Abstract
Background
Third-generation cephalosporin-resistant Gram-negatives (3GCR-GN) and vancomycin-resistant enterococci (VRE) are common causes of multi-drug resistant healthcare-associated infections, for which gut colonisation is considered a prerequisite. However, there remains a key knowledge gap about colonisation and infection dynamics in high-risk settings such as the intensive care unit (ICU), thus hampering infection prevention efforts.
Methods
We performed a three-month prospective genomic survey of infecting and gut-colonising 3GCR-GN and VRE among patients admitted to an Australian ICU. Bacteria were isolated from rectal swabs (n = 287 and n = 103 patients ≤2 and > 2 days from admission, respectively) and diagnostic clinical specimens between Dec 2013 and March 2014. Isolates were subjected to Illumina whole-genome sequencing (n = 127 3GCR-GN, n = 41 VRE). Multi-locus sequence types (STs) and antimicrobial resistance determinants were identified from de novo assemblies. Twenty-three isolates were selected for sequencing on the Oxford Nanopore MinION device to generate completed reference genomes (one for each ST isolated from ≥2 patients). Single nucleotide variants (SNVs) were identified by read mapping and variant calling against these references.
Results
Among 287 patients screened on admission, 17.4 and 8.4% were colonised by 3GCR-GN and VRE, respectively. Escherichia coli was the most common species (n = 36 episodes, 58.1%) and the most common cause of 3GCR-GN infection. Only two VRE infections were identified. The rate of infection among patients colonised with E. coli was low, but higher than those who were not colonised on admission (n = 2/33, 6% vs n = 4/254, 2%, respectively, p = 0.3). While few patients were colonised with 3GCR- Klebsiella pneumoniae or Pseudomonas aeruginosa on admission (n = 4), all such patients developed infections with the colonising strain. Genomic analyses revealed 10 putative nosocomial transmission clusters (≤20 SNVs for 3GCR-GN, ≤3 SNVs for VRE): four VRE, six 3GCR-GN, with epidemiologically linked clusters accounting for 21 and 6% of episodes, respectively (OR 4.3, p = 0.02).
Conclusions
3GCR-E. coli and VRE were the most common gut colonisers. E. coli was the most common cause of 3GCR-GN infection, but other 3GCR-GN species showed greater risk for infection in colonised patients. Larger studies are warranted to elucidate the relative risks of different colonisers and guide the use of screening in ICU infection control.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-021-06386-z.
Keywords: Antimicrobial resistance (AMR), Colonisation, Transmission, Genomic surveillance, Intensive care
Introduction
Healthcare associated infections (HAI) result in considerable morbidity and mortality, with the total burden in high income countries exceeding that of influenza, tuberculosis and other major communicable diseases combined [1]. The emergence and spread of antimicrobial resistant (AMR) organisms, including difficult-to-treat multi-drug resistant (MDR) strains, has further exacerbated this problem [2], and as a consequence several MDR healthcare-associated bacterial pathogens are now recognised by the World Health Organization (WHO) as urgent threats to public health [3]. Among the WHO’s top priorities are carbapenem-resistant Gram-negatives (GNs, specifically Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa) as well as vancomycin-resistant enterococci (VRE).
In Australia, it is estimated that one in ten hospitalised patients suffers an HAI, and the most common MDR organisms are VRE, methicillin-resistant Staphylococcus aureus and extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae [4]. Fortunately carbapenem-resistance has so far remained rare, although the prevalence has been slowly increasing [5]. Notably, Australia suffers a particularly high rate of vancomycin resistance among enterococcal infections (~ 45% of Enterococcus faecium bacteremias [5]), primarily due to the endemic spread of healthcare-associated E. faecium sequence types (STs) 17, ST80 and ST796 [5–8].
Asymptomatic gut colonisation is considered a key risk factor for enterococcal, Enterobacteriaceae and other GN infections [9–13] and in the case of VRE has been associated with longer ICU stay and treatment costs [12, 14]. A number of hospitals have implemented rectal screening programs to identify patients colonised with VRE and/or carbapenemase-producing Enterobacteriaceae in high-risk wards such as intensive care units (ICUs), oncology and hematology wards [7, 8, 15, 16]. However, few facilities conduct routine screening for ESBL-Enterobacteriaceae or other high-risk carbapenem-susceptible GNs in part due to a lack of evidence to inform the interpretation of these results and to appropriately target infection control measures. For example, only a small number of studies have investigated the risks associated with ESBL-Enterobacteriaceae colonisation at a species-specific level [9, 11, 12, 17–20], but there is emerging evidence to suggest important variation, both in terms of the risk of infection and the risk of transmission in the hospital setting. We propose that a better understanding of these risks, as well as the broader colonisation and infection dynamics of AMR organisms in these settings, will facilitate better targeting of infection prevention and control practices.
Here we report a genome-resolved snapshot of high-risk AMR bacteria from rectal swabs and clinical specimens collected in an Australian ICU over a three-month period. We focus on GN organisms resistant to third-generation cephalosporins (including ESBL-Enterobacteriaceae) in addition to VRE. We leverage the genome data to perform high-resolution analysis of these bacterial populations to; i) compare infecting and colonising organisms isolated form the same patient; ii) characterise population diversity at the strain level; and iii) identify putative nosocomial transmission clusters.
Methods
Recruitment and specimen collection
The Alfred Hospital, Melbourne, Australia is a 350-bed tertiary referral hospital including a 45-bed ICU providing care for general medical and surgical patients plus specialist cardiac and trauma services. We conducted a prospective surveillance study for rectal colonisation of third generation cephalosporin-resistant GN (3GCR-GN) organisms and VRE in patients admitted to the ICU from 21 Dec 2013 to 7 April 2014. This study was approved by the Alfred Health Human Research Ethics Committee, Project numbers #550/12 (19 February 2013) and #526/13 (10 December 2013). Patients aged ≥18 years and considered by nursing staff as likely to remain in the ICU > 2 days were eligible for inclusion. The requirement for informed consent to participate was waived by the same ethics committee (Alfred Health Human Research Ethics Committee) as screening for AMR organisms is considered an infection control and surveillance activity, however patients were given the option to refuse screening. Rectal swabs were collected at time of recruitment (day 0–2 of ICU admission) and subsequently each 5–7 days during ICU stay. Information on age, sex, dates of hospital and ICU admission/s, surgery in the last 30 days, and antibiotic treatment in the last 7 days were extracted from hospital records when swabs were taken. Dates of discharge and/or death were extracted from hospital records at the conclusion of the study. All clinical isolates recovered from ICU patients and identified as 3GCR-GN or VRE by the diagnostic laboratory as part of routine care were also stored for inclusion.
