Abstract
During development, oligodendrocytes in the central nervous system extend a multitude of processes that wrap axons with myelin. The highly polarized oligodendrocytes generate myelin sheaths on many different axons, which are far removed from the cell body. Neurons use RNA binding proteins to transport, stabilize, and locally translate mRNA in distal domains of neurons. Local synthesis of synaptic proteins during neurodevelopment facilitates the rapid structural and functional changes underlying neural plasticity and avoids extensive protein transport. We hypothesize that RNA binding proteins also regulate local mRNA regulation in oligodendrocytes to promote myelin sheath growth. Fragile X mental retardation protein (FMRP), an RNA binding protein that plays essential roles in the growth and maturation of neurons, is also expressed in oligodendrocytes. To determine whether oligodendrocytes require FMRP for myelin sheath development, we examined fmr1−/− mutant zebrafish and drove FMR1 expression specifically in oligodendrocytes. We found oligodendrocytes in fmr1−/− mutants developed myelin sheaths of diminished length, a phenotype that can be autonomously rescued in oligodendrocytes with FMR1 expression. Myelin basic protein (Mbp), an essential myelin protein, was reduced in myelin tracts of fmr1−/− mutants, but loss of FMRP function did not impact the localization of mbpa transcript in myelin. Finally, expression of FMR1-I304N, a missense allele that abrogates FMRP association with ribosomes, failed to rescue fmr1−/− mutant sheath growth and induced short myelin sheaths in oligodendrocytes of wild-type larvae. Taken together, these data suggest that FMRP promotes sheath growth through local regulation of translation.
Keywords: fragile X mental retardation protein, messenger RNA, myelin, oligodendrocyte, RNA binding protein, spinal cord, zebrafish
1 |. INTRODUCTION
Oligodendroglia in the central nervous system simultaneously wrap numerous axons in a highly dynamic ensheathment process that takes place at a distance from the nucleus (Kirby et al., 2006). Mature oligodendrocytes can ultimately form more than a dozen stabilized myelin sheaths per cell, with variability in the lengths of individual myelin internodes (Almeida, Czopka, Ffrench-Constant, & Lyons, 2011; Murtie, Macklin, & Corfas, 2007). How can these complex cells coordinate so many independent axonal interactions from a distance? Subcellular localization of mRNA represents a powerful mechanism to achieve on demand, localized protein production (Steward & Worley, 2001). In the nervous system, this phenomenon allows cells to achieve remarkable plasticity, despite physical separation from the cell body (Feuge, Scharkowski, Michaelsen-Preusse, & Korte, 2019; Martin et al., 2009). Oligodendrocytes express an array of RNA binding proteins (RBPs) (Dobson, Zhou, Flint, & Armstrong, 2008; Ebersole, Chen, Justice, & Artzt, 1996; Thomas et al., 2005; Wang et al., 2004), which may serve to regulate mRNA at active sites of ensheathment to meet independent protein requirements in each myelin sheath.
Fragile X syndrome (FXS), the most common heritable form of autism spectrum disorder (Harris et al., 2008), is caused by transcriptional silencing of the FMR1 gene, which encodes the RBP fragile X mental retardation protein (FMRP) (Sutcliffe et al., 1992). FXS can be comorbid with epilepsy, attention deficit/hyperactivity disorder, and anxiety (Berry-Kravis, 2002; Cordeiro, Ballinger, Hagerman, & Hessl, 2010; Nolin et al., 2003; Sullivan et al., 2006), and FMRP plays prominent roles in a wide range of neural functions, including neural plasticity (Huber, Gallagher, Warren, & Bear, 2002) and synaptic development (Dictenberg, Swanger, Antar, Singer, & Bassell, 2008; Doll, Vita, & Broadie, 2017; Pan, Zhang, Woodruff, & Broadie, 2004). Importantly, longitudinal neuroimaging has documented region-specific white matter abnormalities in individuals with FXS (Hoeft et al., 2010; Swanson et al., 2018), thereby also implicating oligodendrocytes in the disease state.
We hypothesize that FMRP regulates mRNA in oligodendrocytes to facilitate myelin sheath growth. As an RBP, FMRP has been implicated in mRNA stabilization (Zalfa et al., 2007), transport (Kao, Aldridge, Weiler, & Greenough, 2010), and activity-dependent translation (Dictenberg et al., 2008; Todd, Mack, & Malter, 2003) of critical synaptic genes in neurons, functions that could also facilitate local protein synthesis in highly polarized oligodendrocytes. Glial cells express FMRP during early brain development (Giampetruzzi, Carson, & Barbarese, 2013; Wang et al., 2004), and Fmr1 knockout mice show delayed cortical myelination during early postnatal stages (Pacey et al., 2013). However, the relative glial contribution of FMRP in myelination remains unclear. We used zebrafish (Danio rerio) to explore roles for FMRP in the early developmental myelination of axons, utilizing cell type-specific methods to visualize and manipulate FMR1 and associated target genes in vivo, finding crucial FMRP requirements in myelin sheath growth and regulation of myelin genes.
2 |. MATERIALS AND METHODS
2.1 |. Zebrafish lines and husbandry
The Institutional Animal Care and Use Committee at the University of Colorado School of Medicine approved all animal work, which is in compliance with U.S. National Research Council’s Guide for the Care and Use of Laboratory Animals, the U.S. Public Health Service’s Policy on Humane Care and Use of Laboratory Animals, and Guide for the Care and Use of Laboratory Animals. Larvae were raised at 28.5°C in embryo medium and staged as hours or days post fertilization (hpf/dpf) according to morphological criteria (Kimmel, Ballard, Kimmel, Ullmann, & Schilling, 1995). Zebrafish lines used in this study included fmr1hu2787 (den Broeder et al., 2009), Tg(sox10:mRFP)vu234 (Kucenas et al., 2008), Tg(sox10:tagRFP)co26 (Blasky, Pan, Moens, & Appel, 2014), Tg(mbp: EGFP-CAAX)co58 (Yergert, Hines, & Appel, 2019), and Tg(olig2:EGFP)vu12 (Shin, Park, Topczewska, Madwsley, & Appel, 2003). All other constructs were delivered by transient transgenesis to facilitate sparse labeling and single cell analysis. Genotyping for fmr1hu2787 was performed as previously described (Ng, Yang, & Lu, 2013).
2.2 |. Plasmid construction
Multisite Gateway cloning was utilized to generate Tol2 expression plasmids, which were injected into single cell embryos along with Tol2 mRNA to produce transient transgenic animals. Human FMR1 and FMR1-I304N were amplified from pFRT-TODestFLAGHAhFMRPiso1, a gift from Thomas Tuschl (Ascano et al., 2012), using the following attB primers (annealed sequence in lowercase): forward, GGGGACAAGTT TGTACAAAAAAGCAGGCTTAatggaggagctg; reverse, GGGGACCACT TTGTACAAGAAAGCTGGGTTgggtactccattcacgagtggt. We generated pME-FMR1 and pME-FMR1-I304N entry clones via binding protein recombination with pDONR221 backbone. Middle entry clones were then cloned behind either sox10 or myrf promoters, either in frame with enhanced green fluorescent protein (EGFP) to generate a fusion protein or with an internal ribosome entry site (IRES) and cysteine-aliphatic amino acid-X (CAAX) prenylation motif (IRES-EGFP-CAAX) to label cell membranes with a distinct peptide.
Entry clones were LR-recombined with either the pDEST-Tol2-CG2 destination vector (green heart marker; for pEXPR-myrf:FMR1-IRES-EGFP-CAAX and pEXPR-myrf:FMR1-I304N-IRES-EGFP-CAAX) or the pDEST-Tol2-pA2 vector for pEXPR-sox10:FMR1-EGFP.
Published plasmids are p3E-7.2sox10 (Mathews et al., 2014), p5E-myrf, p3E-IRES-EGFP-CAAX, pEXPR-tol2-mbp:EGFP-CAAX (Hughes & Appel, 2019), and p3E-EGFP (Kwan et al., 2007).
2.3 |. Imaging and analysis
With the exception of single molecule fluorescent in situ hybridization (smFISH) and IHC, live larvae were imaged in all experiments. Larvae were embedded laterally in 1.2% low-melt agarose containing 0.4% tricaine for immobilization. We acquired images on a Zeiss CellObserver SD 25 spinning disk confocal system for time-lapse microscopy and cell counts (Carl Zeiss, Oberkochen, Germany) or a Zeiss LSM 880 for all other experiments (Carl Zeiss). Images were captured with Zen software (Carl Zeiss), then processed and analyzed using Fiji/ImageJ or Zen Blue (Carl Zeiss).
2.4 |. smFISH probe design
mbpa smFISH probes were designed using the Stellaris RNA FISH Probe Designer tool by entering the zebrafish mbpa cDNA sequences obtained from Ensemble Genome Browser from transcript mbpa-206 (GRCz11). Probes with highly repetitive sequences were removed. The probes were ordered with a CAL Fluor Red 610 Dye. Probes were resuspended in Tris-EDTA, pH 8.0, and stored at a stock concentration of 12.5 μM at −20°C (Table 1).
TABLE 1.
