The H3K36me2 histone modification pathway is an acquired dependency in H3K27M mutant diffuse intrinsic pontine glioma (DIPG).
Abstract
Histone H3K27M is a driving mutation in diffuse intrinsic pontine glioma (DIPG), a deadly pediatric brain tumor. H3K27M reshapes the epigenome through a global inhibition of PRC2 catalytic activity and displacement of H3K27me2/3, promoting oncogenesis of DIPG. As a consequence, a histone modification H3K36me2, antagonistic to H3K27me2/3, is aberrantly elevated. Here, we investigate the role of H3K36me2 in H3K27M-DIPG by tackling its upstream catalyzing enzymes (writers) and downstream binding factors (readers). We determine that NSD1 and NSD2 are the key writers for H3K36me2. Loss of NSD1/2 in H3K27M-DIPG impedes cellular proliferation and tumorigenesis by disrupting tumor-promoting transcriptional programs. Further, we demonstrate that LEDGF and HDGF2 are the main readers mediating the protumorigenic effects downstream of NSD1/2-H3K36me2. Treatment with a chemically modified peptide mimicking endogenous H3K36me2 dislodges LEDGF/HDGF2 from chromatin and specifically inhibits the proliferation of H3K27M-DIPG. Our results indicate a functional pathway of NSD1/2-H3K36me2-LEDGF/HDGF2 as an acquired dependency in H3K27M-DIPG.
INTRODUCTION
H3K27M is a driver mutation in 80% of diffuse midline glioma (DMG), a malignant, treatment-resistant brain tumor that includes diffuse intrinsic pontine glioma (DIPG), arising from the pons as well as other gliomas in the thalamus and spinal cord (1–3). Patients affected by this disease typically range from 5 to 7 years old and have a 5-year survival rate of less than 2% with an average postdiagnostic survival of 9 months (4, 5). The hallmark of H3K27M DMG/DIPG is a global loss in chromatin-associated di- and trimethylated lysine-27 of histone H3 (H3K27me2/me3) (6, 7). H3K27me1/me2/me3 is catalyzed solely by the polycomb repressive complex 2 (PRC2) (8, 9). Notably, PRC2 is allosterically stimulated by its own catalytic product, H3K27me3, fostering a positive feedback loop (10, 11). This mechanism by which PRC2 “writes” and “reads” H3K27me3 is central to the spreading and formation of extensive H3K27me3-chromatin domains, which provide the platform for chromatin compaction and thus repression (12). These H3K27me3-chromatin domains are also inherited upon DNA replication (13). The inheritance of H3K27me3 together with the PRC2 “write-read” mechanism can fully restore H3K27me3-chromatin domains upon DNA replication (13). This unique property of PRC2 points to its critical role in propagating a particular cellular identity. However, as reported by our laboratory and others, H3K27M inhibits PRC2 catalysis of H3K27me2/me3 in multiple ways (6, 14). H3K27M preferentially binds to the allosterically activated state of PRC2, thereby hindering this crucial feedback mechanism leading to a global loss of H3K27me3 (14). This phenomenon not only disrupts the formation of extensive H3K27me3-repressive domains but also is expected to impact their inheritance. Expectedly, H3K27M-mediated dysregulation of this critical epigenetic state fosters genomic derepression and aberrant activation of inappropriate genes that potentially cooperate with other genetic mutations in driving early oncogenesis during tumor evolution.