Bacterial culture and sequencing
Gut colonising 3GCR-GN and VRE were identified by culture on ceftazidime and chromID VRE agar plates (BioMérieux, Marcy L’Etoile, France), respectively. Presumptive 3GCR-GN and VRE colonies were subjected to species identification via matrix-assisted laser desorption ionization time-of-flight analysis with a Vitek MS (BioMérieux). Clinical isolates were collected and identified via standard diagnostic protocols [10]. Genomic DNA was extracted and sequenced on the Illumina platform. Twenty-three isolates were selected for sequencing on the Oxford Nanopore MinION platform as described previously [21]. See Supplementary Methods, Additional File 1 for full details.
Genomic analyses
Genomes were assembled using Unicycler v0.4.7 [22] and subjected to quality control (Supplementary Methods, Additional File 1). Species were identified using Kraken v1.0 [23] and multi-locus sequence types (STs) were identified from assemblies using mlst (github.com/tseemann/mlst) where schemes were available (accessed from pubmlst.org [24]).
AMR genes were identified from final assemblies using Kleborate (https://github.com/katholt/Kleborate) [25] which performs nucleotide and protein BLAST against a curated version of the CARD database, and results were interpreted in a species-specific manner (i.e. to distinguish intrinsic from acquired genes). These data were used to calculate the predicted number of acquired AMR classes per isolate (defined for each species as described in [26], Supplementary Methods, Additional File 1). MDR was defined as predicted acquired resistance for ≥3 drug classes [26].
Single nucleotide variants (SNVs) were identified by mapping Illumina reads to the relevant completed chromosomal reference genomes (ST-specific, Supplementary Table 1, Additional File 2) using Bowtie2 v2.2.9 [27] and SAMtools v1.9 [28], as implemented in the RedDog pipeline (github.com/katholt/RedDog) [29].
Statistical analyses
Statistical analyses were performed using R v3.6.3 [30] and the tidyverse package [31]. Fisher’s Exact test and proportion test were used to test for differences in count data in two-by-two contingency tables (proportion test was applied where any single count was ≤5, otherwise Fisher’s Exact test was used). The Wilcoxon rank sum test was used to test for differences in age distributions, wherein the data did not fit a normal or other distribution appropriate for parametric testing. Univariate and multivariate logistic regression models were used to test risk factors for 3GCR-GN and VRE colonisation at ICU admission.
Results
During the study period there were 716 patients admitted to the ICU, 31 (4.3%) patients had one or more 3GCR-GN infections (n = 41 isolates), and two (0.3%) had VRE infections (n = 2 isolates, both E. faecium, Fig. 1). Of 430 patients eligible for participation in rectal screening, 66 declined to participate and 311 contributed one or more rectal swabs (72.3% of eligible non-refusers). The majority of participants were swabbed within the first 2 days of ICU admission (baseline screening swabs, n = 287, 92.3%), including 79 with ≥1 additional follow-up swab (Fig. 1). Participants with baseline swabs were 64.5% male (n = 185), aged 18–93 years (median 57 years; IQR, 44–71 years, Supplementary Table 2, Additional File 1), and the majority (n = 218, 76.0%) were known to have had recent healthcare exposure prior to ICU admission (surgical procedure within the last 30 days, transferred from another ward with first admission > 2 days prior, or transferred from another hospital).
Fig. 1.
Prevalence of 3GCR-GN and VRE gut colonisation on admission to the ICU. Prevalence of third generation cephalosporin-resistant Gram-negative (3GCR-GN) organisms (A) and VRE (vancomycin-resistant enterococci) organisms (C) among males and females for whom rectal swabs were collected within two days of ICU admission. Age distributions are shown for culture-confirmed 3GCR-GN (B) and VRE (D) carriers and non-carriers, stratified by sex. ns, non-significant using Fisher’s Exact Test (A, C) or Wilcoxon Rank Sum test (B, D); *, p < 0.05
Third generation cephalosporin-resistant (3GCR) GN organisms were isolated from baseline screening swabs of 50 patients (17.4%, n = 56 isolates), while VRE were isolated from 24 (8.4%, n = 24 E. faecium isolates, no E. faecalis were identified; Supplementary Figure 1, Additional File 1). Co-colonisation with 3GCR-GN and VRE was identified in 12 patients (4.2%), indicating a significant association between these organisms (OR 5.6, 95% CI 2.2–15.5, p = 0.0001 using Fisher’s Exact Test). There were no significant differences in colonisation rates between males and females, and no association between age and colonisation status, with the exception of VRE in females (median age 73.5 years amongst carriers versus 55.5 years amongst non-carriers, p = 0.04 using Wilcoxon Rank Sum test; Fig. 1). Neither surgery within the last 30 days, recent healthcare exposure (defined as above), or antibiotic treatment within the last 7 days, were significantly associated with 3GCR-GN or VRE colonisation at baseline (using univariate and multivariate logistic regression, with age and sex as covariates; Supplementary Table 3, Additional File 1).
A total of 138 rectal swabs were collected from 103 patients between 3 days and 12 weeks after ICU admission (Supplementary Table 4, Supplementary Figure 2, Additional File 1). Twenty-five swabs (18.1%) from 18 patients (17.5%) were positive for ≥1 3GCR-GN organism (n = 30 isolates); 15 swabs (10.9%) from ten patients (9.7%) were positive for VRE (n = 15 isolates). Notably, 3GCR-GN organisms were cultured from 15/61 patients (24.6%) who were culture-negative for 3GCR-GN colonisation at baseline, and VRE were cultured from 13/75 (17.3%) patients who were culture-negative for VRE colonisation at baseline, consistent with acquisition and/or overgrowth of the organisms during ICU stay.
