Abstract
ATP synthase is essential in aerobic energy metabolism, and the rotary catalytic mechanism is one of the core concepts to understand the energetic functions of ATP synthase. Disulfide bonds formed by oxidizing a pair of cysteine mutations halted the rotation of the γ subunit in two critical conformations, the ATP-waiting dwell (αE284C/γQ274C) and the catalytic dwell (αE284C/γL276C). Tryptophan fluorescence was used to measure the nucleotide binding affinities for MgATP, MgADP and MgADP-AlF4 (a transition state analog) to wild-type and mutant F1 under reducing and oxidizing conditions. In the reduced state, αE284C/γL276C F1 showed a wild-type-like nucleotide binding pattern; after oxidation to lock the enzyme in the catalytic dwell state, the nucleotide binding parameters remained unchanged. In contrast, αE284C/γQ274C F1 showed significant differences in the affinities of the oxidized versus the reduced state. Locking the enzyme in the ATP-waiting dwell reduced nucleotide binding affinities of all three catalytic sites. Most importantly, the affinity of the low affinity site was reduced to such an extent that it could no longer be detected in the binding assay (Kd > 5 mM). The results of the present study allow to present a model for the catalytic mechanism of ATP synthase under consideration of the nucleotide affinity changes during a 360° cycle of the rotor.
Keywords: ATP synthase, Disulfide crosslink, Nucleotide binding, Catalytic mechanism, Tryptophan fluorescence, Rotational catalysis
1. Introduction
ATP synthase is structurally and functionally conserved among species. ATP synthesis through oxidative phosphorylation or photophosphorylation plays a critical role in the energy metabolism of animals, plants and many microbes [1]. The prototype enzyme, ATP synthase from Escherichia coli consists of eight different subunits: three are found in the membrane-embedded Fo subcomplex (ab2c10), and five are found in the water-soluble F1 subcomplex (α3β3γδε) [2]. ATP synthase couples ATP synthesis/hydrolysis and the movement of protons across the membrane. The conversion of electrochemical energy (proton gradient) to chemical energy (ATP) occurs in ATP synthase through its unique rotary catalytic mechanism, with mechanical energy as intermediate [3]. Protons flowing down the electrochemical gradient push the rotor complex (c10γε) to spin. Rotation of the γ subunit within the catalytic hexamer (α3β3) changes the conformation of three catalytic binding sites on the β subunits to synthesize ATP from ADP and inorganic phosphate (Pi) [4]. In bacteria, under certain physiological conditions the enzyme can run in reverse, hydrolyzing ATP to build up a proton gradient which the bacterium needs for nutrient uptake and locomotion [5]. For further reviews see Refs. [6–10].
According to Boyer’s “binding change” principles, in ATP synthesis, the energy obtained from the proton gradient is used to reduce the affinity of MgATP as well as to increase the affinity for substrates, but not for the chemical synthesis step itself [11]. The rotational angle of the γ subunit determines the conformation of each of the three β subunits, which have been named βTP (AMP-PNP bound), βDP (ADP bound) and βE (empty) according to the nucleotide occupancy in the original crystal structure, with the nucleotide-bound sites in the “closed” conformation and the empty site in the “open” conformation [12]. The catalytic binding sites located in these β subunits have vastly different affinities to Mg2+-nucleotides (thus named high, medium or low affinity site) [13]; a “half-closed” conformation (βHC) is proposed to be the intermediate state between open and closed conformation [14]. The position of γ determines which site has high, medium, or low affinity at any given point of time. Given that three MgATP molecules are synthesized or hydrolyzed during a full revolution of the γ subunit, each 120° rotation step is identical [15]. Viewing from the ATP hydrolysis direction, a 120° step can be further resolved into an 80° substep for MgATP binding and a 40° substep for release of products MgADP and Pi [6,16–18]. The pause before the 80° substep is named “ATP-waiting dwell”, and the time of this dwell is dependent on the MgATP concentration (< 0.1 ms under MgATP saturation). A lag time has been observed before the 40° substep as well; during this time, MgATP is hydrolyzed. This “catalytic dwell” lasts approximately 2 ms and it is independent of the MgATP concentration [19].
The energy necessary to drive rotation in hydrolysis direction is not provided by MgATP hydrolysis itself, but by changes in the binding affinities of substrate and products [1,4,20,21]. Mapping the profile of nucleotide binding affinities as a function of the rotational angle of the γ subunit is essential for complete understanding of the rotary catalytic mechanism. In this study, we use tryptophan fluorescence to measure nucleotide binding affinities after locking the enzyme by disulfide crosslinking [22] in the two most important conformations: in the ATP-waiting dwell and in the catalytic dwell. The outcome will give further insights into the molecular basis of the catalytic mechanism of ATP synthase.
2. Materials and Methods
2.1. Strains
E. coli strain DH5α (New England BioLabs) was used as the competent cell for site-directed mutagenesis. Bearing no intrinsic ATP synthase operon, E. coli strain DK8 (Δ(atpB-atpC), tetracycline resistant) [23] was used to express ATP synthase from the respective plasmid.
2.2. Construction of plasmids
Plasmid pSN6 (pBR322 derivative, atpB-atpC, βY331W, ampicillin resistant) [24] was used as the template throughout this study and is referred to as WT (“wild-type”) in the following text. Mutations were introduced by site-directed mutagenesis strategy (oligonucleotides from Integrated DNA Technology, listed in Supplementary Material Table S1; Q5® High Fidelity DNA Polymerase from New England BioLabs). A plasmid with desired mutation was verified by restriction pattern analysis and DNA sequencing, followed by transformation into strain DK8 for further assays.
2.3. Growth yield assay
A single colony of DK8 strain expressing WT or mutant ATP synthase was inoculated in lysogeny broth (LB) medium with 100 μg/ml ampicillin and grown to late exponential phase. A further 1:500 inoculation was made into medium containing 8 mM succinate or 4 mM glucose [25]. Cells grew aerobically at 30 °C until saturation. Growth yield was quantified from turbidity of the liquid culture by measuring its absorbance at 590 nm. Each mutant was assessed at least in triplicate.
2.4. Inverted membrane vesicle preparations
Cells grew aerobically at 30 °C and then were harvested at late exponential phase. Buffers used in experiments were prepared as previously described [26]. Resuspended cells were lysed mechanically by two passes through a homogenizer (EmulsiFlex-C3; Avestin, Ottawa, Ontario, Canada) at 15 kpsi [27]; after removal of cell debris by ultracentrifugation, membrane vesicles were pelleted from the supernatant and purified. For each strain expressing WT or mutant ATP synthase, at least two biological replicates were prepared.
2.5. F1 subcomplex protein purification
The water-soluble F1 subcomplex of WT and mutants was purified as previously described [28]. In brief, membrane vesicles were washed three times in the buffer of low ionic strength, causing the F1 subcomplex to dissociate from the membrane. F1 was precipitated using polyethylene glycol 6000 (PEG, 10% final concentration). Resuspended F1 was purified by anion-exchange chromatography on Whatman DE52 cellulose with a stepwise elution with Na2SO4. F1 was concentrated by ultrafiltration and further purified by size exclusion chromatography on Sephacryl S-300 HR. Fractions with high specific ATPase activity were pooled and concentrated. F1 was stored at – 80 °C.
2.6. Protein concentration assay
The Bradford method was used to determine protein concentrations, with bovine serum albumin as standard [29]. All samples were measured at least in duplicate.
