Abstract
Potato cyst nematodes (PCN) from the genus Globodera spp. cause major losses in the potato (Solanum tuberosum) industry worldwide. Despite their importance, at present little is known about the status of this plant pathogen in cultivated potatoes in Colombia. In this study, a total of 589 samples collected from 75 geographic localities in nine potato producing regions of Colombia (Cundinamarca, Boyacá, Antioquia, Nariño, Santander, Norte de Santander, Tolima, Caldas and Cauca) were assayed for the presence of potato cyst nematodes. Fifty-seven percent of samples tested positive for PCN. Based on phylogenetic analysis of the internal transcribed spacer region (ITS1-5.8S-ITS2) of the rRNA gene and D2-D3 expansion segments of the 28S rRNA gene, all populations but one were identified as Globodera pallida. Sequences of G. pallida from Colombia formed a monophyletic group closely related to Peruvian populations, with the lowest average number of nucleotide substitutions per site (Dxy = 0.002) and net nucleotide substitutions per site (Da = 0.001), when compared to G. pallida populations from Europe, South and North America. A single sample formed a well-supported subclade along with G. rostochiensis and G. tabacum from Japan, USA and Argentina. To our knowledge this is the first comprehensive survey of Globodera populations from Colombia that includes genetic data. Our findings on species diversity and phylogenetic relationships of Globodera populations from Colombia may help elucidate the status and distribution of Globodera species, and lead to the development of accurate management strategies for the potato cyst nematodes.
Introduction
The cyst nematodes, Globodera Skarbilovich, 1959, are one of the most limiting plant parasitic nematodes around the world [1]. Within the genus, thirteen species have been identified, of which G. rostochiensis, G. pallida, G. ellingtonae, and G. tabacum are important for agriculture [2]. The potato cyst nematodes (PCN), Globodera rostochiensis (golden or yellow potato cyst nematode) and Globodera pallida (pale potato cyst nematode) cause major losses in potato (Solanum tuberosum L.) crops [3], and are also considered as official control pests in many countries [4]. These species cause damage to the potato plants, by penetrating and feeding into the root tissue, which causes nutritional and water deficiency that is expressed in chlorosis and wilting of the leaves, and may also cause low growth, dwarfism and proliferation of small lateral roots that lead to yield reduction [4]. If PCN species are left uncontrolled they may reduce potato yield up to 80% [5,6], representing major economic losses in the potato industry worldwide.
Identification of Globodera species based on morphological characterization of the perineal area of cysts (e.g. distance from vulva and anus and Granek’s ratio) and some characters of the second stage juvenile (e.g. stylet length and stylet knob shape) [4,7] may be ambiguous. Morphometric measurements of these characters often show overlap among species, making morphological identification of cyst nematodes time consuming and difficult, especially when differentiating G. pallida from G. rostochiensis and G. tabacum species complex (G. tabacum tabacum, G. tabacum solanacearum and G. tabacum virginiae) [8,9]. Therefore, molecular diagnosis is a necessary and recommended complement to identify cyst nematode species [4].
For plant-parasitic nematodes, molecular diagnostics not only improve speed and accuracy of nematode identification, but also have allowed a better understanding of the biology of nematodes as agricultural pests [10]. The genomic regions more often used to study phylogenetic relationships for plant-parasitic nematodes include DNA fragments from the 28S ribosomal DNA (rDNA), internal transcribed spacer (ITS), as well as mitochondrial DNA (mtDNA) [2,10–15]. Ribosomal genes exhibit enough conserved inter-specific neutral genetic variation as to inform species delimitation without being prone to marker saturation [15–18]. For cyst nematodes identification, although several methods have been used, DNA-based approaches have shown to be more accurate to separate G. pallida from G. rostochiensis and other Globodera species and, ribosomal regions have also shown to be useful markers to distinguish species within the genus [12,17,19–21]. For new occurrences of Globodera spp., sequencing of DNA fragments is also recommended, especially for regions where genetic data has not been reported before and for PCN species that may not follow a typical profile [17,22]. For Globodera species from Colombia, genetic information including validation of currently available diagnostic DNA markers and molecular phylogenetics have not been documented.
In Colombia, G. pallida was first identified based on morphological characters in 1970 in Cumbal (municipality located in the Nariño department), at the south west extreme of the country [23]. In 1971, the species was regulated under the authority of The Instituto Colombiano Agropecuario (ICA) and listed as quarantine pest, limiting the access to export potato seeds from Nariño and its neighbor department, Cauca, to other potato producing regions of Colombia. In 1983, Nieto [24] conducted an intensive PCN survey and reported G. pallida in other municipalities of Nariño (Túquerres, Pupiales, Ipiales, Gualmatán, Sapuyes, among others), as well as in Cauca (Totoró, Cajibío, Silvia, Popayán, Páez, among others), with an average of 50–80 cysts/100 g of soil in Nariño and 9–10 cysts/100 g of soil in Cauca. The authors also sampled the nematode in Cundinamarca and Boyacá, the main potato producing departments in Colombia, and other minor producing potato regions such as Caldas, Tolima, Valle del Cauca, Santander and Norte de Santander, but only reported the presence of PCN in Nariño and Cauca. In 2004, the species was no longer listed as an official control pest. Yet, in a survey conducted from 2011 to 2012, PCN was reported in 12 out of 14 sampled fields in Tunja, Samacá and Ventaquemada (municipalities of Boyacá department) and Tausa, Tabio and Zipaquirá (municipalities of Cundinamarca department), although population densities were not registered [25]. Therefore, PCN was considered as a re-emerging pathogen in 2012 by the Federación Colombiana de Productores de Papa (Colombian Federation of Potato Producers–FEDEPAPA), and ICA [25].
To obtain better knowledge about Globodera spp. associated with potato crops in Colombia, it is necessary to develop DNA sequence information to better characterize populations from different geographic regions and to understand their distribution patterns. This information will also serve as a foundation to the design of effective control measures that require fast and accurate identification of species, and it is a crucial factor when searching for possible sources of host-plant resistance as well as for other management strategies. Therefore, the objectives of this study were to: i) survey the Globodera spp. populations detected in cultivated potatoes in Colombia; ii) carry out a molecular characterization of these Globodera populations based on sequences of the ITS1 of rRNA, partial 18S rRNA and, D2-D3 expansion segments of the 28S nuclear ribosomal RNA gene; and iii) study the phylogenetic relationships of Globodera spp. from Colombia by comparison with previously published molecular data of populations from other regions of the world.
Materials and methods
Ethics statement
Nematode sampling was performed under a collection permit granted by the Autoridad Nacional de Licencias Ambientales (ANLA) [Colombian National Authority Environmental Permits]: “Permit for collecting specimens of wild species of the biological diversity for non-commercial scientific research purposes”, resolution No. 1466, expedited on December 3, 2014.
Nematode populations and sampling
From 2013 to 2017, an extensive survey was conducted throughout the main commercial potato producing regions of Colombia. A total of 589 sampling sites were selected in 75 geographic localities using a stratified sampling strategy. The strata were defined as the departments (country subdivisions) with the highest potato growing area reported in Colombia [26], for a total of nine departments sampled: Cundinamarca, Boyacá, Antioquia, Nariño, Santander, Norte de Santander, Tolima, Caldas and Cauca (Fig 1, Table 1). At each department, the number of fields sampled per municipality was proportional to the potato area planted and fields at each municipality were selected based on established potato crops in pre-flowering and flowering stages (Fig 1, Table 1). Soil samples at each field were collected from within rows, at roughly equal intervals in a line transect pattern across an area of 10,000 m2 or less. A soil sample consisted of 60 soil cores (1.5 cm in diameter by 5 cm deep) taken near the root of the plants. Infected roots and surrounding soil of samples collected from each field were pooled into one composite sample. Samples were placed into plastic bags, transported to the laboratory of microbiology at the Corporación Colombiana de Investigación Agropecuaria (AGROSAVIA), Tibaitatá Research Center, in Mosquera, Cundinamarca, and stored at 4°C until processing. Cyst nematodes were extracted from soil samples using the Fenwick’s method [27], and cyst individuals per 100 cm3 of soil were counted and morphologically identified using the keys by Handoo et al. and Golden [7,28]. Population density was calculated as the average number of cysts per 100 cm3 of soil, population range as the minimum and maximum densities of cysts per 100 cm3 of soil; prevalence (%) was estimated by the formula: number of samples with cysts/ total number of samples collected)*100. Additionally, a viability test was performed by randomly selecting 10 cyst per population that were crushed using a Huijsman’s homogenizer [29] to release eggs and juveniles, alive eggs and j1 were counted under the stereoscope and viability percentage was calculated per population.