List of smFISH probes targeted to mbpa transcript
| Probe name | Sequence 5′ → 3′ | Probe name | Sequence 5′ → 3′ |
|---|---|---|---|
| Probe 1 | CTTTGGATTGAGCGGAGAAG | Probe 13 | AATCTTCAACCTGGGAGAAA |
| Probe 2 | GTCCAGACTGTAGACCACTG | Probe 14 | GATCTCGCTCTCCACCCAAA |
| Probe 3 | CAGATCAACACCTAGAATGG | Probe 15 | CTGGAGCACCATCTTCTGAG |
| Probe 4 | CTCTGGACAAAACCCCTTCG | Probe 16 | CTTCTCCAAGCAGGAAAACA |
| Probe 5 | TGTCCTGGATCAAATCAGCA | Probe 17 | GAGATGGAAGAGAGTGAAAT |
| Probe 6 | TTCTTCGGAGGAGACAAGAA | Probe 18 | CGGGAAGCAAAAACTTGAGA |
| Probe 7 | AGAGACCCCACCACTCTT | Probe 19 | ATGTCTGCCCTACAGACTCA |
| Probe 8 | TCGTGCATTTCTTCAGGAGC | Probe 20 | GTCGCAGCGAGTTTAACAGA |
| Probe 9 | TCGTGCATTTCTTCAGGAGC | Probe 21 | ACATTGGCCATCTTCGCTTC |
| Probe 10 | TCGAGGTGGAGAGAACTATT | Probe 22 | AGATAGAGATACAATCCAAG |
| Probe 11 | CATTAGATCGCCACAGAGAC | Probe 23 | TCTGTTGCTACATGCCTGCA |
| Probe 12 | AAGGGAACAGAACACACTTT | Probe 24 | TTACAGAAGCACGTGTTGAC |
2.5 |. smFISH experimental procedure
The smFISH protocol was adapted from three published protocols (Hauptmann & Gerster, 2000; Lyubimova et al., 2013; Oka & Sato, 2015). First, larvae were sorted for olig2:EGFP expression and fixed overnight (O/N) in 4% paraformaldehyde at 4°C. Larvae were embedded laterally in 1.5% agar, 5% sucrose blocks, and transferred to a 30% sucrose solution O/N at 4°C. Blocks were frozen on dry ice and sectioned with a Leica cryostat into 20 μm thick sections and placed on microscope slides (Fisherbrand cat no.: 12–550-15). Slides were not allowed to dry more than 5 min before adding 4% paraformaldehyde to fix the tissue at room temperature (RT) for 10–20 min. The slides were quickly rinsed with 1× phosphate buffered saline (PBS) twice. The tissue was permeabilized with 70% cold ethanol at −20°C for 2 hr. Parafilm was placed over tissue to prevent evaporation at all incubation steps. The tissue was rehydrated with wash buffer (10% deionized (DI) formamide, 2× saline-sodium citrate (SSC) in molecular grade water, 1 ml DI formamide, 1 ml 20× SSC) for 5 min at RT. From this point on, care was taken to protect the tissue and probes from light as much as possible. Hybridization buffer was made: 2× SSC, 10% DI formamide, 25 mg/ml tRNA, 50 mg/ml bovine serum albumin, 200 mM ribonucleoside vanadyl complex in diethyl pyrocarbonate water. Aliquots were made and stored at −20°C. Probe was added to hybridization buffer 1:100 for a final concentration of 125 nM. As a control, slides were incubated in hybridization buffer with no probe. Slides were quickly rinsed with fresh wash buffer followed by two wash steps at 37°C for 30 min. 4’6’-diamidino-2-phenylindole (DAPI) was added at 1:1000 concentration in wash buffer to the tissue for 7 min at RT. Slides were quickly rinsed twice with wash buffer. Finally, slides were mounted with Vectashield mounting media (Vector Laboratories, Burlingame, CA) and a No. 1 coverslip and sealed with nail polish. All slides were stored and protected from light at 4°C.
2.6 |. smFISH microscopy
Images of smFISH experiments were obtained using a Zeiss LSM 880 with Airyscan confocal microscope and a Plan-Apochromat 63×, 1.4 numerical aperture oil immersion objective. The acquisition light path used Diode 405, Argon 488, HeNe 594 lasers, 405 beam splitter and 488/594 beam splitters, and Airyscan super resolution detector. Imaging was performed using Zeiss Zen Black software and parameters included: 1024 × 1024 frame size, 1.03 μs pixel dwell time, 16-bit depth, 2.4× zoom. 1.8% 488 laser power, 5% 594 laser power, 0.5% 405 laser power, 705 gain, and z intervals of 0.3 μm. All images were taken in the hindbrain of zebrafish larvae. Myelin segments were selected for imaging based on expression of olig2:EGFP and Quasar-610 fluorescence. Post-image processing was performed using Airyscan Processing set to 6.8.
2.7 |. Immunohistochemistry
Four days post fertilization Tg(sox10:mRFP) larvae were fixed in 4% paraformaldehyde/1× PBS, rocking O/N at 4°C. Larvae were washed 3 × 5 min in 0.1% Tween/1× PBS (PBSTw), rocking at RT. For antigen retrieval, larvae were placed in 150 mM Tris pH 9 for 5 min at RT and then 15 min at 75°C. Larvae were rinsed in 1× PBS/0.1% Tween, then embedded in 1.5% agar/30% sucrose and immersed in 30% sucrose O/N. Blocks were frozen on dry ice and 20 μm sagittal sections were taken with a cryostat microtome and collected on polarized slides. Slides were mounted in Sequenza racks (Thermo Scientific, Waltham, MA), washed 3 × 5 min in 0.1% Triton-X 100/1× PBS (PBSTx), blocked 1 hr in 2% goat serum/2% bovine serum albumin/PBSTx and then placed in primary antibody (in block) O/N: rabbit α-Mbp (1:200; Kucenas, Wang, Knapik, & Appel, 2009). Sections were washed 1.5 hr in PBSTx, and then incubated 2 hr at RT in secondary antibody (in block): AlexaFluor 488 goat α-rabbit. Sections were washed for 1 hr in PBSTx, incubated with DAPI (1:2000 in PBSTx) for 5 min, washed 3 × 5 min in PBSTx, then mounted in Vectashield (Vector Laboratories).
2.8 |. Immunohistochemistry microscopy
Images of immunohistochemistry experiments were obtained with the same settings as smFISH, along with the following changes: 512 × 512 frame size, 0.85 μs pixel dwell time, 16-bit depth, 1.8× zoom. Line averaging was set to 2, 6.5% 488 laser power, 4.5–5.5% 594 laser power, 1% 405 laser power, 619 gain, and z intervals of 0.144 μm. All images of single cells were taken in the hindbrain of zebrafish larvae. Cells were selected for imaging based on sox10:mRFP fluorescence. Post-image processing was performed using Airyscan Processing set to 1.0.
2.9 |. Western blot
At 4 dpf, 20 wild-type and fmr1−/− mutant larvae were anesthetized in 0.1% tricaine, tail-clipped for genotyping, and stored at −80°C in 5 μL RIPA buffer + complete mini protease inhibitor cocktail (Roche) + phosphatase inhibitor cocktail set II (Calbiochem). Larvae were thawed on ice and homogenized using a 0.1 ml mortar and pestle (Electron Microscopy Sciences, Hatfield, PA). Lysates were placed on a rocker for 30 min at 4°C, then sonicated. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Scientific). Samples were then diluted in RIPA buffer + Trident 6× Laemmli SDS (GeneTex) + 15% 2-mercaptoethanol (Sigma, St. Louis, MO), and stored at −20°C. Samples were thawed on ice, boiled for 5 min at 95°C, and centrifuged for 10 min at 4°C. A volume of 20 μg protein was loaded into Mini-Protean TGX Precast Gels (BioRad) along with 5 μL Precision Plus Protein Kaleidescope Prestained Protein Standards (BioRad) in 1× Running Buffer (BioRad, Hercules, CA). Gels were electrophoresed at 125 V for 45 min, and transferred to immobilon fluorescent polyvinylidene fluoride (PVDF) membranes (Millipore, Burlington, MA). Blots were blocked in 5% bovine serum albumin/Tris-buffered saline, 0.1%Tween (BSA/TBST) for 1 hr and incubated with Mbp (1:1000; Kucenas et al., 2009) and β-Tubulin (AA2, 1:5000; Sigma) O/N at 4°C. Blots were then visualized using LiCor infrared secondary antibodies (1:10,000; LiCor, Lincoln, NE), with band intensity (Mbp:β-Tubulin loading control) calculated using ImageStudioLite software (LiCor). Due to the difficulty resolving low molecular weight Mbp isoforms, the 14 and 18.5 kDa bands were combined for quantification.
2.10 |. Reverse transcriptase polymerase chain reaction
RNA was isolated from pooled 4 dpf wild-type or fmr1−/− larvae and treated with 500–1,000 μL of Trizol and snap frozen. All RNA isolation steps were performed on ice and in a 4°C centrifuge at 18,078g. Larvae were thawed on ice and homogenized with a 23 g needle. A volume of 200 μL chloroform was added followed by 15 sec of shaking and centrifugation for 10 min. The aqueous layer was transferred to a new tube, combined with an equal volume of cold 100% isopropanol along with 2 μL GlycoBlue (Thermo Fisher). Samples were incubated at −20°C for 20 min, centrifuged 10 min, and the supernatant was transferred to a new tube. To wash the pellet, 200 μL cold 70% ethanol was added, followed by centrifugation for 5 min. The wash step was repeated. The pellet was dried at RT and RNA was resuspended in 20 μL molecular grade water. RNA abundance was quantified using a Nanodrop spectrophotometer (Thermo Fisher).
To synthesize cDNA, we used the iScript Reverse Transcription Supermix for reverse transcriptase quantitative polymerase chain reaction (PCR) (BioRad) which uses random hexamer primers to synthesize cDNA. We followed manufacturers protocol and completed the reaction by adding 50 μL of molecular grade water and storing samples at −20°C.
cDNA from each biological replicate (four biological replicates) was PCR amplified with primers targeting mbpa and rpl13. Primers used were mbpa forward 5′-AAGGGAAAGAGACCCCACCAC-3′, mbpa reverse 5′-TCAGAAGATGGTGCTCCAGCGT-3′, rpl13 forward 5′-AGATCCGCA GACGTAAGGCC-3′, rpl13 reverse 5′-CTCCTCCTCAGTACTGTCTCCC-3′. PCR product was run on a 1% agarose gel with GeneRuler 100 bp DNA ladder (ThermoFisher) and imaged. Fiji software was used to measure the mean grey values of all bands and background. Background was subtracted from all mbpa and rpl13 grey values. All mbpa values were divided by the loading control, rpl13, values from the same biological replicate to calculate the ratio of mbpa:rpl13.