2.7. Western blot
The relative expression amount of membrane-bound ATP synthase was quantified through Western blot [30] with anti-γ antibody (a kind gift from Drs. Toshiharu Suzuki and Masasuke Yoshida, Japan Science and Technology Agency, Tokyo). Band intensity was evaluated by ImageJ software (National Institutes of Health). Two biological replicates were assessed.
2.8. Disulfide bond crosslinking and reduction
The disulfide bond was formed between two cysteine residues by 1 mM CuSO4 oxidation for one hour at 25 °C; to reduce a disulfide bond, protein samples were treated with 1 mM (or 4 mM, in the recovery assay) dithiothreitol (DTT) for one hour at 25 °C [31]. The concentration of membrane protein as well as the F1 samples was adjusted to 1 mg/ml to ensure the efficient oxidation or reduction.
2.9. ATPase activity assay
ATPase activities were assessed at 30 °C in a medium containing 50 mM Tris/H2SO4, 4 mM MgSO4 and 10 mM ATP, pH 8.0. To trigger ATP hydrolysis, inverted membranes (10 μg/ml) or purified F1 proteins (5 μg/ml) were added into the medium. To terminate the reaction, sodium dodecyl sulfate (SDS) was added to a final concentration of 5% (w/v). Inorganic phosphate released from the reaction was quantified colorimetrically by measuring the absorbance at 700 nm [32]. Each protein sample was assayed at least in duplicate.
2.10. Proton pumping assay
Inverted membrane vesicles (100 μg) were vigorously suspended in 2.0 ml proton pumping medium containing 10 mM HEPES, 300 mM KCl, 5 mM MgCl2, 1 μg/ml valinomycin and 1 μM acridine orange, pH 7.5, at 25 °C. The acridine orange fluorescence intensity was measured at an emission wavelength of 530 nm by using an excitation wavelength of 460 nm. To initiate proton pumping, 1 mM ATP was added; 5 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to dissipate the proton gradient [33]. Each sample was assayed at least in duplicate.
2.11. Nucleotide binding assay
As F1 is purified and stored in buffer containing MgATP, it may have nucleotides bound to the catalytic sites [34]. To remove them, F1 samples were subjected to chromatography with Sephadex G-50 directly before the nucleotide binding assay [35]. Nucleotide binding to the catalytic sites was measured using the fluorescence of βY331W. This technique is described in detail in [13,28], addressing issues such as the precautions taken to keep ATP hydrolysis in MgATP binding experiments to a minimum (see also Supplementary Material) and justification of the assumption that all three catalytic sites contribute equally to the total fluorescence signal. Using a spectrofluorometer type FL3–22 (Jobin-Yvon/Horiba), the excitation wavelength was set to 295 nm, and the intensity at 350 nm from emission spectra was recorded [13]. The assay solution consisted of 50 mM Tris/H2SO4 (pH 8.0), 40 nM of F1 and 2.5 mM MgSO4. ATP or ADP was added from 30 nM to 5 mM. For titration with the transition state analog MgADP-AlF4, the assay solution contained in addition 1 mM AlCl3 and 10 mM NaF [36,37]. It should be noted that binding of MgADP-AlF4 is reversible, with a half-life of inhibition of approximately 100 h [37]. WT F1 (βY331W) was used as positive control. True wild-type F1 (βY331) was used as reference to correct for the inner filter effects at high concentration of nucleotides [28]. At least three assays were done for each data point. To calculate the nucleotide binding affinities, data collected were fit to the following four equations.
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
| (Eq. 4) |
In these equations: Q is the quenching of fluorescence intensity, and Qsat is the quenching of fluorescence intensity when all three catalytic binding sites are occupied. As an exception, as titrations of the αE284C/γQ274C mutant in the oxidized state did not reach the same degree of quenching as the reduced state, Qsat of the reduced state was also used for the oxidized state. F0 is the initial fluorescence intensity of proteins, and Fexp is the fluorescence intensity upon nucleotide titration. [L]0 is the total nucleotide concentration (with an excess of Mg2+, all ATP or ADP was assumed to be in form of a Mg-nucleotide complex), [L]Free is the free nucleotide concentration (the concentration of Mg-nucleotide not bound to the enzyme), which can be calculated based on the total ligand concentration [L]0, the total concentration of F1, [E]0 (40 nM in this study), and the number of occupied catalytic binding sites, n (Eq. 3). Kd1, Kd2 and Kd3 are the dissociation constants of the three catalytic sites.
2.12. Miscellaneous
SigmaPlot (Systat Software Inc.) was used for non-linear least-squares regression.
3. Results
3.1. Functional characterization of cysteine mutants
A previous study [22] had shown that an αE284C/γL276C disulfide bond halts rotation in the catalytic dwell, while an αE284C/γQ274C bond freezes the enzyme in the ATP-waiting dwell. Thus, we introduced the single mutations αE284C, γQ274C and γL276C and the double mutations αE284C/γQ274C and αE284C/γL276C into E. coli ATP synthase. The mutant enzymes were characterized initially by growth yield in medium containing limiting glucose or succinate as sole carbon source to study the effect of the mutations on ATP synthesis; the effect on ATP hydrolysis was determined using ATPase activity and ATP-driven proton pumping of membrane preparations (Table 1). For the in vitro assays (ATPase, proton pumping; Table 1, columns 5 and 8), the membranes were pretreated with 1 mM DTT for one hour at room temperature to reduce any disulfide bond possibly formed by spontaneous oxidation.
Table 1.
Functional characterization of mutants used in the present study
| Strain | Growth Yield | ATP Synthase Amount | ATPase Activity | ATP-driven H+ Pumping | |||
|---|---|---|---|---|---|---|---|
| 8 mM Succinate | 4 mM Glucose | ||||||
| % | % | % | unit/mg protein | % | |||
| Reduced | Oxidized | Ox/Red | |||||
| WT (βY331W) | 100 | 100 | 100 | 1.05 (0.04) | 1.01 (0.03) | 0.99 (0.03) | 100 |
| + αE284C | 68 (4) | 91 (3) | 137 (21) | 0.17 (0.02) | 0.14 (0.01) | 0.15 (0.02) | 45 (3) |
| + γQ274C | 104 (4) | 97 (2) | 122 (17) | 0.80 (0.05) | 0.74 (0.04) | 0.78 (0.04) | 97 (2) |
| + γL276C | 102 (3) | 99 (2) | 117 (15) | 0.79 (0.03) | 0.72 (0.04) | 0.75 (0.05) | 102 (3) |
| + αE284C/γQ274C | 62 (3) | 85 (4) | 113 (12) | 0.13 (0.02) | 0.05 (0.01) | 0.12 (0.02) | 26 (4) |
| + αE284C/γL276C | 72 (4) | 88 (3) | 115 (17) | 0.12 (0.02) | 0.04 (0.01) | 0.12 (0.02) | 42 (4) |
| pUC18 | < 1 | 29 (1) | 0 | < 0.01 | < 0.01 | < 0.01 | < 1 |
All strains generated in this study are based on pSN6/DK8 background (labeled “WT”) and carried the βY331W mutation. Growth yield in limiting succinate was assessed from the turbidity of cell culture, measured as absorbance at 590 nm. Inverted membrane vesicles isolated from WT and mutant strains were used for the ATPase activity and ATP driven H+ pumping measurements. The amount of membrane-bound ATP synthase was determined by Western blot using anti-γ antibody. Before enzymatic assays, membrane protein samples were treated with 1 mM DTT (the Reduced group), 1 mM CuSO4 (the Oxidized group) at room temperature for 1 h in order to reduce or form a disulfide bond. Samples in the Ox/Red group were treated with 1 mM CuSO4 for 1 h and then 4 mM DTT for 1 h at room temperature. This group was served to clarify whether the formation of disulfide bond is reversible. ATPase activities were quantified by the amount of inorganic phosphate released at 30 °C; 1 unit is defined as 1 μmol inorganic phosphate released per minute in the reaction. ATP-driven proton pumping ability was assayed as described in Materials and Methods. All values were normalized against WT, except for the membrane ATPase activity. Standard deviations are shown in parentheses. Strain pUC18/DK8 served as negative control, expressing no ATP synthase.