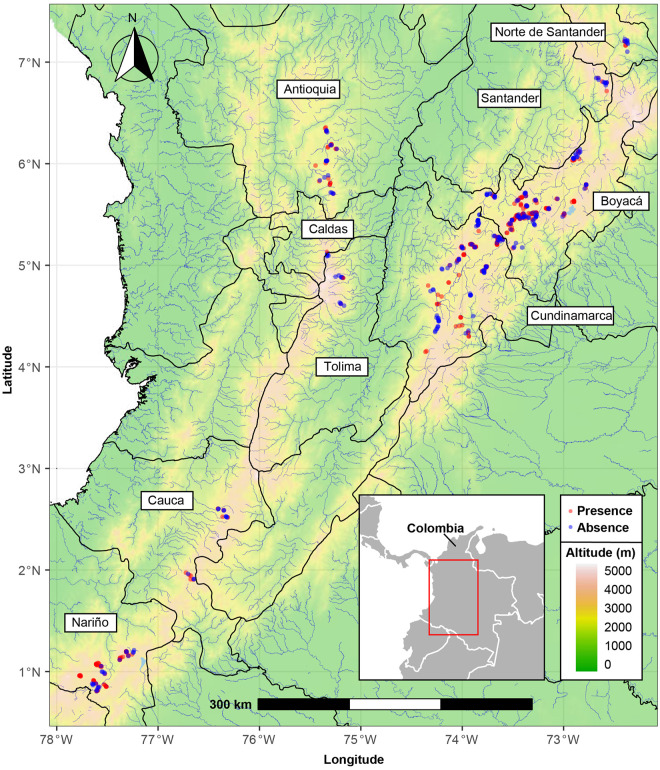
Fig 1. Map of the Colombian Andes showing sampling sites of PCN associated with potato crops in Colombia.
Black lines represent department limits, blue lines represent rivers and lakes. Colors according to elevation map. Red dots mark the position of the sample sites that tested positive for PCN and blue dots mark the position of the sampling sites that tested negative for PCN. This figure has been adapted from a base map and data from OpenStreetMap and OpenStreetMap Foundation, original copyright 2020. ©OpenStreetMap contributors. Please notice that this figure is similar but not identical to the original image and is therefore for illustrative purposes only.
Table 1. Globodera species, density levels, and prevalence (%) in cultivated potato crops from the main growing regions in Colombia.
| Department | Municipality | Species | # of samples collected | # of samples with Globodera spp. | Prevalencea (%) | Averageb/100 cm3 soil | Population rangec/100 cm3 soil | Viabilityd average (%) | Potato variety |
|---|---|---|---|---|---|---|---|---|---|
| Nariño | Túquerres | G. pallida | 12 | 12 | 2.04 | 97.3 | 1–345 | 60 | Diacol Capiro |
| Nariño | Ipiales | G. pallida | 10 | 5 | 0.85 | 9.1 | 2–45 | 41 | Diacol Capiro |
| Nariño | Pasto | G. pallida | 15 | 9 | 1.53 | 14.7 | 1–64 | 60 | Diacol Capiro |
| Nariño | Yacuanquer | G. pallida | 2 | 2 | 0.34 | 3.5 | 3–4 | 45 | Criolla |
| Nariño | Tangua | G. pallida | 5 | 4 | 0.68 | 11 | 1–34 | 55 | Diacol Capiro |
| Nariño | Ospina | G. pallida | 5 | 4 | 0.68 | 47.4 | 5–93 | 38 | Diacol Capiro |
| Nariño | Iles | G. pallida | 5 | 1 | 0.17 | 16 | 1–80 | 59 | Diacol Capiro |
| Nariño | Guachucal | G. pallida | 7 | 7 | 1.19 | 21.3 | 1–84 | 63 | Betina |
| Nariño | Pupiales | G. pallida | 11 | 6 | 1.02 | 5.9 | 1–29 | 30 | Diacol Capiro |
| Nariño | Córdoba | G. pallida | 7 | 7 | 1.19 | 9.4 | 1–27 | 44 | Pastusa Suprema |
| Cundinamarca | Chipaque | G. pallida | 4 | 4 | 0.68 | 68.3 | 2–168 | 51 | Pastusa Suprema |
| Cundinamarca | Madrid | G. pallida | 5 | 2 | 0.34 | 1.8 | 2–7 | 63 | Diacol Capiro |
| Cundinamarca | Pasca | - | 6 | 0 | - | - | - | - | Pastusa Suprema |
| Cundinamarca | Sesquilé | G. pallida | 9 | 5 | 0.85 | 10.2 | 1–25 | 62 | Pastusa Suprema |
| Cundinamarca | Subachoque | G. pallida | 5 | 1 | 0.17 | 4 | 0–20 | 52 | Pastusa Suprema |
| Cundinamarca | Tausa | G. pallida | 25 | 17 | 2.89 | 84.2 | 1–1327 | 51 | Pastusa Suprema |
| Cundinamarca | Ubaque | G. pallida | 4 | 2 | 0.34 | 1.8 | 3–4 | 58 | Superior |
| Cundinamarca | Ubaté | G. pallida | 3 | 3 | 0.51 | 443.3 | 268–556 | 51 | Parda Pastusa |
| Cundinamarca | Une | G. pallida | 3 | 3 | 0.51 | 3.3 | 1–5 | 72 | Criolla |
| Cundinamarca | Villapinzón | G. pallida | 29 | 18 | 3.06 | 23.4 | 1–475 | 74 | Pastusa Suprema |
| Cundinamarca | Zipaquirá | G. pallida | 9 | 5 | 0.85 | 2 | 2–10 | 64 | Pastusa Suprema |
| Cundinamarca | Cajicá | G. pallida | 1 | 1 | 0.17 | 1 | 0–1 | 80 | Diacol Capiro |
| Cundinamarca | Tenjo | G. pallida | 2 | 2 | 0.34 | 72.5 | 50–95 | 76 | Diacol Capiro |
| Cundinamarca | Sibaté | - | 8 | 0 | - | - | - | - | Criolla |
| Cundinamarca | Soacha | G. pallida | 3 | 3 | 0.51 | 1 | 0–1 | 76 | Diacol Capiro |
| Cundinamarca | Cogua | G. pallida | 4 | 4 | 0.68 | 1.5 | 1–2 | 73 | Betina |
| Cundinamarca | San Bernardo | G. pallida | 2 | 2 | 0.34 | 2.5 | 2–3 | 51 | Criolla |
| Cundinamarca | Fosca | G. pallida | 4 | 2 | 0.34 | 2 | 0–5 | 69 | Pastusa Suprema |
| Cundinamarca | Lenguazaque | G. pallida | 3 | 1 | 0.17 | 1.7 | 0 | 60 | Parda Pastusa |
| Cundinamarca | Simijaca | - | 2 | 0 | - | - | - | - | Pastusa Suprema |
| Cundinamarca | Susa | G. pallida | 15 | 1 | 0.17 | 2.7 | 1–125 | 55 | Pastusa Suprema |
| Cundinamarca | Guatavita | G. pallida | 11 | 4 | 0.68 | 11.8 | 1–125 | 55 | Pastusa Suprema |
| Cundinamarca | La Calera | - | 5 | 0 | - | - | - | - | Pastusa Suprema |
| Cundinamarca | Chocontá | Globodera sp. | 8 | 7 | 1.19 | 43 | 1–93 | 70 | Pastusa Suprema |
| Boyacá | Tota | G. pallida | 6 | 4 | 0.68 | 4.7 | 2–19 | 58 | Parda Pastusa |
| Boyacá | Toca | G. pallida | 10 | 7 | 1.19 | 27.3 | 5–122 | 51 | Tocarreña |
| Boyacá | Tunja | G. pallida | 25 | 17 | 2.89 | 47.3 | 1–411 | 60 | Diacol Capiro |
| Boyacá | Chíquiza | G. pallida | 11 | 9 | 1.53 | 47 | 1–142 | 23 | Betina |
| Boyacá | Úmbita | G. pallida | 5 | 1 | 0.17 | 2 | 0–2 | 20 | Tocarreña |
| Boyacá | Samacá | G. pallida | 29 | 22 | 3.74 | 53.2 | 1–429 | 24 | Diacol Capiro |
| Boyacá | Oicatá | G. pallida | 6 | 4 | 0.68 | 7.7 | 1–36 | 24 | Diacol Capiro |
| Boyacá | Sogamoso | G. pallida | 8 | 8 | 1.36 | 107.5 | 1–305 | 38 | Parda Pastusa |
| Boyacá | Siachoque | G. pallida | 27 | 20 | 3.40 | 64 | 1–364 | 54 | Parda Pastusa |
| Boyacá | Arcabuco | G. pallida | 10 | 5 | 0.85 | 122.5 | 1–873 | 23 | Parda Pastusa |
| Boyacá | Ventaquemada | G. pallida | 15 | 12 | 2.04 | 7.9 | 1–73 | 53 | Pastusa Suprema |
| Boyacá | Motavita | G. pallida | 8 | 4 | 0.68 | 2.6 | 1–14 | 54 | Betina |
| Boyacá | Sora | G. pallida | 3 | 3 | 0.51 | 185 | 101–286 | 60 | Diacol Capiro |
| Boyacá | Saboyá | G. pallida | 16 | 5 | 0.85 | 1.4 | 1–15 | 36 | Parda Pastusa |
| Boyacá | Soracá | G. pallida | 9 | 8 | 1.36 | 84.7 | 1–306 | 24 | Diacol Capiro |
| Boyacá | Boyacá | G. pallida | 3 | 1 | 0.17 | 13.7 | 1–41 | 35 | Diacol Capiro |
| Boyacá | Viracachá | G. pallida | 3 | 2 | 0.34 | 1.3 | 1–2 | 32 | Rubí |