Among the 127 3GCR-GN and 41 VRE isolates cultured from ICU patients, 112 (88.2%) and 41 (100%) respectively were successfully revived and sequenced (Supplementary Methods, Supplementary Figure 1, Additional File 1). We combined clinical and genome data to identify distinct colonisation and infection episodes, defined as unique combinations of species and ST (derived from whole-genome sequences (WGS)), per patient and specimen type. This identified 33 3GCR-GN and two VRE infection episodes (Table 1). The latter included one urinary tract infection for which the patient was prescribed amoxycillin (as recommended for treatment of VRE in the urinary tract, where a high concentration of amoxycillin can be achieved) and one respiratory infection treated with teicoplanin. 3GCR-GN infections were mainly respiratory (n = 19, 57.6%), wound (n = 5, 15.2%) and bloodstream (n = 4, 12.1%) infections, and the most common agents were Escherichia coli (10 episodes, 30.3%) and Klebsiella pneumoniae (6 episodes, 18.2%; Table 1, Fig. 2). E. coli was also the most common 3GCR-GN gut coloniser (36/62 3GCR-GN colonisation episodes, 58%), followed by Enterobacter hormaechei, K. pneumoniae, Klebsiella aerogenes, and Klebsiella oxytoca (four episodes, 6.5%, each, Table 1). Thirty-three unique VRE colonisation episodes were identified. Overall, 88 patients were colonised and/or infected with 3GCR-GN and/or VRE. Most (62, 70.5%) experienced a single episode of either 3GCR-GN (n = 61) or VRE (n = 1), however 17 patients (20.2%) had two episodes (16 patients with two different species/STs, 13 with one VRE and one 3GCR-GN species) and nine patients (10.1%) had ≥3 episodes (seven with ≥3 different species/STs, including five with VRE).
Table 1.
Colonisation and infection episodes by species
| Species | Total colonisation episodes (n baseline) | Infection episodes | |||
|---|---|---|---|---|---|
| Total | Pt colonised with same strain (n baseline) | Pt colonised at baseline (%) | Pt not colonised at baseline (%) | ||
| VRE | |||||
| Enterococcus faecium | 33 (25) | 2 | 0 | 0 | 1 (0.4%) |
| 3GC-resistant GN | |||||
| Escherichia coli | 36 (33) | 10 | 1 (0) | 2 (6%) | 4 (2%) |
| Klebsiella pneumoniae | 4 (2) | 7 | 3 (2) | 2 (100%) | 3 (1%)* |
| Pseudomonas aeruginosa | 3 (2) | 2 | 1 (1) | 2 (100%) | 0 (0%)* |
| Enterobacter hormaechei | 4 (3) | 3 | 0 | 0 | 1 |
| Klebsiella aerogenes | 4 (1) | 3 | 0 | 0 | 1 |
| Enterobacter asburiae | 0 | 2 | 0 | 0 | 1 |
| Stenotrophomonas maltophilia | 0 | 2 | 0 | 0 | 1 |
| Serratia marcescens | 1 (0) | 2 | 0 | 0 | 0 |
| Achromobacter xylosoxidans | 0 | 1 | 0 | 0 | 1 |
| Burkholderia cenocepacia | 0 | 1 | 0 | 0 | 1 |
| Citrobacter freundii | 2 (2) | 1 | 0 | 0 | 0 |
| Citrobacter portucalensis | 1 (1) | 0 | 0 | 0 | 0 |
| Enterobacter kobei | 1 (0) | 0 | 0 | 0 | 0 |
| Escherichia marmotae | 1 (1) | 0 | 0 | 0 | 0 |
| Klebsiella oxytoca | 4 (4) | 0 | 0 | 0 | 0 |
| Morganella morganii | 1 (0) | 0 | 0 | 0 | 0 |
* p < 0.00005 for association between colonisation at baseline and infection with same species (proportion test). Distinct episodes of colonisation and infection were defined as those with a unique combination of patient ID, specimen type, species and multi-locus sequence type. Colonisation episodes are expressed as total detected and number detected at baseline (within first 2 days after ICU admission). Strains were defined on the basis of chromosomal SNVs (≤7 SNVs between isolates). Pt patient, VRE vancomycin-resistant enterococci
Fig. 2.
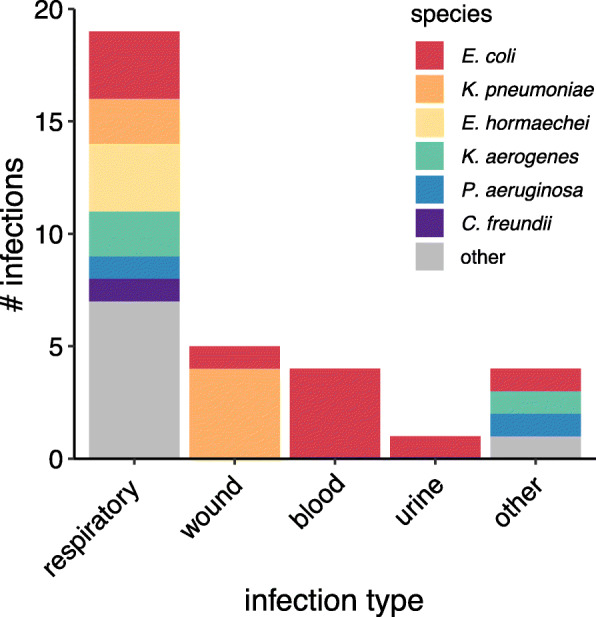
Infections caused by 3GCR-GN organisms. Bar chart shows the number of distinct respiratory, wound, blood, urine and other infections by species as indicated in the legend. Distinct infections were defined as those represented by unique combinations of species, multi-locus sequence type, patient and specimen type
The majority of 3GCR-GN colonisation and infection isolates were predicted to be MDR (see Methods): n = 44/61 (72%) distinct colonisation episodes and n = 18/29 (62%) distinct infection episodes. MDR was particularly common among E. coli (n = 44 episodes, 96%), K. pneumoniae (n = 9, 90%) and Serratia marcescens (n = 3, 100%). WGS identified acquired ESBL genes in 20 representative 3GCR-GN colonisation (32.3%) and 12 representative 3GCR-GN infection (36.4%) isolates. These were mostly MDR K. pneumoniae carrying blaCTX-M-15 (n = 8) or MDR E. coli carrying diverse blaCTX-M genes (n = 19, including n = 6 blaCTX-M-14, n = 5 blaCTX-M-15, n = 3 blaCTX-M-27, n = 3 blaCTX-M-62, n = 2 blaCTX-M-24-like). Carbapenemase genes were rare: two patients had wound or tissue infections with K. pneumoniae ST231 carrying blaOXA-48, one of whom was also colonised by K. pneumoniae ST231 with blaOXA-48. A third patient was colonised with S. marcescens carrying blaIMP-4. One of the patients with K. pneumoniae ST231 wound / tissue infections received a course of meropenem (this patient also had 3GCR P. aeruginosa and 3GCR Burkholderia cenocepacia infections), while the second received meropenem, ciprofloxacin and tigecycline. The S. marcescens rectal colonisation episode was not treated.