Mutation γQ274C or γL276C did not significantly affect the functions of ATP synthase. Mutation αE284C resulted in moderate growth yield (62 – 72% of WT), impaired proton pumping ability (26 – 45% of WT) and low ATPase activity of membrane preparations (< 20% of WT). The behavior of the double mutants essentially reflected the effects observed with the single mutants (Table 1). The impairments of the αE284C mutant were not caused by reduced expression or reduced oligomeric stability (see the Western blot results in column 4 of Table 1). The amino acid residues at position γ274 are not conserved, with charged (K and R), polar (T, N and Q) or small (G) residues (Supplementary Material Table S2); a cysteine in this position does not appear to cause a problem. Position γ276 shows large aliphatic hydrophobic residues (I, L) in most species; replacement by a cysteine appears to be well tolerated. The glutamate residue at position α284 is highly conserved, which might explain the impairment of enzyme function in the αE284C mutant. Unexpectedly, after isolation of the F1-ATPase subcomplex, the double mutants containing the αE284C mutation had WT-like ATPase activities (see 3.2. and Table 2).
Table 2.
ATPase activities of purified F1 samples
| F1 Samples | No Treatment | 1 mM DTT | 1 mM CuSO4 |
|---|---|---|---|
| unit/mg protein | |||
| WT (βY331W) | 6.0 (0.4) | 5.9 (0.2) | 5.7 (0.4) |
| + αE284C/γQ274C | 5.1 (0.3) | 5.0 (0.3) | 0.2 (< 0.1) |
| + αE284C/γL276C | 4.0 (0.2) | 3.9 (0.2) | 0.1 (< 0.1) |
F1 samples were treated for 1 h with either 1 mM DTT or 1 mM CuSO4 to reduce or oxidize the disulfide bond between α and γ subunits. All treatments were done at 25 °C. Experimental procedure of ATPase activity assay was as described in the legend of Table 1. Standard deviations are shown in parentheses.
3.2. The disulfide bond formed between subunits α and γ abolishes enzymatic activity by stopping rotation
F1-ATPase was purified from WT, αE284C/γQ274C, and αE284C/γL276C enzymes. The ATPase activity of these samples was assayed directly after preparation without any further treatment, after reduction of disulfide bonds with 1 mM DTT, or after oxidation of disulfide bonds with 1 mM CuSO4. With no treatment or after reduction by DTT, all three enzymes showed ATPase activities of 4 – 6 units/mg protein. Oxidized by 1 mM CuSO4, the ATPase activity of WT F1 remained high (5.7 unit/mg protein); however, with the α-γ disulfide crosslink formed, ATPase activities of mutant F1 with double cysteine substitutions almost dropped to zero (Table 2). With a disulfide crosslink formed upon oxidation, in the cysteine double mutants, αE284C/γQ274C and αE284C/γL276C, ATP hydrolysis was abolished since the γ subunit could no longer rotate; the residual ATPase activities might be due to a marginal amount of enzyme with no α-γ disulfide bond or due ATP hydrolysis by the uni-site mechanism [38]. Furthermore, the data show that even in absence of treatment with DTT, the disulfide bonds between α and γ are in the reduced state.
The experimental data obtained with the WT enzyme confirmed that the treatment conditions for oxidation or reduction used in this study did not affect the hydrolytic capability. Still, the remote possibility remained that one of the introduced cysteine residues by itself might cause inhibition of the enzyme upon oxidation. We tested therefore the impact of oxidation on membrane ATP synthase samples with single cysteine mutations (Table 1, column 6), and they did not show any significant impairment compared to the reduced sample (Table 1, column 5). As observed for isolated F1, the double mutants were impaired due to formation of an α-γ disulfide crosslink. Full ATPase activity could be recovered by removal of the crosslink by re-reduction using an excess of DTT (Table 1, column 7). It should be noted that reductive removal of the crosslink also resulted in resuming rotation [22], a clear indication of enzymatic activity. These findings demonstrate that oxidation of the cysteines is reversible and does not cause any damage to the enzyme.
3.3. Nucleotide binding affinities of the WT and cysteine double mutant F1
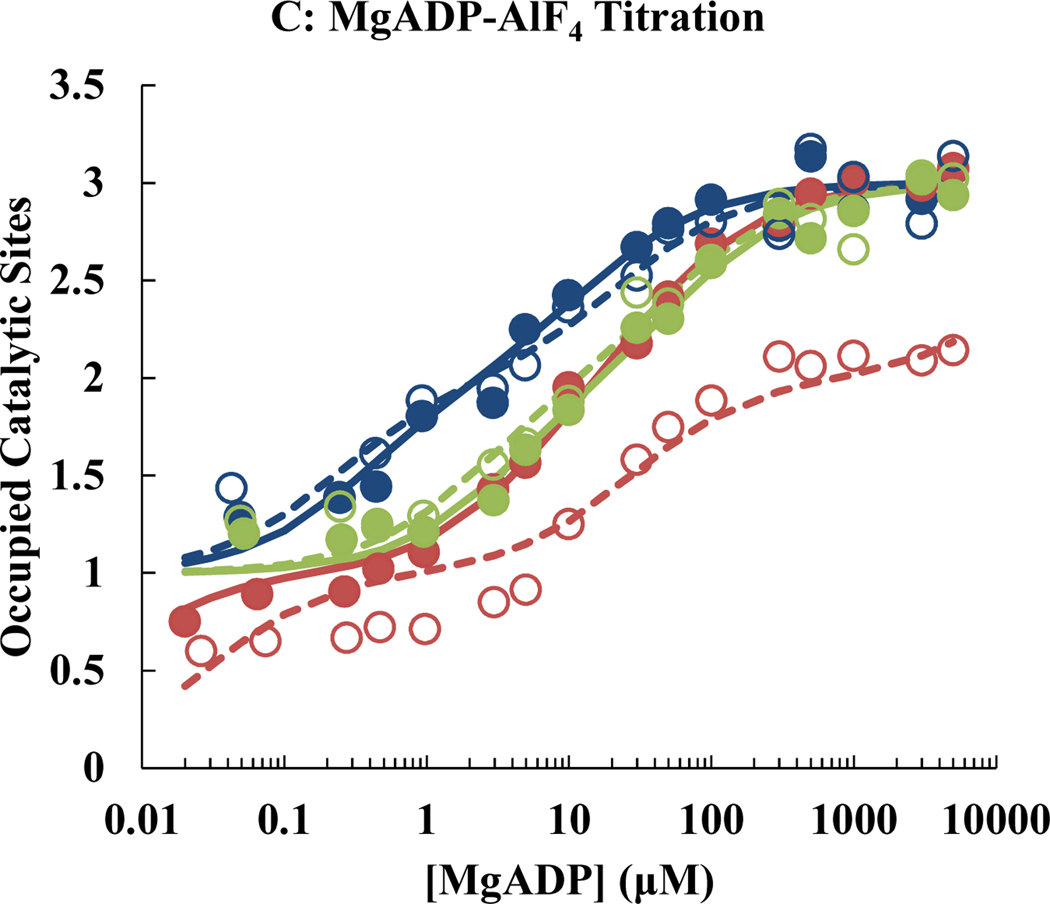
Following reduction or oxidation treatment, WT and mutant F1 samples were titrated with MgATP, MgADP or MgADP-AlF4 (a transition state analog). Quenching of fluorescence intensities of the tryptophan probe (βY331W) reflects the occupancy of the catalytic binding site [13]. The results are summarized in Fig. 1 and Table 3. For WT F1, neither reduction nor oxidation treatment significantly affected the binding parameters of the catalytic sites. For comparison, previous studies [39,40] with untreated enzyme gave values for Kd1, Kd2, and Kd3, of 0.02 μM, 1.5 μM, and 29 μM for MgATP; 0.04 μM, 6.0 μM, and 42 μM for MgADP; and < 1 nM, 0.06 μM, and 40 μM, for MgADP-AlF4.
