Abstract
Mantle cell lymphoma (MCL) is considered one of the most aggressive lymphoid tumors. However, it sometimes displays indolent behavior in patients and might not necessitate treatment at diagnosis; this has been described as “smoldering MCL” (SMCL). There are significant differences in the diagnosis, prognosis, molecular mechanisms and treatments of indolent MCL and classical MCL. In this review, we discuss the progress in understanding the molecular mechanism of indolent MCL to provide insights into the genomic nature of this entity. Reported findings of molecular features of indolent MCL include a low Ki-67 index, CD200 positivity, a low frequency of mutations in TP53, a lack of SOX11, normal arrangement and expression of MYC, IGHV mutations, differences from classical MCL by L-MCL16 assays and MCL35 assays, an unmutated P16 status, few defects in ATM, no NOTCH1/2 mutation, Amp 11q gene mutation, no chr9 deletion, microRNA upregulation/downregulation, and low expression of several genes that have been valued in recent years (SPEN, SMARCA4, RANBP2, KMT2C, NSD2, CARD11, FBXW7, BIRC3, KMT2D, CELSR3, TRAF2, MAP3K14, HNRNPH1, Del 9p and/or Del 9q, SP140 and PCDH10). Based on the above molecular characteristics, we may distinguish indolent MCL from classical MCL. If so, indolent MCL will not be overtreated, whereas the treatment of classical MCL will not be delayed.
Keywords: Smoldering mantle cell lymphoma, Indolent, Molecular features
Background
Mantle cell lymphoma (MCL) is a mature B-cell neoplasm that accounts for 5% to 10% of all lymphomas [1, 2]. MCL represents a subtype of non-Hodgkin lymphoma with a poor prognosis and is considered to be one of the most aggressive lymphoid tumors [2–5]. However, some display indolent behavior in patients that might not necessitate treatment at diagnosis [3]. In recent years, it has become increasingly clear that MCL is more heterogeneous than we initially thought [6]. In the 2017 WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, MCL was classified into indolent leukemic non-nodal MCL or classical MCL. Classical MCL is characterized by unmutated/minimally mutated IGHV and mostly SOX11 + , whereas indolent leukemic non-nodal MCL is defined as MCL in which the patient presents with mutated IGHV and mostly SOX11– and peripheral blood, bone marrow, and sometimes splenic involvement but without significant adenopathy. Of note, in situ mantle cell neoplasia, a new name for in situ MCL, is also an indolent type of MCL, reflecting its low clinical risk (Table 1) [7]. However, we found that the significance of IGHV mutations and the expression level of SOX11 in MCL are controversial [8]. In addition, there are many terms used to describe indolent MCL, such as leukemic non-nodal MCL, early-stage MCL, limited-stage MCL, low-risk MCL, and small-MCL [9–15]. After reviewing a large number of previous studies, we recommend that all indolent MCL presentations be unified under the title “smoldering mantle cell lymphoma (SMCL)”. Our previously proposed definition of SMCL was as follows: lack of B symptoms, Ki-67 expression less than 30%, low MCL-International Prognostic Index (MIPI) score, normal serum LDH and b2-microglobulin levels, PET/CT with the SUVmax < 6, maximum tumor diameter less than 3 cm, spleen size < 20 cm, with some particular immunophenotype, such as CD5 (−) and CD38 (−), markedly increased CD23 positive lymphocyte proportions (compared to usual negative CD23), kappa light chain restriction (compared to typical lambda light chain restriction), high expression of CD200, without C-myc, NOTCH1/2 and TP53 mutation, nonblastoid/pleomorphic histology, and no tumor growth on reevaluation every 3 months, at least 6 months [16]. In recent years, various techniques, such as polymerase chain reaction (PCR), Sanger sequencing, and next-generation sequencing (NGS) approaches, including whole-exome sequencing (WES), whole-genome sequencing (WGS), and targeted panels, have revealed mutations with prognostic significance in MCL [2]. For example, TP53 mutations are associated with blastoid morphology, and CD200 positivity portends an indolent clinical course [1, 17]. However, there is no systematic analysis representative of the molecular features of indolent MCL. Therefore, in this review, we discuss the molecular characteristics and genomic landscape of indolent MCL to distinguish indolent MCL from classical MCL.
Table 1.
2017 WHO classification of mantle cell lymphoma
| Mantle cell lymphoma (MCL) | |
|---|---|
| Classical MCL | Mostly SOX11 ( +) |
| Unmutated/minimally mutated IGHV | |
| Indolent leukemic non-nodal MCLa | Mostly SOX11 (−) |
| Mutated IGHV | |
| In situ mantle cell neoplasia | New name for in situ MCL, reflecting low clinical risk |
aIndolent leukemic non-nodal MCL with peripheral blood, bone marrow, and sometimes splenic involvement, may become more aggressive
The difference in the Ki-67 index between indolent MCL and classical MCL
A low Ki-67 index may imply a more indolent form of MCL [18]. Kimura et al. also indicated that Ki-67 expression in small-MCL was lower than that in classical MCL [15]. Ki-67 is a nuclear protein involved in the regulation of cell proliferation [19]. It is not only the most frequently used determinant of cell proliferation in clinical practice but also a predictor of overall survival (OS) [6]. The higher the Ki-67 index, the shorter the time it takes for MCL to transform into blastic and/or pleomorphic MCL. However, its prognostic value in lymphoma is still inconclusive [19]. Some studies have shown that high Ki-67 expression is associated with poo survival rates, while others have shown no strong associations. Vose et al. demonstrated that a high Ki-67 index is associated with MCL subtypes that have a poor prognosis, such as the blastoid variant subtype [18]. Using Ki-67 positivity cutoffs of < 10%, 10–30% and > 30% of cells, Determann et al. demonstrated 3-year OS rates of 93%, 74% and 66% in MCL patients [6]. Moreover, Gallo et al. reported that the Ki-67 index was lower in leukemic non-nodal MCL (average 2%) than in classical MCL (40%) and aggressive MCL (76%), conferring a better prognosis [20]. Therefore, the proliferative activity reflected by the Ki-67 index has become a strong and independent variable for predicting survival in MCL patients. Hoster et al. suggested that the use of the Ki-67 index is superior to cytology and growth pattern as prognostic factors in MCL [21]. Except for MIPI, the Ki-67 index remains the only routinely available independent prognostic factor. However, the Ki-67 index was already accepted and incorporated into the MIPI in 2017 [22]. The modified combination of the Ki-67 index and MIPI showed a refined risk stratification, reflecting their strong complementary prognostic effects. Furthermore, in MCL, the Ki-67 index increases over time, and the only determinant of recurrence risk found was a Ki-67 level of > 30% [23]. In the study of Chakhachiro et al., of 11 patients with a Ki-67 level > 30%, seven experienced disease recurrence within the first 3 years, while only 3 of 16 patients with a Ki-67 level ≤ 30% experienced relapse. However, Medani et al. indicated that it is easier to perform precise counts and accurately evaluate proliferation indices via immunohistochemistry for phosphohistone H3 (PHH3), a reliable mitosis-specific marker in MCL, than via the Ki-67 index [24]. Moreover, Schrader and colleagues found that during early G1 phase, Ki-67 is undetectable, whereas minichromosome maintenance protein 6 (MCM6) is expressed throughout the entire G1 phase. Therefore, high MCM6 expression is a prognostic marker superior to the Ki-67 index in MCL because MCM6 may indicate early G1-phase arrest [25]. Another issue with the Ki-67 index is interobserver variability, making it difficult to determine the best cutoff values across laboratories [6]. Thus, the Ki-67 index plays an important role in the prognosis of MCL and the classification of subtypes [26]. Given that the Ki-67 index of indolent MCL is very low, Ki-67 may be an independent prognostic factor.
CD200 positivity in indolent MCL
CD200 expression in MCL indicates a unique subgroup. This subgroup is frequently accompanied by IGHV mutations and SOX11 negativity and portends an indolent clinical course [9, 10]. CD200, formerly known as OX-2, is a transmembrane type Ia glycoprotein expressed on thymocytes, activated B cells, T cells, dendritic cells, endothelial cells, and neurons. CD200 transmits inhibitory signals, resulting in the suppression of T-cell-mediated immune activation. CD200 and its ligand CD200R play important roles in the regulation of antitumor activity [27–30]. Recent reports have proven that CD200 immunophenotyping is useful in the differential diagnosis of B-cell neoplasms. CD200 is uncommonly expressed in classical MCL, but it has been noted that CD200 is frequently expressed in indolent MCL. Compared with patients with CD200-negative MCL, those with CD200 + MCL are more likely to have non-nodal leukemic presentation characterized by the loss of lymphadenopathy or extranodal and/or gastrointestinal tract disease [10, 31]. Therefore, CD200 expression is useful for identifying patients with MCL who may have an indolent course [30]. In addition, CD23-positive MCL is more often associated with CD200 positivity and weak SOX11 expression. Recent data have suggested that patients with CD23 + MCL are significantly more likely to have leukemic non-nodal presentation than those with CD23-negative MCL (42% vs 11%) [32]. Kelemen et al. showed that the frequency of bone marrow (BM) and peripheral blood involvement in patients with CD23 + MCL was similar, but extranodal nonmedullary disease was more common. In other words, although patients with CD23-positive MCL have a leukemic presentation similar to CLL, their prognosis is better than that of patients with CD23-negative MCL. In contrast, Saksena and colleagues indicated that patients with CD23 + MCL were more frequently in stage 4 disease with BM involvement and an elevated leukocyte count. In conclusion, indolent MCL is generally CD200 + , CD23 + , and SOX11-negative and has a leukemic presentation with features similar to those of CLL/SLL. In earlier studies, CD200 was proven to be a useful marker for distinguishing CLL from MCL via flow cytometry [27]. However, several recent studies have demonstrated that the IgH-cyclin D1 rearrangement is necessary for the differential diagnosis between MCL and CLL/SLL [30, 33]. CD200 is also a reliable auxiliary marker for classic prognostic factors [29]. The assessment of CD200 is helpful for distinguishing most cases of monoclonal asymptomatic lymphocytosis and cyclin D1–positive (MALD1) indolent MCL from classical MCL to avoid overdiagnosis and unnecessary treatment [17]. Thus, CD200 is frequently expressed in indolent MCL and is associated with CD23 and SOX11.