Most VRE isolates representing distinct episodes (n = 32, 91%) carried the vanB vancomycin resistance operon and the remainder (n = 3, 9%) the vanA operon, which confers resistance to vancomycin plus teicoplanin [32]. Additionally, 32 of these isolates (91%) were predicted to be MDR due to the presence of genes conferring resistance to tetracyclines and/or high-level resistance to gentamicin, streptomycin and/or streptogramins.
We assessed species-specific infection rates amongst patients testing culture-positive at baseline screening, versus those testing culture-negative, for all species with ≥2 infections identified amongst patients with baseline screening data (E. coli, K. pneumoniae, P. aeruginosa). Infection rates were higher amongst baseline-culture-positive patients, although the difference was statistically significant only for K. pneumoniae and P. aeruginosa (Table 1). There were few patients for whom WGS data were available for isolates of the same species detected in baseline screening and infections (two patients for each species). For n = 2/2 such cases with K. pneumoniae (ST231, ST323) and n = 1/2 with P. aeruginosa (ST357), the WGS data confirmed the infections were caused by the colonising strain (0–7 pairwise SNVs), whereas both E. coli infections were caused by different strains to those detected on baseline screening swabs (different STs; Table 1, Supplementary Results, Additional File 1). A further two patients tested culture-negative on baseline screening, but subsequently tested positive on follow-up screening for the same strain that was isolated from their diagnostic specimens (K. pneumoniae ST323, E. coli ST393; Supplementary Results, Additional File 1).
Finally, we assessed strain level diversity and evidence for nosocomial transmission using WGS and patient data. The isolates were diverse, with 62 STs identified amongst 3GCR-GN (24 amongst E. coli alone, including n = 6 ST131 episodes, n = 4 ST10 and n = 3 episodes each of ST38, ST69, ST176, ST648 and ST963) and 8 amongst VRE (including n = 14 ST796, n = 10 ST17 and n = 5 ST203 episodes). Most 3GCR-GN STs (n = 48, 77%) were unique to a single patient; however, ten E. coli STs (42%), two K. pneumoniae (40%), one K. oyxtoca (33%), one Enterobacter hormachaei (17%) and five VRE STs (63%) were found in multiple patients (Fig. 3a). We defined probable strain transmission events on the basis of chromosomal SNVs (see Methods, Fig. 3b-c). Using a threshold of ≤20 SNVs for transmission of 3GCR-GN (based on recent studies [10, 15, 16, 33]) we identified six putative 3GCR-GN transmission clusters each involving 2–3 patients (maximum 8 SNVs, Table 2). With a single exception, all patients within clusters were epidemiologically linked (overlapping ICU stays or ≤ 14 days between stays). Assuming one patient in each epidemiologically linked cluster was the index patient, the data suggest only six (6.3%) 3GCR-GN episodes resulted from recent nosocomial transmission (n = 2, 6.1% of infections; n = 4, 6.5% of colonisation). We also identified K. pneumoniae ST323 isolates from two patients that differed by 45–47 SNVs, notably many fewer than the majority of comparisons (Fig. 3a) and within the range expected for K. pneumoniae subject to wider circulation in the healthcare network rather than those subject to community-transmission [33]. Consistent with this, we have previously shown that K. pneumoniae ST323 were circulating more broadly within the hospital during the study period [34].
Fig. 3.
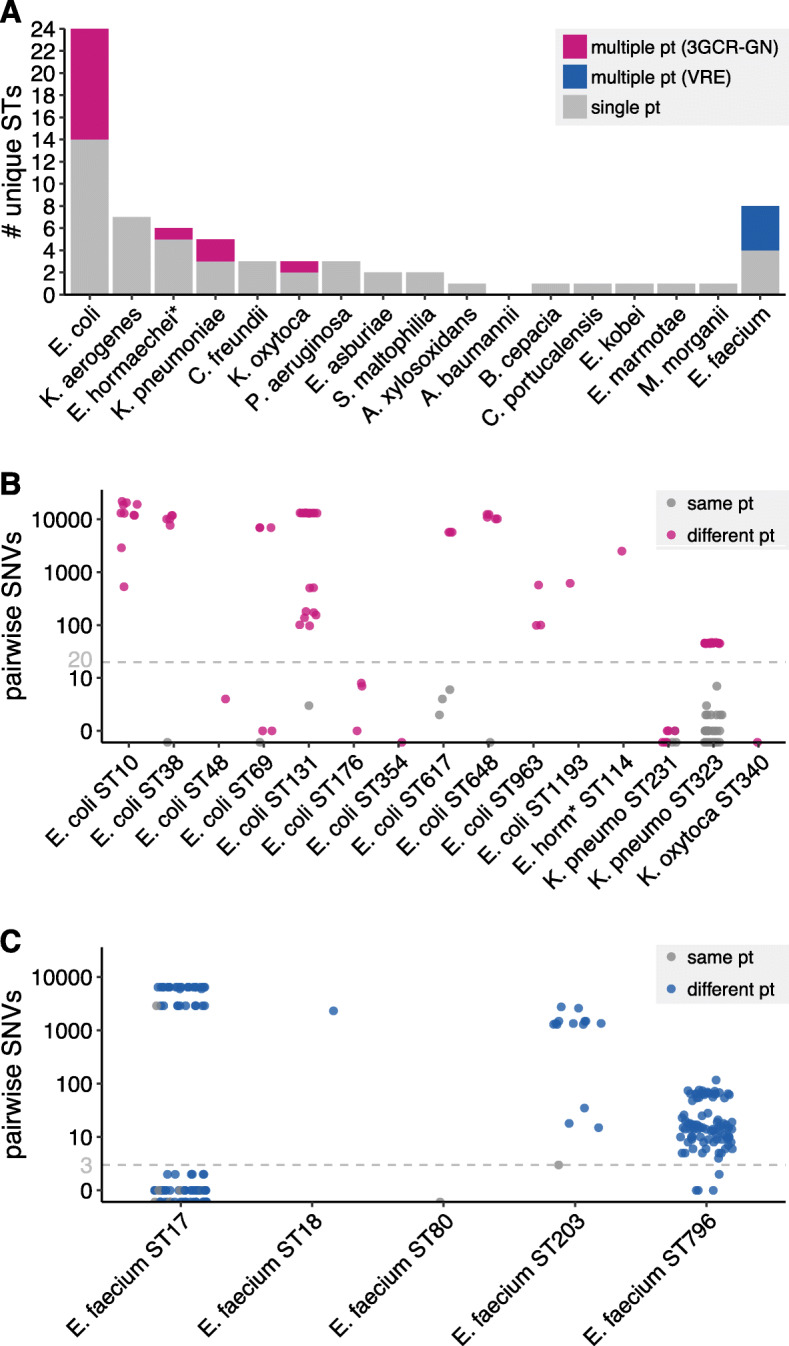
3GCR-GN and VRE strain diversity. (A) Bar plot showing the number of unique multi-locus sequence types (STs) identified for each third-generation cephalosporin-resistant Gram-negative (3GCR-GN) species plus vancomycin-resistant E. faecium (VRE) Bars are coloured as indicated in the legend. (B, C) Pairwise single nucleotide variants (SNVs) for pairs of isolates assigned to the same ST for 3GCR-GN species (B) and VRE (C). Grey points indicate pairs of isolates from the same patient (pt), pink and blue points indicate pairs of isolates from different patients for 3GCR-GN and VRE isolates, respectively. Grey dashed lines indicate the SNV thresholds used to define putative transmissions (n ≤ 20 SNVs for 3GCR-GN and n ≤ 3 SNVs for VRE, see Results for details). *E. hormaechei multi-locus sequence typing was performed using the Enterobacter cloacae scheme, which covers both species
Table 2.