3 |. QUANTIFICATION AND STATISTICAL ANALYSIS
3.1 |. smFISH quantification
All quantification was performed in ImageJ Fiji using a custom script created by Karlie Fedder (available upon request). First, z intervals were selected for myelin tracts using the “Make Substack” feature in Fiji. Substacks of myelin tracts in the hindbrain included 13 steps with an interval of 0.3 μm. Each substack was maximum z-projected. Background was subtracted using a 2.5 rolling ball. The image was then thresholded by taking 3 SDs above the mean fluorescence intensity. Puncta were analyzed using the “Analyze Particles” feature with a size of 0.01-Infinity and circularity of 0.00–1.00. Using the maximum projection of the olig2:EGFP channel, a region of interest (ROI) was drawn around myelin segments using the rectangle tool to draw a square of 100 × 100 pixels (4.39 × 4.39 μm). All thresholded puncta were inspected to ensure single molecules were selected. Occasionally, threshold puncta fell on the border of the ROI and these were excluded from measurements. mbpa transcripts are highly expressed and counting individual puncta was not consistently reliable. Therefore, to measure each puncta, we overlaid the thresholded image on the maximum z projected image and calculated the integrated density (area × average fluorescence intensity) using the “IntDen” measurement.
3.2 |. Immunohistochemistry quantification
First, z intervals were selected for hindbrain myelin tracts using the “Z Project” feature in Fiji. Substacks of myelin tracts in the hindbrain included 20 steps with an interval of 0.144 μm. Each substack was projected via “Sum Slices.” Fluorescence channels were split and saved as 16-bit images. Next, the membrane-bound red fluorescent protein (mRFP) channel was duplicated and automatically thresholded using the Otsu method. The wand tracing tool was used to select myelin tracts and intensity measurements were taken by redirecting to the Mbp channel (Analyze, Set Measurements, Redirect). Following quantification of the entire dataset, individual values were normalized to average wildt-ype Mbp expression levels.
3.3 |. Statistics
All statistics were performed in Graphpad Prism (version 8). Outliers were identified using a ROUT with Q = 1%. Normality was assessed with a D’Agostino and Pearson omnibus test. For two groups, unpaired comparisons were made using either unpaired two-tailed t tests (for normal distributions) or Mann–Whitney tests (abnormal distributions). For multiple comparisons, an analysis of variance was followed with Tukey’s individual comparisons to each mean for normal distributions or Kruskal–Wallis test was followed with Dunn’s individual comparisons to each mean for abnormal distributions.
4 |. RESULTS
4.1 |. FMRP is localized within nascent myelin sheaths
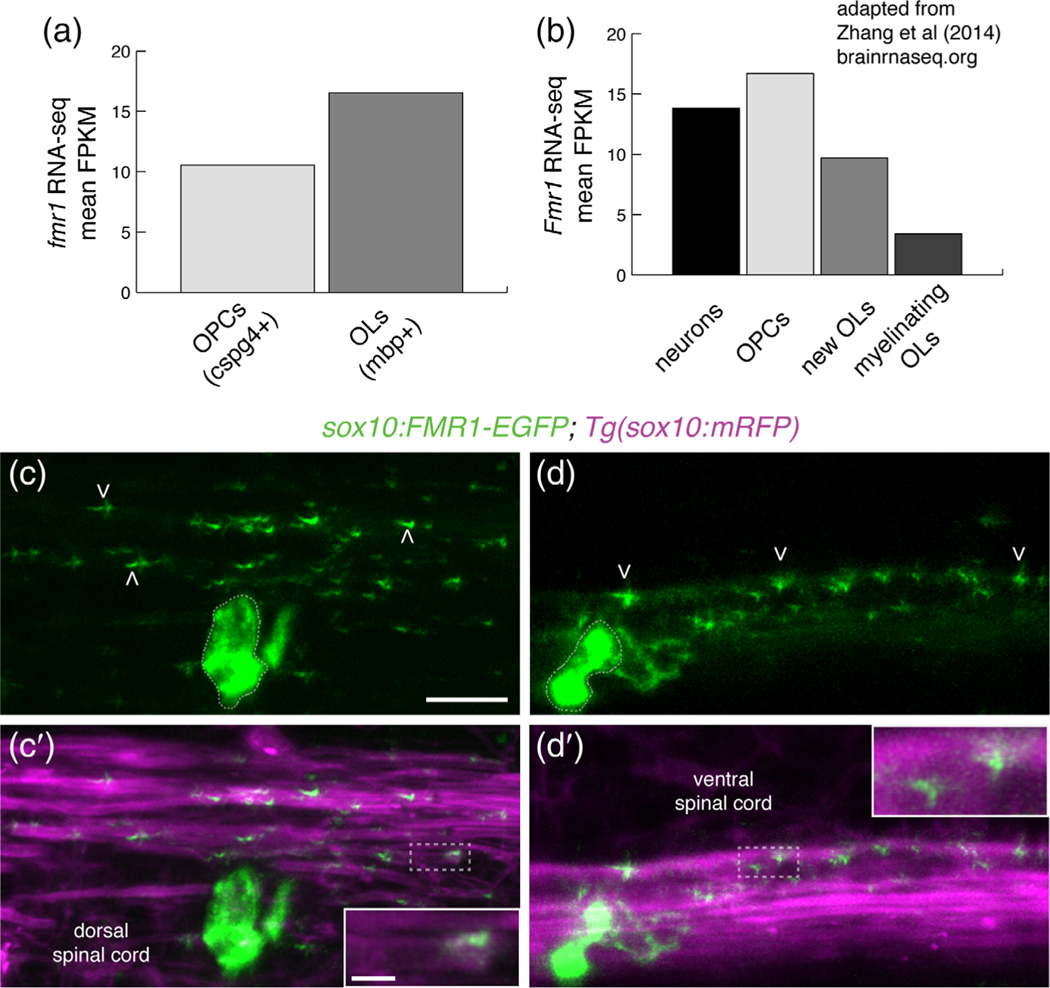
Although the bulk of FMRP studies have focused on neuronal development and function, oligodendrocyte lineage cells also express FMRP (Giampetruzzi et al., 2013; Wang et al., 2004). Our recent RNA-Seq study (Ravanelli et al., 2018) revealed that zebrafish oligodendrocyte precursor cells (OPCs) and myelinating oligodendrocytes express fmr1 (Figure 1a) and RNA-Seq analysis of cells isolated from mouse brain (Zhang et al., 2014) indicated that OPCs and newly formed oligodendrocytes express Fmr1 at levels comparable to neurons (Figure 1b). In order to achieve distal regulation of target mRNAs, FMRP must be localized to myelin sheaths. To determine the subcellular localization of FMRP in live zebrafish larvae, we used Tol2-based transgenesis to transiently express a human FMR1-EGFP fusion protein behind sox10 regulatory elements, which drive expression in OPCs and oligodendrocytes (Mathews et al., 2014). We injected sox10:FMR1-EGFP into 1-cell transgenic sox10:mRFP embryos expressing the oligodendrocyte membrane reporter sox10:mRFP (Kucenas et al., 2008) and subsequently examined individual cells at 4 dpf. In oligodendrocytes of the dorsal and ventral spinal cord, we noted bright FMR1-EGFP expression in cell bodies and more punctate expression in myelin sheaths (Figure 1c,d), including the terminal ends of RFP+ myelin sheaths (Figure 1c′,d′, inset). These data demonstrate subcellular localization of FMRP in myelin sheaths during early developmental myelination.
FIGURE 1.