Abnormal expression of TP53
Mutations and deletions of the tumor protein p53 (TP53) gene are the most frequent genetic alterations detected in human tumors, although they are rather less common in lymphomas [34]. However, acquisition of the TP53 mutation has been proven to be one of the characteristic markers of MCL. Indeed, in MCL, 26% of cases contain TP53 mutations/deletions [35]. TP53 functions mainly as a transcription factor, phosphorylating at multiple sites, and responds to a large number of cellular stresses, such as cell cycle control, DNA repair, senescence, cell metabolism and apoptosis [36]. The phosphorylation and stabilization of TP53 induced by DNA damage is an obstacle to tumorigenesis. Recent studies have shown the poor prognostic impact of TP53 gene aberrations on MCL patients, including those with the indolent MCL subtype [5, 37, 38]. TP53 mutation and TP53 deletion are both associated with significant reductions in OS in patients with indolent MCL [39]. Conversely, the frequency of TP53 mutations has been reported to be much lower in the indolent variants of MCL [1]. However, TP53 mutations may be clonal or subclonal. The seemingly indolent MCL may contain subclonal TP53 mutations [9]. Several methods can be used to identify TP53 aberrations in clinical samples: (i) a negative feedback mechanism between MDM2 and TP53; (ii) DNA sequencing; (iii) functional assays, especially FASAY (functional analyses of separated alleles in yeast); and (iv) FISH (fluorescence in situ hybridization) [40]. In one study, Streich et al. demonstrated that the majority of TP53 mutations (75%) were associated with deletion of the chromosome arm 17p. Moreover, all TP53-mutated cases show strong TP53 expression by immunohistochemistry [41]. These studies also indicated that TP53 mutations were associated with blastoid morphology and a decreased response to chemotherapy. However, another study showed that the deletion of 17p may not have a prognostic impact [39]. Dong et al. found that TP53 mutations were correlated with a mutated IGHV status and CD38 negativity [42]. In a univariate analysis, TP53 mutations were identified as important predictors of survival, but they were insufficient to be used as independent prognostic factors in patients in the advanced stage. Zlamalikova et al. found 20 cases with loss of the TP53 locus, half of which harbored a concurrent TP53 mutation [34]. Compared to TP53 deletions, TP53 mutations are associated with significantly worse outcomes [5]. Similarly, in chronic lymphocytic leukemia (CLL), > 70% of patients with TP53 deletion also carry TP53 mutation. Therefore, there is a rather weak association between TP53 allelic deletion and TP53 mutation [34]. However, some scholars suggest a lack of prognostic significance for TP53 aberrations in patients with indolent MCL. This may be partly due to the frequent subclonal nature of these mutations in this variant [9]. Eskelund et al. showed the prognostic significance of TP53 mutations but not TP53 deletions. One possible explanation for this difference is that TP53 mutations destroy the function of the entire TP53 protein tetramer, and deletions only reduce the amount of transcription, so they may have little effect on protein function [43]. Furthermore, McCall et al. presented a rare case of CD5-negative non-nodal MCL accompanied by TP53 mutation/17p deletion, but the patient still achieved long progression-free survival (PFS) [44]. Some studies have indicated that the higher the MCL risk group, the higher the percentage of patients with > 50% TP53 expression, and the expression of TP53 is related to the outcome of MCL independent of the MIPI and Ki-67 level [14, 45]. However, some studies have suggested that stratifying patients by proliferation to separate them into low-, intermediate- and high-risk groups is more effective than stratifying them by any single driver mutation [45, 46]. Interestingly, Lin et al. showed no significant difference in prognosis between patients with and without TP53 alterations who received reduced-intensity or nonmyeloablative allogeneic hematopoietic cell transplantation, providing a beneficial treatment modality for these high-risk patients for the first time [47].
Overall, indolent MCL carries almost no TP53 mutation/deletion. However, a small portion of indolent MCL may harbor subclonal TP53 mutations. TP53 mutation and TP53 deletion are both associated with a poor prognosis independent of the MIPI, and TP53 mutations are associated with significantly worse outcomes than TP53 deletions. Moreover, TP53 mutations are correlated with CD38 negativity and hypermutated IGHV.
The expression level of SOX11
The clinical features of MCL are closely related to the expression level of SOX11. Leukemic mantle cell lymphoma limited to the blood and bone marrow is characterized by a lack of SOX11, mild-moderate lymphocytosis, and interstitial low-level bone marrow involvement [48]. SOX11, a member of the SOXC family, is a single-exon gene and a high-mobility transcription factor [3, 6]. The related transcription factors SOX11, SOX12 and SOX4 compete for the same target genes, and their developmental necessity is evident from single-gene knockout studies indicating that mice without SOX11 and SOX4 expression cannot survive because of heart outflow tract malformations [49]. SOX11 is considered an oncogene that can induce cell proliferation, enforce the expression of PAX5 and inhibit terminal B-cell differentiation into plasma cells via BCL6 and PRDM1 [50]. In addition, SOX11 mediates the expression of platelet-derived growth factor alpha (PDGFA), focal adhesion kinase (FAK) and C-X-C motif chemokine receptor 4 (CXCR4), which promote angiogenesis, metastasis and tumor cell migration, respectively [51]. MRQ-58 has been applied in the study of SOX11 and B cell lymphoma because it has been proven to be the most sensitive marker and does not cross-react with the SOX4 protein, which has high amino acid sequence similarity with SOX11 [52]. SOX11 is not expressed in normal lymphocytes but is widely expressed in MCL [6]. Previous studies have identified several direct targets of SOX11 in MCL, including DBN1, HIG2, SETMAR and WNT signaling molecules. More recently, an elegant integrative analysis of the epigenome in primary MCL demonstrated a distant regulatory element 675 kb downstream from the SOX11 gene that may influence transcriptional activity at the SOX11 promoter [53]. The expression of the SOX11 gene is high in classical MCL and almost absent in indolent MCL [4, 18]. Agata et al. found fewer chromosomal aberrations and more hypermutated immunoglobulin receptor genes in SOX11-negative MCL patients. The SOX11 promoter region was heterogeneously methylated in SOX11-negative primary MCL cases [49]. Moreover, SOX11 is a highly specific marker for both CCND1-positive and CCND1-negative MCL [15]. Variants with a low frequency of SOX11 negativity and IGHV-mutated genes are more indolent and associated with better prognosis. It is useful to confirm SOX11 negativity with hypermutated IGHV to identify a clearly indolent disease [54, 55]. Sílvia et al. demonstrated that SOX11 knockdown reduced engrafted tumor growth in vivo, which is consistent with the indolent clinical course of human SOX11-negative MCL [38]. However, the significance of SOX11 negativity in indolent MCL is still controversial [4]. A retrospective analysis of 186 MCL patients indicated no significant correlation between the absence of SOX11 and the prognosis of MCL [56]. In some studies, patients with SOX11-negative MCL even had a worse prognosis than those with SOX11-positive MCL [39, 57]. There are many factors that could have led to this conclusion: (i) the inclusion of patients with critical SOX11 expression levels, (ii) SOX11-negative cases could correspond to a progressive or transformed stage and generalized lymphadenopathy, (iii) technical difficulties, (iv) the lack of international guidelines for classifying MCL into treatment groups according to clinical behavior (indolent versus classical MCL), (v) the use of heterogeneously treated patients as a basis for the prognostic analysis of SOX11, (vi) potential cross-reactivity of the polyclonal reagents used, (vii) and other confounding factors, such as the presence of TP53 alterations [58]. A highly sensitive and specific in situ hybridization assay for the quantification of SOX11 mRNA in MCL revealed a close correlation between TP53 and negative/low SOX11 expression levels [50, 59]. SOX11-negative cases may represent indolent MCL that obtains TP53 mutations [60]. That is, SOX11-negative MCL initially has an indolent course but becomes aggressive when TP53 mutation is acquired [61]. Consequently, the expression of SOX11 alone should not be used to define aggressiveness. Christian et al. suggested that in both SOX11-negative and SOX11-positive subtypes, the stage of MCL lesions in situ was similar [62]. The expression of SOX11 in “in situ” mantle cell neoplasias suggested that the upregulation of this transcription factor was an early event in MCL [63]. Therefore, SOX11 should not be considered a mere “prognostic parameter” in MCL but rather a marker helping to distinguish the two subsets. However, Nygren et al. showed that a number of indolent MCLs express SOX11 and that in SOX11( +) MCL, indolent disease cannot be ruled out [64]. SOX11 has always been one of the controversial topics of research on indolent MCL. Venera et al. confirmed that SOX11 downregulated hypoxia-inducible gene 2 (HIG-2) at the protein level. HIG-2 knockdown leads to reduced levels of SOX11 [65]. This discovery may provide new clues for the treatment of MCL.
Overall, the expression of the SOX11 gene is almost absent in indolent MCL. SOX11 negativity indicates a better prognosis. However, the significance of SOX11 negativity in indolent MCL is still inconclusive.