Putative nosocomial transmission clusters
| # | Organism | Cluster size (n pt) | Pairwise SNVs | Days between pt. stays | Episodes attributed to transmission (n total (n very likely)) c | |
|---|---|---|---|---|---|---|
| Genomic a | Genomic + epi b | |||||
| 1 | E. coli ST48 | 2 | 2 | 4 | 8 | 1 (0) |
| 2 | E. coli ST69 | 2 | 2 | 1 | 0 | 1 (1) |
| 3 | E. coli ST176 | 3 | 2 | 1–8 | 0–31 | 1 (1) |
| 4 | E. coli ST354 | 2 | 2 | 0 | 6 | 1 (0) |
| 5 | K. pneumoniae ST231 | 2 | 2 | 0–2 | 0 | 1 (1) |
| 6 | K. oxytoca ST340 | 2 | 2 | 0 | 0 | 1 (1) |
| 7 | E. faecium ST17 | 2 | 2 | 1 | 11 | 1 (0) |
| 8 | E. faecium ST17 | 7 | 7 | 0–2 | 0 | 6 (6) |
| 9 | E. faecium ST796 | 2 | 2 | 1 | 1 | 1 (0) |
| 10 | E. faecium ST796 | 3 | 0 | 1–2 | 20–62 | 0 (0) |
Pt, patient; SNVs, single nucleotide variants
aPatient cluster identified by genomic analysis; ≤ 20 single nucleotide variants between isolates (Gram-negative species), or ≤ 3 single nucleotide variants between isolates (E. faecium)
bPatient cluster identified by genomic analysis (as defined above) and supported by epidemiological data (≤ 14 days between patient ICU stays or overlapping stays)
cNumber of colonisation and infection episodes likely or very likely attributed to transmission; likely, patient was part of the genomic cluster (as defined above) with ≤14 days between the patient ICU stay and that of another patient in the genomic cluster; very likely, patient was part of the genomic cluster (as defined above) and ICU stay overlapped with ≥1 other patient in the genomic cluster
Interpretation of VRE pairwise SNVs is more complex because several healthcare-associated lineages are known to be circulating in Melbourne and the regional hospital network. Consequently, it is not uncommon to find near-identical isolates in patients from different wards, hospitals or cities without epidemiological links [7, 8, 15]. We therefore used a more conservative threshold of ≤3 SNVs to define putative recent VRE transmissions (based on empirical distribution, see Supplementary Figure 3), which identified four clusters of 2–7 patients each (Table 2). Two clusters were ST17 (both epidemiologically linked clusters, separated by ≥2900 SNVs) and two were the locally emerged ST796 [35, 36] (clusters separated by 4–6 SNVs, one was epidemiologically linked). Assuming one patient in each epidemiologically linked cluster is the index patient, we estimate eight (28.6%) VRE episodes resulted from recent transmission (n = 1 infection and n = 7, 21.2% of colonisation episodes). This should be considered a conservative estimate considering the difficulty in distinguishing clusters, particularly for ST796. Nevertheless, the transmission burden is significantly greater than that estimated for 3GCR-GN organisms (OR = 4.3, 95% CI 1.2–16.6, p = 0.02 using Fisher’s Exact test).
Discussion
We have presented a holistic investigation of 3GCR-GN and VRE colonising and infecting patients in an Australian ICU over a three-month duration. Approximately 17% patients were colonised by at least one 3GCR-GN at ICU admission. Just under 40% of these isolates were detected as ESBL-Enterobacteriaceae, hence our data are comparable to reports from Europe where 7–13% of ICU-admitted patients were colonised with these organisms [17, 19, 37], and indicate a lower prevalence of ESBL-Enterobacteriaceae colonisation than reported among patients admitted to ICUs in China (32%) [38] and Thailand (62%) [39]. Additionally, 9% of patients in our study were colonised with VRE at ICU admission; a similar prevalence to that reported recently for patients in a hospital network in Singapore [40], higher than that reported for patients admitted to an ICU in Sri Lanka (2%) [41], and lower than reported in Brazil (15%) [42] and India (31%) [43]. Notably, the VRE colonisation prevalence was lower than the 17.5% (95% CI 13.7–21.9) indicated in a previous point-prevalence survey of patients in our hospital [44].
E. coli was the dominant colonising and infecting organism, and has been previously reported as the most common cause of GN blood-stream infections in Australia [5]. These findings are consistent with the notion of colonisation as a prerequisite for disease and the logic of scales, whereby the most common colonisers have more opportunities to cause infection and therefore contribute the greatest burden of disease. Our data indicated a higher prevalence of infections among patients colonised at admission with 3GCR E. coli, K. pneumoniae or P. aeruginosa than those who were not colonised. However, there were differences in the rate of infection; just 6% for E. coli, versus 100% (n = 2/2 each) for K. pneumoniae and P. aeruginosa. While these data must be interpreted with caution due to the small sample size, we note that the trends are consistent with previous reports [9, 17] including a large study of 2386 patients colonised by ESBL-K. pneumoniae or ESBL-E. coli, which found that the former were twice as likely as the latter to develop an infection with the same organism (6.8% versus 3.2% [9]).
At the time of this study our hospital infection prevention and control (IPC) policy stated that all patients known to have a clinical VRE and/or carbapenemase-producing Enterobacteriaceae (metallo-beta-lactamases only) infection or colonisation were isolated and subject to contact precautions during their hospital admission. Nevertheless, our WGS analysis identified four putative VRE transmission clusters (including three that were epidemiologically linked) and indicated that 21.2% of VRE episodes were attributable to transmission. In contrast, we estimated that just 6.3% of 3GCR-GN episodes were attributable to transmission (21.2% VRE vs 6.3% 3GCR-GN, OR 4.3, p = 0.0209), resulting from six putative 3GCR-GN transmission clusters (all epidemiologically linked). However, no specific IPC policies were applied for patients known to be colonised or infected with 3GCR-GN organisms beyond standard care.