Previously, we and others reported that H3K27M DIPG cells exhibit elevated levels of another chromatin-associated histone posttranslational modification, demethylated lysine-36 of histone H3 (H3K36me2) (14–16). While H3K36me2 is antagonistic to the catalysis of H3K27me2/me3 (17, 18), it remains unclear as to whether the elevated levels of H3K36me2 in DIPG arise through the increased levels of transcription in these cells or letup from the antagonistic effects of H3K27me2/me3. The role of these elevated levels of H3K36me2 in promoting tumorigenesis is also unclear. Five mammalian lysine methyltransferases can “write,” i.e., catalyze, histone H3K36 methylation, generating H3K36me1 and H3K36me2: NSD1, NSD2, NSD3, ASH1L, and SETD2 (19–21). Among these enzymes, only SETD2 can further convert H3K36me2 to H3K36me3 (21), the latter being closely associated with transcribed gene bodies and with the recruitment of RNA splicing factors through direct protein-protein interactions (22). However, unlike H3K36me3, the distribution of H3K36me2 is less restricted and appears to be generally associated with euchromatic regions (23). Both H3K36me2 and H3K36me3 are recognized by downstream proteins, “readers,” that contain one or more methyl-lysine reading Pro-Trp-Trp-Pro (PWWP) domains (24). Among these readers, we previously demonstrated that LEDGF and HDGF2 are associated with all H3K36me2/me3-decorated genomic regions and facilitate RNA polymerase II–dependent transcription by relieving the nucleosomal barrier, functionally resembling the FACT complex (25). Other PWWP-containing readers, such as DNMT3A and DNMT3B, have been shown to localize to intergenic regions and to regulate DNA methylation patterns (26). Here, we investigate the role of H3K36me2 by tackling its writers and readers to ascertain the contribution of any of these writer-reader modules in establishing dependency on H3K36me2 in H3K27M DIPG cells.
RESULTS
NSD1 and NSD2 are responsible for global H3K36me2 levels and required for H3K27M-DIPG proliferation
To probe the functional role of the aberrant elevation of H3K36me2 in H3K27M-DIPG cells, we first engineered a doxycycline-inducible H3K36M construct in DIPG4 cells comprising a H3K27M mutation. H3K36M is a general inhibitor of H3K36 methyltransferases, and its expression in DIPG4 cells effectively reduced endogenous H3K36me2/3 levels and impeded cell proliferation by an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assay (Fig. 1A). To ascertain the writers that are responsible for H3K36me2 catalysis in the context of DIPG, we adopted a small interference RNA (siRNA)–based approach by knocking down NSD1 alone and in conjunction with each of the remaining H3K36 methyltransferases in DIPG4 cells. Knockdown (KD) of NSD1 alone in DIPG4 had a moderate effect on endogenous H3K36me2 levels, and co-KD of NSD2 manifested an additional effect, whereas co-KD of NSD3, ASH1L, or SETD2 was ineffectual (Fig. 1B, left). Consistent with the literature, SETD2 is the only mammalian enzyme responsible for H3K36me3. While co-KD of NSD1 and NSD2 had a substantial impact on H3K36me2 levels, additional KD of NSD3, ASH1L, or SETD2 was ineffectual (Fig. 1B, right). We further extended our analysis to other DIPG cell lines, including DIPG6 (H3K27M), DIPG13 (H3K27M), DIPG38 (H3K27M), DIPG10 [H3–wild type (WT)], and a cortex glioma cell line, pcGBM2 (H3-WT) (Fig. 1C and figs. S1, A and B). While single KD of NSD1 in pcGBM2 and KD of NSD2 in DIPG13 rendered a major effect on H3K36me2 levels, their co-KD demonstrated a more substantial effect on eliminating endogenous H3K36me2 levels (Fig. 1C and fig. S1B). Intriguingly, co-KD of NSD1 and NSD2 appeared to strongly reduce the proliferation of three H3K27M DIPG cell lines analyzed (DIPG6, DIPG13, and DIPG38) (Fig. 1D and fig. S1C), but a considerably milder effect was observed in H3-WT cells, including DIPG10 or human embryonic kidney 293T (HEK293T) cells (Fig. 1D and fig. S1D).
Fig. 1. NSD1/2 and LEDGF/HDGF2 are required for H3K27M-DIPG cell proliferation.