Fragile X mental retardation protein (FMRP) is localized within nascent myelin sheaths. (a) Zebrafish fmr1 expression levels (fragments per kilobase of transcript per million mapped reads; FPKM) from RNA-Seq of FAC-sorted cspg4+ olig2+ OPCs and mbpa+ olig2+ oligodendrocytes (Ravanelli et al., 2018). (b) Murine Fmr1 expression (FPKM) from RNA-Seq of neurons and oligodendrocyte lineage cells (Adapted from Zhang et al., 2014; brainseq.org). (c-c′) Lateral images of a living 4 dpf Tg(sox10:mRFP) transgenic larva transiently expressing sox10: FMR1-EGFP, a human FMR1-EGFP fusion construct, in an oligodendrocyte in the dorsal spinal cord. FMR1-EGFP expression is highest in the cell body (dashed outline), with dimmer, punctate expression noted in sox10:mRFP+ myelin sheaths of the dorsal spinal cord (arrowheads), including the terminal ends of sheaths (c′, inset). (d,d′) An oligodendrocyte in the ventral spinal cord transiently expressing FMR1-EGFP in a single myelin sheath on a large diameter Mauthner axon. The fusion protein is widespread in the cell soma (dashed outline) and punctate throughout the myelin sheath (arrowheads). FMR1-EGFP co-localizes with the sox10:mRFP+ myelin membrane (d′ inset). Wide scale bar = 10 μm, inset scale bar = 2 μm [Color figure can be viewed at wileyonlinelibrary.com]
4.2 |. Global loss of FMRP leads to reduced myelin sheath growth and dynamics
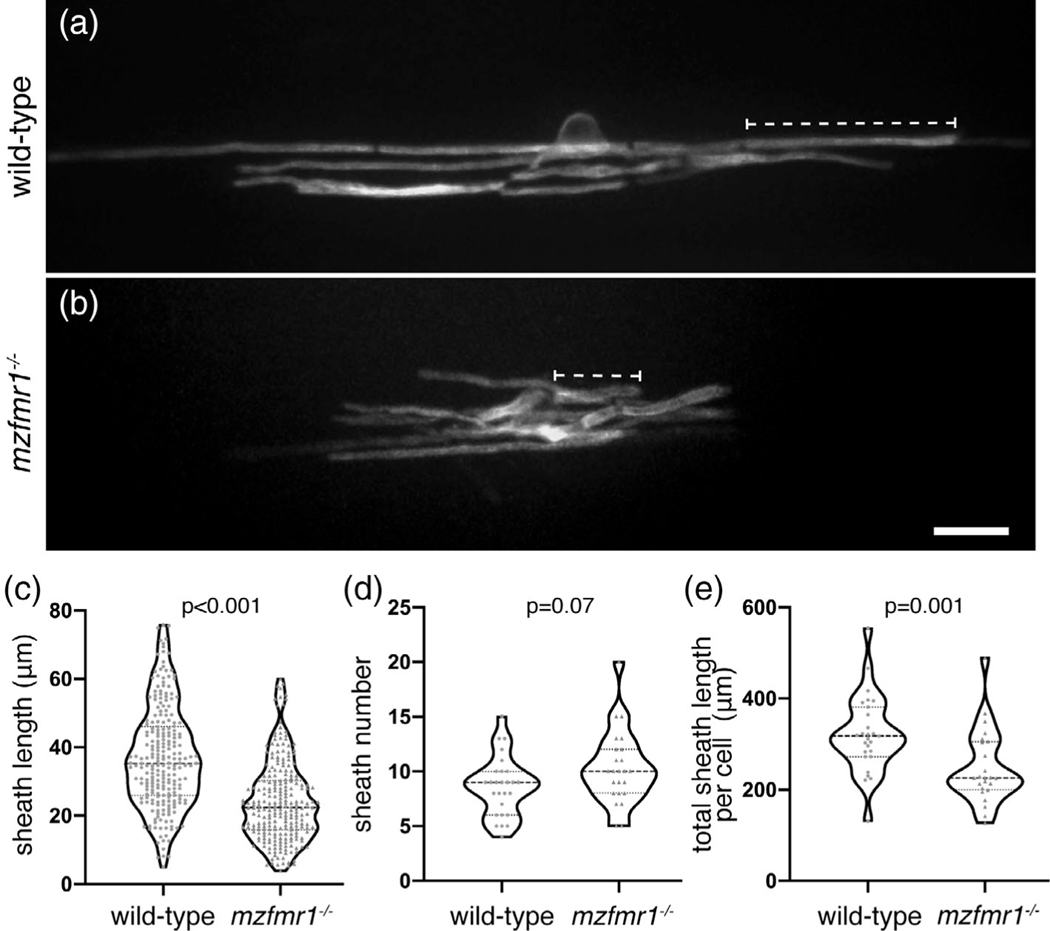
FMRP-deficient neurons display prominent abnormalities in synaptic structure, with immature dendritic spines representing the hallmark phenotype in FXS patients (Irwin, Galvez, & Greenough, 2000). Does FMRP also promote the maturation of myelin sheaths during development? With evidence of subcellular localization of FMRP within developing myelin sheaths, we next examined requirements for FMRP in oligodendrocyte development. Larvae homozygous for the loss-of-function fmr1hu2787 allele do not display any obvious physical defects, and grow into viable, fertile adults (den Broeder et al., 2009). To examine individual oligodendrocytes in the developing spinal cord, we mosaically expressed an oligodendrocyte-specific membrane tethered reporter, mbpa:EGFP-CAAX, which distinctly labels myelin sheaths (Hughes & Appel, 2019). We injected mbpa:EGFP-CAAX into 1-cell wild-type and maternal-zygotic fmr1hu278 (mzfmr1−/−) embryos, in which any potential maternal contribution of FMRP is absent. We then quantified the length of individual myelin sheaths at 4 dpf, finding that sheaths in mzfmr1−/− loss-of-function larvae were reduced ∼35% in length compared to wild-type (Figure 2c). Oligodendrocytes in mzfmr1−/− larvae formed approximately the same total number of sheaths per cell as wild-type (Figure 2d). Taken together, the cumulative sheath length per cell was dramatically reduced in the absence of FMRP (Figure 2e). With prominent deficits in overall myelin sheath growth at 4 dpf, these data suggest that FMRP promotes the growth of myelin sheaths.
FIGURE 2.
Global loss of FMRP leads to reduced myelin sheath length. Lateral images of oligodendrocytes in the spinal cord of living wild-type (a) and mzfmr1−/− (b) larvae labeled by mbp:EGFP-CAAX. The mbp:EGFP-CAAX reporter distinctly labels individual myelin sheaths (brackets). Average sheath length (c), sheaths per cell (d), and total cumulative sheath length (e) in wild-type and mzfmr1−/− loss-of-function mutants. Violin plot centerlines represent median values and hinges represent 25th and 75th percentiles, with all data points shown. Wild-type: n = 234 sheaths, 27 cells; mzfmr1−/−: n = 247 sheaths, 24 cells. Significance determined by Mann–Whitney tests. Scale bar = 10 μm
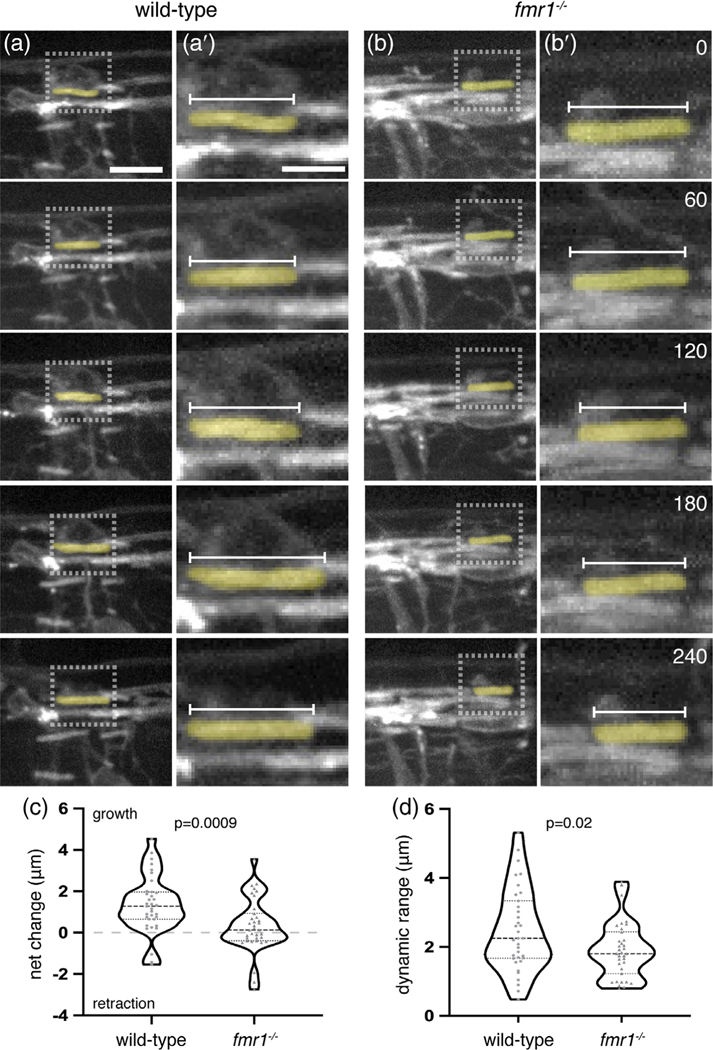
The short myelin sheaths noted in fmr1−/− larvae could result from deficient growth or from sheath instability and excess retraction. We examined these possibilities by capturing time-lapse images of the developing spinal cord at 3 dpf, when sheath development is highly dynamic in newly differentiated oligodendrocytes (Figure 3a,b). We recorded three-dimensional images of transgenic wild-type and fmr1−/− loss-of-function larvae expressing sox10:mRFP every 15 min for 4 hr. We then quantified the length of individual myelin sheaths at each time point and analyzed the overall growth or retraction (t240 − t0) as well as the dynamic range (maximum value – minimum value), the latter a measure of overall capacity for growth or retraction. During this 4-hr imaging window, we found that wild-type myelin sheaths gained an average of 1.34 ± 0.23 μm in length, whereas fmr1−/− mutant sheaths displayed very little positive growth (0.33 ± 0.22 μm; Figure 3c). In addition, wild-type sheaths were highly dynamic, with an overall average range of 2.47 ± 0.2 μm. In contrast, fmr1−/− sheaths displayed an average range of 1.9 ± 0.14 μm, a 23% reduction compared to wild-type (Figure 3d). In summary, these data indicate an oligodendrocyte requirement for FMRP in the highly dynamic phase of initial myelin sheath growth, with reduced sheath growth and dynamic capacity in fmr1−/− mutants.
FIGURE 3.