Fig. 1.
Titration of WT and mutant F1 with nucleotides. Data for WT are shown in blue, those for the αE284C/γQ274C mutant in red, and those for the αE284C/γL276C mutant in green. Filled circles represent the experimental data of F1 in absence of α-γ crosslink; open circles represent the experimental data of F1 in presence of α-γ crosslink. Each data point represents the average of at least three independent assays. Solid lines are the least-squares best fit curves of the filled circle sets, whereas dashed lines are the least-squares best fit curves of the open circle sets. The curves were fitted as described in Materials and Methods, using a linear x-axis. Conversion to a log scale is only for illustration purposes, to allow presentation of binding events with vastly different affinities in a single figure.
Table 3.
Nucleotide dissociation constants of WT and mutant F1
| WT (βY331W) | + αE284C/γQ274C | + αE284C/γL276C | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MgATP (μM) | Kd1 | Kd2 | Kd3 | Kd1 | Kd2 | Kd3 | Kd1 | Kd2 | Kd3 |
| Reduction | < 0.01 | 6.0 (2.5) | 34 (17) | 0.66 (0.17) | 15 (5.5) | 93 (44) | < 0.01 | 10 (3.5) | 81 (35) |
| R2 = 0.9614 | R2 = 0.9755 | R2 = 0.9554 | |||||||
| Oxidation | < 0.01 | 7.6 (2.9) | 58 (28) | 3.0 (0.68) | 432 (105) | > 5000# | < 0.01 | 8.0 (2.2) | 140 (56) |
| R2 = 0.9628 | R2 = 0.9207 | R2 = 0.9580 | |||||||
| MgADP (μM) | Kd1 | Kd2 | Kd3 | Kd1 | Kd2 | Kd3 | Kd1 | Kd2 | Kd3 |
| Reduction | 0.022 (0.0049) | 11 (12) | 16 (19) | 0.20 (0.041) | 19 (5.7) | 120 (47) | 0.025 (0.0035) | 7.3 (1.1) | 88 (18) |
| R2 = 0.9843 | R2 = 0.9859 | R2 = 0.9891 | |||||||
| Oxidation | 0.024 (0.0050) | 12 (7.5) | 24 (16) | 1.3 (0.34) | 160 (42) | > 5000# | 0.026 (0.0057) | 3.6 (0.84) | 76 (25) |
| R2 = 0.9854 | R2 = 0.9408 | R2 = 0.9713 | |||||||
| MgADP-AlF4 (μM) | Kd1 | Kd2 | Kd3 | Kd1 | Kd2 | Kd3 | Kd1 | Kd2 | Kd3 |
| Reduction | < 0.01 | 0.39 (0.13) | 12 (4.9) | < 0.01 | 5.7 (1.2) | 42 (11) | < 0.01 | 3.9 (0.98) | 89 (30) |
| R2 = 0.9289 | R2 = 0.9874 | R2 = 0.9665 | |||||||
| Oxidation | < 0.01 | 0.22 (0.072) | 27 (11) | 0.035 (0.010) | 36 (9.9) | > 5000# | < 0.01 | 2.6 (0.82) | 83 (36) |
| R2 = 0.9278 | R2 = 0.9207 | R2 = 0.9375 | |||||||
Nucleotide binding to the catalytic sites was measured at 25 °C by monitoring the fluorescence signal of residue βY331W. Nucleotide dissociation constants (Kd) were calculated by fitting theoretical curves to the experimental data by using a three-site binding model. The coefficient of determination (R2) for each regression is listed. The method applied was unable to resolve Kd values lower than 0.01 μM with confidence. In the underlying titrations, each data point was measured at least three times. The least-square fits to determine binding constants and statistical parameters were based on these individual data points (not on the average of each condition shown in Fig. 1). Standard errors for the Kd values are shown in parentheses.
This number was too high to be determined through current experimental method (Kd3 > 5 mM).
In the reduced state, the αE284C/γL276C double mutant F1 exhibited a nucleotide binding pattern similar to WT. The affinities of site 2 for the transition state analog MgADP-AlF4 and of site 3 for all tested nucleotides appeared to be slightly reduced by the mutations. More importantly, after oxidation to crosslink the rotor and the stator in the catalytic dwell state, the nucleotide binding parameters remained unchanged.
The αE284C/γQ274C double mutant had an overall reduced affinity for nucleotides. This seems to be mostly due to the γQ274C mutation and not due to αE284C, as it was not observed for the αE284C/γL276C mutant to a similar extent. Still, in the reduced state αE284C/γQ274C F1 showed the typical nucleotide binding pattern with three sites of different affinities. The αE284C/γQ274C mutant was the only enzyme investigated here that exhibited pronounced difference in nucleotide binding behavior between reduced and oxidized state. In the oxidized state, the affinities of the catalytic sites were so far reduced that at the highest MgATP concentration used (5 mM), the quench of βY331W fluorescence was only about 2/3 of the value observed in the reduced state. Thus, locking of γ in the ATP-binding dwell results in the “loss” of a catalytic site, or, more precisely, Kd3 becomes > 5 mM for all nucleotides tested. Furthermore, the affinities of sites 1 and 2 were reduced by factors of 5 – 29 in the oxidized versus the reduced state.
4. Discussion
4.1. Nucleotide binding pattern as a function of the rotational angle of γ
A complete description of the catalytic mechanism of ATP synthase will have to include an analysis of the affinity changes for substrates and products at each of the three catalytic binding sites during rotation; the affinities of the catalytic sites are determined by rotational angle of the central γ subunit. In the present study, we determined the nucleotide binding affinities of the catalytic sites in the two most important conformations in the catalytic cycle, the ATP-binding dwell and the catalytic dwell. For this purpose, we made use of two cysteine double mutants, αE284C/γL276C and αE284C/γQ274C, which upon oxidation to form a disulfide bond lock the enzyme in the catalytic dwell and the ATP-binding dwell, respectively [22], and we measured the nucleotide binding affinities using the fluorescence of a tryptophan introduced at position β331 as signal [13,28]. The most important result of this study was the finding that locking the enzyme in the ATP-binding dwell gave an overall reduction in nucleotide binding affinities; the affinity of site 3 (the lowest-affinity site) was reduced so far that it could not be detected anymore with the technique used (Kd3 > 5 mM).
