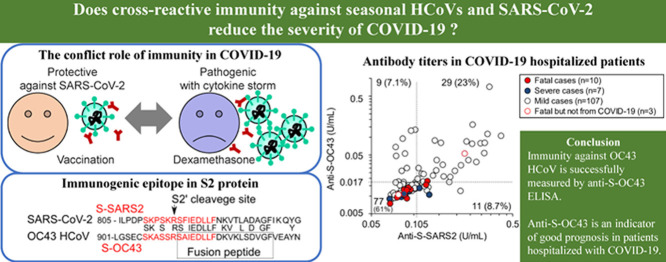
Abstract
Background
The effects of high-intensity immunity on coronavirus disease 2019 (COVID-19) remain unclear. Antibodies against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) are preferentially induced in inpatients with COVID-19 compared with outpatients with milder disease, and immunosuppression is the standard therapy for severe cases. This study investigated the relationship between cross-reactive antibody production against seasonal human coronavirus and the clinical course of COVID-19.
Methods
Among the immunogenic epitopes of SARS-CoV-2, conserved peptides in human coronavirus OC43 were searched and synthesized. Enzyme-linked immunosorbent assay was designed to detect antibodies against synthesized peptides. Antibody titres against S2ʹ cleavage site epitopes near fusion peptides of SARS-CoV-2 and OC43 were determined in the sera of 126 inpatients with COVID-19. The correlation between antibody titres and clinical data was analysed.
Results
Inpatients with COVID-19 who produced antibodies against OC43 did not develop severe or fatal pneumonia. Antibody titres against the corresponding epitope of SARS-CoV-2 did not differ between inpatients with severe and mild COVID-19. Antibody titres against the OC43 epitope increased more than those against SARS-CoV-2 during the first 2 weeks of COVID-19.
Conclusions
Immunity to seasonal human coronavirus OC43 effectively enhanced recovery from COVID-19. Detecting cross-reactive antibodies to OC43 may help to predict prognosis for patients with COVID-19.
Keywords: SARS-CoV-2, Antibody, Fusion peptide, S2ʹ cleavage site, Cross-reactivity
Graphical abstract
Introduction
An important characteristic of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is its variations. During the ongoing COVID-19 pandemic, the numbers of confirmed cases and deaths differ widely between countries, with rates more than 10 times higher in North America and Europe than in East Asia as of March 2021 (Wiersinga et al., 2020; World Health Organization, 2021). The severity of COVID-19 also varies considerably. Most infected cases have no symptoms or recover with mild, self-limiting symptoms, while approximately 5% of infected patients develop deadly pneumonia (Wiersinga et al., 2020). Acquired immunity to other human coronaviruses (HCoVs) may affect the morbidity and mortality of COVID-19. HCoVs such as OC43, HKU-1, NL63 and 229E are less pathogenic than their relative, SARS-CoV-2, and cause the seasonal common cold in the general population (Killerby et al., 2018). Memory T and B lymphocytes against seasonal HCoVs can be re-activated by SARS-CoV-2 infection, producing cross-reactive antibodies (Grifoni et al., 2020; Le Bert et al., 2020; Mateus et al., 2020; Secchi et al., 2020; Shrock et al., 2020). The difference in immunological memory against seasonal HCoVs is one possible explanation for the variations in COVID-19.
A critical question is whether cross-reactive immunity against SARS-CoV-2 and seasonal HCoVs reduces the severity of COVID-19. This issue is important for predicting the effectiveness and safety of SARS-CoV-2 vaccines. The origin of vaccination by Jenner was based on the medical use of cross-reactive immunity triggered by a related virus with low toxicity. Antibodies against the spike (S) protein or the receptor binding domain in the S1 protein of SARS-CoV-2 are detected in patients with COVID-19, and the antibodies may protect the host through their neutralizing activity (Brouwer et al., 2020; Chi et al., 2020; Hassan et al., 2020; Ju et al., 2020; Shi et al., 2020). In contrast, severe pneumonia in patients with COVID-19 is amplified by an excessive immune response called a ‘cytokine storm’ (Moore and June, 2020; Woodruff et al., 2020). Immunosuppression by corticosteroids or interleukin-6 blockade is a reliable therapy in severe cases (Guaraldi et al., 2020; Horby et al., 2021). Antibodies against SARS-CoV-2 have been detected more frequently in patients hospitalized with COVID-19 than in milder cases (Long et al., 2020; Shrock et al., 2020; Wiersinga et al., 2020; Garcia-Beltran et al., 2021). High amounts of immune complexes or activated lymphocytes may induce excessive inflammation. Some studies have proposed antibody-dependent enhancement of infection with suboptimal antibodies (Lee et al., 2020; Garcia-Beltran et al., 2021). One of the most serious concerns is the possibility that immunity activated by vaccines or seasonal HCoVs may increase the severity of COVID-19 owing to excessive inflammation when the host fails to prevent infection with SARS-CoV-2.