| Boyacá | Ciénega | - | 4 | 0 | - | - | - | - | Pastusa Suprema |
| Boyacá | Mongua | G. pallida | 2 | 1 | 0.17 | 0.5 | 0–1 | 38 | ICA Única |
| Boyacá | Firavitoba | G. pallida | 3 | 2 | 0.34 | 1 | 1–2 | 44 | Parda Pastusa |
| Boyacá | Gámeza | G. pallida | 5 | 2 | 0.34 | 0.8 | 1–2 | 20 | Tocarreña |
| Boyacá | Belén | G. pallida | 11 | 7 | 1.19 | 122.3 | 1–757 | 40 | Parda Pastusa |
| Boyacá | Tutazá | G. pallida | 12 | 3 | 0.51 | 6.9 | 1–78 | 49 | ICA Única |
| Cauca | San Sebastián | G. pallida | 8 | 5 | 0.85 | 4.1 | 1–13 | 48 | Parda Pastusa |
| Cauca | Silvia | G. pallida | 6 | 2 | 0.34 | 0.3 | 0–1 | 47 | Criolla |
| Cauca | Totoró | G. pallida | 6 | 2 | 0.34 | 4.3 | 1–23 | 51 | Criolla |
| Antioquia | San Vicente | G. pallida | 9 | 5 | 0.85 | 7.11 | 2–24 | 40 | Criolla |
| Antioquia | La Unión | G. pallida | 9 | 3 | 0.51 | 6 | 1–51 | 32 | Diacol Capiro |
| Antioquia | Sonsón | G. pallida | 9 | 5 | 0.85 | 7.5 | 1–29 | 16 | Criolla |
| Antioquia | La Ceja | G. pallida | 1 | 1 | 0.17 | 7 | 7–7 | 20 | Criolla |
| Antioquia | Abejorral | G. pallida | 6 | 3 | 0.51 | 2.17 | 1–7 | 15 | Criolla |
| Antioquia | Marinilla | G. pallida | 7 | 5 | 0.85 | 5.71 | 1–26 | 20 | Criolla |
| Antioquia | Santuario | G. pallida | 3 | 2 | 0.34 | 1.33 | 1–3 | 20 | ICA Nevada |
| Norte de Santander | Pamplona | G. pallida | 8 | 2 | 0.34 | 0.9 | 1–4 | 37 | Criolla |
| Norte de Santander | Cácota | G. pallida | 4 | 2 | 0.34 | 1 | 1–2 | 16 | Criolla |
| Santander | Concepción | G. pallida | 8 | 2 | 0.34 | 0.9 | 1–6 | 20 | Parda Pastusa |
| Santander | Carcasí | G. pallida | 2 | 1 | 0.17 | 0.5 | 0–1 | 36 | Parda Pastusa |
| Santander | Cerrito | G. pallida | 4 | 1 | 0.17 | 1 | 1–4 | 31 | ICA Única |
| Caldas | Manizales | G. pallida | 7 | 1 | 0.17 | 0.43 | 1–3 | 15 | Parda Pastusa |
| Tolima | Anzoátegui | G. pallida | 5 | 1 | 0.17 | 0.4 | 1–2 | 29 | Pastusa Suprema |
| Tolima | Murillo | G. pallida | 7 | 1 | 0.17 | 0.2 | 0–1 | 19 | Parda Pastusa |
a Prevalence: (Number of samples with cysts/ total number of samples collected)*100.
b Population density: Average number of cysts per 100 cm3 of soil.
c Population range: Minimum and maximum densities of cysts per 100 cm3 of soil.
d Viability: Average of cysts with viable eggs per population.
DNA extraction, polymerase chain reaction and sequencing
For molecular characterization, from the departments that showed the highest cyst nematode densities, soil samples taken per municipality were pooled according to proximity distance (1 km and 5 km range), resulting in one to two samples per municipality, for a total of 26 populations analyzed (Table 1). From each pooled population, DNA was extracted from individual cysts or juveniles using the "Sigma Extract-N-Amp Kit (XNAT2)" kit (Sigma, St. Louis, MO) according to the protocol reported by Ma et al. [30] at AGROSAVIA, La Selva Research Center, Rionegro, Antioquia. The number of individuals analyzed per population depended upon the cyst nematode density present in each soil sample. DNA was then stored at -20°C until used.
PCR amplification of two genomic regions were performed using 12.5 μl of the Extract-N-AmpTM Tissue PCR kit (Sigma), 1 μl of each primer, 4 μl of DNA and water to complete a volume of 25 μl. The rDNA primers used for PCR and DNA sequencing are listed in Table 2. The ITS region of ribosomal DNA was amplified using 94° C for 2.5 min for initial denaturation, followed by 40 cycles at 94° C for 1 min, 55° C for 1 min, 72° C for 2 min, and a final extension of 72° C for 5 minutes. For the 28S region, initial denaturation was 94° C for 5 min, 40 cycles of 94° C for 30 sec, 58° C for 30 sec, 72° C for 1 min, and a final extension of 72° C for 10 min [31]. The products were loaded on a 1.5% agarose gel and visualized using gel red (Biotium, San Francisco, CA). Sanger sequencing of the amplicons was performed in both directions by CorpoGen (Bogotá, Colombia).
Table 2. Primers used for polymerase chain reaction and DNA sequencing of Globodera spp. individuals recovered from cultivated potatoes in Colombia.
| Primer | Marker | Sequence (5’ to 3’) | Reference |
|---|---|---|---|
| F194 | ITS | CGTAACAAGGTAGCTGTAG | [32] |
| F195 | ITS | TCCTCCGCTAAATGATATG | [33] |
| D2A | 28S | ACAAGTACCGTGAGGGAAAGTTG | [33] |
| D3B | 28S | TCGGAAGGAACCAGCTACTA | [34] |
28S = Large ribosomal RNA subunit and ITS = internal transcribed spacer 1 and 2 including 5.8S rRNA.
Sequence alignment and phylogenetic analyses
Resulting sequences were assembled in Sequencher® software version 5.1 (Gene Codes Corporation, Ann Arbor, MI USA) and manually reviewed for base calling errors. Partial 28S rRNA and ITS1-2 + 5.8S rRNA gene sequences from G. pallida, G. mexicana, G. rostochiensis, G. tabacum, G. ellingtonae, and G. artemisiae, were retrieved from GenBank nucleotide database and included in the alignment (Table 3) [8,19,21,35–43]. Sequences of Punctodera punctata and P. chalcoensis also obtained from GenBank (AF274416.1, DQ328699.1.1, AY090885.1), were used as outgroup taxa for both gene regions. After that, sequence alignments were performed using Clustal W [44] and manually edited using BioEdit v 7.2 [45]. To remove ambiguous regions in the alignment the program Gblocks v.0.91b was used with the standard settings [46]. Newly generated sequences for both gene regions were deposited in GenBank (Table 3).
