Abstract
Chronic obstructive pulmonary disease (COPD) is a common inflammatory disease associated with restricted lung airflow. Quantitative computed tomography (CT)-based bronchial measures are popularly used in COPD-related studies, which require both airway segmentation and anatomical branch labeling. This paper presents an algorithm for anatomical labeling of human airway tree branches using a novel two-step machine learning and hierarchical features. Anatomical labeling of airway branches allows standardized spatial referencing of airway phenotypes in large population-based studies. State-of-the-art anatomical labeling methods are associated with mandatory manual reviewing and correction for mislabeled branches—a time-consuming process susceptible to inter-observer variability. The new method is fully automated, and it uses hierarchical branch-level features from the current as well as ancestral and descendant branches. During the first machine learning step, it differentiates candidate anatomical branches from insignificant topological branches, often, responsible for variations in airway branching patterns. The second step is designed for lung lobe-based classification of anatomical labels for valid candidate branches. The machine learning classifiers has been designed, trained, and validated using total lung capacity (TLC) CT scans (n = 350) from the Iowa cohort of the nationwide COPDGene study during their baseline visits. One hundred TLC CT scans were used for training and validation, and a different set of 250 scans were used for testing and evaluative experiments. The new method achieved labeling accuracies of 98.4, 97.2, 92.3, 93.4, and 94.1% in the right upper, right middle, right lower, left upper, and left lower lobe, respectively, and an overall accuracy of 95.9%. For five clinically significant segmental branches, the method has achieved an accuracy of 95.2%.
Keywords: Airway tree, computed tomography, airway branch labeling, centerline analysis, neural network
1. INTRODUCTION
Chronic obstructive pulmonary disease (COPD) is an inflammatory disease that causes restricted lung airflow. COPD is the fourth leading cause of death in the United States of America affecting over 328 million people worldwide.1,2 Quantitative computed tomography (CT)-based measures of bronchial morphology are popularly used in COPD-related research and clinical studies exploring the pathophysiology and mechanism of the disease occurrence and progression.3-6 For example, Smith et al. observed that, when the same anatomic airway segments are evaluated across subjects, airway walls, on average, are thinner in smokers with COPD compared with normal non-smokers.7 The important precursory steps for quantitative CT-based analysis of bronchial morphology are, first, segmentation of the airway lumen tree, followed by labeling of major anatomical branch segments in the extracted airway tree. Anatomical labeling of airway branches enables standardized spatial referencing and matching of airway phenotypes among individuals in large population-based studies.
Several computerized airway tree labeling algorithms are available in literature. For example, Mori et al. presented a rule-based classification method.8 This method suffers from errors related to false branching and branching variability beyond the finite rule-base, and, once, an error is committed, it propagates through the tree triggering additional errors. Kitaoka et al. developed a mathematical reference model of human airway tree with geometric and topologic features representing individual branches and inter-branch relationships and used this model for matching and anatomical branch labeling for test cases.9 This approach was further extended where a reference model was generated from a large set of human airway tree data mitigating the effects of anatomical branching variabilities, and, also, skeletal pruning was added to reduce false branching.10 Feragen et al. used the notion of geometric tree-space to map individual airway trees on to the tree-space and use geodesic distance analysis for matching and anatomical labeling.11,12 Other researchers have used statistical modeling of individual anatomical branches from a training set and determine branch labels in a test set using breadth-first search and probability maximization.13,14 However, these methods still need manual intervention to achieve an accuracy suitable for large research and clinical studies.
In this paper, we present an automated algorithm for anatomical labeling of human airway tree branches using a novel two-step machine learning and hierarchical features. The method uses geometric and topologic features of the current as well as ancestral and descendant generations through a series of neural network (NN)-based machine learning classifiers. During the first machine learning step, candidate anatomical branches are differentiated from insignificant topological branches, often, responsible for variations in airway branching patterns. The second machine learning step is designed for lung lobe-based classification of anatomical labels for valid candidate branches.