This study assessed the clinical effect of immunity against seasonal HCoVs on COVID-19. Antibody titres against the most immunogenic epitope near the fusion peptide in the S2 protein (Shrock et al., 2020), shared by SARS-CoV-2 and OC43 HCoV, were measured, and the clinical outcomes of patients hospitalized for COVID-19 (excluding mild, non-hospitalized cases) were compared, mainly using blood samples collected within 2 weeks of diagnosis.
Materials and methods
Database search for cross-reactive epitopes of SARS-CoV-2 and OC43 HCoV
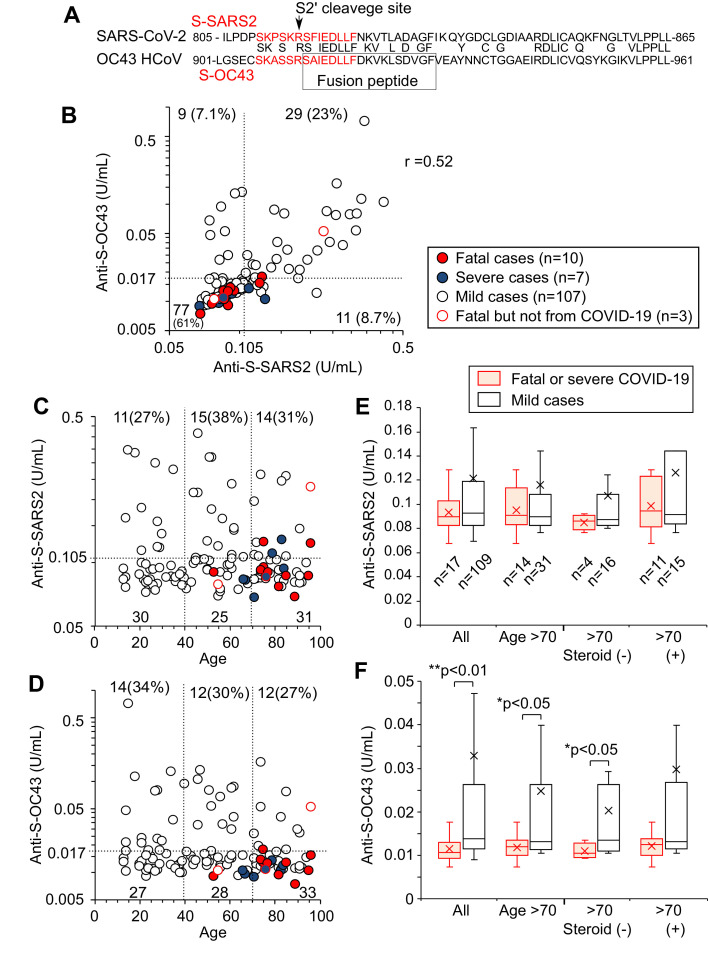
For 61 peptide sequences of epitope candidates of SARS-CoV-2 listed by Wang et al. (2020), blastp in NCBI (https://blast.ncbi.nlm.nih.gov) was used to search for similar peptides in OC43 HCoV by activating ‘Align two or more sequences’. AAR01012.1–AAR012020.1, listed in AY391777.1, were used for protein sequences of OC43 HCoV. The sequences of S2 protein of SARS-CoV-2 (QHD43416.1) and OC43 (AAR01015.1) were aligned with blastp in NCBI. Figure 1 A shows the functional domains in S2 protein known as the ‘fusion peptide’ (Madu et al., 2009) and the ‘predicted S2ʹ cleavage site’ (Benton et al., 2020).
Figure 1.
Antibody titres against S-OC43 but not S-SARS2 are low in patients with severe coronavirus disease 2019 (COVID-19). (A) Part of the protein sequence of S2 proteins. SARS-CoV-2 S2 protein (from 805th to 865th amino acid) and OC43 HCoV S2 protein (from 901st to 961st amino acid) were aligned based on homology, indicating the shared amino acids. Peptide sequences of S-SARS2 and S-OC43 are shown in red letters. The functional domains known as the S2′ cleavage site and the fusion peptide are also shown. (B) Antibody titres against S-SARS2 and S-OC43 in each patient. Antibody titres were measured for 126 patients hospitalized with COVID-19 using sera drawn 1–2 weeks after diagnosis. Red and blue circles indicate fatal and severe cases, respectively. Dotted lines indicate the antibody titres of sera used for dilution of standard, meaning the lower detection limits. Median values of replicated tests are shown. The numbers of cases in the fractions and correlation coefficient are inserted. (C,D) Antibody titres were plotted against age. The number of cases in the area is also shown. (E,F) Titres of anti-S-SARS2 (E) and anti-S-OC43 (F) were compared between severe or fatal cases (closed markers) and mild hospitalized cases (open markers) among all populations, cases aged >70 years, and cases aged >70 years with or without systemic corticosteroid treatment. The number of cases in each group is shown in (E). Boxes indicate 75th, 50th and 25th percentiles. Crosses indicate the mean. Statistical significance was tested using Mann–Whitney U-test.