MYC rearrangement or overexpression
Velden and colleagues suggested that B-cell prolymphocytic leukemia is a specific subtype of MCL [66]. Interestingly, patients with prolymphocytic leukemia-like MCL with MYC amplification, no expression of CD38, and loss of TP53 showed prolonged survival outcomes similar to those with indolent leukemic MCL. MYC (8q24) is a critical global transcription factor that regulates 10–15% of all human genes and controls many cellular functions, including the cell cycle, survival, cell growth, apoptosis and metabolism [67, 68]. Contrary to the research of Velden et al., most studies have demonstrated that overexpression/structural changes of MYC in MCL are related to a more aggressive course, a higher MIPI and a worse prognosis [69, 70]. Similar to MYC rearrangements, IGH-BCL2 is also commonly observed in MCLs, and approximately 60% are double-hit lymphomas [67]. MCL with MYC rearrangement is characterized by p53 expression, a high proliferation rate and a complex karyotype [71]. Patients with high MYC or p53 expression have significantly shorter OS and progression-free survival (PFS) [72]. Furthermore, alterations in TP53 and MYC add prognostic information to somatic gene copy number alteration (CNA), enhancing the prognostic correlation of these individual changes observed in previous studies [1]. Choe et al. suggested MYC overexpression to be a predictor of a poor prognosis in MCL [72]. However, Wang and colleagues indicated that MYC rearrangement rather than extra MYC copies is an independent prognostic factor in patients with MCL [67]. Interestingly, there is a close relationship between MYC and miRNAs. MiR-34a was associated with poor outcomes in two independent series of leukemic and nodal MCLs and correlated with high expression of the MYC oncogene [73]. MYC plays an important role in intrinsic ibrutinib resistance in MCL, probably by inhibiting miR16-1 and miR15a, two tumor suppressor miRNAs involved in MCL pathogenesis. The status of MYC could also predict the response to BTKi [67].
Thus, the role of MYC amplification is currently unclear, with this aberrancy reported in studies on both indolent and aggressive MCLs.
IGHV mutations
As early as 2003, Orchard et al. suggested that mutated immunoglobulin heavy chain variable region (IGHV) genes might help identify indolent MCL [74]. In recent years, some cases of MCL have been characterized by IGHV mutations, and they are truly associated with an indolent disease course [57, 59]. The typical clinical presentation of patients with IGHV mutations comprises leukemic non-nodal CLL-like, including splenomegaly, a Ki-67 proliferation fraction < 10%, a low tumor burden and noncomplex karyotypes [55]. The identified VH rearrangements in indolent MCL were VH1, VH2, VH3 and VH4 [3]. Ferna`ndez et al. compared the disease behavior of indolent MCL with that of classical MCL and showed that 70% of patients with indolent disease carried hypermutated IGHV, and of those mutations, 50% harbored a VH4 mutation [8]. If IGHV genes have been highly mutated, the clinical course is indolent, with involvement of the peripheral blood, and treatment is not necessary [6]. However, the clinical significance of IGHV mutations in MCL is controversial. Some series have shown prolonged patient survival with hypermutations, but most studies have suggested no clear survival benefit [8]. In some instances, the IGHV gene mutation status has been associated with significant reductions in OS [39]. Moreover, the response to chemotherapy and 5-year survival are negatively correlated with the degree of IGHV gene mutation [75].
Thus, most patients with indolent MCL carry IGHV mutations. However, the significance of hypermutated IGHV in patient survival is still conflicting.
L-MCL16 assay and MCL35 assay
In a recent study, Guillem et al. developed a novel gene expression assay that could identify classical MCL and leukemic non-nodal MCL— the L-MCL16 assay [4]. The L-MCL16 assay includes 16 genes (13 genes that are upregulated in classical MCL and 3 novel genes that are upregulated in non-nodal MCL: CD200, SLAMF1 and BTLA). This novel assay was performed in an independent cohort of 70 leukemic MCL patients and suggested that non-nodal MCL patients predicted by the L-MCL16 assay had significant biological and clinical differences compared to classical MCL patients at diagnosis. Although the L-MCL16 assay recognized three subgroups, high, standard and low proliferation, of classical MCL and leukemic non-nodal MCL, the predictive ability was not well correlated, as previously found in nodal samples. However, their findings emphasized the value of the L-MCL16 assay for the biological determination of indolent MCL. Although the Ki-67 score can be used as a prognostic factor of MCL, Ki-67 staining and interpretation are affected by considerable interlaboratory and interobserver variability [14]. Gene expression analyses are very accurate; however, they are carried out on fresh frozen tissue, which is not easy to obtain for most patients. Therefore, the MCL35 assay, a new identification method, was developed. The MCL35 assay is a proliferation detection method based on the expression of Nanostring-based RNA. It is composed of 35 different genes, 18 of which are housekeeping genes and 17 of which are related to proliferation [76]. The MCL35 assay translated the proliferation characteristics derived from studies on MCL into a test suitable for routinely available formalin-fixed paraffin-embedded (FFPE) biopsies. Scott et al. demonstrated that the MCL35 assay produced gene expression levels of sufficient quality to specify an assay score and risk group in 108 FFPE biopsies from 110 samples (98%). The MCL35 assay, as a continuous score, stratified patients into high-risk, standard-risk and low-risk groups in a training set containing 47 biopsies, with different OS times (medians of 1.1 years, 2.6 years, and 8.6 years, respectively). The MCL35 assay has been shown to have good reproducibility in different centers. Furthermore, the prognostic ability of the assay was validated in younger patients for whom there was a plan to undergo autohematopoietic stem cell transplantation (ASCT). However, Holte et al. suggested that the MCL35 assay cannot separate the standard-risk group from the low-risk group. Furthermore, the MCL35 assay might identify a subgroup with a terrible outcome despite treatment with very active therapy. Moreover, recent studies have suggested that the MCL35 assay is independent of the MIPI but is strongly associated with the Ki-67 index and MCL cytology (classical versus pleomorphic/blastic types). The MCL35 assay was able to identify the three risk groups within the patient subgroup with a Ki-67 index ≥ 30%, and the MCL35 assay subsumed the prognostic power of the Ki-67 PI in pairwise multivariable analysis. Therefore, the MCL35 assay showed stronger prognostic power than the Ki-67 index. However, inversely, some scholars have pointed out that the outcomes of patients stratified according to the Ki-67 score (≥ vs. < 30%) alone is better than the outcomes predicted by the MCL35 assay [77]. Hilka et al. also suggested shortcomings of the MCL35 assay, such as being established only for biopsies from involved lymph nodes with a high tumor load (≥ 60%), poor fixation of the FFPE sample and failure from unknown reasons (10% of MCL specimens). Intriguingly, a recent study showed that the MCL35 assay together with the MIPI and Ki-67 could not predict outcome in nodal samples from patients with indolent MCL who adopted an attitude of waiting.
Therefore, the L-MCL16 assay and MCL35 assay may be used to screen out indolent MCL from all variants of MCL. However, their prognostic value remains unclear.
P16 aberrations
Deletions of the P16 gene are closely associated with aggressive MCL subtypes, such as the blastoid variant of MCL, with a poor prognosis [18, 78]. P16, also known as INK4a or CDKN2A, is a tumor suppressor gene [79]. The P16 gene is frequently silenced in various types of lymphoma, and this silencing is usually caused by the hypermethylation of CpG islands in its promoter but may also be achieved through the activation of Bmi-1. Streich et al. indicated that 7% of MCL cases in their study harbored codeletions of TP53 and CDKN2A [41]. Of note, the combination of both aberrations represents significantly more adverse prognostic effects than the isolated aberration of either gene [5]. Moreover, the prognostic effects of p16 and TP53 deletions were not related to the proliferation marker Ki-67 [80]. Importantly, patients with concurrent TP53 mutation/deletion and P16 deletion might benefit from innovative treatments, such as Bruton’s tyrosine kinase inhibitors (BTKi). Thus, the expression level of P16 is normal or higher in indolent MCL.
ATM defects
The frequency of defects in ataxia telangiectasia mutated (ATM) gene has been reflected to be much lower in the indolent variants of MCL [9]. The ATM gene encodes a 370-kDa ATM protein, which is a member of the PI-3 protein kinase family and is essential for DNA processing, the cell cycle and telomere length regulation [81]. Notably, 11q deletion and/or ATM mutation, in cooperation with cyclin D1, consistently represent the most frequent oncogenic event in MCL [82]. This response is closely related to the activation (stabilization) of TP53 mediated by ATM. Indeed, in MCL, 26% of cases contain TP53 mutation/deletion, 56% contain ATM alteration and 10% contain both genetic changes [35]. Mareckova and colleagues revealed the mutual exclusivity of ATM and TP53 mutations in MCL [82]. Due to limited information on the clinical impact of ATM mutations on MCL and although ATM has been repeatedly proven to be the most commonly mutated gene, followed by CDKN2A and TP53, the latest publications consistently show no statistically significant difference in the OS of patients with wild-type vs. mutated ATM, which is in sharp contrast to TP53 defects [3, 83]. Nevertheless, Delfau et al. reported that somatic gene copy number alterations in MYC, CDK2, ATM, MDM2 and CDK4 had no prognostic value [80]. Of note, however, non-nodal leukemic MCLs with aberrations in ATM are more aggressive [61]. In the report of Bea et al., ATM mutations were found in more than half of tumors with SOX11 positivity but not in those with SOX11 negativity [84]. ATM was present at similar allelic frequencies in the two subclones of different cases, indicating that it represents early events. Furthermore, interestingly, the loss of ATM may actually contribute to the radiosensitivity of MCL cells [85].
The frequency of defects in ATM is much lower in indolent MCL. Furthermore, indolent MCLs with aberrations in ATM are more aggressive. However, some recent studies have suggested that ATM has little prognostic value.