Fragile X mental retardation protein (FMRP) promotes dynamic myelin sheath growth. sox10: mRFP+ myelin sheaths in the dorsal spinal cord of transgenic Tg(sox10:mRFP) wild-type (a) and fmr1−/− mutant (b) larvae at 3 dpf. Lateral three-dimensional images of the spinal cord were captured every 15 min for 4 hr (each row represents an hourly time point; scale bar = 10 μm). (a′,b′) High magnification detail of individual sox10:mRFP+ myelin sheaths, inset in (a) and (b) (scale bar = 5 μm). Numbers in top right corners denote time elapsed from initial time point. (c) Change in average length of individual myelin sheaths (t240 − t0; wild-type = 35 sheaths, eight larvae; fmr1−/− = 35 sheaths, seven larvae; Mann–Whitney test). Positive values indicate sheath growth and negative values denote retraction from the initial time point. (d) Dynamic range of individual myelin sheaths (maximum value − minimum value; unpaired t test) [Color figure can be viewed at wileyonlinelibrary.com]
4.3 |. FMRP autonomously promotes myelin sheath growth
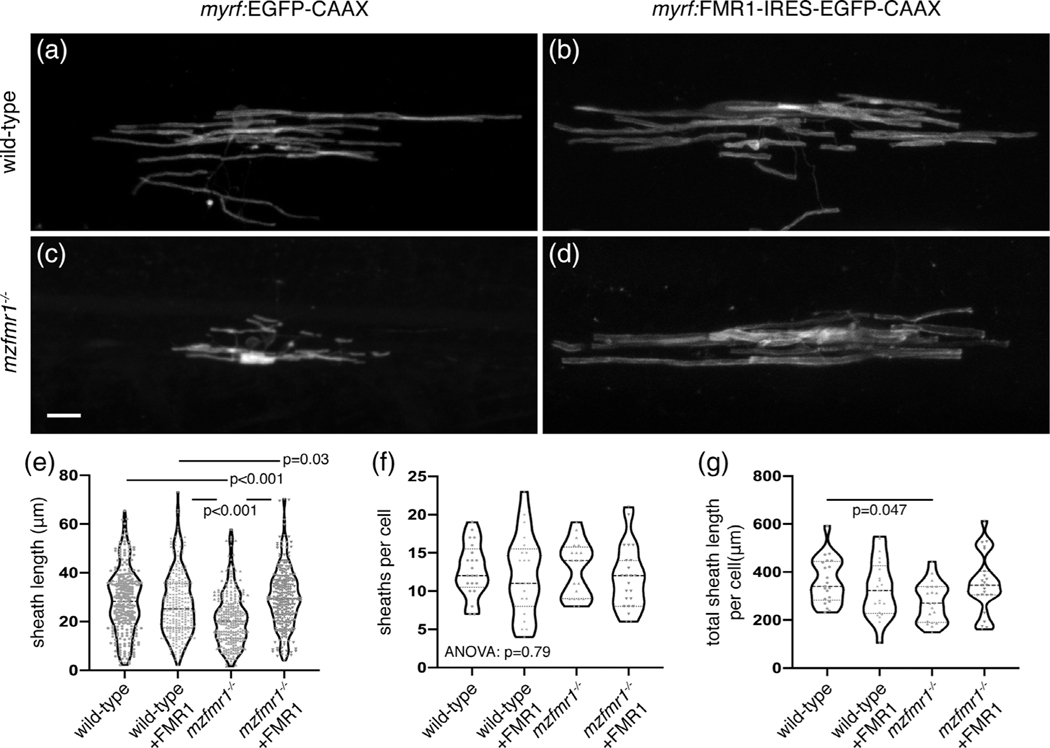
Because fmr1hu2787 mutants globally lack FMRP, deficient sheath growth could stem from dysfunction in neurons or oligodendrocytes. To test whether FMRP functions autonomously to promote myelin sheath growth, we transiently expressed human FMR1 using myrf regulatory DNA, which drives expression in differentiated oligodendrocytes (Hornig et al., 2013). In contrast to the fusion protein encoded by sox10:FMR1-EGFP presented in Figure 1, FMR1 and EGFP-CAAX are produced as distinct peptides from the myrf:FMR1-IRES-EGFP-CAAX construct. We injected control myrf:EGFP-CAAX and FMR1 plasmids in both wild-type and mzfmr1−/− loss-of-function embryos at the one-cell stage, and quantified myelin sheaths at 4 dpf. Similar to results shown in Figure 2, oligodendrocytes in mzfmr1−/− larvae expressing the control myrf:EGFP-CAAX reporter developed sheaths of diminished length compared to the wild-type control group (Figure 4a,c). Expression of FMR1-IRES-EGFP-CAAX in wild-type did not impact sheath development (Figure 4b). However, targeted FMR1-IRES-EGFP-CAAX expression in oligodendrocytes of mzfmr1−/− larvae restored individual and cumulative myelin sheath length (Figure 4d,e,g) without impacting the average number of sheaths per cell (Figure 4f). These results demonstrate an intrinsic oligodendrocyte-requirement for FMRP in sheath growth.
FIGURE 4.
Fragile X mental retardation protein (FMRP) autonomously promotes myelin sheath growth. Images of living wild-type (a) and mzfmr1−/− loss-of-function mutant (b) larvae expressing myrf:EGFP-CAAX in oligodendrocytes. Oligodendrocyte-targeted expression of FMR1-IRES-EGFP-CAAX in wild-type (c) and mzfmr1−/− mutant (d) larvae. Average sheath length (e), sheaths per cell (f), and cumulative sheath length (g) in each condition. Wild-type control: 268 sheaths, 21 cells; wild-type+FMR1: 255 sheaths, 21 cells; mzfmr1−/−: 257 sheaths, 20 cells; mzfmr1−/−+FMR1: 270 sheaths, 23 cells. Significance determined by Kruskal-Wallis test with Dunn’s multiple comparisons test for average sheath length, analysis of variance (ANOVA) for sheath number, and ANOVA with Tukey’s test for and cumulative sheath length. Scale bar = 10 μm
4.4 |. Loss of FMRP function leads to reduced membrane-associated Mbp protein expression but does not impact oligodendrocyte quantity
Oligodendrocytes in fmr1−/− mutants develop diminished myelin sheaths, which may suggest that FMRP regulates the expression of myelin proteins. We focused on Myelin basic protein (Mbp), encoded by mRNA that previous studies have shown to be a target of FMRP (Li, 2001; Wang et al., 2004). Although FMRP can repress translation of Mbp mRNA in vitro (Li, 2001; Wang et al., 2004), Mbp expression was reduced in early postnatal cerebellum of Fmr1 knockout mice (Pacey et al., 2013). We performed western blots to detect Mbp protein in whole larva lysates, and found comparable levels of the 21.5 kDa isoform, which is primarily cytoplasmic and nuclear-localized (Allinquant, Staugaitis, D’Urso, & Colman, 1991), in wild-type and mutant larvae. By contrast, mzfmr1−/− mutant larvae had less of the 14 and 18.5 kDa isoforms (Figure S1a,b), which localize to plasma membrane of oligodendrocytes in rodents (Allinquant et al., 1991). To investigate whether these isoform-specific changes in Mbp levels could result from changes in mbpa RNA splicing, we performed RT-PCR to detect mbpa mRNA splice variants (Figure S1c). This approach revealed no differences in the amount of four splice variants between wild-type and mutant larvae (Figure S1d,e), suggesting that FMRP does not regulate Mbp production by regulating RNA splicing.
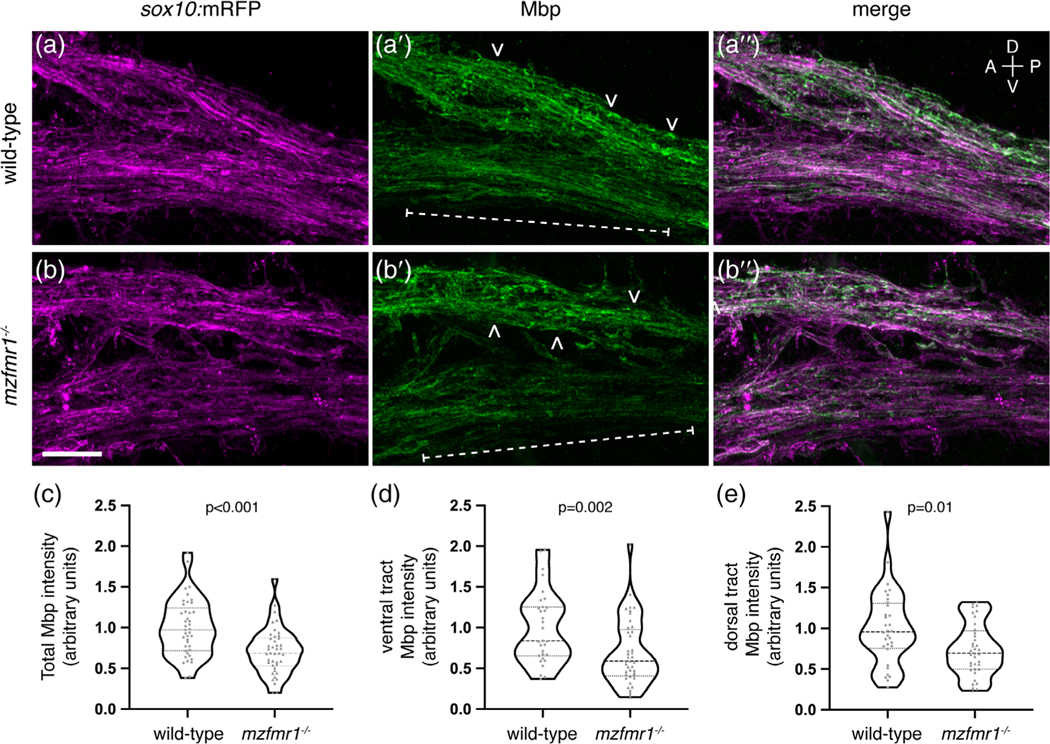
To further test whether FMRP regulates production of membrane-associated Mbp, we labeled sagittal sections of transgenic larvae expressing the membrane reporter sox10:mRFP with antibody against Mbp, focusing on the dense myelin tracts of the hindbrain (Figure 5a,b). Overall, we found that Mbp protein was reduced by ∼29% in fmr1−/− myelin tracts at 4 dpf (Figure 5c). Interestingly, we also noted a regional difference in Mbp expression, in that the ventral myelin tract was most profoundly impacted by the loss of FMRP (brackets; Figure 5d). In contrast, Mbp expression was more comparable in the dorsal myelin tract (arrowheads; Figure 5e). These results demonstrate reduced myelin-localized Mbp expression in fmr1−/− larvae, which suggests a crucial role for FMRP in the synthesis of an essential myelin membrane protein isoform.
FIGURE 5.