Clinical subjects in the first epidemic period
By June 2020, 66 adults had been diagnosed with SARS-CoV-2 infection at Nozaki Tokushukai Hospital in Osaka, Japan using quantitative reverse transcription polymerase chain reaction (qRT-PCR). Residual blood samples were available for 49 patients, including 34 hospitalized patients. The median age was 51 years (range 15–92 years). Among the 34 hospitalized patients, 25 (74%) had pneumonia detected on computed tomography (CT). One hundred and fifty-two residual blood samples, collected on 8–10 April 2020, from patients whose tests showed they were not infected with SARS-CoV-2 were used as SARS-CoV-2-negative samples.
Clinical subjects in the second epidemic period
From July to October 2020, 126 patients with COVID-19 were hospitalized at Nozaki Tokushukai Hospital. Residual blood samples drawn during hospitalization were available from all patients at some time points. Blood samples were available for 122 cases within 7 days of diagnosis, and for 106 cases from >7 days after diagnosis. The median age of these 126 patients was 55.5 years (range 13–96 years). Thirteen patients died during the observation period lasting until November 2020. Among them, 10 patients died from COVID-19, including two patients who died shortly after discharge from the infectious disease ward (median day 13, 3–28 days after diagnosis). The other three patients died from causes other than COVID-19, and were excluded from the fatal COVID-19 cases. Although the hospital did not have an intensive care unit for patients with COVID-19, seven cases were classified as severe COVID-19 according to the respiratory conditions and CT findings. The cause of death and the classification of severity were determined by physicians who were blinded to the results of this study. Clinical data can be seen in Table 2. The mean sampling day of the analysed bloods of patients hospitalized with fatal, severe or mild COVID-19 was 8.4, 11.7 and 9.7 days after diagnosis, respectively.
Table 2.
Clinical data for severe cases receiving antibodies and cases who died from other causes during the second epidemic period.
| Index | Age | Sex | Pneumonia | Max O2(L/min) | Treatmenta | Days to death | Days of qRT-PCR(+)b | Anti-S-SARS2c | Anti-S-OC43c | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 90s | F | Non | 0 | - | 42 | 14 | - | ++ | - | ++ |
| 13 | 90s | F | Bilateral | 10 | S | 3 | NA | + | NA | - | NA |
| 14 | 70s | F | Bilateral | 15 | S,F,Na | 10 | >9 | - | + | - | + |
| 15 | 80s | M | Bilateral | 15 | S,Na | 0 (<14) | - | + | - | - | |
| 16 | 70s | M | Bilateral | MV | S,F,Ni | 17 | NA | + | NA | - | |
| 17 | 70s | M | Bilateral | 8 | - | 86 | 4 | - | - | - | - |
| 18 | 50s | F | Bilateral | 4 | S,Na | 11 | 0 (<8) | - | - | - | - |
qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; MV, mechanical ventitlation.
Of the hospitalized patients infected with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) from July to October 2020, five severe or fatal cases induced detectable levels of anti-S-SARS2 (Cases 12–16). Three cases who died from causes other than coronavirus disease 2019 are also listed (Cases 12, 17 and 18). Pneumonia was evaluated with chest X-ray imaging in Case 13, and with computed tomography in the other cases. Maximum flow rates of oxygen inhalation required during hospitalization are shown. MV was required from day 2 to day 9.
Case 12 was discharged from the infectious disease ward on day 28 but died on day 42 from senility. Pneumonia was not detected on repeated computed tomography. Case 17 recovered from severe COVID-19 pneumonia and was discharged on day 31, but was rehospitalized due to respiratory failure on day 82 and died on day 86 when SARS-CoV-2 was negative on qRT-PCR. Case 18 was hospitalized due to hydrocephalus and arteriovenous malformation. SARS-CoV-2 infection was diagnosed by screening qRT-PCR test at hospitalization. Her respiratory condition recovered, but she died from brain damage. Cases 12 and 18 were classified as mild COVID-19.
Treatments for pneumonia other than antibiotics are listed. S, systemic corticosteroids; F, favipiravir; Na, nafamostat; Ni, nintedanib.
Days of qRT-PCR(+) indicate days from diagnosis until the last day of a positive result on qRT-PCR for SARS-CoV-2.
Antibody titres of blood drawn within 1 week of diagnosis (left) and later (right) are shown. ++, antibody titre is higher than 0.21 and 0.105 U/mL for anti-S-SARS2; +, antibody titre is higher than 0.034 and 0.017 U/mL for anti-S-OC43, respectively.