2. METHODS
The new method relies on leveraging knowledge of the overall airway branching structure to compartmentalize the labeling process. The airway tree structure and the 32 anatomical segments of interest are illustrated in Figure 1. Starting at the trachea, the left and right main bronchi (LMB and RMB) lead to the left and right lungs. Beneath the LMB and RMB, bronchi lead to the left and right upper lobes, right middle lobe, and the left and right lower lobes. In the right lung, the right upper lobe (RUL) branch leads to the upper lobe while the Bronchus Intermedius (BronchInt) feeds the middle and lower lobes. For the first three generations, the airway branching pattern is standard, and branching variabilities are mostly observed beyond this depth. Moreover, such variabilities of bronchopulmonary segments are only local phenomena. An example of such variability is trifurcations of RB1, RB2, and RB3 versus one bifurcation leading to RB1 shortly followed by another bifurcation leading to RB2 and RB3 branches.
Figure 1.
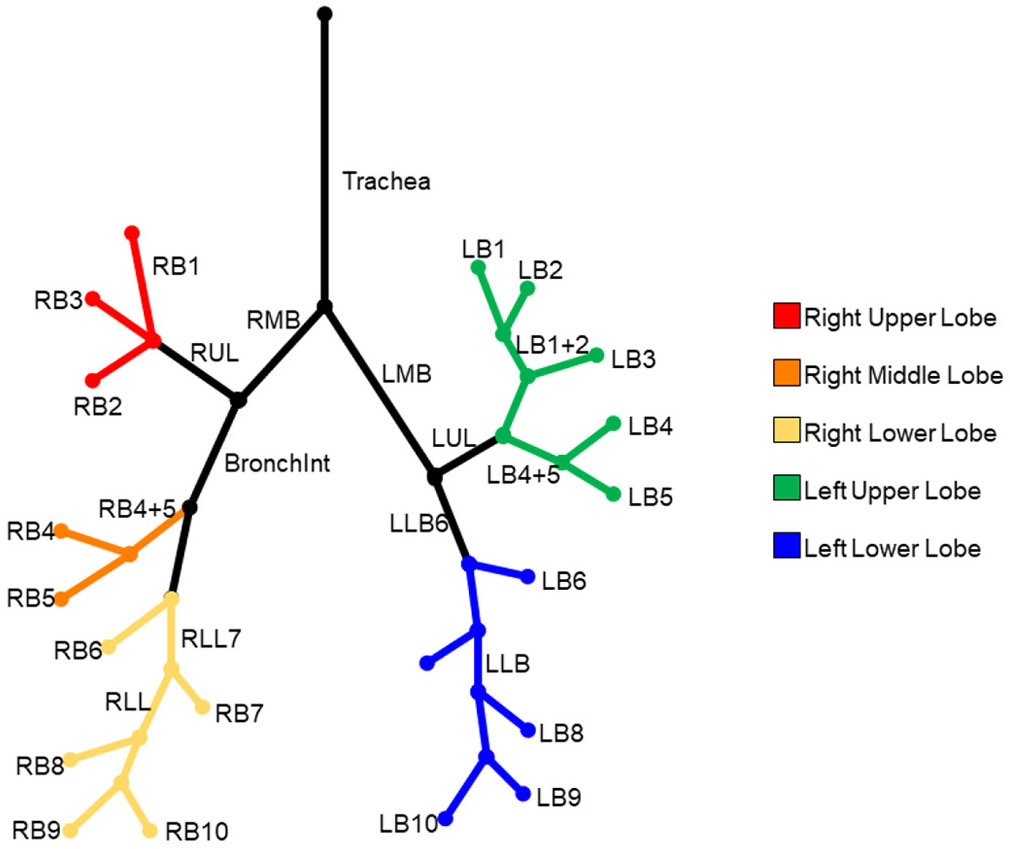
Graphical description of topological and spatial relationship among 32 anatomical human bronchi up to the segmental level. Segmental branches for different lobes are color-coded.