NOTCH1/2 mutations
The frequency of mutations in the NOTCH1 gene has been reported to be much lower in the indolent variants of MCL [9]. Other studies have indicated that indolent MCL lacks NOTCH1/2, MYC and TP53 mutations [16]. In other words, NOTCH1 and/or NOTCH2 mutations have been related to poor outcomes [43]. Jain et al. suggested that patients with variants of aggressive histology MCL (AH-MCL) frequently exhibited CCND1, NOTCH1, and SMARCA4 (SWI/SNF) gene mutations [86]. Moreover, the presence of NOTCH2, UBR5, and NOTCH3 mutations was exclusive to AH-MCL. Zhou and colleagues believe that MYC rearrangement together with NOTCH2 mutation contributes to the aggressive subtype switch from CD19 + CD10– cells to CD19 + CD10 + cells [87]. Interestingly, NOTCH2 mutation is a substitute for NOTCH1 mutation [84]. However, NOTCH1 was not identified as an independent prognostic factor in a multivariate model with TP53 mutations [46]. In conclusion, 9.5% of MCL patients present NOTCH1/2 mutations, which can be used to identify a subset of tumors with more aggressive biological and clinical features, including those with blastoid/pleomorphic morphology. Indolent MCLs express no or low NOTCH1/2.
Several genes that have been valued in recent years
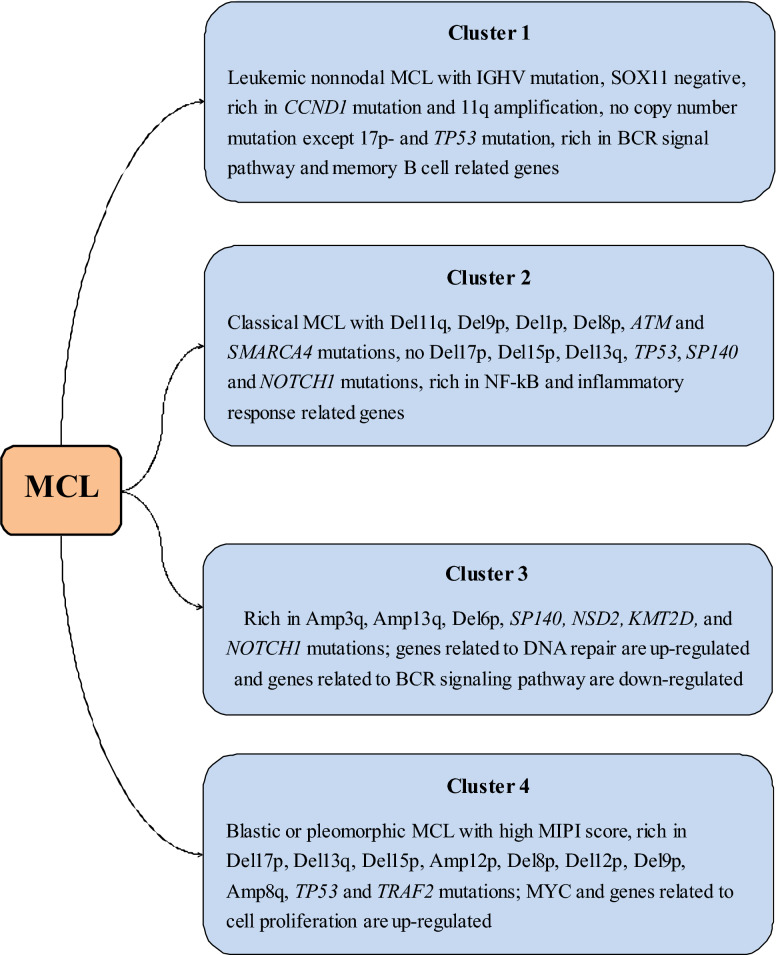
Compared to those with a low Ki-67 level, AH-MCL patients with Ki-67 ≥ 50% have exclusive mutations in TP53, RANBP2, SMARCA4, KMT2C, SPEN, and NSD2 (WHSC1) [86, 88]. The study group of Sakhdari et al. included twenty-one (81%) patients with nodal MCLs and 5 (19%) patients with leukemic variant MCLs. In their study, TP53, CARD11, ATM, FBXW7, SPEN, NOTCH1 and BIRC3 were mutated to varying degrees, and most mutations were clonal in nature [9]. Among these mutated genes, SMARCA4 and BCL2 were altered only after progression, while TP53, KMT2D (MLL2), CDKN2A, CCND1, CELSR3, NOTCH2 and ATM were altered 2–4 times more frequently after progression [89]. Zhao et al. demonstrated that in the progression of MCL, there was clonal evolution of novel SMARCA4 and KMT2C/D mutations. However, Jain et al. did not observe BIRC3, MAP3K14 and TRAF2 mutations at any time point in their experiment [90]. Yang and colleagues suggested that after the MIPI and MIPI-c score, TP53 and WHSC1 mutations were the most significant prognostic factors in MCL, and all patients benefited from WHSC1 and TP53 mutations at diagnosis [91]. KMT2D mutations are associated with an increased risk of progression and death [92]. By adding KMT2D mutations and TP53 disruption to the MIPI-c backbone, Ferrero et al. derived a new prognostic indicator, the “MIPI-genetic”. The “MIPI-g” improved the discrimination ability of the model, defining low-risk, intermediate-risk and high-risk patients (4-year PFS: 72%, 42.2%, and 11.5%, respectively; 4-year OS: 94.5%, 65.8%, and 44.9%, respectively). They also demonstrated that among patients with TP53 disruption, those with KMT2D mutations had a poor outcome. Some researchers described the genetic spectrum of 134 MCL patients, and they suggested that ATM and KMT2D mutations were more frequent in patients without IGHV mutations. Abnormal SP140 expression and PCDH10 mutation independently imply shorter PFS and OS. Del 9p and/or Del 9q, the most common variants, were found in 40% of the 134 patients and were significantly associated with a poor prognosis. In contrast, abnormalities in the Amp 11q gene are associated with prolonged PFS and OS. The researchers further divided the samples into four groups: no chr9 deletion, major Del 9q, major Del 9p and massive chr9 deletion. They found that the prognosis of the group without chr9 deletion was significantly better than that of the other three groups, suggesting that tumor suppressor genes may be located on the part of chr9 that is deleted [93]. Recently, a new classification of MCL that can predict prognosis was proposed (Fig. 1). The altered splicing of HNRNPH1 was related to inferior outcomes in MCL patients [46]. All patients with WHSC1, MLL2 and UBR5 mutations responded to therapy. Conversely, alterations in NOTCH1, CCND1 and SMARCA4 occurred only in nonresponders [94]. The deletion of SMARCA4 results in transcriptional changes that enable cells to survive a therapeutic challenge, and this is reflected in the fact that SMARCA4 mutations are enriched in patients with primary progression. Interestingly, mutations in CARD11 and BIRC3 were previously associated with ibrutinib resistance and regulation of the activation of NF-κB, but this did not rule out the response to the combination of ibrutinib and venetoclax. Overexpression of CARD11 mutants was demonstrated to endow resistance to the NF-κB inhibitor lenalidomide and the BCR inhibitor ibrutinib [95]. Moreover, mutations in the SWI–SNF chromatin-remodeling complex confer resistance to venetoclax and ibrutinib. Last, in future analyses, changes in the mutational status from baseline samples to samples at disease progression and current mutations of interest in MCL should be taken into account [2].
Fig. 1.
New classification of MCL
In conclusion, the frequency of mutations in SPEN, SMARCA4 (SWI/SNF), RANBP2, KMT2C, NSD2 (WHSC1), Del 9p and/or Del 9q, CARD11, FBXW7, SP140, BIRC3, KMT2D (MLL2), PCDH10, CELSR3 and HNRNPH1 is much lower in indolent MCL. Conversely, Amp 11q gene mutations without chr9 deletion have a better prognosis.
Genes that lack data
Indolent MCL is characterized by kappa light chain restriction. In contrast, a more typical lambda light chain restriction is presented in cMCL [48, 96]. The incidence of frameshift Bax mutations is low in indolent and mantle cell lymphomas, which indicates that microsatellite instability (MSI) is not a feature of the pathogenesis of these lymphomas [97]. Petrakis et al. showed a close positive correlation between the expression of CD34 and SOX11 [59]. However, we still cannot draw a conclusion on the correlation between SOX11 expression and CD83 expression in MCL. The levels of cell membrane CD83 (mCD83) in MCL patients are significantly elevated. The expression of mCD83 is mainly limited to lymphocytes, neutrophils and activated DCs. The CD83 promoter contains NF-κB-binding sites. CD83 expression is correlated with canonical NF-κB activation in MCL [98, 99]. Moreover, indolent MCL shows low or no expression of genes in the high-mobility group (HMG) [100]. Importantly, poly(ADP-ribose) polymerase 1 (PARP1) protein expression is related to a progressive course of indolent MCL and shortened OS. Gallo and colleagues indicated that PARP1 should be included in initial diagnostic studies as a negative predictor [20]. PARP1 is a nuclear protein involved in DNA repair and the maintenance of genomic stability and is overexpressed in a number of aggressive cancers. As reported, through caspase-9–3-7-PARP signaling, SOX11 silencing promotes proliferation and inhibits the apoptosis of MCL cells and desensitizes MCL cells to bortezomib [101]. At diagnosis, the peripheral blood absolute monocyte count (AMC) is an independent prognostic factor for OS in MCL patients, suggesting its ability to predict outcomes in addition to the MIPI [102]. Jain et al. pointed out that the downregulation of BACH2 was related to an increase in proliferation, and patients with cluster 1 methylation patterns with somatic mutations had less aggressive disease [103]. Activation of the Akt pathway has been found in most blastoid MCLs, and the loss of PTEN promotes this process. Additionally, antiapoptotic Mcl-1 overexpression is significantly related to blastoid MCL and TP53 mutations.
Indolent MCL is characterized by kappa light chain restriction, a low incidence of frameshift Bax mutations, low expression of CD34, low or no expression of HMG, low PARP1 protein content, upregulation of BACH2, inhibition of the Akt pathway and low expression of Mcl-1.