Enzyme-linked immunosorbent assay using the different peptides of OC43 and SARS-CoV-2
Peptides (S-SARS2, S-OC43, Hel and NSP13 listed in Table S1, see online supplementary material) were synthesized and purified using high-performance liquid chromatography at >50% purity by Eurofins Genomics K.K. (Tokyo, Japan). Dry peptides were dissolved in water at a concentration of 2 mM, except for NSP13. NSP13 was dissolved in water with a 10% volume of acetate. A 96-well enzyme-linked immunosorbent assay (ELISA) plate (IWAKI, AGC Techno Glass, Shizuoka, JAPAN) was coated with 1 µM peptide diluted in 50 mM carbonate buffer (pH 9.6) at 4°C overnight. After washing with PBST (0.05% Tween-20 in phosphate buffered saline) three times, plates were blocked with blocking buffer (1% bovine serum albumin in PBST) for 1 h. After washing with PBST, the plates were incubated with patient serum diluted in blocking buffer for 1 h. After washing with PBST, 0.4 µg/mL anti-human IgG(H+L) conjugated with horseradish peroxidase (Promega, Fitchburg, WI, USA) in blocking buffer was added and incubated for 30 min. After washing with PBST five times, 50 µL of TMB substrate solution (Thermo Fisher Scientific, Waltham, MA, USA) was added. The reaction was stopped by adding 150 µL sulfuric acid after 10 min of incubation. Optical density (OD) values were measured at 450 nm using a microplate reader (Sunrise, Tecan, Männedorf, Switzerland). For standard antibody titration, sera from Cases 2 and 3 drawn on day 35 were mixed in a 1:1 ratio, and the mixture was defined to be 1 U/mL. The mixture was serially diluted with serum from a healthy donor which had 0.105 U/mL of anti-S-SARS2 and 0.017 U/mL of anti-S-OC43. Using 10% serum diluted with blocking buffer, the antibody concentration in each sample was determined by referring to a fitting curve made on the same ELISA plate.
Results
Search for conserved epitopes between SARS-CoV-2 and OC43 HCoV
A published database was searched for immunogenic epitopes to detect antibodies against OC43 HCoV in ELISA. Among 61 epitope candidates listed by Wang et al. (2020), four epitope sequences of SARS-CoV-2 were found to be completely matched to sequences in Orf1ab of OC43 HCoV, as expected from the high homology of their Orf1ab genes (Zhou et al., 2020). Two regions of OC43 HCoV were selected: 5802–5815 in helicase (Hel) and 6920–6934 in NSP-13; these included the matched epitope and had one amino acid difference from their counterparts in SARS-CoV-2 (Table S1, see online supplementary material). In the S protein, an epitope candidate of SARS-CoV-2, 816-SFIED-820, was found to share sequence with OC43 HCoV, except for 817-F. Two peptides, S-OC43 (906–919 of OC43 HCoV) and S-SARS2 (810–823 of SARS-CoV-2) were synthesized (Table S1, see online supplementary material). This domain in the S2 protein partially overlaps with the fusion peptide (Madu et al., 2009), and includes the predicted S2ʹ cleavage site (Benton et al., 2020) (Figure 1A). Interestingly, lists of the most immunogenic epitopes that were searched using unbiased methods by five independent groups commonly include this S2ʹ cleavage site epitope (Farrera-Soler et al., 2020; Poh et al., 2020; Shrock et al., 2020; Wang et al., 2020; Yi et al., 2020).
Antibodies against the S2 peptides were detected in COVID-19 and non-COVID-19 cases
In order to detect the prevalence of anti-OC43 humoral immunity in normal populations, ELISA was performed with four peptides of OC43 and SARS-CoV-2 (Table S1, see online supplementary material) using blood samples from 152 uninfected cases, drawn in April 2020 at the same time as the qRT-PCR test for SARS-CoV-2 infection. Among them, six cases (3.9%) showed higher OD values at 450 nm (OD450) against S-SARS2 than the thresholds. Regarding S-OC43, 12 cases (7.9%) were positive, of which four cases were also positive for S-SARS2 (Figure S1, see online supplementary material). The other two cases were positive for anti-S-SARS2 alone. Regarding Hel and NSP13, 22 and 77 cases were higher than the thresholds, respectively. The OD450 values were slightly higher in almost all samples in wells coated with NSP13 or Hel than in those without peptides. This implies a wide prevalence of immunity against HCoVs, as reported by Grifoni et al. (2020), but the possibility that the result reflected characteristics of the coating peptides could not be ruled out. The OD450 values for S-SARS2 and S-OC43 were clearly high in four cases, and the average values were lower than those of the other peptides. These results indicate that approximately 8% of uninfected people have humoral immunity against OC43 HCoV which is detectable with anti-S-OC43 ELISA, and half of them have cross-reactive antibodies against S-SARS2.
Next, blood samples of SARS-CoV-2-infected cases were tested to assess the effect of SARS-CoV-2 infection on antibody titres. During the first epidemic period, until June 2020, blood samples were collected from 49 patients who were diagnosed with SARS-CoV-2 infection at Nozaki Tokushukai Hospital. Among them, seven cases, including five cases whose samples were taken >1 week after diagnosis, had anti-S-SARS2 (Table 1 and Figure S2A, see online supplementary material). All seven cases with anti-S-SARS2 were also positive for anti-S-OC43, and six cases had anti-NSP13. In the first epidemic period, patients with COVID-19 were hospitalized for a long period until repeated qRT-PCR tests became negative. Among the seven cases with a high titre of anti-S-SARS2, SARS-CoV-2 was detected in four cases for a long duration (>1 month), and six cases suffered from pneumonia (Table 1 and Figure S2B, see online supplementay material). Seven patients were rehospitalized due to mild symptoms and positive results on re-examination of SARS-CoV-2 qRT-PCR, including Cases 1 and 5. These findings indicate that cross-reactive antibodies against S-SARS2 and S-OC43 can be detected in hospitalized patients with COVID-19 pneumonia, but the contribution to recovery is unclear.
Table 1.