Loss of fragile X mental retardation protein (FMRP) function leads to reduced Mbp protein expression. Sagittal sections through the hindbrain of Tg(sox10:mRFP) wild-type (a-a″) and mzfmr1−/− loss-of-function mutant (b-b″) larvae, labeled with antibody against Mbp. Mbp strongly co-localizes with sox10:mRFP+ myelin sheaths in the hindbrain. Mbp expression was quantified in both ventral (brackets) and dorsal myelin tracts (arrowheads). (c) Cumulative normalized quantification of Mbp intensity (wild-type n = 41 sections, 30 larvae; mzfmr1−/− n = 42 sections, 30 larvae; unpaired t test). (d) Normalized Mbp intensity in ventral hindbrain myelin tracts (wild-type n = 31 sections, 26 larvae; mzfmr1−/− n = 41 sections, 28 larvae; Mann–Whitney test). (e) Normalized Mbp intensity in dorsal hindbrain myelin tracts (wild-type n = 32 sections, 26 larvae; mzfmr1−/− n = 37 sections, 28 larvae; Mann–Whitney test). mRNA abundance in mzfmr1−/− mutants was normalized to wild-type. Scale bar = 10 μm. A = anterior, P = posterior, D = dorsal, V = ventral [Color figure can be viewed at wileyonlinelibrary.com]
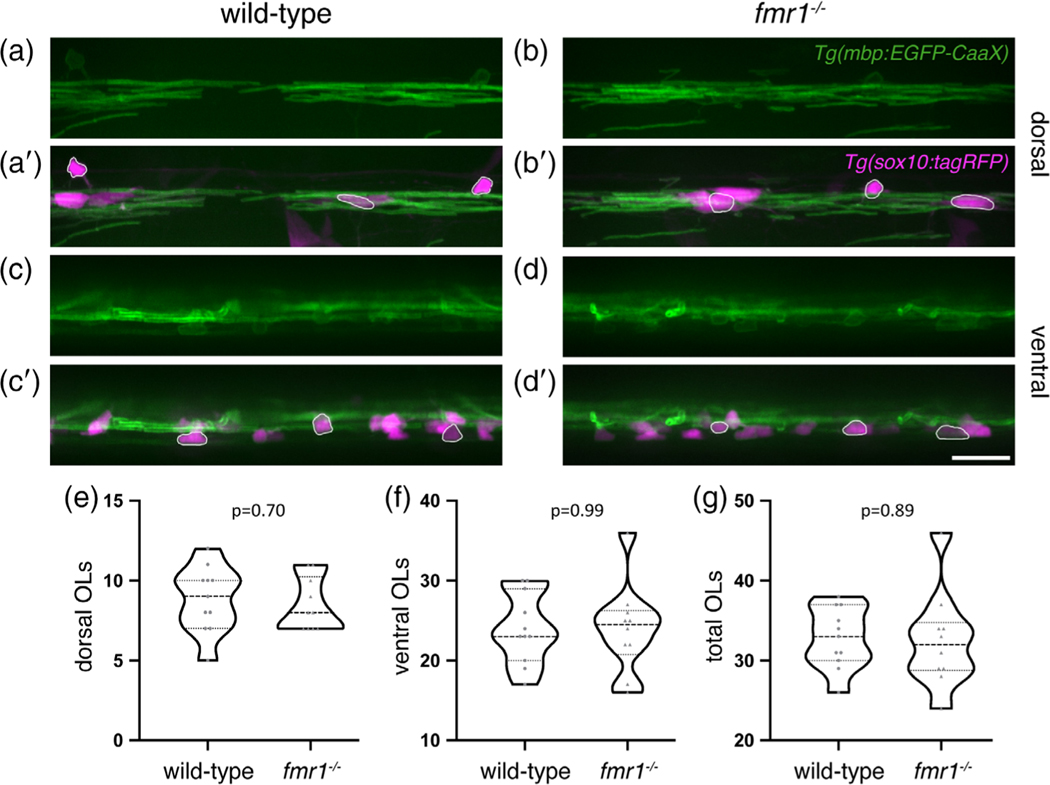
Reduced Mbp abundance in fmr1−/− myelin tracts could stem from deficient Mbp production in individual oligodendrocytes or from a reduction in the total number of cells. To investigate the latter possibility, we quantified the number of mature oligodendrocytes in the spinal cord of transgenic wild-type and fmr1−/− larvae expressing mbpa:EGFP-CAAX, and sox10:tagRFP, a cytosolic reporter of the oligodendrocyte lineage (Blasky et al., 2014) (Figure 6). We quantified the number of mbpa+/sox10+ cells in both the dorsal (Figure 6a,b′) and ventral (Figure 6c,d′) regions of the cord, finding comparable numbers between genotypes (Figure 6e,f). The cumulative number of mbpa+/sox10+ cells was also unaffected by FMRP loss (Figure 6g). These data indicate that FMRP does not regulate the quantity of oligodendrocytes in the developing spinal cord.
FIGURE 6.
Fragile X mental retardation protein (FMRP) does not affect oligodendrocyte quantity in the spinal cord. Lateral images of living transgenic Tg(mbp-EGFP-CAAX); Tg (sox10:tagRFP) wild-type and fmr1−/− larvae at 4 dpf. mbp+/sox10+ oligodendrocytes in the dorsal (a-b′) and ventral (c-d′) spinal cord (outlined examples in merged images). Quantification of dorsal (e), ventral (f), and cumulative oligodendrocytes (g) (wild-type n = 11 larvae; fmr1−/− n = 10 larvae). Statistical significance assessed using unpaired t tests. Scale bar = 20 μm; OL, oligodendrocyte [Color figure can be viewed at wileyonlinelibrary.com]
4.5 |. Loss of FMRP does not affect mbpa mRNA abundance in myelin tracts
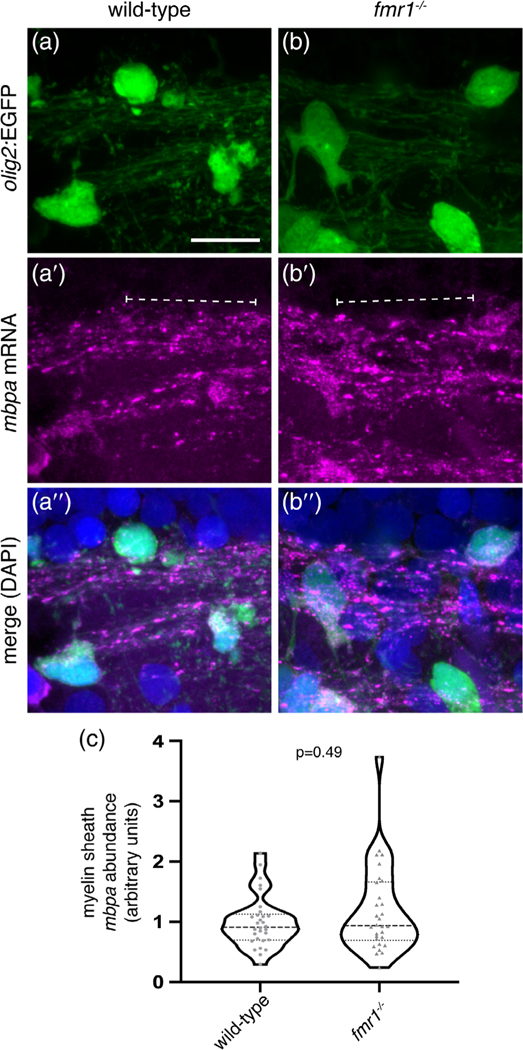
Because FMRP does not regulate the number of mature oligodendrocytes, reduced levels of Mbp could be due to faulty localization of mbp mRNA or from diminished Mbp translation. Because FMRP regulates subcellular localization of synaptic mRNAs (Dictenberg et al., 2008), it could play a similar role for mbp mRNA in oligodendrocytes. To test this possibility, we performed smFISH (Femino, Fay, Fogarty, & Singer, 1998; Raj, van den Bogaard, Rifkin, van Oudenaarden, & Tyagi, 2008) on 4 dpf wild-type and fmr1−/− Tg(olig2:EGFP) larvae. We quantified transcripts expressed by mbpa, a zebrafish ortholog of mouse Mbp, in individual olig2:EGFP+ myelin tracts in the hindbrain, which contained robust levels of mbpa (Figure 7a,b). We found that wild-type and fmr1−/− larvae contained comparable levels of mbpa transcript (brackets; Figure 7c). These data indicate that FMRP does not affect mbpa abundance in myelin tracts, the presumptive site of Mbp translation (Colman, Kreibich, Frey, & Sabatini, 1982).
FIGURE 7.