The putative VRE transmission clusters involved ST17 and ST796, which are considered healthcare-adapted lineages and are widely distributed in Australia [5–8, 36, 45]. While four of the six 3GCR-GN transmission clusters involved E. coli, these included just four of 24 distinct E. coli STs, and did not include the globally distributed ST131, ST10 or ST38. This is consistent with a recent report showing that these E. coli STs were commonly identified from patients in other Melbourne hospitals, but rarely associated with nosocomial transmission [15]. Hence our data adds to the growing evidence base [15, 18–20, 46] that ESBL-E. coli are mainly spread in the community setting.
While the risk of ESBL-E. coli transmission in hospitals may be low, there is emerging evidence that the risk may be 2–4 times greater for other ESBL-Enterobacteriaceae such as K. pneumoniae and Enterobacter sp. [19, 20]. In our study, 2/11 (18.2%) ESBL-K. pneumoniae episodes were attributed to recent transmission, with one additional episode attributed to broader transmission within the hospital as reported previously [34]; however, a larger sample size is required to make a definitive comparison.
The inclusion of contemporaneous rectal screening and clinical isolates is a key strength of this study, which in combination with the breadth of species sampling and use of WGS, has allowed high-resolution analysis of the dynamics of AMR organisms in the ICU. Notably, the majority of putative transmissions we identified would have been missed if not for the inclusion of rectal screening. However, the study also has several limitations: firstly, we were not able to screen all eligible patients, nor to follow our patients beyond the three-month study period, thus we likely underestimate the true burdens of infection and transmission. Secondly, due to its short duration this study was underpowered for the assessment of patient risk factors (e.g. recent surgery or antibiotic use), which we expect to impact the risk of infection and onwards transmission. Thirdly, our focus on the ICU as a high-risk setting for HAI fails to capture the additional burden of 3GCR-GN and VRE in other wards. Finally, as noted above our assessment of infection risk among patients colonised with 3GCR-GN species other than E. coli was limited due to sample size. Nevertheless, in combination with the small number of existing reports our results highlight and contrast several key features of 3GCR-GN and VRE dynamics, and the importance of understanding species-specific risk(s).
Conclusions
Our data show that 3GCR-GN and VRE causing gut colonisation and infections in the ICU are highly diverse, and that the most common AMR colonisers (VRE and E. coli) do not necessarily pose the greatest risk to patients (in terms of incident infection, or onward transmission) in comparison to other gut colonising species. A more detailed assessment of species-specific risks (especially attack rate and risk of transmission) is needed to guide the efficient use of rectal screening programs and targeted approaches to IPC aimed at limiting the MDR disease burden.
Supplementary Information
Additional file 1. Supplementary Methods, Supplementary Results, Supplementary Tables 2-6, Supplementary Figures 1-3.
Additional file 2: Supplementary Table 1. Specimen and genotype information of isolates included in this study.
Acknowledgements
We gratefully acknowledge the contribution and support of Janine Roney, Mellissa Bryant, Jennifer Williams, and Noelene Browne at the Alfred Hospital, and the sequencing team at the Wellcome Trust Sanger Institute. We also acknowledge the curators of the relevant MLST schemes and the developers of the PubMLST website (https://pubmlst.org/), Keith Jolley and Martin Maiden. The development of that website was funded by the Wellcome Trust.
Abbreviations
- ICU
Intensive care unit
- IPC
Infection prevention and control
- MDR
Multi-drug resistant
- AMR
Antimicrobial resistant
- 3GCR
Third generation cephalosporin resistant
- GN
Gram-negative
- VRE
Vancomycin resistant enterococci
- ST
Sequence type
- SNV
Single nucleotide variant
Authors’ contributions
AWJJ and KEH conceived and designed the study, and obtained funding. MM, NFP, JSG, KMW, DVP, SAM, IJA, NM, DWS collected and processed isolates and/or clinical data. LMJ and CLG performed DNA extractions and/or generated sequence data. KLW, JH and KEH designed data analyses. KLW, JH, RRW and KEH performed data analyses. KLW, JH, AWJJ and KEH drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Health and Medical Research Council of Australia (Project Grant 1043822, Investigator Grant APP1176192 to KLW), the Viertel Charitable Foundation of Australia (Senior Medical Research Fellowship to KEH.) and this work was supported, in whole or in part, by the Bill & Melinda Gates Foundation (OPP1175797). Under the grant conditions of the Bill and Melinda Gates Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version that might arise from this submission. The funders played no role in the design of the study, the collection, analysis or interpretation of data, or writing the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article (and its additional files and/or available via public data repositories: Bacterial isolate metadata and genotypes are listed in Supplementary Table 1, Additional File 2. Illumina read data are publicly available via the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under project accessions PRJEB6891 and PRJNA646837, and hybrid genome assemblies are publicly available via GenBank (https://www.ncbi.nlm.nih.gov/genbank/); accessions are listed in Supplementary Table 1, Additional File 2.
Declarations
Ethics approval and consent to participate
This study was approved by the Alfred Health Human Research Ethics Committee, Project numbers #550/12 (19 February 2013) and #526/13 (10 December 2013), and the requirement for informed consent to participate was waived by the same ethics committee as screening for AMR organisms is considered an infection control and surveillance activity. All methods were carried out in accordance with the relevant local guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kelly L. Wyres and Jane Hawkey contributed equally to this work.