Development of pneumonia in patients infected with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) treated with antibodies during the first epidemic period.
| Index | Age | Sex | Pneumonia | Max O2(L/min) | Treat-menta | Days ofqRT-PCR(+)b | Anti-S-SARS2c | Anti-S-OC43c | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 30s | M | Bilateral | 0 | B | 39 | NA | ++ | NA | ++ |
| 2 | 50s | F | Bilateral | 1 | F,B | 44 | ++ | ++ | ++ | ++ |
| 3 | 20s | M | Bilateral | 1 | S,F | 40 | NA | ++ | NA | ++ |
| 4 | 50s | F | Bilateral | 0 | B | 8 | NA | ++ | NA | + |
| 5 | 60s | M | Left | 0 | B | 33 | NA | + | NA | + |
| 6 | 40s | F | Left | 0 | B | 14 | NA | + | NA | + |
| 7 | 10s | F | Non | 0 | - | NA (<47) | + | NA | + | NA |
| 8 | 40s | M | Bilateral | 1 | F | 30 | - | - | - | - |
| 9 | 20s | F | Bilateral | 1 | F,I | 36 | - | - | - | ++ |
| 10 | 50s | F | Bilateral | 0 | - | NA | - | NA | - | NA |
| 11 | 70s | M | Bilateral | 15 | - | NA | - | NA | - | NA |
qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
Among 49 patients infected with SARS-CoV-2 before June 2020, Cases 1–8 had the highest amounts of anti-S-SARS2 and anti-S-OC43, Case 9 had the highest amount of anti-Hel, Case 10 had the highest amount of anti-NSP13, and Case 11 died 8 days after diagnosis. Cases 3, 7 and 10 were family members. Pneumonia was evaluated with computed tomography. Maximum flow rates of oxygen inhalation required during hospitalization are shown.
Treatments for pneumonia other than antibiotics are listed. B, baloxavir marboxil; F, favipiravir; S, systemic corticosteroids; I, inhaled corticosteroids.
Days of qRT-PCR(+) indicate days from diagnosis until the last day of a positive result on qRT-PCR for SARS-CoV-2; NA, qRT-PCR testing was not repeated at the study hospital.
The results for antibody detection are shown for blood drawn at diagnosis (left) and at least 20 days later (right). ++, antibody was detected at 1/100 dilution; +, antibody was detected at 1/10 dilution; NA, blood samples were not available.
Low antibody titres against OC43 S2 peptide in patients with severe COVID-19
In order to reveal the effect of cross-reactive immunity on the clinical outcomes of COVID-19, this study focused on the titres of antibodies against S-SARS2 and S-OC43 in patients hospitalized with COVID-19, and analysed the correlation between antibody titres and the severity of each case in the next epidemic period. Blood samples were collected from 126 hospitalized patients with COVID-19 during the second epidemic period, from July to October 2020. The majority were patients with mild or moderate pneumonia, or patients with a mild or asymptomatic infection who were at high risk due to old age or comorbidities. Antibody titres against S-SARS2 and S-OC43 were determined by comparing OD450 values with those of the serum mixture of Cases 2 and 3 (defined as 1 U/mL) diluted in serum from a healthy donor that contained 0.105 U/mL of anti-S-SARS2 and 0.017 U/mL of anti-S-OC43. When blood samples drawn 1–2 weeks after diagnosis (median day 9, range 1–17 days) were tested, 40 (32%) and 38 (30%) cases had anti-S-SARS2 and anti-S-OC43 above the detection thresholds, respectively (Figure 1B). Antibody titres against S-SARS2 and S-OC43 were correlated (r=0.52), while nine (7.1%) cases had anti-S-OC43 alone at high levels. These results suggest that antibodies against S-OC43 are not always cross-reactive with S-SARS2, but the immune state producing anti-S-OC43 may easily induce generation of antibodies reactive to S-SARS2.
Among 126 cases, 10 patients (shown as red circles in Figure 1) died during the observation period (median day 13, range 3–28 days after diagnosis), excluding three patients who died from causes other than COVID-19 on days 11, 42 and 86 after diagnosis (Table 2 ). Seven cases, including Case 17 who recovered from COVID-19 pneumonia but died on day 86, were classified as severe COVID-19 with a broad area of pneumonia on CT scan and poor respiratory condition (shown as blue circles, Figure 1B–D). Classification was determined by physicians who were blinded to the antibody titre results. Case 12, a female patient in her 90s with high antibody titres, did not suffer from COVID-19 pneumonia, but died, possibly from senility, 42 days after diagnosis (shown by an open red circle). In contrast, Cases 13 and 14, with low levels of anti-S-SARS2, died of COVID-19 pneumonia 3 and 10 days after diagnosis, respectively (Table 2). Antibody titres against S-SARS2 and S-OC43 were compared between 17 fatal or severe patients (closed circles in Figure 1B) and 109 mild inpatients with COVID-19 (open circles in Figure 1B). Interestingly, the titres of anti-S-OC43 were significantly higher in less severe cases than in fatal or severe cases (P<0.01, Mann–Whitney U-test), while those of anti-S-SARS2 did not differ (P>0.1, Mann–Whitney U-test) (Figure 1E,F).