It is highly unlikely that formation of the αE284C/γQ274C disulfide bond per se would lead to complete fluorescence quenching of βY331W at the third catalytic site. The three tryptophan residues in position β331 have a relatively high quantum yield (0.22) [28]. Complete quenching of the fluorescence requires a drastic change in their environment; formation of a π-π stacking complex between the tryptophan indole group and the adenine base of bound nucleotide provides such a change, which is the basis of the technique used here to study nucleotide binding to the catalytic sites [13,28]. The crystal structure of the E. coli enzyme [41] and those of the mitochondrial enzyme [12,14] show that, except for the absence or presence of nucleotide, all three βY331 residues are in a very similar environment. Furthermore, except for the nucleotide, there is no other possible known quenching group nearby, such as the amide groups of glutamine and asparagine, the carboxyl groups of glutamic and aspartic acids, a lysine ε-amino group, a tyrosine phenol, a cysteine sulfhydryl, a histidine imidazole or a disulfide bond [42–44]. Thus, all fluorescence responses of βY331W (or the lack thereof) have to be attributed to solely to the presence (or absence) of bound nucleotide, and not to any other independent quenching phenomenon.
4.2. Integration into mechanistic model
The new findings can be integrated into the previously-proposed mechanism for ATP hydrolysis by ATP synthase [15] as follows (Fig. 2). As in the previous version, the revised scheme takes into account (a) that in time-average, all three catalytic sites of E. coli ATP synthase have to be occupied by nucleotide to achieve significant catalysis rates [13,28,45]; (b) that under substrate saturation the MgATP to MgADP distribution is 1:2 [46]; (c) that the high-affinity site is the catalytically active site [47]; (d) that under substrate saturation virtually all enzyme molecules are in the catalytic dwell [19,48]; and (e) that the βTP site is the high-affinity site [15]. While the last point is not universally accepted (for a different point of view, see e.g. [8]), the results of the present study strengthen the argument for the assignment of the βTP site as the high-affinity site. Fluorescence resonance energy transfer experiments showed that residue γ262 in the C-terminal helix of γ was much closer to the high-affinity site than residue γ5 in the N-terminal helix [15]. In the crystal structures [12,14], residue γ262 is much closer to the βTP site than γ5. The present study confirms that the distance measurements were actually performed in an F1 conformation similar to that seen in the crystal structures (see part 4.4.).
Fig. 2.
Catalytic site affinity changes in the rotary mechanism of ATP synthase. (A) Revised catalytic mechanism [15] incorporating the results of the present study. For details, see part 4.2. of the Discussion. It should be noted that the red arrow symbolizes the changes in the rotational angle of γ; it is not associated with a specific structural feature of γ. The hypothetical intermediate conformation βIM during closing of β might or might not be the same as the half-closed conformation βHC seen in crystal structures [14,41]. (B) Catalytic site affinity as a function of the rotational angle of γ. For each specified angle, the approximate range of experimentally-determined affinities is given (for details, see part 4.2. of the Discussion). The letters indicate the symbols for the different affinities of the catalytic sites used in Fig. 2A (H, high; M, medium; L, low; O, open site; asterisk, ATP-waiting dwell; no asterisk, catalytic dwell). The red horizontal arrow and the red L** indicate that γ probably has to undergo thermal rotational fluctuations in the positive direction before MgATP can bind effectively (see part 4.4. of the Discussion; see also state B in Fig. 2A).
The revised rotary mechanism presented in Fig. 2A additionally takes into consideration the findings of the present study, (i) the observation that, unless the enzyme is locked in the ATP-binding dwell, the nucleotide binding affinities measured by the tryptophan fluorescence assay represent the enzyme in the catalytic dwell, and (ii) the newly-determined affinities for the ATP-binding dwell. Fig. 2B shows the changes in nucleotide binding affinity during the progress of catalysis. The Kd values for the ATP-binding dwell (the sites are labeled with an asterisk) are corrected for the affinity decrease due to the γQ274C mutation itself, even without oxidation.
After release of the reaction products from the previous catalytic cycle, the enzyme is in the ATP-waiting dwell (defined as 0°), with an empty low-affinity site (state A in Fig. 2B). The respective β subunit is in the open conformation with a Kd for MgATP of > 5 mM. At saturating MgATP concentrations, MgATP is immediately bound to this site (state B), which provides the energy to close the conformation of the β subunit, which in turn drives a rotation of γ by 80°, converting the low-affinity site into the high-affinity site (for further discussion, see Chapter 4.4. below). The enzyme is now in the 80° catalytic dwell and MgATP is bound very tightly (Kd(MgATP) < 10 nM by tryptophan fluorescence ([13,28] and this study); uni-site experiments gave a number of 0.2 nM [49]) (state C). MgATP is hydrolyzed at the end of the catalytic dwell [19] (state C→D); MgATP hydrolysis might actually trigger the subsequent 40° rotation step [50]. At 120° the enzyme is again in the ATP-waiting dwell (state E). Compared to the 80° catalytic dwell, the affinity is reduced by five- to seven-fold (Kd1(ox)/Kd1(red) for αE284C/γQ274C), which would give Kd values between 1 and 50 nM for the WT enzyme. After the next 80° rotation step, at 200°, γ is back in the catalytic dwell with a Kd in the range of 1 – 10 μM ([13,28] and this study). Continuing rotation reduces the affinity further (Fig. 2B), finally reaching a Kd of > 5 mM at 360° and allowing release of the reaction product MgADP. The techniques developed in our laboratory do not give us any insight into the timing of Pi release. There is considerable evidence that Pi is released during the 40° substep (state D→E) [51,52], as suggested in the scheme in Fig. 2A. However, the sequence of MgADP and Pi release is strictly hypothetical, as arguments for either scenario have been presented [51–53]. For an overview of residues involved in phosphate binding, see [54].
4.3. Comparison with literature data
The present study is not the first attempt to quantify the γ-angle dependence of the affinity of the catalytic sites. Two previous studies [55,56] measured on- and off-rates in single molecule experiments with the enzyme of Geobacillus stearothermophilus (formerly known as Bacillus sp. PS3) to calculate affinities; both agree with our results that the affinity of a nucleotide, newly-bound in the ATP-waiting dwell, will increase with the rotation angle of γ. Watanabe et al. [55] measured the affinity for MgATP at angles between – 30° and + 30° around the ATP-waiting dwell and found a continuous increase in affinity, with a Kd of around 200 nM at – 30° (equivalent to 330°) to 10 nM at 0° to 1 nM at 30°. The numbers raise the question of which binding site is monitored in these experiments; the highest ATP concentration used was 200 nM. MgATP hydrolysis at physiological rates requires that all three catalytic sites are participating. The Km for MgATP hydrolysis in this enzyme is at least 7 μM [57]; at 200 nM ATP, less than 3% of all enzyme molecules will work in the physiological mode. Adachi et al. [56] used Cy3-ATP and Cy3-ADP as ligands in a rather ambitious project trying to cover all angles between 0° and 360°; in the end, there were some gaps. Similar to our results, the affinity for Cy3-ATP increased from 0° to 80° (Kd 1 μM and 30 nM, respectively) and then dropped off. In contrast to our observation that the affinity is lowest at 0° (360°), the Cy3 nucleotides reach minimum affinity at 320° (Kd 30 – 40 μM). Another difference is the range of affinity changes. For Cy3 nucleotides it is 3 – 4 orders of magnitude; our study shows nearly twice as much. It is not quite clear if the Cy3-nucleotides are true representatives of the natural ligands/substrates; the fluorescent “Cy3” attachment is actually larger than the nucleotide. The authors state that the Cy3-attachment reduces on- and off-binding rates [56], but they do not give a Km for bulk Cy3-ATP hydrolysis.