Table 3. Details of cyst nematode populations included in the molecular and phylogenetic studies from cultivated potatoes in Colombia and reported in other studies.
| Globodera species | Location (Municipality, Department) | Specimen code | Accession number 28S | Accession number ITS |
|---|---|---|---|---|
| Globodera pallida | La Unión, AN* | A1A | MH389939 | MH389979 |
| Globodera pallida | La Unión, AN | A1B | MH389938 | MH389978 |
| Globodera pallida | La Unión, AN | A1C | - | MH389977 |
| Globodera pallida | Ventaquemada, BO | B2A | MH389946 | MH389986 |
| Globodera pallida | Ventaquemada, BO | B2B | MH389945 | MH389985 |
| Globodera pallida | Ventaquemada, BO | B2C | MH389944 | MH389984 |
| Globodera pallida | Ventaquemada, BO | B2D | MH389943 | MH389983 |
| Globodera pallida | Arcabuco, BO | B3D | MH389942 | MH389982 |
| Globodera pallida | Chíquiza, BO | B4D | MH389937 | - |
| Globodera pallida | Tunja, BO | B5D | - | MH389976 |
| Globodera pallida | Toca, BO | B6D | MH389936 | MH389975 |
| Globodera pallida | Sogamoso, BO | B7A | MH389935 | MH389974 |
| Globodera pallida | Sogamoso, BO | B7B | MH389934 | - |
| Globodera pallida | Sogamoso, BO | B7D | MH389933 | - |
| Globodera pallida | Sora, BO | B8A | MH389932 | MH389973 |
| Globodera pallida | Sora, BO | B8C | - | MH389972 |
| Globodera pallida | Sora, BO | B8D | - | MH389971 |
| Globodera pallida | Samacá, BO | B9D | MH389941 | MH389981 |
| Globodera pallida | Soracá, BO | B11A | MH389931 | - |
| Globodera pallida | Soracá, BO | B11B | MH389930 | - |
| Globodera pallida | Soracá, BO | B11C | MH389929 | - |
| Globodera pallida | Soracá, BO | B12A | MH389940 | MH389980 |
| Globodera pallida | Susa, CU | C1A | MH389928 | MH389970 |
| Globodera pallida | Susa, CU | C1B | MH389927 | - |
| Globodera pallida | Susa, CU | C1D | - | MH389969 |
| Globodera pallida | Guatavita, CU | C2B | MH389926 | MH389968 |
| Globodera pallida | Guatavita, CU | C2C | MH389925 | MH389967 |
| Globodera pallida | Guatavita, CU | C3C | MH389923 | MH389965 |
| Globodera pallida | Guatavita, CU | C3D | MH389922 | - |
| Globodera pallida | Ubaté, CU | C4C | MH389921 | - |
| Globodera pallida | Ubaté, CU | C4D | MH389920 | - |
| Globodera pallida | Tausa, CU | C5A | MH389919 | MH389964 |
| Globodera pallida | Tausa, CU | C5B | MH389918 | - |
| Globodera pallida | Subachoque, CU | C6B | MH389917 | - |
| Globodera pallida | Sesquilé, CU | C8A | MH389916 | - |
| Globodera pallida | Sesquilé, CU | C8C | MH389915 | MH389963 |
| Globodera pallida | Sesquilé, CU | C8D | - | MH389962 |
| Globodera pallida | Cajicá, CU | C10D | - | MH389961 |
| Globodera pallida | Villapinzón, CU | C11A | MH389914 | MH389960 |
| Globodera pallida | Villapinzón, CU | C11D | MH389913 | MH389959 |
| Globodera pallida | Túquerres, NA | N2A | MH389949 | MH389990 |
| Globodera pallida | Túquerres, NA | N2B | MH389948 | MH389989 |
| Globodera pallida | Túquerres, NA | N2C | - | MH389988 |
| Globodera pallida | Túquerres, NA | N2D | MH389947 | MH389987 |
| Globodera pallida | Guachucal, NA | N4A | MH389912 | - |
| Globodera pallida | Guachucal, NA | N4B | MH389911 | - |
| Globodera pallida | Guachucal, NA | N4C | MH389910 | MH389958 |
| Globodera pallida | Guachucal, NA | N4D | MH389909 | - |
| Globodera pallida | Belén, NA | N5A | - | MH389957 |
| Globodera pallida | Belén, NA | N5B | - | MH389956 |
| Globodera pallida | Belén, NA | N5C | MH389908 | MH389955 |
| Globodera pallida | Belén, NA | N5D | MH389907 | MH389954 |
| Globodera pallida | Ospina, NA | N8B | - | MH389953 |
| Globodera pallida | Ospina, NA | N8C | MH389906 | MH389952 |
| Globodera pallida | Ospina, NA | N8D | MH389905 | MH389951 |
| Globodera pallida | Ipiales, NA | N9A | MH389904 | MH389950 |
| Globodera sp | Chocontá, CU | C3A | MH389924 | MH389966 |
| Globodera pallida | Peru | - | - | GU084813.1 |
| Globodera pallida | Peru | - | - | GU084805.1 |
| Globodera pallida | Chile | - | - | GU084800.1 |
| Globodera pallida | Ukraine | - | - | AJ606687.1 |
| Globodera pallida | England | - | - | DQ847110.1 |
| Globodera pallida | Poland | - | - | EU855119.1 |
| Globodera pallida | Peru | - | - | GU084804.1 |
| Globodera pallida | Peru | - | - | GU084806.1 |
| Globodera pallida | Peru | - | - | HQ670269.1 |
| Globodera mexicana | Mexico | - | - | EU006707.1 |
| Globodera mexicana | Mexico | - | - | EU006708.1 |
| Globodera rostochiensis | USA | - | - | EF153839.1 |
| Globodera rostochiensis | Australia | - | - | EF622524.1 |
| Globodera rostochiensis | Canada | - | - | FJ212166.1 |
| Globodera rostochiensis | England | - | - | EF153840.1 |
| Globodera rostochiensis | Bolivia | - | - | GU084809.1 |
| Globodera tabacum | Japan | - | - | AB207272.1 |
| Globodera tabacum | USA | - | - | GQ294525.1 |
| Globodera tabacum | USA | - | - | DQ847112.1 |
| Globodera tabacum | Argentina | - | - | DQ097515.2 |
| Globodera ellingtonae | USA | - | - | GQ896543.1 |
| Globodera ellingtonae | Chile | - | - | GU084808.1 |
| Globodera ellingtonae | USA | - | - | DQ097514.2 |
| Punctodera punctata | Belgium | - | - | AF274416.1 |
| Punctodera chalcoensis | Mexico | - | - | AY090885.1 |
| Globodera pallida | France | - | KJ409636.1 | - |
| Globodera pallida | Slovakia | - | KJ409626.1 | - |
| Globodera pallida | England | - | JN712219.1 | - |
| Globodera pallida | Chile | - | JN712220.1 | - |
| Globodera pallida | France | - | GU338021.1 | - |
| Globodera ellingtonae | USA | - | JN712217.1 | - |
| Globodera tabacum | USA | - | GQ294492.1 | - |
| Globodera rostochiensis | Canada | - | JN712223.1 | - |
| Globodera rostochiensis | Slovakia | - | KJ409625.1 | - |
| Globodera artemisiae | Hungary | - | KU845472.1 | - |
28S = Large ribosomal RNA subunit and ITS = internal transcribed spacer 1 and 2 including 5.8S rRNA.
Phylogenetic relationships among partial sequences of the 28S rRNA and Internal Transcribed Spacer 1 and 2 plus 5.8S rRNA genes were inferred using Bayesian Inference (BI) and Maximum Likelihood (ML) methods. For both gene regions, PartitionFinder v.2.0 [47] was used to select the best-fit partitioning scheme and models of evolution for phylogenetic analyses according to the Greedy algorithm using the Bayesian Information Criterion (BIC). The sequence partition for 28S rRNA gene was a single partition with HKY substitution model [48] for each of the 3 positions of the codon. The best sequence partition for ITS1-2 + 5.8S rRNA gene internal transcribed spacer 1 and 2 including the 5.8S rRNA region was a partition that included the first and second position of the codon with a K80 model [49] substitution model, and a second partition that consisted on the third codon position under the K80 with proportions of invariable sites (K80+I). Bayesian Inference analyses were performed using MrBayes v.3.1.2 [50], with five independent runs of four Markov chains for 1 x 106 generations and default heating values, sampling every 100 generations with 2500 samples discarded as burn-in after checking for convergence. Clades were considered strongly supported when values were > 0.95 [51]. For Maximum likelihood analyses the software GARLI v.2.0 [52] was used. Bootstrap support for trees was generated with 1,000 replicate searches and summarized in a consensus tree using SumTrees [53], clades were considered as well/strongly supported when bootstrap was >70%. In addition, in order to characterize the genetic divergence between cyst nematodes from Colombia and Globodera species already reported, the average number of nucleotide substitution per site (Dxy), net nucleotide substitutions per site (Da) and number of fixed differences (Fd) among genetic groups were computed using DnaSP v.6.12.03 [54].