The overall strategy of our labeling algorithm is to label the first three generations of the airway tree, referred to as confirmed airway branches, using simple branch features. Once the first three generations of branch segments are identified, entry points to each of the five lung lobes are located and a two-stage classification strategy is used. During the first stage, anatomical branches are identified, which are fed to the second stage for classifying bronchopulmonary segments in individual lobes.
2.1. Skeletonization and detection of confirmed airway branches
The method starts with leakage-free segmented airway lumen tree as the input, which we obtained from chest CT scans using our previously published iterative multi-parametric freeze-and-grow algorithm.15,16 This method combines deep learning and conventional image processing approaches to iteratively segment the airway tree volume from TLC chest CT scans. First, a deep learning network is used to generate a voxel-level lumen likelihood map from the CT image. Next, it iteratively segments the airway lumen volume from the voxel-level lumen likelihood map starting at a conservative likelihood threshold and iteratively captures finer branches by progressively relaxing its value and correcting for segmentation leakages. Next, the centerline representation of the airway lumen tree is computed using a minimum cost path-based skeletonization algorithm developed at out laboratory17 This method generates the curve skeleton of airway tree by iteratively growing new skeletal branches computed as a minimum-cost path. The meaningfulness of a skeletal branch is defined by its global context and scales. It is necessary to remove spurious skeletal branches from the airway tree centerline, since they introduce critical challenges for computerized airway labeling due to erroneous branching patterns and branch features. A local scale-based method is developed and applied to prune spurious centerline branches while preserving true one. The basic idea of this step is that the for valid airway tree centerline branches the ratio of skeletal depth to local scale will be higher. Let p be a skeletal voxel immediately following a junction. The skeletal subtree Sp emanating from p is pruned if depth(p)/scale(p) is smaller than a threshold, where scale(p) is the local airway lumen radius, computed as the largest distance transform value in the 26-connected neighborhood of p, and depth(p) is the geodesic distance from p to the farthest voxel in Sp. After pruning, the airway tree centerline is rotated and translated such that the trachea aligned with the image z-axis with its root at the origin. The first step of our method identifies the seven anatomical airway centerline branches (Trachea, RMB, LMB, RUL, LUL, BronchInt, and LLB6 in Figure 1) in the first three generations including the trachea using their tree generation and relative coordinate positions. These branches are used to identify the entry points into each of the five lung lobes.
2.2. Anatomical branch classification
The first classification step is aimed to differentiate among valid anatomic branches from insignificant topological branches, often, responsible for variations in airway branching patterns. A fully connected NN with three hidden layers consisting of 128, 256, and 512 neurons was trained to compute validity likelihood of topological branches. Rectified linear unit (ReLu) activation function is used for hidden layers, while sigmoid activation is used for the output layer to determine the likelihood that a given branch is an anatomical branch. A binary cross entropy loss function is used and a likelihood threshold of 0.3 is used for anatomical branches. The threshold value was determined by taking the mean minus three times standard deviation of anatomic branches in the training dataset.
Lobe-specific NN classifiers are trained for labeling anatomical branches in individual lobes. For example, consider the right upper lobe. All first classifier-cleared anatomical branches beyond the branch RUL are used to train a multi-class NN. During the training process, a class label of 1, 2, and 3 are assigned for RB1, RB2, RB3 branches, respectively; class label 0 is assigned to all anatomical branches other than RB1, RB2, RB3. For this classifier, a fully connected NN with three hidden layers consisting of 48, 96, and 192 neurons each with rectified linear (ReLu) activation is used with batch size of 8. A softmax activation function is used for the output layer to predict class output. A weighted categorical cross entropy loss function is used, where the weights of each output class are calculated as one minus the fraction of samples of that class in the training dataset. At runtime, anatomical branches in a given lobe are fed to the lobe-specific classifier and a branch with the maximum likelihood for a given target anatomical branch is labeled as that target branch. If the maximum likelihood for a given target branch is below a threshold among all input anatomical branches, then that specific target branch is considered as missing.