Contributor Information
Kelly L. Wyres, Email: kelly.wyres@monash.edu
Adam W. J. Jenney, Email: a.jenney@alfred.org.au
References
- 1.Cassini A, Plachouras D, Eckmanns T, Abu Sin M, Blank HP, Ducomble T, Haller S, Harder T, Klingeberg A, Sixtensson M, Velasco E, Weiß B, Kramarz P, Monnet DL, Kretzschmar ME, Suetens C. Burden of six healthcare-associated infections on European population health: estimating incidence-based disability-adjusted life years through a population prevalence-based modelling study. PLoS Med. 2016;13(10):e1002150. doi: 10.1371/journal.pmed.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, Ouakrim DA, Oliveira TC, Struelens MJ, Suetens C, Monnet DL, Strauss R, Mertens K, Struyf T, Catry B, Latour K, Ivanov IN, Dobreva EG, Tambic Andraševic A, Soprek S, Budimir A, Paphitou N, Žemlicková H, Schytte Olsen S, Wolff Sönksen U, Märtin P, Ivanova M, Lyytikäinen O, Jalava J, Coignard B, Eckmanns T, Abu Sin M, Haller S, Daikos GL, Gikas A, Tsiodras S, Kontopidou F, Tóth Á, Hajdu Á, Guólaugsson Ó, Kristinsson KG, Murchan S, Burns K, Pezzotti P, Gagliotti C, Dumpis U, Liuimiene A, Perrin M, Borg MA, de Greeff SC, Monen JCM, Koek MBG, Elstrøm P, Zabicka D, Deptula A, Hryniewicz W, Caniça M, Nogueira PJ, Fernandes PA, Manageiro V, Popescu GA, Serban RI, Schréterová E, Litvová S, Štefkovicová M, Kolman J, Klavs I, Korošec A, Aracil B, Asensio A, Pérez-Vázquez M, Billström H, Larsson S, Reilly JS, Johnson A, Hopkins S. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization . Global priority list of antibiotic-resistant bacteria to guide research, discovery, and devlopment of new antibiotics. 2017. [Google Scholar]
- 4.Russo PL, Stewardson AJ, Cheng AC, Bucknall T, Mitchell BG. The prevalence of healthcare associated infections among adult inpatients at nineteen large Australian acute-care public hospitals: a point prevalence survey. Antimicrob Resist Infect Control. 2019;8(1):114. doi: 10.1186/s13756-019-0570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coombs G, Bell J, Daley D, Collignon P, Cooley L, Gottlieb T, et al. Australian group on antimicrobial resistance Sepsis outcome programs; 2018 report. Sydney: Australian Commisssion on Safety and Quality in Health Care; 2019.
- 6.Gorrie C, Higgs C, Carter G, Stinear TP, Howden B. Genomics of vancomycin-resistant Enterococcus faecium. Microb Genomics. 2019;5(7). 10.1099/mgen.0.000283. [DOI] [PMC free article] [PubMed]
- 7.Lee RS, Gonçalves Da Silva A, Baines SL, Strachan J, Ballard S, Carter GP, et al. The changing landscape of vancomycin-resistant Enterococcus faecium in Australia: a population-level genomic study. J Antimicrob Chemother. 2018;73(12):3268–78. 10.1093/jac/dky331. [DOI] [PubMed]
- 8.van Hal SJ, Ip CLC, Ansari MA, Wilson DJ, Espedido BA, Jensen SO, et al. Evolutionary dynamics of Enterococcus faecium reveals complex genomic relationships between isolates with independent emergence of vancomycin resistance. Microb Genomics. 2016;2:e000048. [DOI] [PMC free article] [PubMed]
- 9.Denkel LA, Maechler F, Schwab F, Kola A, Weber A, Pfäfflin F, et al. Infections caused by extended-spectrum beta-lactamase-producing Enterobacterales after rectal colonisation with ESBL-producing Escherichia coli or Klebsiella pneumoniae. Clin Microbiol Infect. 2019;S1198-743X:30627–30625. doi: 10.1016/j.cmi.2019.11.025. [DOI] [PubMed] [Google Scholar]
- 10.Gorrie CL, Wick RR, Edwards DJ, Strugnell RA, Pratt N, Garlick J, et al. Gastrointestinal carriage is a major reservoir of K. pneumoniae infection in intensive care patients. Clin Infect Dis. 2017;65(2):208–215. doi: 10.1093/cid/cix270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Souverein D, Euser SM, Herpers BL, Kluytmans J, Rossen JWA, Den Boer JW. Association between rectal colonization with highly resistant Gram-negative rods (HR-GNRs) and subsequent infection with HR-GNRs in clinical patients: a one year historical cohort study. PLoS One. 2019;14(1):e0211016. 10.1371/journal.pone.0211016. [DOI] [PMC free article] [PubMed]
- 12.Freedberg DE, Zhou MJ, Cohen ME, Annavajhala MK, Khan S, Moscoso D, et al. Pathogen colonization of the gastrointestinal microbiome at intensive care unit admission and risk for subsequent death or infection. Intensive Care Med. 2018;44(8):1203–1211. doi: 10.1007/s00134-018-5268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boldt AC, Schwab F, Rohde AM, Kola A, Bui MT, Märtin N, et al. Admission prevalence of colonization with third-generation cephalosporin-resistant Enterobacteriaceae and subsequent infection rates in a German university hospital. PLoS One. 2018;13:e0201548. doi: 10.1371/journal.pone.0201548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung E, Byun S, Lee H, Moon SY, Lee H. Vancomycin-resistant enterococcus colonization in the intensive care unit: clinical outcomes and attributable costs of hospitalization. Am J Infect Control. 2014;42(10):1062–1066. doi: 10.1016/j.ajic.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Sherry NL, Lee RS, Gorrie CL, Kwong JC, Stuart RL, Korman T, Marshall C, Higgs C, Chan HT, Graham M, Johnson PDR, Leroi MJ, Reed C, Richards MJ, Slavin MA, Worth LJ, Howden BP, Grayson ML, Controlling Superbugs Study Group Pilot study of a combined genomic and epidemiologic surveillance program for hospital-acquired multidrug-resistant pathogens across multiple hospital networks in Australia. Infect Control Hosp Epidemiol. 2021;42(5):573–581. doi: 10.1017/ice.2020.1253. [DOI] [PubMed] [Google Scholar]
- 16.Sherry NL, Lane CR, Kwong JC, Schultz M, Sait M, Stevens K, et al. Genomics for molecular epidemiology and detecting transmission of carbapenemase-producing Enterobacterales in Victoria, Australia, 2012-2016. J Clin Microbiol. 2019;57:1–12. doi: 10.1128/JCM.00573-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Razazi K, Mekontso Dessap A, Carteaux G, Jansen C, Decousser JW, de Prost N, Brun-Buisson C. Frequency, associated factors and outcome of multi-drug-resistant intensive care unit-acquired pneumonia among patients colonized with extended-spectrum β-lactamase-producing Enterobacteriaceae. Ann Intensive Care. 2017;7(1):61. doi: 10.1186/s13613-017-0283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prevel R, Boyer A, M’Zali F, Cockenpot T, Lasheras A, Dubois V, Gruson D. Extended spectrum beta-lactamase producing Enterobacterales faecal carriage in a medical intensive care unit: low rates of cross-transmission and infection. Antimicrob Resist Infect Control. 2019;8(1):112. doi: 10.1186/s13756-019-0572-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurieva T, Dautzenberg MJD, Gniadkowski M, Derde LPG, Bonten MJM, Bootsma MCJ. The transmissibility of antibiotic-resistant Enterobacteriaceae in intensive care units. Clin Infect Dis. 2018;66(4):489–493. doi: 10.1093/cid/cix825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilty M, Betsch BY, Bögli-Stuber K, Heiniger N, Stadler M, Küffer M, Kronenberg A, Rohrer C, Aebi S, Endimiani A, Droz S, Mühlemann K. Transmission dynamics of extended-spectrum beta-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin Infect Dis. 2012;55(7):967–975. doi: 10.1093/cid/cis581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wick RR, Judd LM, Gorrie CL, Holt KE. Completing bacterial genome assemblies with multiplex MinION sequencing. MGen. 2017;3(10):e000132 [DOI] [PMC free article] [PubMed]