Results
Field survey
Of the 589 potato fields sampled, cyst nematodes were detected in 355 fields distributed in 69 municipalities of Colombia, with densities ranging from 1 to 1,327 cysts per 100 cm3 of soil (Table 1). The predominant species was G. pallida (Fig 2), identified in 51% of the fields sampled in Cundinamarca, 63.3% in Boyacá, 72.2% in Nariño, 54% in Antioquia, 45% in Cauca, 33% in Norte de Santander, 29% in Santander, 17% in Tolima and 14% in Caldas, with densities ranging from 1 to 1327 cysts/100 cm3 of soil, 1 to 873 cysts/100 cm3 of soil, 1 to 345 cysts/100 cm3 of soil, 1 to 51 cysts/100 cm3 of soil, 1 to 23 cysts/100 cm3 of soil, 1 to 4 cysts/100 cm3 of soil, 1 to 6 cysts/100 cm3 of soil, 1 to 3 cysts/100 cm3 of soil and 1 to 2 cysts/100 cm3 of soil, respectively. Among municipalities, the highest mean densities were detected in Ubaté (443.3 cysts/100 cm3 of soil), followed by Sora, Arcabuco, Belén, Sogamoso, Túquerres and Tausa (185, 122.5, 122.3, 107.5, 97.3 and 84,2 cysts/100 cm3 of soil, respectively). Cyst nematodes were not found in Pasca, Sibaté, Simijaca, and La Calera in Cundinamarca, nor in Ciénega in Boyacá (Table 1). All samples positive for PCN showed cysts with viable eggs, and viability percentage ranged from 15 to 80%. Boyacá and Cundinamarca were the departments that had in average the higher viability (Table 1).
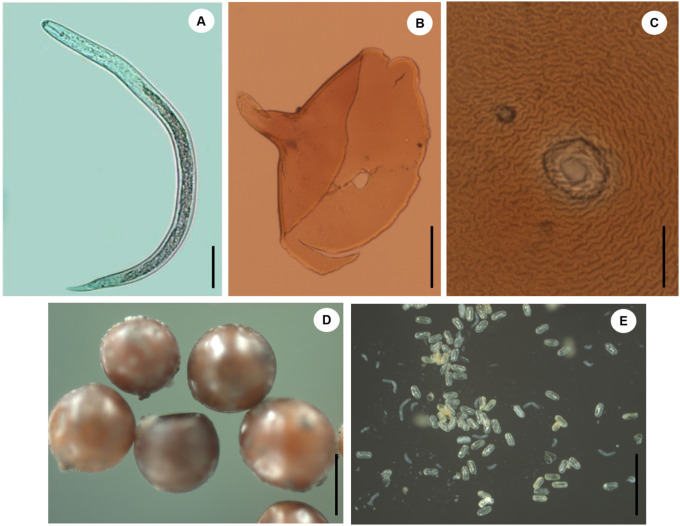
Fig 2. Light micrographs of Globodera pallida from Colombia (Population B2: Ventaquemada, Boyacá).
A) Second-stage juvenile. B) Anterior region. C) Cysts. D) Eggs. Scale bars: A-C = 50 μm; D, E = 100 μm.
Given host-based grouping, PCN was detected in samples taken from varieties of Solanum tuberosum Group Andigena such as Diacol Capiro (107 out of 158 samples), Betina (24 out of 30 samples), Pastusa Suprema (81 out of 167 samples), Parda Pastusa (66 out of 105 samples), Tocarreña (10 out of 20 samples), Rubí (2 out of 3 samples), ICA Única (4 out of 16 samples), ICA Nevada (2 out of 3 samples), Superior (2 out of 10) and, from varieties of Solanum tuberosum Group Phureja such as Criolla variety (32 out of 69 samples) (Table 1). A morphologically different species (under description), was only detected in one field in Chocontá (Cundinamarca) on Suprema variety (Table 1).
Molecular analysis
The amplification of D2-D3 expansion segments of 28S rRNA and internal transcribed spacer 1, 2 including the 5.8S rRNA yielded single fragments of 609 and 848 bp, respectively. Forty-two new D2-D3 of 28S rRNA gene sequences and twenty-eight new internal transcribed spacer 1 and 2 including the 5.8S rRNA were obtained in the present study (Table 2).
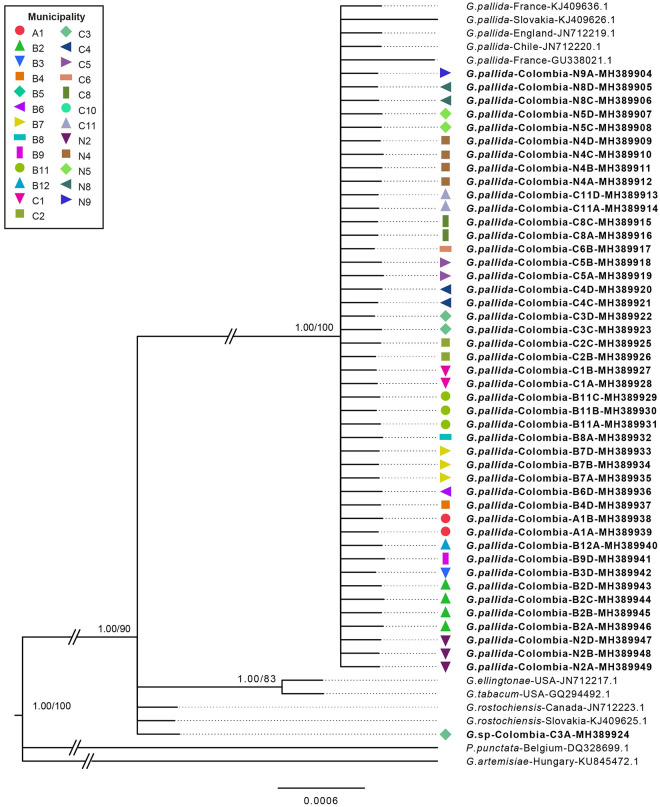
Phylogenetic relationships inferred from analyses of D2-D3 expansion segments of 28S rRNA of a multiple-edited alignment (57 sequences), showed two well supported major clades based on BI and ML inferences (PP = 1.00, BP = 90) (Fig 3). A highly supported clade (i) (PP = 1.00, BP = 90), was formed by sequences of G. pallida from France, Chile, England, Slovakia and all but one cyst nematode sequence obtained in this study from Colombia. The second major clade, Clade (ii) grouped three species, G. ellingtonae and G. tabacum that formed a well-supported subclade (PP = 1.00, BP = 83) clearly separated from a politomy formed by a single cyst nematode sample (G. sp) from Colombia and G. rostochiensis from Canada and Slovakia (Fig 3).
Fig 3. Phylogenetic relationships within the genus Globodera.
Bayesian 50% majority rule consensus trees as inferred from D2–D3 expansion segments of 28S rRNA as a single partition with HKY model. Node-support values: Left value posterior probability BI shown if >95%, right value bootstrap from ML analysis shown only if >70%. Newly obtained sequences in this study are in bold.
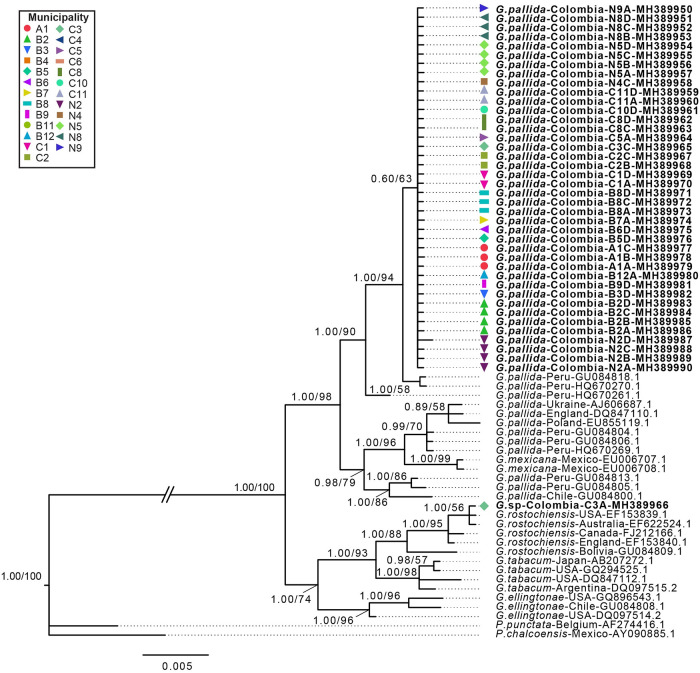
The 50% majority-rule BI consensus tree of the alignment generated for the 69 sequences of the region conformed by the internal transcribed spacer 1 and 2 including the 5.8S rRNA regions, showed two well supported major clades (PP = 1.00/ BP = 100) that were consistent with the findings based on 28S rRNA phylogeny (Fig 4). Clade (i) was formed by G. pallida and G. mexicana, and Clade (ii) was formed by G. rostochiensis, G. tabacum, G. ellingtonae and one sequence from Colombia. In Clade (i) two sequences of G. pallida from Peru along with the rest of sequences from Colombia formed a well-supported subclade (PP = 1.00, BP = 90) that was clearly separated (PP = 1.00, BP = 94) from one sequence of G. pallida from Peru. The sister clade of this sub-clade was formed by other sequences of G. pallida from Peru and European countries such as Ukraine, England and Poland, that were separated with high support (PP = 1.00, BP = 96) from sequences of G. mexicana and moderately support (PP = 1.00, BP = 79) of G. pallida from Chile and Peru. In Clade (ii) a single sequence from Colombia formed a well-supported subclade (PP = 1.00, BP = 93) along with G. rostochiensis from USA, Australia, Canada, England and Bolivia that was related with G. tabacum from Japan, USA and Argentina. This subclade formed a sister clade with G. ellingtonae from USA and Chile with high support (PP = 1.00, BP = 74).