For both classifiers, hierarchical geometric and topologic features are used from the candidate branch, its parent and grandparent, and all sibling and immediate children branches. For each of these branches, the following features are computed and used for training—(1) branch generation number, (2) branch position relative to the tree root, (3) Euclidean branch length, (4) projected branch length along each image coordinate axis, (5) branch angles with each image coordinate axis, and (6) the number of siblings and immediate children resulting a feature length of 90. All feature values are scaled between −1 and 1.
3. EXPERIMENTAL RESULTS
The new airway branch labeling algorithm was trained, validated, and tested using chest CT scans at TLC of three hundred and fifty subjects (age (years): [45 80] ([Min Max]), 65.1±7.5 (mean±std.); 168 female; smoking (pack-year): 48.5±26.8; COPD GOLD status (N): GOLD −1 (31); GOLD 0 (173); GOLD 1 (45), GOLD 2 (68), GOLD 3 (20), GOLD 4 (13)) from the Iowa cohort of the COPDGene study5 at their baseline visits. CT scans were acquired on a Siemens Sensation 64 scanner (Forchheim, Germany) at the University of Iowa Comprehensive Lung Imaging Center (I-CLIC) research CT facility. The following CT parameters were used: 120 kV, 110 effective mAs, pitch of 1.0; and the images were reconstructed on 512×512 matrices using the standard B35 body kernel with 0.75 mm slice thickness, 0.5 mm slice spacing, and approximately 0.62±0.06 mm in-plane pixel size, slices: [493 808] 675.6±51.5. This study was approved by the University of Iowa Institutional Review Board. The dataset was randomly partitioned into one hundred training scans, further partitioned into eighty and twenty training and validation scans, respectively, and two hundred fifty scans for testing and evaluative purposes. The reference anatomical branch labeling was obtained by manual annotation based on airway masks and their centerlines using an ITK-provided graphical user interface and editing tools18 following the standardized bronchopulmonary anatomy up to the segmental level.19,20
Two sets of training samples were generated for the two-step classifiers as described in the method section. For the Step 1 classifier, a training sample vector was generated for each airway centerline topological branch. A total of 27,521 training samples were generated for this step from the one hundred chest CT scans. For the Step 2 classifier of a given lobe, a training sample vector was generated for each possible anatomical branch within the specific lobe after clearance by the Step 1 classifier. A total of 18,194 and 4,551 samples were generated over the five lung lobes for training and validation, respectively. For all classifiers, the network was trained for 200 epochs using Adam optimization algorithm21 and a learning rate of 1×10−4. Step 1 classifier took 2 hours and 14 minutes for training on an Intel Core i9-79000X CPU using a NVIDIA GeForce GTX 1080 Ti graphics card. On an average a lob-specific Step 2 classifier required 37 minutes for training on the same machine.
Results of intermediate steps of our airway labeling algorithm are presented in Figure 2. As observed in Figure 2(c), the airway tree centerline includes no apparent spurious branches, while visually valid branches are preserved after our pruning step. As shown in Figure 2(d), the method successfully identifies the first seven anatomical bronchi in the first three generation. Results of anatomical labeling of segmental bronchi are presented in Figures 2(e) and (f). In these figures, red, orange, yellow, green, and blue used to denote segmental bronchi in the right upper, right middle, right lower, left upper, and left lower lung lobes. Results of anatomical labeling of segmental bronchi in right and left lungs, shown in Figures 2(e) and (f), respectively, are correct as visually confirmed by an expert using an ITK-based graphical user interface with reference and computed labeled airway masks.
Figure 2.