4.4. Open questions
The results of the present study leave some open questions. Most importantly, how can the open binding site with a Kd of > 5 mM efficiently bind the substrate MgATP when the Km for MgATP hydrolysis is around 30 μM [13]. Here, thermal fluctuations appear to play a significant role [22,58,59]. Sielaff et al. [22] show that the fluctuations are actually larger in the ATP-waiting dwell and in the catalytic dwell during rotation than in the enzyme stalled by disulfide crosslinking. At some point in time, the γ subunit will be at a forward angle sufficiently large to increase the affinity of the open binding site so far that it can bind the substrate effectively (indicated by the red L** in Fig. 2A state B and in Fig. 2B).
Another open problem is the role of the ATP-waiting dwell. Sielaff et al. [22] demonstrated that nucleotide (MgADP) saturation gives an enzyme with γ in the position of the catalytic dwell. The results of the present study show that in absence of nucleotides on the catalytic sites γ must also be in the catalytic dwell position, as locking the enzyme in this position by disulfide crosslinking of the αE284C/γL276C mutant had no effect on MgADP binding to the first (high affinity) site. In contrast, locking the enzyme in the ATP-waiting dwell with the αE284C/γQ274C mutant resulted in a significantly reduced affinity. This finding agrees with the observation that in the crystal structure of the yeast enzyme in absence of nucleotide [60] the portion of γ within the α3β3 cylinder is in essentially the same position as in the structures containing nucleotides [8,12]. The position of γ in the crystal structure bears more resemblance to the catalytic dwell than to the ATP-waiting dwell [22,61]. Thus, it seems that the ATP-waiting dwell just exists as an intermediate in the catalytic cycle. This agrees with the observation that the catalytic dwell position is thermodynamically favored [62].
4.5. Catalytic properties of the αE284C mutation
Finally, all mutants containing the αE284C exhibited a peculiar behavior that deserves a comment although it has little to do with the main topic of the present study. The ATPase activity of purified F1 from the αE284C/γQ274C or αE284C/γL276C double mutants in the reduced state was comparable to the WT enzyme (Table 2); however, the ATPase activity of membrane-bound αE284C/γQ274C or αE284C/γL276C ATP synthase in inverted membrane vesicles reached less than 20% of the WT activity (Table 1). As Table 1 shows, the αE284C mutation is the reason for this discrepancy, as the γQ274C and γL276C mutations by themselves did not significantly impair ATP synthase activity. We could exclude assembly problems or oligomeric instability as cause of the impairment (Table 1). Furthermore, the αE284C mutation did not cause uncoupling between ATP hydrolysis and proton translocation; uncoupling would have resulted in high membrane ATPase activity, but reduced ATP-driven proton pumping [25,63]. A number of deletion mutants in the DELSEED-loop have been described which combine high activity of the isolated F1 with impaired ATP-driven proton pumping and decreased ATP synthesis rates as holoenzyme in membrane preparations [64–66]. The reason for this impairment seems to be that shortening of the DELSEED-loop reduces the rotational torque by half [64]. However, these mutants show high membrane ATPase activity. A possible explanation for the behavior of the αE284C mutant could be a further reduction in torque, allowing the enzyme to rotate an isolated γ subunit at full speed, but not an γεc10 rotor subcomplex integrated in the membrane. It should be noted that there is no simple structural explanation for such a far-reaching consequence of a point mutation.
Supplementary Material
Highlights.
The affinity of the catalytic sites of ATP synthase depends on the rotational angle of γ.
Disulfide bonds lock the enzyme into the two most important conformations.
Nucleotide binding affinities were measured at the catalytic and ATP-waiting dwell.
Locking γ into the ATP-binding dwell results in the “loss” of a catalytic site.
Our observations support the tri-site catalytic mechanism of ATP synthase in E. coli.
Acknowledgments
The authors thank Drs. Mary E. Anderson, Richard D. Sheardy and Hendrik Sielaff for the helpful discussions and their input into this project.
Funding
This work was supported in part by the Robert A. Welch Foundation (m-0200) to the Department of Chemistry and Biochemistry, Texas Woman’s University and supported in part by the Texas Woman’s University Teaching and Research Grant for Equipment and Technology to MEA and YL. This work was also supported in part by National Institutes of Health grant GM071462 (including ARRA Administrative Supplement) to JW.
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Weber J, Structural biology: Toward the ATP synthase mechanism, Nat. Chem. Biol 6 (2010) 794–795. 10.1038/nchembio.458. [DOI] [PubMed] [Google Scholar]
- [2].Guo H, Suzuki T, Rubinstein JL, Structure of a bacterial ATP synthase, Elife. 8 (2019) e43128. 10.7554/eLife.43128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Junge W, Sielaff H, Engelbrecht S, Torque generation and elastic power transmission in the rotary FoF1-ATPase, Nature. 459 (2009) 364–370. 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- [4].Nakamoto RK, Baylis Scanlon JA, Al-Shawi MK, The rotary mechanism of the ATP synthase, Arch. Biochem. Biophys 476 (2008) 43–50. 10.1016/j.abb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yoshida M, Muneyuki E, Hisabori T, ATP synthase - A marvellous rotary engine of the cell, Nat. Rev. Mol. Cell Biol 2 (2001) 669–677. 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- [6].Senior AE, Nadanaciva S, Weber J, The molecular mechanism of ATP synthesis by F1Fo-ATP synthase, Biochim. Biophys. Acta - Bioenerg 1553 (2002) 188–211. 10.1016/S0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- [7].Weber J, Senior AE, ATP synthesis driven by proton transport in F1Fo-ATP synthase, FEBS Lett. 545 (2003) 61–70. https://www.ncbi.nlm.nih.gov/pubmed/12788493. [DOI] [PubMed] [Google Scholar]
- [8].Walker JE, The ATP synthase: The understood, the uncertain and the unknown, Biochem. Soc. Trans 41 (2013) 1–16. 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- [9].Kühlbrandt W, Structure and mechanisms of F-Type ATP synthases, Annu. Rev. Biochem 88 (2019) 515–549. 10.1146/annurev-biochem-013118-110903. [DOI] [PubMed] [Google Scholar]
- [10].Sielaff H, Yanagisawa S, Frasch WD, Junge W, Börsch M, Structural asymmetry and kinetic limping of single rotary F-ATP synthases, Molecules. 24 (2019) 504–528. 10.3390/molecules24030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Boyer PD, A perspective of the binding change mechanism for ATP synthesis, FASEB J. 3 (1989) 2164–2178. 10.1096/fasebj.3.10.2526771. [DOI] [PubMed] [Google Scholar]