The expression of microRNAs in indolent MCL
Javeed et al. suggested that a set of stroma-associated microRNAs might define a more indolent group of MCL [104]. MicroRNAs (miRNAs) are noncoding single-stranded RNA molecules composed of 21–23 nucleotides that bind to the 3′-untranslated region (UTR) of the target gene messenger RNA (mRNA) [105, 106]. MiRNAs do not encode proteins, but they go through the process from a primary transcript to a precursor miRNA and then to a mature miRNA [107]. MiRNAs are involved in the posttranscriptional regulation of gene expression and important biological processes, including oncogenesis [108]. Studies have shown that miRNAs are frequently deregulated in a variety of human malignancies [107]. Therefore, miRNA expression may predict outcomes in both solid tumors and hematologic malignancies. Previously, the abnormal expression of miRNAs was frequently observed in various types of lymphomas, suggesting their potential benefits in clinical diagnosis [109]. Zhao et al. demonstrated that patients with significantly downregulated miR-29 had short survival and that downregulation of miR-29 together with cyclin D1 had a synergistic effect in the pathogenesis of MCL [107]. Additionally, Navarro et al. suggested that 7 miRNAs showed prognostic significance independent of the IGHV status and SOX11 expression, and among the top 3 were miR-708 and miR-455-5p/3p [73]. Thus, miR-708 and miR-455-5p/3p may well distinguish between IGHV mutated/wild-type and SOX11 positivity/negativity in MCL. This study also suggested that the expression level of miR-181a/c had an effect on leukemic MCL, mainly by downregulating the expression of ATM, and the CD40 signaling pathway was identified as an important target for the differentially expressed miRNAs between unmutated MCL and mutated MCL. Goswami et al. demonstrated that overexpression of miR-20b was associated with a worse prognosis and high proliferation in MCL, as well as low expression of miR-29. Here, compared with miR-127-3p, the expression of miR-615-3p could better distinguish the different prognostic groups in MCL [108]. MiR-223 is a miRNA that has been well studied. MiR-223 expression is also decreased in unmutated IGVH MCLs, and these types of lymphoma have an aggressive clinical course [110]. Overexpression of miR-223 decreases the viability and proliferation of MCL cells and promotes apoptosis, which may be due to the downregulation of SOX11 mediated by miR-223 [106]. A database search showed that SOX11 was the hypothetical target for miR-223. Luciferase reporter assays confirmed that posttranscriptional miR-223 suppressed the 3′-untranslated region of wild-type SOX11 but did not inhibit that of mutated SOX11. Therefore, these studies not only suggest that miR-223 is highly suppressed in MCL patients and associated with high-risk clinical features but also prove the tumorigenic role of SOX11 [63]. MiR-132-3p is very similar to miR-223. The higher expression of miR-132-3p predicts prolonged survival in MCL patients, and its direct target is SOX11 [109]. Husby et al. combined the expression levels of miR-18b with new biological MCL International Prognostic Index (MIPI-B) data for the first time. Overexpression of miR-18b significantly downregulated MCL cell proliferation [111]. The study also suggested a number of prognostic miRNAs, including the entire miR-17 ~ 92a cluster [112].
In conclusion, the expression level of miR-20b in indolent MCL is lower than that in classical MCL. MiR-127-3p and miR-615-3p are related to the OS of patients with indolent MCL. Moreover, miR-29, miR-708, miR-455-5p/3p, miR-223, miR-132-3P and miR-18b are highly expressed in indolent MCL.
Conclusions
MCL is one of the most aggressive lymphoid tumors, and the toxicity of chemotherapeutic drugs is unfavorable to every patient. Indolent MCL is a subtype of mantle cell lymphoma. Indolent MCL has a better prognosis than classical MCL. However, the process of “wait and watch” puts these patients at the potential risk of evolution to classical and aggressive MCL [16]. Therefore, a clear understanding of the molecular underpinnings of indolent MCL is necessary.
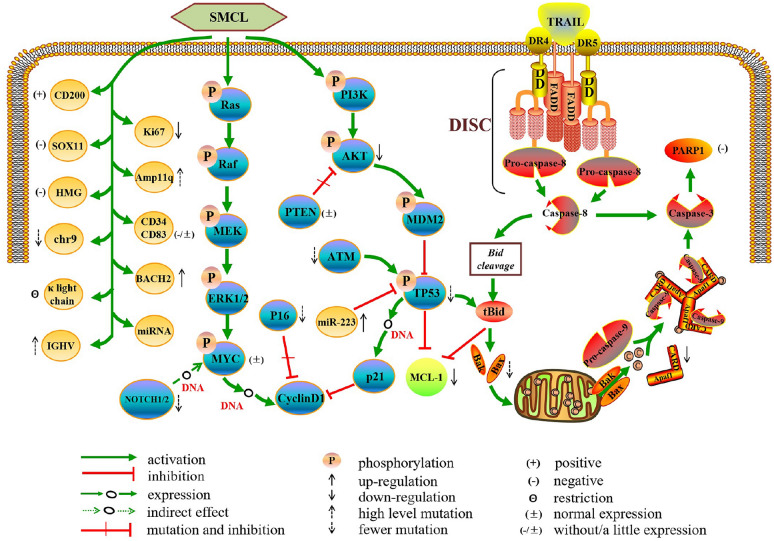
In this review, we summarize and analyze the common molecular characteristics of indolent MCL described in previous studies (Fig. 2). Furthermore, we focus on several genes that have been valued in recent years and some genes that lack data (Table 2). We firmly believe that a more in-depth study of the molecular characteristics may distinguish indolent MCL from classical MCL. If we can clearly distinguish indolent MCL from classical MCL, indolent MCL will not be overtreated, whereas the treatment of classical MCL will not be delayed.
Fig. 2.
Molecular network of indolent MCL
Table 2.
Molecular markers of indolent MCL
| Ki-67 index ≤ 30% |
| CD200 positivity |
| No TP53 mutation/deletion |
| Lack of SOX11 |
| Without rearrangement and overexpression of MYC |
| IGHV mutations |
| L-MCL16 assay and MCL35 assay |
| Unmutated P16 status |
| Without/minimal defects of ATM |
| No NOTCH1/2 mutation |
| Amp 11q gene mutation |
| No chr9 deletion |
| Several genes that have been valued in recent years (all of them are weakly expressed) |
| SPEN |
| SMARCA4 (SWI/SNF) |
| RANBP2 |
| KMT2C |
| NSD2 (WHSC1) |
| CARD11 |
| FBXW7 |
| BIRC3 |
| KMT2D (MLL2) |
| CELSR3 |
| TRAF2 |
| MAP3K14 |
| HNRNPH1 |
| Del 9p and /or Del 9q |
| SP140 |
| PCDH10 |
| Genes that lack data |
| Kappa light chain restriction |
| Almost no incidence of frameshift Bax mutations |
| CD34 (−/ ±), CD83(−/ ±) |
| No expression of HMG |
| PARP1 silencing |
| Up-regulation of BACH2 |
| Cluster 1 methylation pattern with somatic mutation |
| Inhibition of Akt pathway |
| No loss of PTEN |
| Without overexpression of Mcl-1 |
| MicroRNAs upregulation/downregulation |
Acknowledgements
Thanks are due to American Journal Experts (AJE) for assistance with professional language editing.
Abbreviations
- AMC
Absolute monocyte count
- ASCT
Auto-hematopoietic stem cell transplantation
- ATM
Ataxia telangiectasia mutated
- BM
Bone marrow
- BTKi
Bruton’s tyrosine kinase inhibitors
- CLL
Chronic lymphocytic leukemia
- FASAY
Functional analyses of separated alleles in yeast
- FFPE
Formalin-fixed paraffin-embedded
- FISH
Fluorescence in situ hybridization
- HIG-2
Hypoxia-inducible gene 2
- IGHV
Immunoglobulin heavy chain variable region
- LDH
Lactate dehydrogenase
- MCL
Mantle cell lymphoma
- MCM6
Minichromosome maintenance protein 6
- MIPI
MCL-International Prognostic Index
- MSI
Microsatellite instability
- NF-κB
Nuclear factor kappa-B
- OS
Overall survival
- PARP1
Poly (ADP-ribose) polymerase 1
- PET/CT
Positron Emission Tomography/computerized tomography
- PFS
Progression-free survival
- PHH3
Phosphohistone H3
- SMCL
Smoldering mantle cell lymphoma
- SUV
Standard Uptake Value
- TP53
Tumor protein p53
- UTR
3’-Untranslated region
Authors’ contributions
PJ and AD designed this review, collected the related references and drafted the manuscript. HY performed the selection of literature, prepared the figures and revised the manuscript. All authors contributed to this manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LY20H080002), the Wenzhou Municipal Sci-Tech Bureau’s program (Y20180218), and the Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (2019KY452).
Availability of data and materials
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Panruo Jiang and Aakash Desai: Equal contributors
References
- 1.Nadeu F, Martin-Garcia D, Clot G, et al. Genomic and epigenomic insights into the origin, pathogenesis and clinical behavior of mantle cell lymphoma subtypes. Blood. 2020. [DOI] [PMC free article] [PubMed]
- 2.Hill HA, Qi X, Jain P, et al. Genetic mutations and features of mantle cell lymphoma: a systematic review and meta-analysis. Blood Adv. 2020;4(13):2927–2938. doi: 10.1182/bloodadvances.2019001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez V, Salamero O, Espinet B, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70(4):1408–1418. doi: 10.1158/0008-5472.CAN-09-3419. [DOI] [PubMed] [Google Scholar]
- 4.Clot G, Jares P, Gine E, et al. A gene signature that distinguishes conventional and leukemic nonnodal mantle cell lymphoma helps predict outcome. Blood. 2018;132(4):413–422. doi: 10.1182/blood-2018-03-838136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malarikova D, Berkova A, Obr A, et al. Concurrent TP53 and CDKN2A gene aberrations in newly diagnosed mantle cell lymphoma correlate with chemoresistance and call for innovative upfront therapy. Cancers. 2020;12(1(No.2120)):2120. doi: 10.3390/cancers12082120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsi, Eric, D., et al. Indolent mantle cell lymphoma. Leukemia Lymphoma. 2014. [DOI] [PubMed]
- 7.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised Fourth Edition. 2017;Volume 2.
