu X. MiRNA-155 regulates lymphangiogenesis in natural killer/T-cell lymphoma by targeting BRG1. Canc Biol Ther. 2019;20:31–41. doi: 10.1080/15384047.2018.1504721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen H., Huang W., Yang L., Lin C. The PTEN-AKT-mTOR/RICTOR pathway in nasal natural killer cell lymphoma is activated by miR-494-3p via PTEN but inhibited by miR-142-3p via RICTOR. Am J Pathol. 2015;185:1487–1499. doi: 10.1016/j.ajpath.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 86.Watanabe A., Tagawa H., Yamashita J. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- 87.Paik J., Jang J., Jeon Y. MicroRNA-146a downregulates NFκB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Canc Res : an official journal of the American Association for Cancer Research. 2011;17:4761–4771. doi: 10.1158/1078-0432.CCR-11-0494. [DOI] [PubMed] [Google Scholar]
- 88.Go H., Jang J., Kim C., Huh J., Kim P., Jeon Y. Identification of microRNAs modulated by DNA hypomethylating drugs in extranodal NK/T-cell lymphoma. Leuk Lymphoma. 2020;61:66–74. doi: 10.1080/10428194.2019.1654096. [DOI] [PubMed] [Google Scholar]
- 89.Zhang M. miRNA-155 modulates the malignant biological characteristics of NK/T-Cell lymphoma cells by targeting FOXO3a gene. J Huazhong Univ Sci Technol. 2014;34:882–888. doi: 10.1007/s11596-014-1368-z. [DOI] [PubMed] [Google Scholar]
- 90.Wang S., Song W., Wei S. Functional titanium carbide MXenes-loaded entropy-driven RNA explorer for long noncoding RNA PCA3 imaging in live cells. Anal Chem. 2019;91(13):8622–8629. doi: 10.1021/acs.analchem.9b02040. [DOI] [PubMed] [Google Scholar]
- 91.Yoon S. Association of the long non-coding RNA MALAT1 with the polycomb repressive complex pathway in T and NK cell lymphoma. Oncotarget. 2017;8:31305–31317. doi: 10.18632/oncotarget.15453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liang L., Nong L., Zhang S. The downregulation of PRDM1/Blimp-1 is associated with aberrant expression of miR-223 in extranodal NK/T-cell lymphoma, nasal type. J Exp Clin Cancer Res. 2014;33(1) doi: 10.1186/1756-9966-33-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liang L., Zhang Z., Wang Y. The genetic deletion of 6q21 and PRDM1 and clinical implications in extranodal NK/T cell lymphoma, nasal type. Biomed Res Int. 2015;2015 doi: 10.1155/2015/435423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harabuchi Y. Downregulation of miR-15a due to LMP1 promotes cell proliferation and predicts poor prognosis in nasal NK/T-cell lymphoma. Am J Hematol. 2014;89:25–33. doi: 10.1002/ajh.23570. [DOI] [PubMed] [Google Scholar]
- 95.Trotta R., Chen L., Ciarlariello D. miR-155 regulates IFN-γ production in natural killer cells. Blood. 2012;119:3478–3485. doi: 10.1182/blood-2011-12-398099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng Y., Ren J., Zhao J. MicroRNA-155 regulates interferon-γ production in natural killer cells via Tim-3 signalling in chronic hepatitis C virus infection. Immunology. 2015;145:485–497. doi: 10.1111/imm.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang H., Zhang Y., Wu X. Regulation of human natural killer cell IFN-γ production by microRNA-146a via targeting the NF-κB signaling pathway. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu D., Han Q., Hou Z., Zhang C., Zhang J. miR-146a negatively regulates NK cell functions via STAT1 signaling. Cell Mol Immunol. 2017;14:712–720. doi: 10.1038/cmi.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang Y., Jin Z., Dong R. MicroRNA-29b/142-5p contribute to the pathogenesis of biliary atresia by regulating the IFN-γ gene. Cell Death Dis. 2018;9(5) doi: 10.1038/s41419-018-0605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guo H.Q., Huang G.L., Guo C.C., Pu X.X., Lin T.Y. Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis Markers. 2010;29:251–258. doi: 10.3233/DMA-2010-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kitadate A., Ikeda S., Teshima K. MicroRNA-16 mediates the regulation of a senescence-apoptosis switch in cutaneous T-cell and other non-Hodgkin lymphomas. Oncogene. 2016;35:3692–3704. doi: 10.1038/onc.2015.435. [DOI] [PubMed] [Google Scholar]
- 102.Chang T., Yu D., Lee Y. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]