Fig 4. Phylogenetic relationships within the genus Globodera.
Bayesian 50% majority rule consensus trees as inferred from Internal Transcribed Spacer 1 and 2 plus 5.8S rRNA gene with first and second position with K80 substitution model, and a second partition with the third codon under K80+I model. Node-support values: Left value posterior probability BI shown if >95%, right value bootstrap from ML analysis shown only if >50%. Newly obtained sequences in this study are in bold.
Genetic distances among cyst nematodes sequences from Colombia and other nematodes species included in the phylogenetic analyses are summarized in Table 4. Based on the 28S rRNA gene sequences, all sequences from Colombia from Clade (i) had the lowest average number of nucleotide substitutions per site (Nucleotide divergence—Dxy = 0.002) and lowest number of net nucleotide substitutions per site (net genetic distance—Da = 0.001) when compared to G. pallida sequences from the other countries without fixed differences among groups (Table 4). The cyst nematode sequence from Colombia in Clade (ii) had the lowest divergence when compared with G. rostochiensis (Dxy = 0.001, Da = 0.000) without showing any fixed differences among groups (Table 4). In agreement with the 28S rRNA marker, the genetic distances based on the internal transcribed spacer 1 and 2 including the 5.8S rRNA gene sequences of cyst nematodes from Colombia in Clade (i) was lowest when compared with G. pallida (Dxy = 0.014, Da = 0.008), with one fixed substitution (Fig 3, Table 4) and lowest in Clade (ii) when compared with G. rostochiensis (Dxy = 0.003, Da = 0.000) with no fixed differences among groups (Table 4).
Table 4. Gene divergence between potato cyst nematodes from Colombia and other species retrieved from GenBank.
| Marker | Group 1 | Group 2 | Dxy | Da | Fd |
|---|---|---|---|---|---|
| 28S | G. pallida - Colombia | G. pallida | 0.00227 | 0.00116 | 0 |
| G. rostochiensis | 0.01072 | 0.00988 | 4 | ||
| G. ellingtonae | 0.01370 | 0.01243 | 6 | ||
| G. tabacum | 0.01443 | 0.01400 | 6 | ||
| G. artemisiae | 0.02661 | 0.02618 | 15 | ||
| 28S | G. sp - Colombia | G. pallida | 0.01165 | 0.01013 | 6 |
| G. rostochiensis | 0.00125 | 0.00000 | 0 | ||
| G. ellingtonae | 0.00498 | 0.00332 | 2 | ||
| G. tabacum | 0.00415 | 0.00332 | 2 | ||
| G. artemisiae | 0.02076 | 0.01993 | 12 | ||
| ITS | G.pallida - Colombia | G. pallida | 0.01418 | 0.00818 | 1 |
| G. rostochiensis | 0.03188 | 0.02871 | 21 | ||
| G. ellingtonae | 0.02973 | 0.02634 | 18 | ||
| G. tabacum | 0.02721 | 0.02550 | 20 | ||
| G. mexicana | 0.02035 | 0.02026 | 16 | ||
| ITS | G.sp - Colombia | G. pallida | 0.03230 | 0.02600 | 20 |
| G. rostochiensis | 0.00395 | 0.00027 | 0 | ||
| G. ellingtonae | 0.02582 | 0.02189 | 16 | ||
| G. tabacum | 0.01675 | 0.01478 | 12 | ||
| G. mexicana | 0.03289 | 0.03230 | 27 | ||
28S = Large ribosomal RNA subunit and ITS = internal transcribed spacer 1 and 2 including 5.8S rRNA.
Dxy, Da and Fd correspond to average number of nucleotide substitutions per site, the number of net nucleotide substitutions per site, and number of fixed differences between compared groups, respectively.
Discussion
Even when PCN is considered a re-emerging potato pathogen in Colombia, first identified in the department of Nariño in 1970 [23], documented surveys only report PCN in municipalities of Nariño, Cauca, Boyacá and Cundinamarca [24,25]. Our comprehensive study that sampled 75 municipalities in 9 potato producing departments of Colombia found that 60% of the tested samples were positive for PCN (355 out of 589 sampled fields), and that the pathogen is widespread in all Colombian producing potato departments (Cundinamarca, Boyacá, Nariño, Antioquia, Cauca, Norte de Santander, Santander, Tolima and Caldas) (Fig 1), with cysts that contain viable eggs present in all sampled regions.
However, there was variation in population densities among regions. The highest densities were found in Cundinamarca and Boyacá, ranging from 1 to 1,327 cyts/100 g of soil and 1 to 873 cysts/100 g of soil, respectively. Nieto et al. (1983) [24] surveyed these departments in early 1980s, but since detected cysts were empty or contained non-viable eggs, these regions were declared as PCN free. Later, Arciniegas et al (2012) [25], reported PCN in Tunja, Samacá and Ventaquemada in Boyacá and Tausa, Tabio and Zipaquirá in Cundinamarca. In our study, PCN was detected in new municipalities with the highest densities found in Tausa, Ubaté and Villapinzón in Cundinamarca and, in Arcabuco, Belén, Tunja, Samacá, Sogamoso and Sora in Boyacá (Table 1). Boyacá and Cundinamarca are the largest producers of potatoes in Colombia, with 40,724 and 61,322 harvested hectares corresponding to 26% and 39% of the total potato national production in 2018, respectively [26]. Potatoes are the main plant crop grown by farmers and fields are usually planted in monocultures for several continuous cycles. As PCNs are highly specialized, sedentary and obligate endoparasites of solanaceous plants [1,22,55], the constant presence of potato crops in monocultures for several cycles may lead to persistence and increase of this plant pathogen in these two regions overtime. In contrast, in Nariño department, although the nematode was found in all municipalities sampled, PCN densities were lower, from 1 to 345 cysts/100 g of soil, and similar ranges were found by Nieto et al (1983) [24]. Nariño ranks third in production with 31,611 harvested hectares (19,35% of total potato national production), in contrast to Boyacá and Cundinamarca, in this department farmers usually grow different potato cultivars within a field (e.g. Diacol Capiro, Pastusa Suprema, Betina) with a crop rotation scheme usually with non-host plants such as corn, cabbage, lettuce, onion and pastures, and the use of biological microorganisms for the control of other pest problems has also implemented [56], which reduces pesticides use. Similar potato production scheme was observed in its neighbor department, Cauca, and PCN densities decreased in the latter from 23 cysts/100 g of soil [24] to 5,97 cysts/100 of soil in this study. Considering that G. pallida requires a living potato plant to complete its life cycle [6], the management practices implemented in both departments may reflect the reduction of PCN densities in these regions.
Our results also show G. pallida populations have spread into new regions of Colombia. In the departments of Antioquia, Caldas, Tolima, Santander and Norte de Santander, PCN was detected in all municipalities sampled, although with low population levels (5.97 cysts/100 of soil in average in Antioquia, 0.95 cysts/100 of soil in Norte de Santander, 0.8 cysts/100 of soil in Santander, 0.43 cysts/100 of soil in Caldas and 0.3 cysts/100 of soil in Tolima). To our knowledge, this is the first report of the presence of G. pallida in these departments. These regions represent in general, low potato growing areas with low participation in national potato production (2.27% in average in 2018) [26], and were therefore considered before as PCN free. The spread of PCN is mainly caused through tubers, soil or equipment contaminated with cysts [55] and potato seed tubers in these departments frequently come from Boyacá and Cundinamarca, which may allow the dissemination of this plant pathogen into these new regions. Nevertheless, population levels in these departments are low and the extent of PCN is limited, therefore, to maintain low levels and to avoid the spread into new areas, intensive monitoring program for PCN should be implemented in all potato producing regions of Colombia.