Fatality due to COVID-19 is known to be largely affected by age (Wiersinga et al., 2020). Consistent with this, the ratio of severe COVID-19 cases among hospitalized patients was found to be high in older patients (Figure 1C). Plotting antibody titres against age revealed that the antibody titre of anti-S-OC43 was not higher in the elderly population, despite possible exposure to OC43 HCoV at more time points during their life (Figure 1D). This may be due to a reduction in antibody titres after seasonal HCoV pre-exposure for several years (Edridge et al., 2020). When antibody titres were compared between 14 fatal or severe COVID-19 cases and 31 mild cases aged >70 years (Figure 1C-F), the titres of anti-S-OC43 were still significantly higher in mild cases than in severe cases (P<0.05, Mann–Whitney U-test). The titres of anti-S-SARS2 did not differ between the two groups. Immunosuppression by corticosteroid therapy may affect antibody titres (Garcia-Beltran et al., 2021). When antibody titres among patients aged >70 years old who were not treated with systemic corticosteroids were compared, four fatal or severe cases had significantly lower titres of anti-S-OC43 than 16 mild cases (P<0.05, Mann–Whitney U-test); in contrast, there were no significant differences in titres of anti-S-SARS2 between the same groups, or in titres of anti-S-OC43 among cases treated with systemic corticosteroids (Figure 1 E,F, right). While the titres of antibodies against SARS-CoV-2 could be largely affected by high viral loads or the severity of COVID-19, the high titre of anti-S-OC43 was an indicator of good prognosis of COVID-19, with high immunity against OC43 HCoV which potentially cross-reacts with SARS-CoV-2.
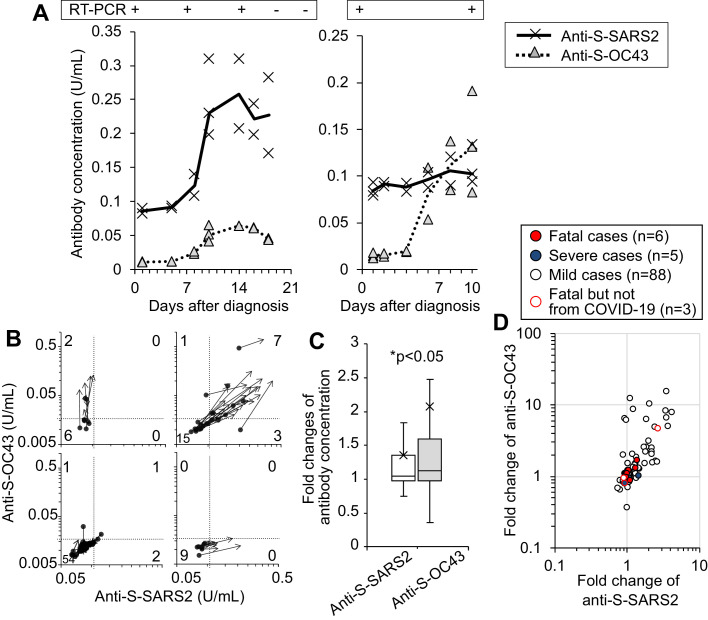
Anti-S-OC43 titre increases before the induction of specific antibody against S-SARS2
To investigate whether the acquired immunity against OC43 HCoV is re-activated by SARS-CoV-2 infection, the kinetics of antibody titres during hospitalization were analysed. For example, the titres of anti-S-SARS2 and anti-S-OC43 in the blood of Case 12 were low at hospitalization and increased three- and six-fold, respectively, 2 weeks after diagnosis, removing the virus successfully (Figure 2 A, left). In another case, a man in his 40s with mild pneumonia without treatment, the titre of anti-S-OC43 increased 8.7-fold, while the titre of anti-S-SARS2 increased 1.2-fold (Figure 2A, right). To confirm the increase in anti-S-OC43 titre with SARS-CoV-2 infection, ELISA was performed using 101 serum samples drawn at hospitalization within 6 days of diagnosis, and the antibody titres were compared with those in blood drawn at least 5 days after the initial sampling (mean interval 9 days, range 5–14 days). Among the 26 cases that produced detectable amounts of anti-S-SARS2 and anti-S-OC43, initial blood samples in 15 (58%) cases contained low amounts of antibodies (Figure 2B, right upper plots). When the ratio of antibody titres in sera drawn at the later time point were compared with those in sera drawn at the initial time point, the median fold changes were >1 for anti-S-SARS2 (median 1.05-fold) and anti-S-OC43 (median 1.12-fold) (Figure 2C). More interestingly, the fold changes in antibody titres were significantly higher for anti-S-OC43 than anti-S-SARS2 among patients hospitalized with COVID-19 (P<0.05, Wilcoxon signed-rank test, Figure 2C,D). Six cases were found to have increased anti-S-OC43 titres but not anti-S-SARS2 titres (Figure 2B,D). These results indicate that patients who have already acquired immunity against S-OC43 have increased anti-S-OC43 titres after SARS-CoV-2 infection, and 75% of these patients successfully generated antibodies reactive to S-SARS2. These results suggest that re-activation of immunological memory against seasonal HCoVs is induced more frequently than new construction of specific antibodies against SARS-CoV-2 during the first 2 weeks of SARS-CoV-2 infection.