- 8.Rule SA, Poplar S, Evans PA, et al. Indolent mantle-cell lymphoma: immunoglobulin variable region heavy chain sequence analysis reveals evidence of disease 10 years prior to symptomatic clinical presentation. J Clin Oncol. 2011;29(15):e437–e439. doi: 10.1200/JCO.2010.31.8006. [DOI] [PubMed] [Google Scholar]
- 9.Sakhdari A, Ok CY, Patel KP, et al. TP53 mutations are common in mantle cell lymphoma, including the indolent leukemic non-nodal variant. Ann Diagn Pathol. 2019;41:38–42. doi: 10.1016/j.anndiagpath.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Hu Z, Sun Y, Schlette EJ, et al. CD200 expression in mantle cell lymphoma identifies a unique subgroup of patients with frequent IGHV mutations, absence of SOX11 expression, and an indolent clinical course. Mod Pathol. 2018;31(2):327–336. doi: 10.1038/modpathol.2017.135. [DOI] [PubMed] [Google Scholar]
- 11.S C, M P, Ajm F, et al. Mantle cell lymphoma. Critical reviews in oncology/hematology. 2020;153:103038. [DOI] [PubMed]
- 12.Dabaja BS, Zelenetz AD, Ng AK, et al. Early-stage mantle cell lymphoma: a retrospective analysis from the International Lymphoma Radiation Oncology Group (ILROG) Ann Oncol. 2017;28(9):2185–2190. doi: 10.1093/annonc/mdx334. [DOI] [PubMed] [Google Scholar]
- 13.Jo JC, Kim SJ, Lee HS, et al. Clinical features and treatment outcomes of limited-stage mantle cell lymphoma: consortium for improving survival of lymphoma report. Ann Hematol. 2020;99(2):223–228. doi: 10.1007/s00277-019-03803-x. [DOI] [PubMed] [Google Scholar]
- 14.Rauert-Wunderlich H, Mottok A, Scott DW, et al. Validation of the MCL35 gene expression proliferation assay in randomized trials of the European Mantle Cell Lymphoma Network. Br J Haematol. 2019;184(4):616–624. doi: 10.1111/bjh.15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura Y, Sato K, Imamura Y, et al. Small cell variant of mantle cell lymphoma is an indolent lymphoma characterized by bone marrow involvement, splenomegaly, and a low Ki-67 index. Cancer Sci. 2011;102(9):1734–1741. doi: 10.1111/j.1349-7006.2011.01988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye H, Desai A, Zeng D, et al. Smoldering mantle cell lymphoma. J Exp Clin Cancer Res. 2017;36(1):185. doi: 10.1186/s13046-017-0652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinet B, Ferrer A, Bellosillo B, et al. Distinction between asymptomatic monoclonal B-cell lymphocytosis with cyclin D1 overexpression and mantle cell lymphoma: from molecular profiling to flow cytometry. Clin Cancer Res. 2014;20(4):1007–1019. doi: 10.1158/1078-0432.CCR-13-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vose JM. Mantle cell lymphoma: 2017 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2017;92(8):806–813. doi: 10.1002/ajh.24797. [DOI] [PubMed] [Google Scholar]
- 19.He X, Chen Z, Fu T, et al. Ki-67 is a valuable prognostic predictor of lymphoma but its utility varies in lymphoma subtypes: evidence from a systematic meta-analysis. BMC Cancer. 2014;14:153. doi: 10.1186/1471-2407-14-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallo M, Cacheux V, Vincent L, et al. Leukemic non-nodal mantle cell lymphomas have a distinct phenotype and are associated with deletion of PARP1 and 13q14. Virchows Arch. 2016;469(6):697–706. doi: 10.1007/s00428-016-2016-8. [DOI] [PubMed] [Google Scholar]
- 21.Hoster E, Rosenwald A, Berger F, et al. Prognostic value of Ki-67 index, cytology, and growth pattern in mantle-cell lymphoma: results from randomized trials of the european mantle cell lymphoma network. J Clin Oncol. 2016;34(12):1386–1394. doi: 10.1200/JCO.2015.63.8387. [DOI] [PubMed] [Google Scholar]
- 22.Hou W, Wei P, Xie J, et al. The degree of overlap between the follicular dendritic cell meshwork and tumor cells in mantle cell lymphoma is associated with prognosis. Pathol Res Practice. 2018;214(4):513–520. doi: 10.1016/j.prp.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Chakhachiro ZI, Saliba RM, Okoroji G, et al. Cytarabine, Ki-67, and SOX11 in patients with mantle cell lymphoma receiving rituximab-containing autologous stem cell transplantation during first remission. Cancer-Am Cancer Soc. 2013;119(18):3318–3325. doi: 10.1002/cncr.28219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medani H, Elshiekh M, Naresh KN. Improving precise counting of mitotic cells in mantle cell lymphoma using phosphohistone H3 (PHH3) antibody. J Clin Pathol. 2020:2020–206956. [DOI] [PubMed]
- 25.Schrader C, Janssen D, Klapper W, et al. Minichromosome maintenance protein 6, a proliferation marker superior to Ki-67 and independent predictor of survival in patients with mantle cell lymphoma. Br J Cancer. 2005;93(8):939–945. doi: 10.1038/sj.bjc.6602795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogt N, Klapper W. Variability in morphology and cell proliferation in sequential biopsies of mantle cell lymphoma at diagnosis and relapse: clinical correlation and insights into disease progression. Histopathology. 2013;62(2):334–342. doi: 10.1111/his.12009. [DOI] [PubMed] [Google Scholar]
- 27.Ting YS, Smith SABC, Brown DA, et al. CD200 is a useful diagnostic marker for identifying atypical chronic lymphocytic leukemia by flow cytometry. Int J Lab Hematol. 2018;40(5):533–539. doi: 10.1111/ijlh.12857. [DOI] [PubMed] [Google Scholar]
- 28.Challagundla P, Medeiros LJ, Kanagal-Shamanna R, et al. Differential expression of CD200 in B-cell neoplasms by flow cytometry can assist in diagnosis, subclassification, and bone marrow staging. Am J Clin Pathol. 2014;142(6):837–844. doi: 10.1309/AJCPBV9ELXC0ECVL. [DOI] [PubMed] [Google Scholar]
- 29.Miao Y, Fan L, Wu Y, et al. Low expression of CD200 predicts shorter time-to-treatment in chronic lymphocytic leukemia. Oncotarget. 2016;7(12):13551–13562. doi: 10.18632/oncotarget.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorigue M, Magnano L, Miljkovic MD, et al. Positive predictive value of CD200 positivity in the differential diagnosis of chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2020;98(5):441–448. doi: 10.1002/cyto.b.21849. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Wang L, Li J, et al. SOX11-negative mantle cell lymphoma: clinicopathologic and prognostic features of 75 patients. Am J Surg Pathol. 2019;43(5):710–716. doi: 10.1097/PAS.0000000000001233. [DOI] [PubMed] [Google Scholar]
- 32.Saksena A, Yin CC, Xu J, et al. CD23 expression in mantle cell lymphoma is associated with CD200 expression, leukemic non-nodal form, and a better prognosis. Hum Pathol. 2019;89:71–80. doi: 10.1016/j.humpath.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Yoshino T, Tanaka T, Sato Y. Differential diagnosis of chronic lymphocytic leukemia/small lymphocytic lymphoma and other indolent lymphomas, including mantle cell lymphoma. J Clin Exp Hematop. 2020. [DOI] [PMC free article] [PubMed]
- 34.Zlamalikova L, Moulis M, Ravcukova B, et al. Complex analysis of the TP53 tumor suppressor in mantle cell and diffuse large B-cell lymphomas. Oncol Rep. 2017;38(4):2535–2542. doi: 10.3892/or.2017.5891. [DOI] [PubMed] [Google Scholar]
- 35.Williamson CT, Kubota E, Hamill JD, et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. Embo Mol Med. 2012;4(6):515–527. doi: 10.1002/emmm.201200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almeida A, Sánchez Morán I, Rodríguez C. Mitochondrial–nuclear p53 trafficking controls neuronal susceptibility in stroke. IUBMB Life. 2021;73(3):582–591. doi: 10.1002/iub.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obr A, Klener P, Furst T, et al. A high TP53 mutation burden is a strong predictor of primary refractory mantle cell lymphoma. Brit J Haematol. 2020. [DOI] [PubMed]
- 38.Bea S, Amador V. Role of SOX11 and Genetic Events Cooperating with Cyclin D1 in Mantle Cell Lymphoma. Curr Oncol Rep. 2017;19(6):43. doi: 10.1007/s11912-017-0598-1. [DOI] [PubMed] [Google Scholar]
- 39.Caballero D, Campo E, Lopez-Guillermo A, et al. Clinical practice guidelines for diagnosis, treatment, and follow-up of patients with mantle cell lymphoma Recommendations from the GEL/TAMO Spanish Cooperative Group. Ann Hematol. 2013;92(9):1151–1179. doi: 10.1007/s00277-013-1783-4. [DOI] [PubMed] [Google Scholar]
- 40.Stefancikova L, Moulis M, Fabian P, et al. Loss of the p53 tumor suppressor activity is associated with negative prognosis of mantle cell lymphoma. Int J Oncol. 2010;36(3):699–706. doi: 10.3892/ijo_00000545. [DOI] [PubMed] [Google Scholar]
- 41.Streich L, Sukhanova M, Lu X, et al. Aggressive morphologic variants of mantle cell lymphoma characterized with high genomic instability showing frequent chromothripsis, CDKN2A/B loss, and TP53 mutations: A multi-institutional study. Genes Chromosomes Cancer. 2020;59(8):484–494. doi: 10.1002/gcc.22849. [DOI] [PubMed] [Google Scholar]