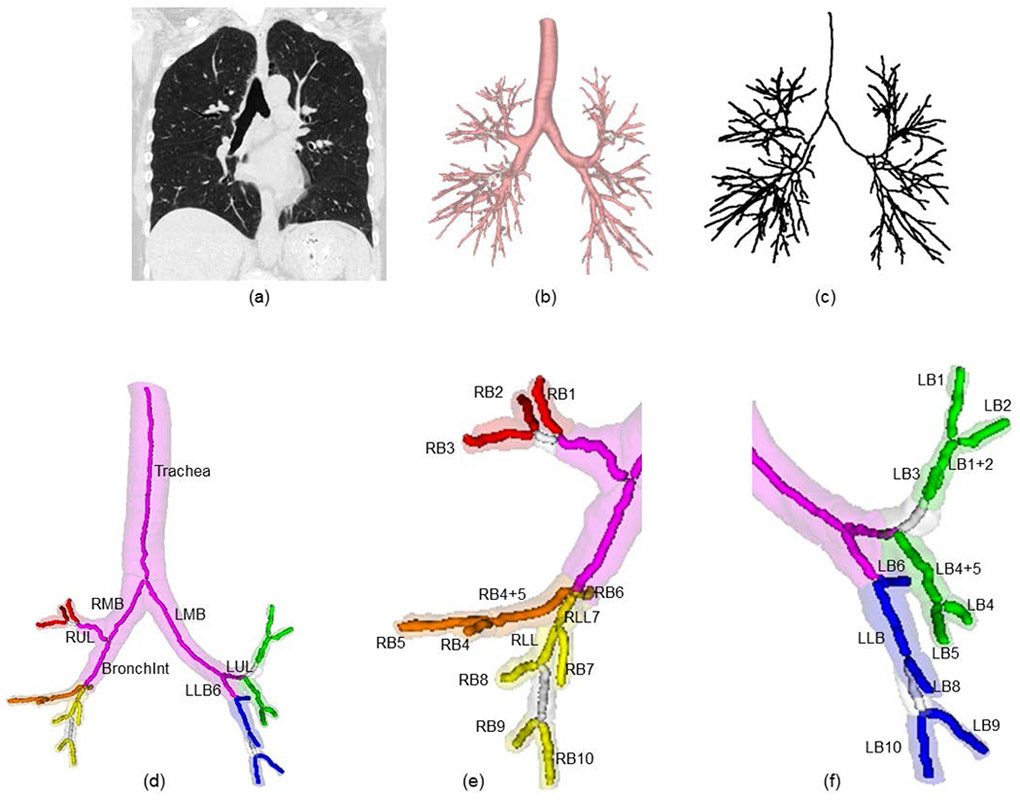
Results of intermediate steps within our anatomical human bronchial labeling. (a) input chest CT image at total lung capacity, (b) segmented airway tree volume computed from (a), (c) airway tree centerline after skeletal pruning computed from (b), (d) anatomical labeling results for the seven branches in the first three generations of the airway tree, results of complete anatomical bronchial labeling at the segmental level with right and left lung results enlarged in (e) and (f), respectively.
For quantitative evaluation, the labeling method was applied on two hundred and fifty TLC CT scans and the results were visually compared with manually annotated labels of 32 anatomical bronchial segments and the labeling accuracy is presented in Table 1. For all test cases, the seven bronchi in the first three generations were correctly identified. The method achieved 98.6, 97.2, 92.3, 93.4, and 94.1% accuracies in labeling segmental bronchi in the right upper, right middle, right lower, left upper, and left lower lobe, respectively, and an overall labeling accuracy of 95.9% was achieved for all 32 anatomical bronchial segments. To simplify the review process and the subsequent use of the many resulting bronchial measures, the radiology center of the Severe Asthma Research Program4 (SARP) has standardized five bronchopulmonary segments, namely, RB1, RB4, RB10, LB1, and LB10. For these five clinically significant bronchi, our labeling method achieved an accuracy of 95.2%.
Table 1.