Figure 2.
Preferential induction of anti-S-OC43 over anti-S-SARS2 after infection with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection. (A) Titres of anti-S-SARS2 and anti-S-OC43 in sera of two cases are plotted against sampling days after diagnosis. The median values are connected with lines. The results of quantitative reverse transcription polymerase chain reaction of SARS-CoV-2 are also shown. The left is Case 12 listed in Table 2. (B) Change in antibody titres over 1–2 weeks after diagnosis of SARS-CoV-2. For 101 cases whose serum samples were available, antibody titres were compared in each case between serum within 6 days of diagnosis (shown as circle) and serum 5–14 days later (shown as arrow). Data are classified in four groups depending on the titres in the later sera. The numbers of cases in each area are inserted in the figures. (C,D) Ratio of antibody titres in sera drawn at the later time point to the initial titres in each patient, shown as box and whisker plots (C) and dot plots (D). Statistical significance was analysed using Wilcoxon's signed-rank test. COVID-19, coronavirus disease 2019.
Discussion
This study showed that the antibody titre against the epitope near the fusion peptide of OC43 HCoV was significantly lower in patients hospitalized with severe or fatal COVID-19 than in patients with mild disease. While 30% of cases had detectable levels of anti-S-OC43 during hospitalization, only one patient among 17 fatal or severe cases induced anti-S-OC43 at a marginal level. In contrast, anti-S-SARS2 was induced in four severe or fatal cases, albeit at low levels. These results suggest that immunity against OC43 HCoV is measured successfully by anti-S-OC43 ELISA, and that the titre is an indicator of good prognosis in patients hospitalized with COVID-19.
Many groups have shown that the neutralizing antibodies and cross-reactive immunity to seasonal HCoVs are induced in patients infected with SARS-CoV-2 (Long et al., 2020; Shrock et al., 2020; Sun et al., 2020; Wiersinga et al., 2020; Garcia-Beltran et al., 2021). Consistent with the present findings, higher titres of antibodies were detected in symptomatic hospitalized cases than in asymptomatic cases, possibly due to their higher viral loads. Consequently, the correlation between antibody titres and good prognosis of COVID-19 was difficult to demonstrate. Higher titres of antibodies against S1 antigens or neutralizing activity were reported to be associated with less severe cases among hospitalized patients (Atyeo et al., 2020; Secchi et al., 2020; Sun et al., 2020; Garcia-Beltran et al., 2021). Following these reports, clinical outcomes were compared among hospitalized patients, excluding mild, non-hospitalized cases of SARS-CoV-2 infection. The results with titres of cross-reactive antibody are more informative than reports based on titre ratios of specific antibody over other anti-SARS-CoV-2 (Atyeo et al., 2020; Garcia-Beltran et al., 2021) in order to assess whether high acquired immunity is protective against COVID-19. For cross-reactive antibody, a group reported that antibody against SARS-CoV-2 S1+S2 antigens was associated with improved survival, and the anti-SARS-CoV-2 titres were correlated with antibody titres against OC43 S2 antigen (Secchi et al., 2020). The larger antigen epitopes or longer duration of blood sampling used in the present study may explain the slight discrepancy (i.e. anti-S-OC43 but not anti-S-SARS2 was correlated with good clinical outcome).
The epitope including the S2ʹ cleavage site near the fusion peptide is the most immunogenic epitope (Farrera-Soler et al., 2020; Poh et al., 2020; Shrock et al., 2020; Wang et al., 2020; Yi et al., 2020). The fusion peptide is known to be highly conserved among various coronaviruses, and mediates the fusion of viral membranes and mammalian cells through its hydrophobic amino acids (Madu et al., 2009; Chan et al., 2013). Antibodies against this domain may have neutralizing activity (Miyoshi-Akiyama et al., 2011; Poh et al., 2020). Although the authors have not evaluated the neutralizing activity or cross-reactivity directly, antibodies against S-OC43 may be acquired during previous exposure to OC43 HCoV, and may interfere with SARS-CoV-2 infection of host cells.
Some improvements are required to clarify the results and to make this ELISA system useful as a clinical test. The antibodies to OC43 HCoV were detectable in <10% of cases in blood drawn at diagnosis or before infection with SARS-CoV-2. A wait of 1–3 weeks was needed until the antibodies increased to detectable levels in 30% of cases (Secchi et al., 2020). The requirement for this delay may cause bias in the availability of blood. One-quarter of patients with fatal disease died within 1 week, and one-quarter of patients were discharged within 1 week. This also explains why hospitalized patients with pneumonia showed higher titres of antibodies than outpatients with milder disease in the results during the first epidemic period. Treatments with a high dose of corticosteroids for severe COVID-19 may affect antibody production (Garcia-Beltran et al., 2021). As many as 76% of severe or fatal cases received systemic corticosteroid treatment, which is a higher proportion than that in mild COVID-19 cases. Although the treatment in either severe or mild cases did not show reduced titres of antibodies in blood samples drawn approximately 10 days after diagnosis, treatments before blood sampling may affect antibody titres. In order to detect cross-reactive immunity in blood drawn at diagnosis, sensitivity should be improved by some modification, such as by increasing the number of epitopes evaluated in order to cover epitopes of the other three seasonal HCoVs.