- [12].Abrahams JP, Leslie AGW, Lutter R, Walker JE, Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria, Nature. 370 (1994) 621–628. 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- [13].Weber J, Wilke-Mounts S, Lee RSF, Grell E, Senior AE, Specific placement of tryptophan in the catalytic sites of Escherichia coli F1-ATPase provides a direct probe of nucleotide binding: Maximal ATP hydrolysis occurs with three sites occupied, J. Biol. Chem 268 (1993) 20126–20133. [PubMed] [Google Scholar]
- [14].Menz RI, Walker JE, Leslie AGW, Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: Implications for the mechanism of rotary catalysis, Cell. 106 (2001) 331–341. 10.1016/S0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- [15].Mao HZ, Weber J, Identification of the βTP site in the x-ray structure of F1-ATPase as the high-affinity catalytic site, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 18478–18483. 10.1073/pnas.0709322104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ueno H, Suzuki T, Kinosita K, Yoshida M, ATP-driven stepwise rotation of FoF1- ATP synthase, Proc. Natl. Acad. Sci. U. S. A 102 (2005) 1333–1338, 10.1073/pnas.0407857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nakanishi-Matsui M, Sekiya M, Nakamoto RK, Futai M, The mechanism of rotating proton pumping ATPases, Biochim. Biophys. Acta Bioenerg 1797 (2010) 1343–1352, 10.1016/j.bbabio.2010.02.014. [DOI] [PubMed] [Google Scholar]
- [18].Okuno D, Iino R, Noji H, Rotation and structure of FoF1-ATP synthase, J. Biochem 149 (2011) 655–664, 10.1093/jb/mvr049. [DOI] [PubMed] [Google Scholar]
- [19].Shimabukuro K, Yasuda R, Muneyuki E, Hara KY, Kinosita K, Yoshida M, Catalysis and rotation of F1 motor: Cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation, Proc. Natl. Acad. Sci. U. S. A 100 (2003) 14731–14736. 10.1073/pnas.2434983100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Oster G, Wang H, Reverse engineering a protein: The mechanochemistry of ATP synthase, Biochim. Biophys. Acta - Bioenerg 1458 (2000) 482–510. 10.1016/S0005-2728(00)00096-7. [DOI] [PubMed] [Google Scholar]
- [21].Gao YQ, Yang W, Karplus M, A structure-based model for the synthesis and hydrolysis of ATP by F1-ATPase, Cell. 123 (2005) 195–205. 10.1016/j.cell.2005.10.001. [DOI] [PubMed] [Google Scholar]
- [22].Sielaff H, Rennekamp H, Engelbrecht S, Junge W, Functional halt positions of rotary FoF1-ATPase correlated with crystal structures, Biophys. J 95 (2008) 4979–4987. 10.1529/biophysj.108.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Klionsky DJ, Brusilow WS, Simoni RD, In vivo evidence for the role of the epsilon subunit as an inhibitor of the proton-translocating ATPase of Escherichia coli, J Bacteriol. 160 (1984) 1055–1060. https://www.ncbi.nlm.nih.gov/pubmed/6238948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ahmad Z, Senior AE, Role of βAsn-243 in the phosphate-binding subdomain of catalytic sites of Escherichia coli F1-ATPase, J. Biol. Chem 279 (2004) 46507–46064. 10.1074/jbc.M407608200. [DOI] [PubMed] [Google Scholar]
- [25].Li Y, Ma X, Weber J, Interaction between γC87 and γR242 residues participates in energy coupling between catalysis and proton translocation in Escherichia coli ATP synthase, Biochim. Biophys. Acta - Bioenerg 1860 (2019) 679–687. 10.1016/j.bbabio.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Senior AE, Fayle DR, Downie JA, Gibson F, Cox GB, Properties of membranes from mutant strains of Escherichia coli in which the β-subunit of the adenosine triphosphatase is abnormal., Biochem. J 180 (1979) 111–118. 10.1042/bj1800111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Steed PR, Zou P, Trone KE, McHaourab HS, Structure and pH-induced structural rearrangements of the putative multidrug efflux pump EmrD in liposomes probed by site-directed spin labeling, Biochemistry. 52 (2013) 7964–7974. 10.1021/bi4012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Weber J, Senior AE, Fluorescent probes applied to catalytic cooperativity in ATP synthase, Methods Enzymol. 390 (2004) 132–152. 10.1016/S0076-6879(04)80006-5. [DOI] [PubMed] [Google Scholar]
- [29].Bradford MM, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem 72 (1976) 248–254. https://www.ncbi.nlm.nih.gov/pubmed/942051. [DOI] [PubMed] [Google Scholar]
- [30].Mnatsakanyan N, Li Y, Weber J, Identification of two segments of the subunit of ATP synthase responsible for the different affinities of the catalytic nucleotide-binding sites, J. Biol. Chem 294 (2019) 1152–1160. 10.1074/jbc.RA118.002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gumbiowski K, Cherepanov D, Müller M, Pänke O, Promto P, Winkler S, Junge W, Engelbrecht S, F-ATPase: Forced full rotation of the rotor despite covalent cross-link with the stator, J. Biol. Chem 276 (2001) 42287–42292. 10.1074/jbc.M106884200. [DOI] [PubMed] [Google Scholar]
- [32].Taussky HH, Shorr E, A simplified method for estimating urinary inorganic phosphate during aluminum gel therapy for phosphatic calculi, J. Urol 69 (1953) 454–455. https://www.ncbi.nlm.nih.gov/pubmed/13035938. [DOI] [PubMed] [Google Scholar]
- [33].Ketchum CJ, Nakamoto RK, A mutation in the Escherichia coli FoF1-ATP synthase rotor, γE208K, perturbs conformational coupling between transport and catalysis, J. Biol. Chem 273 (1998) 22292–22297. https://www.ncbi.nlm.nih.gov/pubmed/9712846. [DOI] [PubMed] [Google Scholar]
- [34].Senior AE, Lee RSF, Al-Shawi MK, Weber J, Catalytic properties of Escherichia coli F1-ATPase depleted of endogenous nucleotides, Arch. Biochem. Biophys 297 (1992) 340–344. 10.1016/0003-9861(92)90682-M. [DOI] [PubMed] [Google Scholar]
- [35].Weber J, Wilke-Mounts S, Senior AE, Cooperativity and stoichiometry of substrate binding to the catalytic sites of Escherichia coli F1-ATPase: Effects of magnesium, inhibitors, and mutation, J. Biol. Chem 269 (1994) 20462–20467. [PubMed] [Google Scholar]
- [36].Nadanaciva S, Weber J, Senior AE, Binding of the transition state analog MgADP-fluoroaluminate to F1-ATPase, J. Biol. Chem 274 (1999) 7052–7058. 10.1074/jbc.274.11.7052. [DOI] [PubMed] [Google Scholar]
- [37].Nadanaciva S, Weber J, Senior AE, New probes of the F1-ATPase catalytic transition state reveal that two of the three catalytic sites can assume a transition state conformation simultaneously, Biochemistry. 39 (2000) 9583–9590. 10.1021/bi000941o. [DOI] [PubMed] [Google Scholar]
- [38].Duncan TM, Senior AE, The defective proton-ATPase of uncD mutants of Escherichia coli. Two mutations which affect the catalytic mechanism, J. Biol. Chem 260 (1985) 4901–4907. [PubMed] [Google Scholar]