Labeling accuracy for 32 different airway anatomical branches from TLC chest CT scans of 250 test subjects from the COPDGene study baseline visits.
| Left Lung | Right Lung | ||
|---|---|---|---|
| Left Upper Lobe | Right Upper Lobe | ||
| LB1+2 | 96.0 | RB1 | 98.6 |
| LB1 | 94.3 | RB2 | 98.6 |
| LB2 | 91.2 | RB3 | 98.1 |
| LB3 | 91.9 | Right Middle Lobe | |
| LB4+5 | 97.8 | RB4+5 | 98.2 |
| LB4 | 91.1 | RB4 | 97.5 |
| LB5 | 91.3 | RB5 | 95.5 |
| Left Lower Lobe | Right Lower Lobe | ||
| LB6 | 98.0 | RB6 | 95.6 |
| LLL | 99.0 | RLL7 | 90.6 |
| LB8 | 92.6 | RB7 | 96.5 |
| LB9 | 87.5 | RLL | 93.5 |
| LB10 | 92.0 | RB8 | 89.4 |
| RB9 | 87.1 | ||
| RB10 | 92.7 | ||
100% labelling accuracy was achieved for Trachea, RMB, LMB, RUL, BronchInt, LUL, LLB6.
4. CONCLUSIONS
In this paper, we have presented an automated anatomical airway branch labeling algorithm using a two-step machine learning and hierarchical features. The method uses an NN-based machine learning approach and hierarchical geometric and topologic features from a candidate branch, its parent and grandparent, all sibling, and immediate children branches. A two-step classification approach has been introduced to compartmentalize and cope with the challenges of anatomic human bronchial labeling and variabilities of airway branching patterns. The first machine learning step differentiates candidate anatomical branches from insignificant topological branches, often, responsible for variations in airway branching patterns. The second step was designed for lung lobe-based classification of bronchopulmonary segments for valid candidate branches. Our experimental results suggest that the method is suitable for automated anatomic labeling of human bronchi up to the segmental level, especially, for the five clinically significant segmental branches.
ACKNOWLEDGEMENTS
This work was supported by the NIH grants 5U01 HL089897, R01 HL112986, R01 HL142042, and S10 OD026960.
REFERENCES
- [1].Kiley JP and Gibbons GH, "COPD National Action Plan: Addressing a Public Health Need Together," Chest, 152, 698–699, 2017. [DOI] [PubMed] [Google Scholar]
- [2].Quaderi SA and Hurst JR, "The unmet global burden of COPD," Glob Health Epidemiol Genom, 3, e4, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Couper D, LaVange LM, Han M, Barr RG, Bleecker E, Hoffman EA, Kanner R, Kleerup E, Martinez FJ, Woodruff PG, Rennard S, and Group SR, "Design of the Subpopulations and Intermediate Outcomes in COPD Study (SPIROMICS)," Thorax, 69, 491–4, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jarjour NN, Erzurum SC, Bleecker ER, Calhoun WJ, Castro M, Comhair SA, Chung KF, Curran-Everett D, Dweik RA, Fain SB, Fitzpatrick AM, Gaston BM, Israel E, Hastie A, Hoffman EA, Holguin F, Levy BD, Meyers DA, Moore WC, Peters SP, Sorkness RL, Teague WG, Wenzel SE, Busse WW, and Program NSAR, "Severe asthma: lessons learned from the National Heart, Lung, and Blood Institute Severe Asthma Research Program," Am. J. Respir. Crit. Care Med, 185, 356–62, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, and Crapo JD, "Genetic epidemiology of COPD (COPDGene) study design," COPD, 7, 32–43, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tan WC, Sin DD, Bourbeau J, Hernandez P, Chapman KR, Cowie R, FitzGerald JM, Marciniuk DD, Maltais F, Buist AS, Road J, Hogg JC, Kirby M, Coxson H, Hague C, Leipsic J, O'Donnell DE, Aaron SD, and Can CCRG, "Characteristics of COPD in never-smokers and ever-smokers in the general population: results from the CanCOLD study," Thorax, 70, 822–9, 2015. [DOI] [PubMed] [Google Scholar]