Loss of fragile X mental retardation protein (FMRP) does not affect mbpa mRNA abundance in myelin tracts. Single molecule fluorescent in situ hybridization to detect mbpa mRNA in the hindbrain of Tg(olig2:EGFP) wild-type (a) and fmr1−/− (b) larvae. Mbpa mRNA (a′,b′; brackets) is localized to olig2:EGFP+ myelin tracts (a″,b″). (c) Normalized quantification of mbpa mRNA abundance in hindbrain myelin tracts (wild-type n = 30 myelin segments, eight larvae; fmr1−/− n = 30 myelin segments, seven larvae). Statistical significance assessed with a Mann–Whitney test. mRNA abundance in fmr1−/− mutants was normalized to wild-type. Scale bar = 10 μm [Color figure can be viewed at wileyonlinelibrary.com]
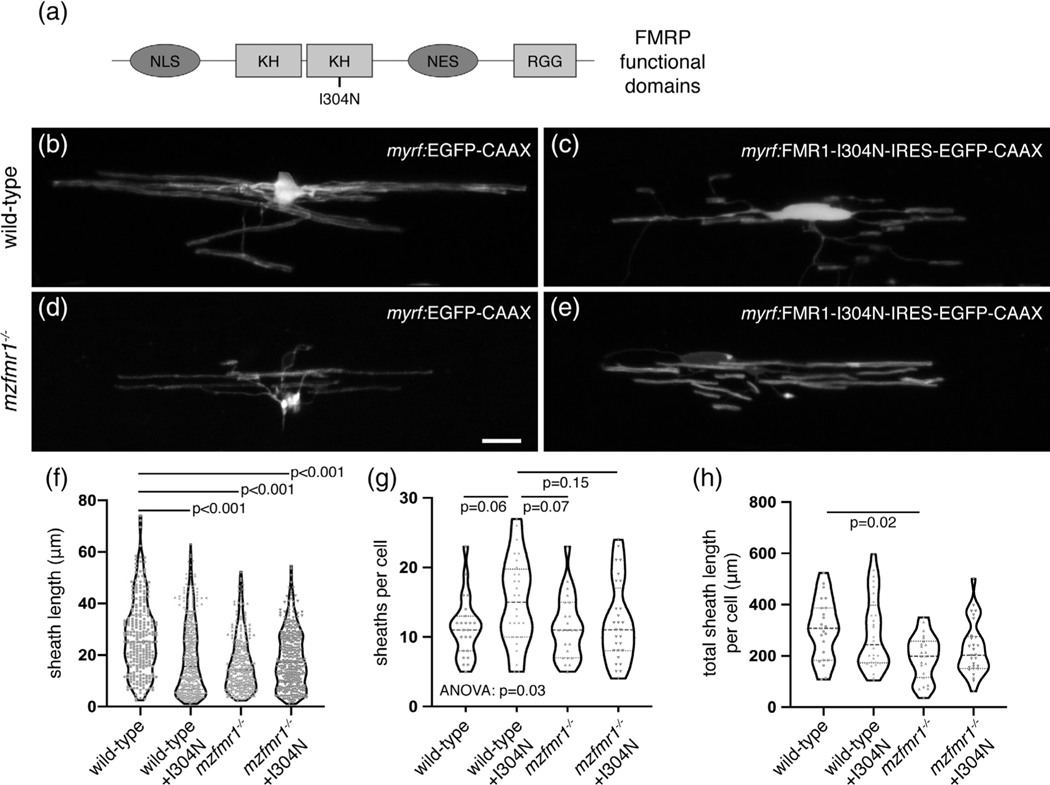
4.6 |. The second KH domain of FMRP is required for myelin sheath growth
FMRP contains several functional domains that mediate nuclear transport, RNA binding, and association in ribonucleoprotein (RNP) complexes (Figure 8a), and all of these functions could contribute to myelin sheath growth. Studies of the missense mutation FMR1-I304N have demonstrated that a hydrophobic region within the second KH domain is crucial for polyribosome association (Feng et al., 1997), FMRP homodimerization, and translational regulation (Laggerbauer, 2001), thereby pinpointing a domain that regulates translation. We therefore tested whether this domain is required for myelin sheath growth by expressing FMR1-I304N in oligodendrocytes (Ascano et al., 2012). We injected control myrf-EGFP-CAAX and myrf-FMR1-I304N-IRES-EGFP-CAAX constructs in wild-type and fmr1−/− one-cell embryos and examined oligodendrocyte morphology at 4 dpf (Figure 8b-e). We found that targeted expression of FMR1-I304N in fmr1−/− mutant oligodendrocytes failed to rescue the length of individual sheaths (Figure 8e,f), but cumulative sheath length was not significantly different than wild-type (p = .11; Figure 8h). Interestingly, expression of FMR1-I304N in wild-type oligodendrocytes led to a ∼30% reduction in individual myelin sheath length compared to controls (Figure 8c,f). Many cells in this group also developed supernumerary short branches (Figure 8g), but FMR1-I304N expression in wild-type did not impact cumulative sheath length (Figure 8h). These results demonstrate that the second KH2 domain of FMRP is essential for the growth of individual myelin sheaths but may not regulate total myelination capacity.
FIGURE 8.
The second KH domain of fragile X mental retardation protein (FMRP) is required for myelin sheath growth. (a) Domain structure of FMRP, including two KH regions and one RGG region (light gray boxes), as well as nuclear localization signal (NLS) and nuclear export signal (NES; dark gray boxes). The I304N missense mutation is located in the second KH domain. Lateral images of living wild-type (b) and mzfmr1−/− loss-of-function mutant (c) larvae expressing myrf:EGFP-CAAX in oligodendrocytes. Oligodendrocyte-targeted expression of FMR1-I304N-IRES-EGFP-CAAX in wildtype (d) and mzfmr1−/− mutant (e) larvae. Average sheath length (f), sheaths per cell (g), and cumulative sheath length (h) in each condition. Wild-type control: 259 sheaths, 23 cells; wild-type+FMR1: 422 sheaths, 28 cells; mzfmr1−/−: 260 sheaths, 23 cells; mzfmr1−/−+FMR1: 335 sheaths, 28 cells. Statistical significance assessed by Kruskal–Wallis test with Dunn’s multiple comparisons test for average sheath length and cumulative sheath length and analysis of variance with Tukey’s test for sheath number. Scale bar = 10 μm
5 |. DISCUSSION
FXS white matter abnormalities have been attributed to primary neuronal dysmorphia or dysfunction (Hoeft et al., 2010). We find that oligodendrocytes in fmr1−/− mutants develop individual myelin sheaths of reduced length as well as a dramatic decrease in total myelin. These phenotypes could result from changes in axon structure or physiology or an autonomous oligodendrocyte requirement for FMRP. Although we cannot completely rule out axonal contributions, our results suggest an intrinsic oligodendrocyte requirement, because targeted expression of FMR1 can rescue myelin sheath growth in fmr1−/− mutant larvae.
FMRP can regulate several crucial steps in the lifecycle of an mRNA, including localization (Dictenberg et al., 2008), stabilization (Zalfa et al., 2007), and translational control (Todd et al., 2003). At subcellular resolution, we show that the developing hindbrain myelin tracts of both wild-type and fmr1−/− mutant larvae contained comparable levels of mbpa mRNA. However, membrane-associated Mbp protein isoform expression was reduced in the absence of FMRP, and consistently, the myelin tracts of fmr1−/− larvae also showed reduced Mbp expression. Taken together, FMRP appears to promote Mbp translation in myelin sheaths, downstream of mRNA localization. Both in vivo studies run contradictory to prior in vitro demonstrations of translational repression of Mbp by FMRP (Wang et al., 2004), though it is important to note that FMRP does not always repress translation of mRNA targets: FMRP appears to promote translation of some N-methyl-D-aspartate (NMDA) receptor subunits, scaffolding proteins, and the immediate early gene Arc, all of which are reduced in prefrontal cortex of Fmr1 knockout mice (Krueger, Osterweil, Chen, Tye, & Bear, 2011). Intriguingly, many of these targets are also expressed in the oligodendrocyte lineage (Zhang et al., 2014), and could contribute to myelin sheath growth and stabilization. FMRP plays various roles in mRNA regulation and translational control and uncovering precise in vivo requirements in developing subcellular compartments remains challenging.
Our future work will investigate the additional functional domains of FMRP to better pinpoint regions that determine localization, stabilization, and translational regulation, along with the specific target sites on mRNAs that are bound by these domains. FMRP may act to stabilize and prevent decay of mbpa, as detected in neuron–glia co-cultures (Zalfa et al., 2007). Association of Mbp mRNA with an additional RBP, Quaking (Qki), appears to localize Mbp mRNA (Li, Zhang, Li, & Feng, 2000), which could explain why myelin-localized mbpa abundance was unaffected by the lack of FMRP. FMRP could also function more directly in translational regulation of sheath-promoting mRNAs within larger RNP complexes (Feng et al., 1997; Zang et al., 2009), as introduction of FMR1-I304N fails to rescue the growth of individual sheaths in fmr1−/− mutants and induces reduced sheath growth in wild-type. Therefore, our data predict that translational control underlies the specific FMRP contribution to individual sheath growth. The precise dissection of FMRP function in mRNA regulation will require careful in vivo examinations of both FMRP binding domains and the requisite sequence motifs on mRNA targets.
Taken together, our experiments indicate a vital role for FMRP in the growth of nascent myelin sheaths. As myelination is influenced by axonal activity (Hines, Ravanelli, Schwindt, Scott, & Appel, 2015; Mensch et al., 2015), we speculate that the short myelin sheaths noted in fmr1−/− mutants are analogous to the immature dendritic spines noted in FXS patients and animal models (Comery et al., 1997; Irwin et al., 2000); FMRP may serve as part of an activity-dependent mechanism that responds to axonal activity and initiates ensheathment. Our time-lapse experiments indicate that FMRP promotes sheath growth and dynamics during the height of axonal ensheathment by oligodendrocytes. These complex processes likely require a host of proteins, many of which may be regulated by FMRP as mRNA targets (Darnell & Klann, 2013). Our future work will explore additional FMRP targets driving sheath growth, including mRNA candidates that encode synaptogenic proteins. FMRP binds dozens of transcripts encoding synaptic proteins (Ascano et al., 2012; Darnell et al., 2011), many of which are also expressed in the oligodendrocyte lineage (Zhang et al., 2014). Interestingly, our laboratory has shown that disruption of synaptic protein function in oligodendrocytes can lead to excess myelin sheaths of diminished length (Hughes & Appel, 2019), which is comparable to FMR1-I304N overexpression (Figure 8). Shared mechanisms of mRNA regulation in both neurons and glial would only expand the essential roles for RBPs in neural circuit development.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Lisbet Finseth for Western blot expertise, Howard Sirotkin for the fmr1hu2787 allele, and Matthew Taliaferro for insightful conversation. This work was supported by US National Institute of Health (NIH) grant R01 NS095679 and a gift from the Gates Frontiers Fund to B.H.A. and NIH grant R21 NS110213 to C.A.D. and B.H.A. The University of Colorado Anschutz Medical Campus Zebrafish Core Facility was supported by NIH grant P30 NS048154.