Molecular identification and phylogenetic relationships of PCN species from Colombia
The molecular phylogeny of PCN populations based on ITS1-5.8S-ITS2 rDNA and 28S D2-D3 regions supported the presence of at least two PCN species in Colombia. Globodera pallida was found in all populations that resulted positive for PCN and, molecular phylogeny based on the ITS1-5.8S-ITS2 rDNA grouped all G. pallida from Colombia in a single clade that was closely related to P5A pathotype strains (GenBank accession number HQ670270.1) and clone La Libertad (GenBank accession number GU084818) (Fig 4). This finding suggests that G. pallida present in Colombia have a different origin than G. pallida present in countries such as Ukraine, England and Poland that cluster as a monophyletic clade with other Peruvian strains, and G. pallida present in Chile that cluster with a different Peruvian strain (Figs 3 and 4). Despite that P5A Peruvian strain has been considered as a different species [12,17], a recent study based on ITS rRNA, COI and cytb mitochondrial regions concluded that all clades within G. pallida belong to a single species [2]. The 28S D2-D3 phylogeny, although with lower level of resolution, also clustered all but one PCN populations from Colombia with G. pallida around the globe as a monophyletic clade (Fig 3). For both gene regions, a single sequence from the C3 population (Chocontá) grouped in a distant clade along with individuals of G. rostochiensis. Genetic distance analyses based on gene regions ITS1-5.8S-ITS2 and D2-D3, were congruent with these findings showing G. pallida from Colombia with the smallest Da (0.8% and 0.12% for ITS rDNA and D2-D3, respectively) and the smallest Dxy (1.41% and 0.23% for ITS rDNA and D2-D3, respectively) when compared with other G. pallida populations. Similarly, genetic distances from C3 population showed the lowest distance when compared with G. rostochiensis (Dxy = 0.001, Da = 0.000).
Therefore, the ITS1-5.8S-ITS2 rDNA and 28S D2-D3 molecular analyses were able to identify with high phylogenetic support G. pallida and G. rostochiensis. Additionally, ITS1-5.8S-ITS2 rDNA phylogenetic resolution supports a northern Peru origin of G. pallida present in Colombia, nevertheless this hypothesis must be further investigated using additional samples and molecular markers, with statistical inference such as model testing and coalescent demographic reconstruction. Although with less taxa included, molecular phylogeny based on 28S D2-D3 gene improved the node support found in previous phylogenies between G. pallida and G.tabacum (i.e. PP = 54 and 72%) (e.g., [9,16]), and the unresolved positions for G. rostochiensis [16]. Taken all together, both DNA markers used in this study showed to be useful to identify Globodera species present in Colombia, with ITS1-5.8S-ITS2 rDNA being more informative in phylogeographic perspective [12].
Conclusions
This study provides new information about the status and prevalence of PCN species associated with cultivated potatoes in the main producing regions of Colombia including for the first time genetic information. Molecular phylogenies with ITS1-5.8S-ITS2 rDNA and D2/D3 28S regions were effective in the identification of G. pallida, the dominant species present in all departments surveyed in this study, and suggest the presence of G. rostochiensis, in one municipality of Cundinamarca, which is currently under description. Considering the presence of PCN species constitute a threat for potato production, intensive sampling and monitoring of this plant pathogen should be conducted to reduce and prevent the spread into new areas. The development of management practices that involves the evaluation of resistant varieties for populations that tested positive for PCN, as well as other practices such as crop rotations, trap crops, biofumigants, biocontrol agents among others that have shown to be effective for other G. pallida populations worldwide, are also a crucial step to reduce population densities of PCN in Colombia.
Acknowledgments
The authors thank the supporting research assistant of Corporación Colombiana de Investigación Agropecuaria, La Selva Research Station, Mario Alonso Mesa, for greenhouse and field work.
Data Availability
All relevant data (including accession numbers) are within the paper.
Funding Statement
This work belongs to the project “Recomendaciones técnicas para el manejo integrado de los problemas fitosanitarios: Globodera pallida, síndrome X, virus PYVV y sus posibles vectores en papa”, with the financial support of the Colombian Ministry of Agriculture (Ministerio de Agricultura y Desarrollo Rural de Colombia).
References
- 1.Jones JT, Haegeman A, Danchin EGJ, Gaur HS, Helder J, Jones MGK, et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol Plant Pathol. 2013;14: 946–961. doi: 10.1111/mpp.12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subbotin SA, Franco J, Knoetze R, Roubtsova T V., Bostock RM, Cid Del Prado Vera I. DNA barcoding, phylogeny and phylogeography of the cyst nematode species from the genus Globodera (Tylenchida: Heteroderidae). Nematology. 2020;22: 269–297. doi: 10.1163/15685411-00003305 [DOI] [Google Scholar]
- 3.Van Riel HR, Mulder A. Potato cyst nematodes (Globodera species) in Western Europe. In: Marks R, Brodie B, editors. Potato cyst nematodes: Biology, distribution and control. Wallingford: CAB International; 1998. pp. 271–298. [Google Scholar]
- 4.O.E.P.P./E.P.P.O. PM 7/40 (4) Globodera rostochiensis and Globodera pallida. EPPO Bull. 2017;47: 174–197. doi: 10.1111/epp.12391 [DOI] [Google Scholar]
- 5.Talavera M, Andreu M, Valor H, Tobar A. Nematodos fitoparásitos en áreas productoras de patata de Motril y Salobreña. Investig Agrar Prod y Protección Veg. 1998;13: 87–95. [Google Scholar]
- 6.Contina JB, Dandurand LM, Knudsen GR. A spatiotemporal analysis and dispersal patterns of the potato cyst nematode Globodera pallida in Idaho. Phytopathology. 2020;110: 379–392. doi: 10.1094/PHYTO-04-19-0113-R [DOI] [PubMed] [Google Scholar]
- 7.Handoo ZA, Carta LK, Skantar AM, Chitwood DJ. Description of Globodera ellingtonae n. sp. (Nematoda: Heteroderidae) from Oregon. J Nematol. 2012;44: 40–57. [PMC free article] [PubMed] [Google Scholar]
- 8.Skantar AM, Handoo ZA, Carta LK, Chitwood DJ. Morphological and molecular identification of Globodera pallida associated with potato in Idaho. J Nematol. 2007;39: 133–144. [PMC free article] [PubMed] [Google Scholar]
- 9.Lax P, Rondan Dueñas JC, Franco-Ponce J, Gardenal CN, Doucet ME. Morphology and DNA sequence data reveal the presence of Globodera ellingtonae in the Andean region. Contrib to Zool. 2014;83: 227–243. doi: 10.1163/18759866-08304002 [DOI] [Google Scholar]
- 10.Subbotin SA, Vierstraete A, De Ley P, Rowe J, Waeyenberge L, Moens M, et al. Phylogenetic relationships within the cyst-forming nematodes (Nematoda, Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Mol Phylogenet Evol. 2001;21: 1–16. doi: 10.1006/mpev.2001.0998 [DOI] [PubMed] [Google Scholar]
- 11.Blaxter ML, De Ley P, Garey JR, Llu LX, Scheldeman P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392: 71–75. doi: 10.1038/32160 [DOI] [PubMed] [Google Scholar]
- 12.Subbotin SA, Prado Vera Del C I, Mundo-Ocampo M, Baldwin JG. Identification, phylogeny and phylogeography of circumfenestrate cyst nematodes (Nematoda: Heteroderidae) as inferred from analysis of ITS-rDNA. Nematology. Nematoda; 2011. pp. 805–824. doi: 10.1163/138855410X552661 [DOI] [Google Scholar]
- 13.Gutiérrez-Gutiérrez C, Cantalapiedra-Navarrete C, Montes-Borrego M, Palomares-Rius JE, Castillo P. Molecular phylogeny of the nematode genus Longidorus (Nematoda: Longidoridae) with description of three new species. Zool J Linn Soc. 2013;167: 473–500. doi: 10.1111/zoj.12019 [DOI] [Google Scholar]
- 14.Ye W, Zeng Y, Kerns J. Molecular characterisation and diagnosis of root-knot nematodes (Meloidogyne spp.) from turfgrasses in North Carolina, USA. PLoS One. 2015;10: 143556. doi: 10.1371/journal.pone.0143556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archidona-Yuste A, Cantalapiedra-Navarrete C, Liébanas G, Rapoport HF, Castillo P, Palomares-Rius JE. Diversity of root-knot nematodes of the genus Meloidogyne Göeldi, 1892 (Nematoda: Meloidogynidae) associated with olive plants and environmental cues regarding their distribution in southern Spain. PLoS One. 2019;13: 1–40. doi: 10.1371/journal.pone.0198236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madani M, Subbotin SA, Ward LJ, Li X, De Boer SH. Molecular characterization of canadian populations of potato cyst nematodes, Globodera rostochiensis and G. pallida using ribosomal nuclear RNA and cytochrome B genes. Can J Plant Pathol. 2010;32: 252–263. doi: 10.1080/07060661003740033 [DOI] [Google Scholar]
- 17.Skantar AM, Handoo ZA, Zasada IA, Ingham RE, Carta LK, Chitwood DJ. Morphological and molecular characterization of Globodera populations from oregon and idaho. Phytopathology. 2011;101: 480–491. doi: 10.1094/PHYTO-01-10-0010 [DOI] [PubMed] [Google Scholar]
- 18.Li X, Maria M, Cai R, Barsalote EM, Peneva V, Zheng J. Distribution of trichodorid species in mainland china with description of Trichodorus hangzhouensis sp. Nov. (nematoda, triplonchida). Zookeys. 2020;2020: 163–189. doi: 10.3897/zookeys.945.50424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subbotin SA, Halford PD, Warry A, Perry RN. Variations in ribosomal DNA sequences and phylogeny of Globodera parasitising solanaceous plants. Nematology. 2000;2: 591–604. doi: 10.1163/156854100509484 [DOI] [Google Scholar]
- 20.Grenier E, Bossis M, Fouville D, Renault L, Mugniéry D. Molecular approaches to the taxonomic position of Peruvian potato cyst nematodes and gene pool similarities in indigenous and imported populations of Globodera. Heredity (Edinb). 2001;86: 277–290. doi: 10.1046/j.1365-2540.2001.00826.x [DOI] [PubMed] [Google Scholar]
- 21.Bulman SR, Marshall JW. Differentiation of Australasian potato cyst nematode (PCN) populations using the polymerase chain reaction (PCR). New Zeal J Crop Hortic Sci. 1997;25: 123–129. doi: 10.1080/01140671.1997.9513998 [DOI] [Google Scholar]
- 22.Grenier E, Fournet S, Petit E, Anthoine G. A cyst nematode “species factory” called the Andes. Nematology. 2010;12: 163–169. doi: 10.1163/138855409X12573393054942 [DOI] [Google Scholar]
- 23.Baeza CA. El nematodo dorado (Heterodera rostochiensis Wol) en Colombia. ICA Ibagué, segunda reunión de Fitopatología y Sanidad Vegetal. 1972. p. 20.