- [7].Smith BM, Hoffman EA, Rabinowitz D, Bleecker E, Christenson S, Couper D, Donohue KM, Han MK, Hansel NN, Kanner RE, Kleerup E, Rennard S, and Barr RG, "Comparison of spatially matched airways reveals thinner airway walls in COPD. The Multi-Ethnic Study of Atherosclerosis (MESA) COPD Study and the Subpopulations and Intermediate Outcomes in COPD Study (SPIROMICS)," Thorax, 69, 987–96, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mori K, Hasegawa J, Suenaga Y, and Toriwaki J, "Automated anatomical labeling of the bronchial branch and its application to the virtual bronchoscopy system," IEEE Trans. Med. Imag, 19, 103–14, 2000. [DOI] [PubMed] [Google Scholar]
- [9].Kitaoka H, Park Y, Tschirren J, Reinhardt J, Sonka M, McLennan G, and Hoffman EA, "Automated nomenclature labeling of the bronchial tree in 3D-CT lung images," Proc. of Med. Imag. Comp. Comp-Assist. Interven. (MICCAI), 1–11, 2002. [Google Scholar]
- [10].Tschirren J, McLennan G, Palagyi K, Hoffman EA, and Sonka M, "Matching and anatomical labeling of human airway tree," IEEE Trans. Med. Imag, 24, 1540–7, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Feragen A, Petersen J, Owen M, Lo P, Thomsen LH, Wille MM, Dirksen A, and de Bruijne M, "A hierarchical scheme for geodesic anatomical labeling of airway trees," Proc. of Med. Imag. Comp. Comp-Assist. Interven. (MICCAI), 147–155, 2012. [DOI] [PubMed] [Google Scholar]
- [12].Feragen A, Petersen J, Owen M, Pechin L, Hohwu Thomsen L, Wille MM, Dirksen A, and de Bruijne M, "Geodesic Atlas-Based Labeling of Anatomical Trees: Application and Evaluation on Airways Extracted From CT," IEEE Trans. Med. Imag, 34, 1212–26, 2015. [DOI] [PubMed] [Google Scholar]
- [13].Lo P, van Rikxoort EM, Goldin J, Abtin F, de Bruijne M, and Brown M, "A bottom-up approach for labeling of human airway trees," Med. Imag. Comp. Comp-Assist Interven. (MICCAI) Int. WS. Pulm. Im. Anal, 2011. [Google Scholar]
- [14].van Ginneken B, Baggerman W, and van Rikxoort EM, "Robust segmentation and anatomical labeling of the airway tree from thoracic CT scans," Proc. of Med. Imag. Comp. Comp-Assist. Interven. (MICCAI), 219–226, 2008. [DOI] [PubMed] [Google Scholar]
- [15].Nadeem SA, Hoffman EA, and Saha PK, "A fully automated CT-based airway segmentation algorithm using deep learning and topological leakage detection and branch augmentation approaches," Proc. of Med Imag: Imag Proc, 10949, 109490C, 2019. [Google Scholar]
- [16].Nadeem SA, Hoffman EA, Sieren JP, and Saha PK, "Topological leakage detection and freeze-and-grow propagation for improved CT-based airway segmentation," Proc. of Proc SPIE Med Imag: Imag Process, 10574, 105741A, 2018. [Google Scholar]
- [17].Jin D, Iyer KS, Chen C, Hoffman EA, and Saha PK, "A Robust and Efficient Curve Skeletonization Algorithm for Tree-Like Objects Using Minimum Cost Paths," Pat Recog Lett, 76, 32–40, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, and Gerig G, "User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability," Neuroimage, 31, 1116–1128, 2006. [DOI] [PubMed] [Google Scholar]
- [19].Boyden EA, Segmental anatomy of the lungs. A study of the patterns of the segmental bronchi and related pulmonary vessels, The Blakiston Division, McGraw-Hill Book Company, 1955. [Google Scholar]
- [20].Sealy WC, Connally SR, and Dalton ML, "Naming the bronchopulmonary segments and the development of pulmonary surgery," Ann Thorac Surg, 55, 184–8, 1993. [DOI] [PubMed] [Google Scholar]
- [21].Kingma DP and Ba J, "Adam: A method for stochastic optimization," arXiv preprint arXiv:1412.6980, 2014. [Google Scholar]