Despite these limitations and the small size of the cohort, these results indicate that high immunity against OC43 HCoV protects patients hospitalized with COVID-19 from fatal exacerbations. Humoral immunity against seasonal HCoVs may be functionally more important than specific antibodies against SARS-CoV-2, which take 1 month to reach a peak (Secchi et al., 2020; Garcia-Beltran et al., 2021), and may be a better indicator of good prognosis. The present results also include important suggestions regarding the SARS-CoV-2 vaccine. The protective function of cross-reactive immunity supports the notion that vaccines are fundamentally safe and effective against COVID-19. A new type of vaccine that activates immunity to seasonal HCoVs may be worth considering to prepare for the possible epidemics of new and existing SARS-CoV-2 variants.
Author contributions
Study design: TY and TS.
Data collection and laboratory experiments: TY, HK and HN.
Data analysis: TY and HN.
Writing: TY and TS.
Declaration of Competing Interest
None declared.
Acknowledgments
Acknowledgements
The authors wish to thank M. Minata for the analysis of clinical data; H. Nakamoto for critical reading of the manuscript; Y. Tanizawa, T. Chishiro, S. Hirayama and other staff at Nozaki Tokushukai Hospital for the management of patients with COVID-19 and samples; and Editage (www.editage.com) for English language editing.
Funding
This work was financially supported by Tokushukai Medical Group.
Ethical approval
This study was approved by the Ethics Committee of Medical Group Tokushukai (TGE01425-002). Blood was collected following informed consent.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2021.07.015.
Appendix. Supplementary materials
References
- Atyeo C, Fischinger S, Zohar T, Slein MD, Burke J, Loos C, et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53 doi: 10.1016/j.immuni.2020.07.020. 524–32e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KH, Chan JF, Tse H, Chen H, Lau CC, Cai JP, et al. Cross-reactive antibodies in convalescent SARS patients' sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J Infect. 2013;67:130–140. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge AWD, Kaczorowska J, Hoste ACR, Bakker M, Klein M, Loens K, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Farrera-Soler L, Daguer JP, Barluenga S, Vadas O, Cohen P, Pagano S, et al. Identification of immunodominant linear epitopes from SARS-CoV-2 patient plasma. PLoS One. 2020;15 doi: 10.1371/journal.pone.0238089. e0238089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran WF, Lam EC, Astudillo MG, Yang D, Miller TE, Feldman J, et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184 doi: 10.1016/j.cell.2020.12.015. 476–88e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181 doi: 10.1016/j.cell.2020.05.015. 1489–501e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaraldi G, Meschiari M, Cozzi-Lepri A, Milic J, Tonelli R, Menozzi M, et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2:e474–e484. doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182 doi: 10.1016/j.cell.2020.06.011. 744–53e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, et al. Dexamethasone in hospitalized patients with Covid-19 – preliminary report. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Killerby ME, Biggs HM, Haynes A, Dahl RM, Mustaquim D, Gerber SI, et al. Human coronavirus circulation in the United States 2014–2017. J Clin Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Lee WS, Wheatley AK, Kent SJ, DeKosky BJ. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Madu IG, Roth SL, Belouzard S, Whittaker GR. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi-Akiyama T, Ishida I, Fukushi M, Yamaguchi K, Matsuoka Y, Ishihara T, et al. Fully human monoclonal antibody directed to proteolytic cleavage site in severe acute respiratory syndrome (SARS) coronavirus S protein neutralizes the virus in a rhesus macaque SARS model. J Infect Dis. 2011;203:1574–1581. doi: 10.1093/infdis/jir084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Poh CM, Carissimo G, Wang B, Amrun SN, Lee CY, Chee RS, et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secchi M, Bazzigaluppi E, Brigatti C, Marzinotto I, Tresoldi C, Rovere-Querini P, et al. COVID-19 survival associates with the immunoglobulin response to the SARS-CoV-2 spike receptor binding domain. J Clin Invest. 2020;130:6366–6378. doi: 10.1172/JCI142804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shrock E, Fujimura E, Kula T, Timms RT, Lee IH, Leng Y, et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370 doi: 10.1126/science.abd4250. eabd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Feng Y, Mo X, Zheng P, Wang Q, Li P, et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect. 2020;9:940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hou X, Wu X, Liang T, Zhang X, Wang D, et al. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Cent Sci. 2020;6:2238–2249. doi: 10.1021/acscentsci.0c00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . WHO; Geneva: 2021. World Health Organization coronavirus dashboard.https://covid19.who.int Available at: [Google Scholar]
- Yi Z, Ling Y, Zhang X, Chen J, Hu K, Wang Y, et al. Functional mapping of B-cell linear epitopes of SARS-CoV-2 in COVID-19 convalescent population. Emerg Microbes Infect. 2020;9:1988–1996. doi: 10.1080/22221751.2020.1815591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.