- 22.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comp Biol. 2017;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;10:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:4188. 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed]
- 26.Magiorakos A, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 27.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NTK, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R Development Core Team R . R: A Language and Environment for Statistical Computing. 2011. [Google Scholar]
- 31.Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen T, Miller E, Bache S, Müller K, Ooms J, Robinson D, Seidel D, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H. Welcome to the Tidyverse. J Open Source Softw. 2019;4(43):1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 32.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37(8):1563–1571. doi: 10.1128/AAC.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4(11):1919–29. [DOI] [PMC free article] [PubMed]
- 34.Gorrie CL, Mirceta M, Wick RR, Judd LM, Wyres KL, Thomson NR, Strugnell RA, Pratt NF, Garlick JS, Watson KM, Hunter PC, McGloughlin SA, Spelman DW, Jenney AWJ, Holt KE. Antimicrobial resistant Klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to acquisition in the referring hospital. Clin Infect Dis. 2018;67(2):161–170. doi: 10.1093/cid/ciy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lister DM, Kotsanas D, Ballard SA, Howden BP, Carse E, Tan K, et al. Outbreak of vanB vancomycin-resistant Enterococcus faecium colonization in a neonatal service. Am J Infect Control. 2015;43(10):1061–5. 10.1016/j.ajic.2015.05.047. [DOI] [PubMed]
- 36.Buultjens AH, Lam MMC, Ballard S, Monk IR, Mahony AA, Grabsch EA, et al. Evolutionary origins of the emergent ST796 clone of vancomycin resistant Enterococcus faecium. PeerJ. 2017;5:e2916. 10.7717/peerj.2916. [DOI] [PMC free article] [PubMed]
- 37.Alves M, Lemire A, Decré D, Margetis D, Bigé N, Pichereau C, Ait-Oufella H, Baudel JL, Offenstadt G, Guidet B, Barbut F, Maury E. Extended-spectrum beta-lactamase - producing Enterobacteriaceae in the intensive care unit: acquisition does not mean cross-transmission. BMC Infect Dis. 2016;16(1):1–9. doi: 10.1186/s12879-016-1489-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X, Li G, Yi L, Li M, Wang J. The epidemiology of multidrug-resistant bacteria colonization and analysis of its risk factors in intensive care unit. Chinese Crit Care Med. 2015;27:667–671. doi: 10.3760/cma.j.issn.2095-4352.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Kiddee A, Assawatheptawee K, Na-Udom A, Boonsawang P, Treebupachatsakul P, Walsh TR, et al. Risk factors for extended-spectrum β-lactamase-producing Enterobacteriaceae carriage in patients admitted to intensive care unit in a tertiary Care Hospital in Thailand. Microb Drug Resist. 2019;25(8):1182–90. 10.1089/mdr.2018.0318. [DOI] [PubMed]
- 40.Tan D, Htun HL, Koh J, Kanagasabai K, Lim JW, Hon PY, et al. Comparative epidemiology of vancomycin-resistant enterococci colonization in an acute-care hospital and its affiliated intermediate- and long-term care facilities in Singapore. Antimicrob Agents Chemother. 2018;62:e01507–e01518. doi: 10.1128/AAC.01507-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kannangara C, Chandrasiri P, Corea EM. Vancomycin resistant enterococcal (VRE) colonization among patients treated in intensive care units at the National Hospital of Sri Lanka, and determination of genotype/s responsible for resistance. Ceylon Med J. 2018;63(4):154–158. doi: 10.4038/cmj.v63i4.8766. [DOI] [PubMed] [Google Scholar]
- 42.Batistão DW da F, Gontijo-Filho PP, Conceição N, de Oliveira AG, Ribas RM. Risk factors for vancomycin-resistant enterococci colonisation in critically ill patients. Mem Inst Oswaldo Cruz. 2012;107(1):57–63. [DOI] [PubMed]
- 43.Banerjee T, Anupurba S, Filgona J, Singh DK. Vancomycin-resistance Enterococcal colonization in hospitalized patients in relation to antibiotic usage in a tertiary Care Hospital of North India. J Lab Physicians. 2015;7(2):108–111. doi: 10.4103/0974-2727.163123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karki S, Houston L, Land G, Bass P, Kehoe R, Borrell S, et al. Prevalence and risk factors for VRE colonisation in a tertiary hospital in Melbourne, Australia: a cross sectional study. Antimicrob Resist Infect Control. 2012;1:31. doi: 10.1186/2047-2994-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howden BP, Holt KE, Lam MMC, Seemann T, Ballard S, Coombs GW, et al. Genomic insights to control the emergence of vancomycin-resistant enterococci. MBio. 2013;4:e00412–e00413. doi: 10.1128/mBio.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukerji S, Stegger M, Truswell AV, Laird T, Jordan D, Abraham RJ, Harb A, Barton M, O’Dea M, Abraham S. Resistance to critically important antimicrobials in Australian silver gulls (Chroicocephalus novaehollandiae) and evidence of anthropogenic origins. J Antimicrob Chemother. 2019;74(9):2566–2574. doi: 10.1093/jac/dkz242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Methods, Supplementary Results, Supplementary Tables 2-6, Supplementary Figures 1-3.
Additional file 2: Supplementary Table 1. Specimen and genotype information of isolates included in this study.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article (and its additional files and/or available via public data repositories: Bacterial isolate metadata and genotypes are listed in Supplementary Table 1, Additional File 2. Illumina read data are publicly available via the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under project accessions PRJEB6891 and PRJNA646837, and hybrid genome assemblies are publicly available via GenBank (https://www.ncbi.nlm.nih.gov/genbank/); accessions are listed in Supplementary Table 1, Additional File 2.