- [39].Senior AE, Weber J, Nadanaciva S, The catalytic transition state in ATP synthase, J. Bioenerg. Biomembr 32 (2000) 523–529. 10.1023/A:1005625326721. [DOI] [PubMed] [Google Scholar]
- [40].Mao HZ, Gray WD, Weber J, Does F1-ATPase have a catalytic site that preferentially binds MgADP?, FEBS Lett. 580 (2006) 4131–4135. 10.1016/j.febslet.2006.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cingolani G, Duncan TM, Structure of the ATP synthase catalytic complex F1 from Escherichia coli in an autoinhibited conformation, Nat. Struct. Mol. Biol 18 (2011) 701–707. 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen Y, Barkley MD, Toward understanding tryptophan fluorescence in proteins, Biochemistry. 37 (1998) 9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- [43].Hennecke J, Sillen A, Huber-Wunderlich M, Engelborghs Y, Glockshuber R, Quenching of tryptophan fluorescence by the active-site disulfide bridge in the DsbA protein from Escherichia coli, Biochemistry. 36 (1997) 6391–6400. doi: 10.1021/bi963017w. [DOI] [PubMed] [Google Scholar]
- [44].Callis PR, Liu T, Quantitative prediction of fluorescence quantum yields for tryptophan in proteins, J. Phys. Chem. B 108 (2004) 4248–4259. doi: 10.1021/jp0310551. [DOI] [Google Scholar]
- [45].Weber J, Senior AE, Bi-site catalysis in F1-ATPase: Does it exist?, J. Biol. Chem 276 (2001) 35422–35428. 10.1074/jbc.M104946200. [DOI] [PubMed] [Google Scholar]
- [46].Weber J, Bowman C, Senior AE, Specific tryptophan substitution in catalytic sites of Escherichia coli F1-ATPase allows differentiation between bound substrate ATP and product ADP in steady-state catalysis, J. Biol. Chem 271 (1996) 18711–18718. 10.1074/jbc.271.31.18711. [DOI] [PubMed] [Google Scholar]
- [47].Baylis Scanlon JA, Al-Shawi MK, Le Nga P, Nakamoto RK, Determination of the partial reactions of rotational catalysis in F1-ATPase, Biochemistry. 46 (2007) 8785–8797. 10.1021/bi700610m. [DOI] [PubMed] [Google Scholar]
- [48].Yasuda R, Noji H, Yoshida M, Kinosita K, Itoh H, Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase, Nature. 410 (2001) 898–904. 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- [49].Weber J, Senior AE, Catalytic mechanism of F1-ATPase, Biochim. Biophys. Acta - Bioenerg 1319 (1997) 19–58. 10.1016/S0005-2728(96)00121-1. [DOI] [PubMed] [Google Scholar]
- [50].Weber J, Senior AE, ATP synthase: What we know about ATP hydrolysis and what we do not know about ATP synthesis, Biochim. Biophys. Acta - Bioenerg 1458 (2000) 300–309. 10.1016/S0005-2728(00)00082-7. [DOI] [PubMed] [Google Scholar]
- [51].Watanabe R, Iino R, Noji H, Phosphate release in F1-ATPase catalytic cycle follows ADP release, Nat. Chem. Biol 6 (2010) 814–820. 10.1038/nchembio.443. [DOI] [PubMed] [Google Scholar]
- [52].Okazaki KI, Hummer G, Phosphate release coupled to rotary motion of F1-ATPase, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 16468–16473. 10.1073/pnas.1305497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bason JV, Montgomery MG, Leslie AGW, Walker JE, How release of phosphate from mammalian F1-ATPase generates a rotary substep, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 6009–6014. 10.1073/pnas.1506465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ahmad Z, Senior AE, Identification of phosphate binding residues of Escherichia coli ATP synthase, J. Bioenerg. Biomembr 37 (2005) 437–440. 10.1007/s10863-005-9486-8. [DOI] [PubMed] [Google Scholar]
- [55].Watanabe R, Okuno D, Sakakihara S, Shimabukuro K, Iino R, Yoshida M, Noji H, Mechanical modulation of catalytic power on F1-ATPase, Nat. Chem. Biol 8 (2012) 86–92. 10.1038/nchembio.715. [DOI] [PubMed] [Google Scholar]
- [56].Adachi K, Oiwa K, Yoshida M, Nishizaka T, Kinosita K, Controlled rotation of the F1-ATPase reveals differential and continuous binding changes for ATP synthesis, Nat. Commun 3 (2012) Article number 1022. 10.1038/ncomms2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shimo-Kon R, Muneyuki E, Sakai H, Adachi K, Yoshida M, Kinosita K, Chemo-mechanical coupling in F1-ATPase revealed by catalytic site occupancy during catalysis, Biophys. J 98 (2010) 1227–1236. 10.1016/j.bpj.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sielaff H, Rennekamp H, Wachter A, Xie H, Hilbers F, Feldbauer K, Dunn SD, Engelbrecht S, Junge W, Domain compliance and elastic power transmission in rotary FoF1-ATPase, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 17760–17765. 10.1073/pnas.0807683105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Adachi K, Oiwa K, Nishizaka T, Furuike S, Noji H, Itoh H, Yoshida M, Kinosita K, Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation, Cell. 130 (2007) 309–321. 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- [60].Kabaleeswaran V, Shen H, Symersky J, Walker JE, Leslie AGW, Mueller DM, Asymmetric structure of the yeast F1 ATPase in the absence of bound nucleotides, J. Biol. Chem 284 (2009) 10546–10551. 10.1074/jbc.M900544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yasuda R, Masaike T, Adachi K, Moji H, Itoh H, Kinosita K, The ATP-waiting conformation of rotating F1-ATPase revealed by single-pair fluorescence resonance energy transfer, Proc. Natl. Acad. Sci. U. S. A 100 (2003) 9314–9318. 10.1073/pnas.1637860100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nam K, Karplus M, Insights into the origin of the high energy-conversion efficiency of F1-ATPase, Proc. Natl. Acad. Sci. U. S. A 116 (2019) 15924–15929. 10.1073/pnas.1906816116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ketchum CJ, Al-Shawi MK, Nakamoto RK, Intergenic suppression of the γM23K uncoupling mutation in FoF1 ATP synthase by βGlu-381 substitutions: the role of the β380DELSEED386 segment in energy coupling, Biochem. J 330 (1998) 707–712. https://www.ncbi.nlm.nih.gov/pubmed/9480879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Usukura E, Suzuki T, Furuike S, Soga N, Saita EI, Hisabori T, Kinosita K, Yoshida M, Torque generation and utilization in motor enzyme FoF1-ATP synthase: Half-torque F1 with short-sized pushrod helix and reduced ATP synthesis by half-torque FoF1, J. Biol. Chem 287 (2012) 1884–1891. 10.1074/jbc.M111.305938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mnatsakanyan N, Kemboi SK, Salas J, Weber J, The β subunit loop that couples catalysis and rotation in ATP synthase has a critical length, J. Biol. Chem 286 (2011) 29788–29796. 10.1074/jbc.M111.254730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mnatsakanyan N, Krishnakumar AM, Suzuki T, Weber J, The role of the βDELSEED-loop of ATP synthase, J. Biol. Chem 284 (2009) 11336–11345. 10.1074/jbc.M900374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.