- 42.Dong H, Zhou L, Fang C, et al. TP53 mutation is not an independent prognostic factor in patients with mantle cell lymphoma at advanced stage. Med Oncol. 2012;29(3):2166–2173. doi: 10.1007/s12032-011-0096-5. [DOI] [PubMed] [Google Scholar]
- 43.Eskelund CW, Dahl C, Hansen JW, et al. TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood. 2017;130(17):1903–1910. doi: 10.1182/blood-2017-04-779736. [DOI] [PubMed] [Google Scholar]
- 44.Mccall RK, Pang CS, Pettenati MJ. Non-Nodal CD5-Negative mantle cell lymphoma with secondary TP53 Deletion. Case Rep Hematol. 2020;2020:9185432. doi: 10.1155/2020/9185432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aukema SM, Hoster E, Rosenwald A, et al. Expression of TP53 is associated with the outcome of MCL independent of MIPI and Ki-67 in trials of the European MCL Network. Blood. 2018;131(4):417–420. doi: 10.1182/blood-2017-07-797019. [DOI] [PubMed] [Google Scholar]
- 46.Pararajalingam P, Coyle KM, Arthur S, et al. Coding and non-coding drivers of mantle cell lymphoma identified through exome and genome sequencing. Blood. 2020. [DOI] [PMC free article] [PubMed]
- 47.Lin RJ, Ho C, Hilden PD, et al. Allogeneic haematopoietic cell transplantation impacts on outcomes of mantle cell lymphoma with TP53 alterations. Brit J Haematol. 2018. [DOI] [PMC free article] [PubMed]
- 48.Ondrejka SL, Lai R, Smith SD, et al. Indolent mantle cell leukemia: a clinicopathological variant characterized by isolated lymphocytosis, interstitial bone marrow involvement, kappa light chain restriction, and good prognosis. Haematologica. 2011;96(8):1121–1127. doi: 10.3324/haematol.2010.036277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wasik AM, Lord M, Wang X, et al. SOXC transcription factors in mantle cell lymphoma: the role of promoter methylation in SOX11 expression. Sci Rep. 2013;3:1400. doi: 10.1038/srep01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Federmann B, Frauenfeld L, Pertsch H, et al. Highly sensitive and specific in situ hybridization assay for quantification of SOX11 mRNA in mantle cell lymphoma reveals association of TP53 mutations with negative and low SOX11 expression. Haematologica. 2020;105(3):754–764. doi: 10.3324/haematol.2019.219543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balsas P, Palomero J, Eguileor Á, et al. SOX11 promotes tumor protective microenvironment interactions through CXCR4 and FAK regulation in mantle cell lymphoma. Blood. 2017;130(4):501–513. doi: 10.1182/blood-2017-04-776740. [DOI] [PubMed] [Google Scholar]
- 52.Wasik AM, Priebe V, Lord M, et al. Flow cytometric analysis of SOX11: a new diagnostic method for distinguishing B-cell chronic lymphocytic leukemia/small lymphocytic lymphoma from mantle cell lymphoma. Leuk Lymphoma. 2015;56(5):1425–1431. doi: 10.3109/10428194.2014.953147. [DOI] [PubMed] [Google Scholar]
- 53.Mohanty A, Sandoval N, Phan A, et al. Regulation of SOX11 expression through CCND1 and STAT3 in mantle cell lymphoma. Blood. 2019;133(4):306–318. doi: 10.1182/blood-2018-05-851667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cencini E, Sicuranza A, Fabbri A, et al. Study of gene polymorphisms as predictors of treatment efficacy and toxicity in patients with indolent non-hodgkin lymphomas and mantle cell lymphoma receiving bendamustine and rituximab. Br J Haematol. 2019;184(2):223–231. doi: 10.1111/bjh.15582. [DOI] [PubMed] [Google Scholar]
- 55.Yoon DH, Cao J, Chen TY, et al. Treatment of mantle cell lymphoma in Asia: a consensus paper from the Asian Lymphoma Study Group. J Hematol Oncol. 2020;13(1):21. doi: 10.1186/s13045-020-00855-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang R, Huo Z, Duan Y, et al. SOX11 inhibits tumor proliferation and promotes cell adhesion mediated-drug resistance via a CD43 dependent manner in mantle cell lymphoma. Leuk Lymphoma. 2020:1–14. [DOI] [PubMed]
- 57.Navarro A, Clot G, Royo C, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res. 2012;72(20):5307–5316. doi: 10.1158/0008-5472.CAN-12-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nordstrom L, Sernbo S, Eden P, et al. SOX11 and TP53 add prognostic information to MIPI in a homogenously treated cohort of mantle cell lymphoma–a Nordic Lymphoma Group study. Br J Haematol. 2014;166(1):98–108. doi: 10.1111/bjh.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Annese T, Ingravallo G, Tamma R, et al. Inflammatory infiltrate and angiogenesis in mantle cell lymphoma. Transl Oncol. 2020;13(3):100744. doi: 10.1016/j.tranon.2020.100744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodrigues JM, Hassan M, Freiburghaus C, et al. p53 is associated with high-risk and pinpoints TP53 missense mutations in mantle cell lymphoma. Br J Haematol. 2020. [DOI] [PMC free article] [PubMed]
- 61.Sander B, Quintanilla-Martinez L, Ott G, et al. Mantle cell lymphoma–a spectrum from indolent to aggressive disease. Virchows Arch. 2016;468(3):245–257. doi: 10.1007/s00428-015-1840-6. [DOI] [PubMed] [Google Scholar]
- 62.Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest. 2012;122(10):3416–3423. doi: 10.1172/JCI61272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beekman R, Amador V, Campo E. SOX11, a key oncogenic factor in mantle cell lymphoma. Curr Opin Hematol. 2018;25(4):299–306. doi: 10.1097/MOH.0000000000000434. [DOI] [PubMed] [Google Scholar]
- 64.Kiel MJ, Smith LB. Transformation of Indolent Mantle Cell Lymphoma to Pleomorphic Mantle Cell Lymphoma: Case Report and Review of Clinical and Morphologic Variants. Arch Pathol Lab Med. 2012;(No.8):871–875. [DOI] [PubMed]
- 65.Kuci V, Nordstrom L, Conrotto P, et al. SOX11 and HIG-2 are cross-regulated and affect growth in mantle cell lymphoma. Leuk Lymphoma. 2016;57(8):1883–1892. doi: 10.3109/10428194.2015.1121257. [DOI] [PubMed] [Google Scholar]
- 66.van der Velden VH, Hoogeveen PG, de Ridder D, et al. B-cell prolymphocytic leukemia: a specific subgroup of mantle cell lymphoma. Blood. 2014;124(3):412–419. doi: 10.1182/blood-2013-10-533869. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, Tang G, Medeiros LJ, et al. MYC rearrangement but not extra MYC copies is an independent prognostic factor in patients with mantle cell lymphoma. Haematologica (Roma). 2020. [DOI] [PMC free article] [PubMed]
- 68.Nguyen L, Papenhausen P, Shao H. The Role of c-MYC in B-Cell Lymphomas: Diagnostic and Molecular Aspects. Genes-Basel. 2017;8(4):116. doi: 10.3390/genes8040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jardin F, Picquenot J, Parmentier F, et al. Detection of gene copy number aberrations in mantle cell lymphoma by a single quantitative multiplex PCR assay: clinicopathological relevance and prognosis value. Brit J Haematol. 2009;146(6):607–618. doi: 10.1111/j.1365-2141.2009.07791.x. [DOI] [PubMed] [Google Scholar]
- 70.Scapinello G, Riva M, Branca A, et al. A case of “double hit” mantle cell lymphoma carrying CCND1 and MYC translocations relapsed/refractory to rituximab bendamustine cytarabine (R-BAC) and ibrutinib. Ann Hematol. 2020. [DOI] [PubMed]
- 71.Hu Z, Medeiros LJ, Chen Z, et al. Mantle Cell Lymphoma With MYC Rearrangement: A Report of 17 Patients. Am J Surg Pathol. 2017;41(2):216. doi: 10.1097/PAS.0000000000000758. [DOI] [PubMed] [Google Scholar]
- 72.Choe J, Yun JY, Na HY, et al. MYC overexpression correlates with MYC amplification or translocation, and is associated with poor prognosis in mantle cell lymphoma. Histopathology. 2016;68(3):442–449. doi: 10.1111/his.12760. [DOI] [PubMed] [Google Scholar]
- 73.Navarro A, Clot G, Prieto M, et al. microRNA expression profiles identify subtypes of mantle cell lymphoma with different clinicobiological characteristics. Clin Cancer Res. 2013;19(12):3121–3129. doi: 10.1158/1078-0432.CCR-12-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orchard J, Garand R, Davis Z, et al. A subset of t(11;14) lymphoma with mantle cell features displays mutated IgVH genes and includes patients with good prognosis, nonnodal disease. Blood. 2003;101(12):4975–4981. doi: 10.1182/blood-2002-06-1864. [DOI] [PubMed] [Google Scholar]
- 75.Li X, Wu N, Li B. A high mutation rate of immunoglobulin heavy chain variable region gene associates with a poor survival and chemotherapy response of mantle cell lymphoma patients. Medicine. 2019;98(22):e15811. doi: 10.1097/MD.0000000000015811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scott DW, Abrisqueta P, Wright GW, et al. New molecular assay for the proliferation signature in mantle cell lymphoma applicable to formalin-fixed paraffin-embedded biopsies. J Clin Oncol. 2017;35(15):1668–1677. doi: 10.1200/JCO.2016.70.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holte H, Beiske K, Boyle M, et al. The MCL35 gene expression proliferation assay predicts high-risk MCL patients in a Norwegian cohort of younger patients given intensive first line therapy. Br J Haematol. 2018;183(2):225–234. doi: 10.1111/bjh.15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rubio-Moscardo F, Climent J, Siebert R, et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005;105(11):4445–4454. doi: 10.1182/blood-2004-10-3907. [DOI] [PubMed] [Google Scholar]