- 24.Nieto L., Varón F., & Dees J. Reconocimiento y distribución del nemátodo quiste de la papa, Globodera pallida Stone, en Colombia. Rev ICA Colomb. 1983;18: 87–94. [Google Scholar]
- 25.Arciniegas N, Caicedo R, Árevalo E. Nematodo dorado. Rev papa. 2012; 33–36. [Google Scholar]
- 26.AGRONET. Red de información y comunicación del sector Agropecuario Colombiano. Anuario estadístico del sector agropecuario. 2020. https://www.agronet.gov.co/Paginas/inicio.aspx%0Ahttp://www.agronet.gov.co..
- 27.Fenwick DW. Methods for the recovery and counting of cysts of Heterodera schachtii from soil. J Helminthol. 1940;18: 155–172. doi: 10.1017/S0022149X00031485 [DOI] [Google Scholar]
- 28.Golden AM. Morphology and Identification of Cyst Nematodes. Lamberti F, Taylor CE, editors. Cyst Nematodes. New York: Plenum Press; 1986. doi: 10.1007/978-1-4613-2251-1_2 [DOI] [Google Scholar]
- 29.Huijsman CA. Veredeling van de aardappel op resistentie tegen Heterodera rostochiensis Wollenweber. Veenman. 1957.
- 30.Ma X, Agudelo P, Mueller JD, Knap HT. Molecular characterization and phylogenetic analysis of Hoplolaimus stephanus. J Nematol. 2011;43: 25–34. [PMC free article] [PubMed] [Google Scholar]
- 31.Nunn GB. Nematode molecular evolution. Ph. D. Thesis, University of Nottingham, Nottingham, UK. 1992.
- 32.Ferris VR, Fer Ris M, Faghihj Amal. Variation in spacer ribosornal DNA in sorne cyst-forming species of plant parasitic nernatodes. Fundam appl Nemalol. 1993;16: 177–184. [Google Scholar]
- 33.Vrain T, Wakarchuk D, Levesque A, Hamilton R. Intraspecific rDNA restriction fragment length polymorphism in the Xiphinema americanum group. Fundam Appl Nematol. 1992;15: 563–573. [Google Scholar]
- 34.Vinet L, Zhedanov A. A “missing” family of classical orthogonal polynomials. Journal of Physics A: Mathematical and Theoretical. University of Nottingham. 2011. doi: 10.1088/1751-8113/44/8/085201 [DOI] [Google Scholar]
- 35.Ferris VR, Miller LI, Faghihi J, Ferris JM. Ribosomal DNA comparisons of Globodera from two continents. J Nematol. 1995;27: 273–283. [PMC free article] [PubMed] [Google Scholar]
- 36.Powers TO, Szalanski AL, Mullin PG, Harris TS, Bertozzi T, Griesbach JA. Identification of seed gall nematodes of agronomic and regulatory concern with PCR-RFLP of ITS1. J Nematol. 2001;33: 191–194. [PMC free article] [PubMed] [Google Scholar]
- 37.Blok VC, Malloch G, Harrower B, Phillips MS, Vrain TC. Intraspecific variation in Ribosomal DNA in populations of the potato cyst nematode Globodera pallida. J Nematol. 1998;30: 262–274. [PMC free article] [PubMed] [Google Scholar]
- 38.Širca S, Urek G. Morphometrical and ribosomal DNA sequence analysis of Globodera rostochiensis and Globodera achilleae from Slovenia. Russ J Nematol. 2004;12: 161–168. [Google Scholar]
- 39.Uehara T, Kushida A, Itou K, Narabu T, Momota Y. Discrimination of three cyst-forming nematodes of the genus Globodera (Nematode: Heteroderidae) from Japan based on PCR-RFLP of ribosomal DNA. Appl Entomol Zool. 2005;40: 537–543. doi: 10.1303/aez.2005.537 [DOI] [Google Scholar]
- 40.Knoetze R, Malan AP, Mouton C. Differentiation of South African potato cyst nematodes (PCN) by analysis of the rDNA internal transcribed spacer region. African Plant Protection. African Plant; 2006. [Google Scholar]
- 41.Plantard O, Picard D, Valette S, Scurrah M, Grenier E, Mugniéry D. Origin and genetic diversity of Western European populations of the potato cyst nematode (Globodera pallida) inferred from mitochondrial sequences and microsatellite loci. Mol Ecol. 2008;17: 2208–2218. doi: 10.1111/j.1365-294X.2008.03718.x [DOI] [PubMed] [Google Scholar]
- 42.Quader M, Nambiar L, Cunnington J. Conventional and real-time PCR-based species identification and diversity of potato cyst nematodes (Globodera spp.) from Victoria, Australia. Nematology. 2008;10: 471–478. doi: 10.1163/156854108784513860 [DOI] [Google Scholar]
- 43.Pylypenko LA, Uehara T, Phillips MS, Sigareva DD, Blok VC. Identification of Globodera rostochiensis and G. pallida in the Ukraine by PCR. Eur J Plant Pathol. 2005;111: 39–46. doi: 10.1007/s10658-004-2732-9 [DOI] [Google Scholar]
- 44.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinforma. 2003; 2–3. [DOI] [PubMed] [Google Scholar]
- 45.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series. 1999. pp. 95–98.10780396 [Google Scholar]
- 46.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17: 540–552. doi: 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 47.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. Partitionfinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2017;34: 772–773. doi: 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- 48.Hasegawa M, Kishino H, Yano T aki. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22: 160–174. doi: 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- 49.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16: 111–120. doi: 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- 50.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19: 1572–1574. doi: 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- 51.Alfaro ME, Zoller S, Lutzoni F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol Evol. 2003;20: 255–266. doi: 10.1093/molbev/msg028 [DOI] [PubMed] [Google Scholar]
- 52.Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas at Austin. 2006.
- 53.Sukumaran J, Holder MT. DendroPy: A Python library for phylogenetic computing. Bioinformatics. 2010;26: 1569–1571. doi: 10.1093/bioinformatics/btq228 [DOI] [PubMed] [Google Scholar]
- 54.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34: 3299–3302. doi: 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- 55.Dandurand LM, Zasada IA, Wang X, Mimee B, De Jong W, Novy R, et al. Current Status of Potato Cyst Nematodes in North America. Annu Rev Phytopathol. 2019;57: 117–133. doi: 10.1146/annurev-phyto-082718-100254 [DOI] [PubMed] [Google Scholar]
- 56.Lucero AM. Manejo integrado de chisas en fincas de minifundio del departamento de Nariño (Colombia). Corpoica Cienc y Tecnol Agropecu. 2006;7: 70. doi: 10.21930/rcta.vol7_num1_art:63 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data (including accession numbers) are within the paper.