Funding information
National Institute of Neurological Disorders and Stroke, Grant/Award Numbers: R01 NS095679, R21 NS110213; Gates Frontiers Fund; US National Institute of Health, Grant/Award Number: P30 NS048154
Footnotes
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
REFERENCES
- Allinquant B, Staugaitis SM, D’Urso D, & Colman DR (1991). The ectopic expression of myelin basic protein isoforms in shiverer oligodendrocytes: Implications for myelinogenesis. The Journal of Cell Biology, 113, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RG, Czopka T, Ffrench-Constant C, & Lyons DA (2011). Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development, 138, 4443–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, … Tuschl T. (2012). FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature, 492, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis E. (2002). Epilepsy in fragile X syndrome. Developmental Medicine and Child Neurology, 44, 724–728. [DOI] [PubMed] [Google Scholar]
- Blasky AJ, Pan L, Moens CB, & Appel B. (2014). Pard3 regulates contact between neural crest cells and the timing of Schwann cell differentiation but is not essential for neural crest migration or myelination. Developmental Dynamics, 243, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman DR, Kreibich G, Frey AB, & Sabatini DD (1982). Synthesis and incorporation of myelin polypeptides into CNS myelin. The Journal of Cell Biology, 95, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, & Greenough WT (1997). Abnormal dendritic spines in fragile X knockout mice: Maturation and pruning deficits. Proceedings of the National Academy of Sciences of the United States of America, 94, 5401–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro L, Ballinger E, Hagerman R, & Hessl D. (2010). Clinical assessment of DSM-IV anxiety disorders in fragile X syndrome: Prevalence and characterization. Journal of Neurodevelopmental Disorders, 3, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, & Klann E. (2013). The translation of translational control by FMRP: Therapeutic targets for FXS. Nature Neuroscience, 16, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, … Darnell RB (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell, 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Broeder MJ, van der Linde H, Brouwer JR, Oostra BA, Willemsen R, & Ketting RF (2009). Generation and characterization of Fmr1 knockout zebrafish. PLoS One, 4, 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, & Bassell GJ (2008). A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Developmental Cell, 14, 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson NR, Zhou Y-X, Flint NC, & Armstrong RC (2008). Musashi1 RNA-binding protein regulates oligodendrocyte lineage cell differentiation and survival. Glia, 56, 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll CA, Vita DJ, & Broadie K. (2017). Fragile X mental retardation protein requirements in activity-dependent critical period neural circuit refinement. Current Biology, 27, 2318–2330.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole TA, Chen Q, Justice MJ, & Artzt K. (1996). The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nature Genetics, 12, 260–265. [DOI] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, & Singer RH (1998). Visualization of single RNA transcripts in situ. Science, 280, 585–590. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, & Warren ST (1997). FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Molecular Cell, 1, 109–118. [DOI] [PubMed] [Google Scholar]
- Feuge J, Scharkowski F, Michaelsen-Preusse K, & Korte M. (2019). FMRP modulates activity-dependent spine plasticity by binding Cofilin1 mRNA and regulating localization and local translation. Cerebral Cortex, 1–13. Available at https://academic.oup.com/cercor/article-abstract/doi/10.1093/cercor/bhz059/5429722/?redirectedFrom=fulltext [DOI] [PubMed] [Google Scholar]
- Giampetruzzi A, Carson JH, & Barbarese E. (2013). FMRP and myelin protein expression in oligodendrocytes. Molecular and Cellular Neurosciences, 56, 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SW, Hessl D, Goodlin-Jones B, Ferranti J, Bacalman S, Barbato I, … Hagerman RJ (2008). Autism profiles of males with fragile X syndrome. American Journal of Mental Retardation, 113, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann G, & Gerster T. (2000). Multicolor whole-mount in situ hybridization. Methods in Molecular Biology, 137, 139–148. [DOI] [PubMed] [Google Scholar]
- Hines JH, Ravanelli AM, Schwindt R, Scott EK, & Appel B. (2015). Neuronal activity biases axon selection for myelination in vivo. Nature Neuroscience, 18, 683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F, Carter JC, Lightbody AA, Cody Hazlett H, Piven J, & Reiss AL (2010). Region-specific alterations in brain development in one- to three-year-old boys with fragile X syndrome. Proceedings of the National Academy of Sciences, 107, 9335–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig J, Fröb F, Vogl MR, Hermans-Borgmeyer I, Tamm ER, & Wegner M. (2013). The transcription factors Sox10 and Myrf define an essential regulatory network module in differentiating oligodendrocytes. PLoS Genetics, 9, e1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, & Bear MF (2002). Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proceedings of the National Academy of Sciences, 99, 7746–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A, & Appel B. (2019). Oligodendrocytes express synaptic proteins that modulate myelin sheath formation. Nature Communications, 10, 4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, & Greenough WT (2000). Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cerebral Cortex, 10, 1038–1044. [DOI] [PubMed] [Google Scholar]
- Kao D-I, Aldridge GM, Weiler IJ, & Greenough WT (2010). Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proceedings of the National Academy of Sciences, 107, 15601–15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, & Schilling TF (1995). Stages of embryonic development of the zebrafish. Developmental Dynamics, 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, & Appel B. (2006). In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nature Neuroscience, 9, 1506–1511. [DOI] [PubMed] [Google Scholar]
- Krueger DD, Osterweil EK, Chen SP, Tye LD, & Bear MF (2011). Cognitive dysfunction and prefrontal synaptic abnormalities in a mouse model of fragile X syndrome. Proceedings of the National Academy of Sciences, 108, 2587–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucenas S, Takada N, Park HC, Woodruff E, Broadie K, & Appel B. (2008). CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nature Neuroscience, 11, 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucenas S, Wang W-D, Knapik EW, & Appel B. (2009). A selective glial barrier at motor axon exit points prevents oligodendrocyte migration from the spinal cord. The Journal of Neuroscience, 29, 15187–15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, … Bin CC (2007). The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Developmental Dynamics, 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B. (2001). Evidence that fragile X mental retardation protein is a negative regulator of translation. Human Molecular Genetics, 10, 329–338. [DOI] [PubMed] [Google Scholar]
- Li Z. (2001). The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Research, 29, 2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Li D, & Feng Y. (2000). Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. The Journal of Neuroscience, 20, 4944–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimova A, Itzkovitz S, Junker JP, Fan ZP, Wu X, & Van Oudenaarden A. (2013). Single-molecule mRNA detection and counting in mammalian tissue. Nature Protocols, 8, 1743–1758. [DOI] [PubMed] [Google Scholar]
- Martin KC, Zhao Y, Kim SM, Wang DO, Sossin WS, Miura SK, & Hwang H. (2009). Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science, 324, 1536–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews ES, Mawdsley DJ, Walker M, Hines JH, Pozzoli M, & Appel B. (2014). Mutation of 3-hydroxy-3-methylglutaryl CoA synthase I reveals requirements for isoprenoid and cholesterol synthesis in oligodendrocyte migration arrest, axon wrapping, and myelin gene expression. The Journal of Neuroscience, 34, 3402–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensch S, Baraban M, Almeida R, Czopka T, Ausborn J, El Manira A, & Lyons DA (2015). Synaptic vesicle release regulates myelin sheath number of individual oligodendrocytes in vivo. Nature Neuroscience, 18, 628–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtie JC, Macklin WB, & Corfas G. (2007). Morphometric analysis of oligodendrocytes in the adult mouse frontal cortex. Journal of Neuroscience Research, 85, 2080–2086. [DOI] [PubMed] [Google Scholar]
- Ng MC, Yang YL, & Lu KT (2013). Behavioral and synaptic circuit features in a zebrafish model of fragile X syndrome. PLoS One, 8, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolin SL, Brown WT, Glicksman A, Houck GE Jr., Gargano AD, Sullivan A, … Sherman SL (2003). Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. American Journal of Human Genetics, 72, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, & Sato TN (2015). Whole-mount single molecule FISH method for zebrafish embryo. Scientific Reports, 5, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacey LKK, Xuan ICY, Guan S, Sussman D, Henkelman RM, Chen Y, … Hampson DR (2013). Delayed myelination in a mouse model of fragile X syndrome. Human Molecular Genetics, 22, 3920–3930. [DOI] [PubMed] [Google Scholar]
- Pan L, Zhang YQ, Woodruff E, & Broadie K. (2004). The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Current Biology, 14, 1863–1870. [DOI] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, & Tyagi S. (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods, 5, 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanelli AM, Kearns CA, Powers RK, Wang Y, Hines JH, Donaldson MJ, & Appel B. (2018). Sequential specification of oligodendrocyte lineage cells by distinct levels of hedgehog and notch signaling. Developmental Biology, 444, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park HC, Topczewska JM, Madwsley DJ, & Appel B. (2003). Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods in Cell Science, 25, 7–14. [DOI] [PubMed] [Google Scholar]
- Steward O, & Worley PF (2001). A cellular mechanism for targeting newly synthesized mRNAs to synaptic sites on dendrites. Proceedings of the National Academy of Sciences, 98, 7062–7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K, Hatton D, Hammer J, Sideris J, Hooper S, Ornstein P, & Bailey D. (2006). ADHD symptoms in children with FXS. American Journal of Medical Genetics - Part A, 140, 2275–2288. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, & Warren ST (1992). DNA methylation represses FMR-1 transcription in fragile X syndrome. Human Molecular Genetics, 1, 397–400. [DOI] [PubMed] [Google Scholar]
- Swanson MR, Wolff JJ, Shen MD, Styner M, Estes A, Gerig G, … Hazlett HC (2018). Development of white matter circuitry in infants with fragile X syndrome. JAMA Psychiatry, 75, 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Tosar LJM, Loschi M, Pasquini JM, Correale J, Kindler S, & Boccaccio GL (2005). Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Molecular Biology of the Cell, 16, 405–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, & Malter JS (2003). The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proceedings of the National Academy of Sciences, 100, 14374–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ku L, Osterhout DJ, Li W, Ahmadian A, Liang Z, & Feng Y. (2004). Developmentally-programmed FMRP expression in oligodendrocytes: A potential role of FMRP in regulating translation in oligodendroglia progenitors. Human Molecular Genetics, 13, 79–89. [DOI] [PubMed] [Google Scholar]
- Yergert KM, Hines JH, & Appel B. (2019). Neuronal activity enhances mRNA localization to myelin sheaths during development. bioRxiv, 1–30. [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, Di Penta A, … Bagni C. (2007). A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nature Neuroscience, 10, 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, … Darnell RB (2009). A mouse model of the human fragile X syndrome I304N mutation. PLoS Genetics, 5, e1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, … Wu JQ (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of Neuroscience, 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.