- 79.Spike BT, Macleod KF. The Rb tumor suppressor in stress responses and hematopoietic homeostasis. Cell Cycle. 2005;4(1):42–45. doi: 10.4161/cc.4.1.1337. [DOI] [PubMed] [Google Scholar]
- 80.Delfau-Larue M, Klapper W, Berger F, et al. High-dose cytarabine does not overcome the adverse prognostic value of CDKN2A and TP53 deletions in mantle cell lymphoma. Blood. 2015;126(5):604–611. doi: 10.1182/blood-2015-02-628792. [DOI] [PubMed] [Google Scholar]
- 81.Stankovic T, Stewart GS, Byrd P, et al. ATM mutations in sporadic lymphoid tumours. Leukemia Lymphoma. 2009;43(8):1563–1571. doi: 10.1080/1042819021000002884. [DOI] [PubMed] [Google Scholar]
- 82.Mareckova A, Malcikova J, Tom N, et al. ATM and TP53 mutations show mutual exclusivity but distinct clinical impact in mantle cell lymphoma patients. Leukemia Lymphoma. 2019;60(6):1420–1428. doi: 10.1080/10428194.2018.1542144. [DOI] [PubMed] [Google Scholar]
- 83.Ny F, Tc G, Dd W, et al. Oligonucleotide microarrays demonstrate the highest frequency of ATM mutations in the mantle cell subtype of lymphoma. Proc Natl Acad Sci. 2003;100(9):5372–5377. doi: 10.1073/pnas.0831102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bea S, Valdes-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci. 2013;110(45):18250–18255. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Filippi AR, Franco P, Galliano M, et al. Peripheral blood complete remission after splenic irradiation in mantle-cell lymphoma with 11q22-23 deletion and ATM inactivation. Radiation Oncol (London, England) 2006;1(1):35. doi: 10.1186/1748-717X-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jain P, Zhang S, Kanagal-Shamanna R, et al. Genomic profiles and clinical outcomes of de novo blastoid/pleomorphic MCL are distinct from those of transformed MCL. Blood Adv. 2020;4(6):1038–1050. doi: 10.1182/bloodadvances.2019001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou J, Hu L, Zuo M, et al. An uncommon case of double-hit mantle cell lymphoma that demonstrates a transformation process. Am J Clin Pathol. 2020;153(1):49–57. doi: 10.1093/ajcp/aqz133. [DOI] [PubMed] [Google Scholar]
- 88.Zhang J, Jima D, Moffitt AB, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123(19):2988–2996. doi: 10.1182/blood-2013-07-517177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao S, Kanagal Shamanna R, Navsaria L, et al. Efficacy of venetoclax in high risk relapsed mantle cell lymphoma (MCL) - outcomes and mutation profile from venetoclax resistant MCL patients. Am J Hematol. 2020;95(6):623–629. doi: 10.1002/ajh.25796. [DOI] [PubMed] [Google Scholar]
- 90.Jain P, Kanagal-Shamanna R, Zhang S, et al. Long-term outcomes and mutation profiling of patients with mantle cell lymphoma (MCL) who discontinued ibrutinib. Brit J Haematol. 2018;183(4):578–587. doi: 10.1111/bjh.15567. [DOI] [PubMed] [Google Scholar]
- 91.Yang P, Zhang W, Wang J, et al. Genomic landscape and prognostic analysis of mantle cell lymphoma. Cancer Gene Ther. 2018;25(5–6):129–140. doi: 10.1038/s41417-018-0022-5. [DOI] [PubMed] [Google Scholar]
- 92.Ferrero S, Rossi D, Rinaldi A, et al. KMT2D mutations and TP53 disruptions are poor prognostic biomarkers in mantle cell lymphoma receiving high-dose therapy: a FIL study. Haematologica. 2020;105(6):1604–1612. doi: 10.3324/haematol.2018.214056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yan Y, Yi S, Jin M, et al. Genomic and Transcriptomic Profiling Reveals Distinct Subsets Associated with Outcomes in Mantle Cell Lymphoma. Blood. 2020;136(Supplement 1):28–29. doi: 10.1182/blood-2020-139955. [DOI] [Google Scholar]
- 94.Agarwal R, Chan Y, Tam CS, et al. Dynamic molecular monitoring reveals that SWI–SNF mutations mediate resistance to ibrutinib plus venetoclax in mantle cell lymphoma. Nat Med. 2019;25(1):119–129. doi: 10.1038/s41591-018-0243-z. [DOI] [PubMed] [Google Scholar]
- 95.Wu C, de Miranda NF, Chen L, et al. Genetic heterogeneity in primary and relapsed mantle cell lymphomas: Impact of recurrent CARD11 mutations. Oncotarget. 2016;7(25):38180–38190. doi: 10.18632/oncotarget.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Furtado M, Rule S. Indolent mantle cell lymphoma. Haematologica. 2011;96(8):1086–1088. doi: 10.3324/haematol.2011.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martini M, D'Alo F, Pierconti F, et al. Bax mutations are an infrequent event in indolent lymphomas and in mantle cell lymphoma. Haematologica. 2000;85(10):1019–1023. [PubMed] [Google Scholar]
- 98.Hock BD, Haring LF, Steinkasserer A, et al. The soluble form of CD83 is present at elevated levels in a number of hematological malignancies. Leukemia Res. 2004;28(3):237–241. doi: 10.1016/S0145-2126(03)00255-8. [DOI] [PubMed] [Google Scholar]
- 99.Ziduo Li EAKL. Targeting CD83 in mantle cell lymphoma with anti-human CD83 antibody. Clinical & translational immunology. 2020;9(7). [DOI] [PMC free article] [PubMed]
- 100.Navarro A, Royo C, Hernandez L, et al. Molecular pathogenesis of mantle cell lymphoma: new perspectives and challenges with clinical implications. Semin Hematol. 2011;48(3):155–165. doi: 10.1053/j.seminhematol.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 101.Yang W, Wang Y, Yu Z, et al. SOX11 regulates the pro-apoptosis signal pathway and predicts a favorable prognosis of mantle cell lymphoma. Int J Hematol. 2017;106(2):212–220. doi: 10.1007/s12185-017-2236-y. [DOI] [PubMed] [Google Scholar]
- 102.Koh YW, Shin S, Park C, et al. Absolute monocyte count predicts overall survival in mantle cell lymphomas: correlation with tumour-associated macrophages. Hematol Oncol. 2014;32(4):178–186. doi: 10.1002/hon.2106. [DOI] [PubMed] [Google Scholar]
- 103.Jain P, Wang M. Blastoid Mantle Cell Lymphoma. Hematol Oncol Clin North Am. 2020;34(5):941–956. doi: 10.1016/j.hoc.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 104.Iqbal J, Shen Y, Liu Y, et al. Genome-wide miRNA profiling of mantle cell lymphoma reveals a distinct subgroup with poor prognosis. Blood. 2012;119(21):4939–4948. doi: 10.1182/blood-2011-07-370122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhu X, Wang J, Zhu M, et al. MicroRNA-506 inhibits the proliferation and invasion of mantle cell lymphoma cells by targeting B7 H3. Biochem Bioph Res Co. 2019;508(4):1067–1073. doi: 10.1016/j.bbrc.2018.12.055. [DOI] [PubMed] [Google Scholar]
- 106.Zhou K, Feng X, Wang Y, et al. miR-223 is repressed and correlates with inferior clinical features in mantle cell lymphoma through targeting SOX11. Exp Hematol. 2018;58:27–34. doi: 10.1016/j.exphem.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 107.H Y, J T, Jq C, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood: The Journal of the American Society of Hematology. 2010;(13):2630–2639. [DOI] [PMC free article] [PubMed]
- 108.Goswami RS, Atenafu EG, Xuan Y, et al. MicroRNA signature obtained from the comparison of aggressive with indolent non-hodgkin lymphomas: potential prognostic value in mantle-cell lymphoma. J Clin Oncol. 2013;31(23):2903–2911. doi: 10.1200/JCO.2012.45.3050. [DOI] [PubMed] [Google Scholar]
- 109.Wu B, Li J, Wang H, et al. MiR-132–3p serves as a tumor suppressor in mantle cell lymphoma via directly targeting SOX11. Naunyn-Schmiedeberg's Archives of Pharmacology. 2020. [DOI] [PubMed]
- 110.Zhou K, Yi S, Yu Z, et al. MicroRNA-223 expression is uniformly down-regulated in B cell lymphoproliferative disorders and is associated with poor survival in patients with chronic lymphocytic leukemia. Leukemia Lymphoma. 2012;53(6):1155–1161. doi: 10.3109/10428194.2011.642303. [DOI] [PubMed] [Google Scholar]
- 111.Husby S, Ralfkiaer U, Garde C, et al. miR-18b overexpression identifies mantle cell lymphoma patients with poor outcome and improves the MIPI-B prognosticator. Blood. 2015;125(17):2669–2677. doi: 10.1182/blood-2014-06-584193. [DOI] [PubMed] [Google Scholar]
- 112.Roisman A, Huamán Garaicoa F, Metrebian F, et al. SOXC and MiR17-92 gene expression profiling defines two subgroups with different clinical outcome in mantle cell lymphoma. Genes Chromosom Cancer. 2016;55(6):531–540. doi: 10.1002/gcc.22355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.