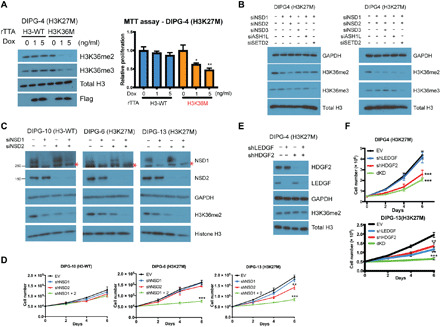
(A) Left: Western blot of H3K36me2, H3K36me2, histone H3, and Flag in DIPG4 (H3K27M) cells expressing doxycycline-inducible, flag-tagged H3-WT or H3K36M constructs. Doxycycline (Dox; 0, 1, or 5 ng/ml) was administrated for an induction of ectopic histones. Cell lysates were harvested 3 days after induction. Right: An MTT assay was conducted for assessing the proliferation of cells treated with respective conditions in the left panel. Cells were seeded 3 days after induction and the MTT assay was conducted 6 days after induction. (B) Western blot of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), H3K36me2, H3K36me3, and histone H3 in DIPG4 cells transfected with indicated siRNAs, including NSD1 and NSD2, NSD3, ASH1L, or SETD2. (C) Western blot of NSD1, NSD2, GAPDH, H3K36me2, and histone H3 in DIPG10 (H3-WT), DIPG6 (H3K27M), and DIPG13 (H3K27M) cells transfected with and without siRNAs against NSD1 and/or NSD2. Asterisks indicate nonspecific bands detected by an anti-NSD1 antibody (Bethyl Laboratories, no. A300-BL715). Cell lysates were harvested 3 days after transfection, and GAPDH was used as a loading control in (B) and (C). (D) Proliferation assays of DIPG10, DIPG6, and DIPG13 cells stably expressing control or shRNAs against NSD1 and/or NSD2. Cell numbers were counted after 2, 4, and 6 days. (E) Western blot of LEDGF, HDGF2, GAPDH, H3K36me2, and histone H3 in DIPG4 cells stably expressing control or shRNAs against LEDGF and/or HDGF2. (F) Proliferation assays of DIPG4 cells used in (E) and DIPG13 cells with the same conditions. rTTA, reverse tetracycline-controlled transactivator; EV, empty vector. *P < 0.05, **P < 0.01, and ***P < 0.001 by Student’s t test.
LEDGF and HDGF2, but not DNMT3A and DNMT3B, are essential for proliferation of H3K27M-DIPG
Having established that NSD1/2 is crucial for the chromatin deposition of H3K36me2 and the proliferation of H3K27M-DIPG cells, we next investigated which readers might mediate the downstream effects. Among the PWWP domain–containing proteins, LEDGF, HDGF2, DNMT3A, and DNMT3B appeared to have preferential binding to H3K36me2/me3 peptides and nucleosomes (24). KD of LEDGF and/or HDGF2 had little impact on H3K36me2 levels in DIPG4 cells (Fig. 1E). Similar to the single KDs of NSD1 and NSD2, a moderate reduction in proliferation was observed in the single KD of HDGF2 in DIPG4 cells and the single KD of LEDGF or HDGF2 in DIPG13 cells, while their co-KD exhibited a far more substantial impact on DIPG4 and DIPG13 cells (Fig. 1F). Further, co-KD of LEDGF and HDGF2 had only a very mild effect on the proliferation of DIPG10 (H3-WT) cells (fig. S1E). On the other hand, neither the single nor the joint KDs of DNMT3A and DNMT3B had a notable impact on cell proliferation (fig. S1F). At the cellular levels, co-KD of NSD1 and NSD2 as well as co-KD of LEDGF and HDGF2 induced a G1/S cell cycle arrest and a reduced expression of proliferation marker gene, Ki67, but had no impact on apoptotic markers, cleaved poly(ADP-ribose) polymerase and caspase 3, in H3K27M-DIPG cells (fig. S1, G and H).
NSD1/2 and LEDGF/HDGF2 facilitate H3K27M-DIPG tumor growth in vivo
Next, we implanted these stable KD cell lines using lentivirus-based short hairpin RNAs (shRNAs) in a xenograft mouse model via an intracranial injection in the cerebral hemisphere. Of note, DIPG4 does not steadily form tumors in the brain in NOD/SCID/IL2γ (NSG) mice although it has been reported to form tumors by flank injections in nude mice (27). As DIPG13 is more dependent on NSD2 for its H3K36me2 levels, KD of NSD2 exhibited reduced tumor size and extended survival of recipient mice relative to control and the NSD1 KD group, and this effect was further amplified in the NSD1 and NSD2 co-KD group (Fig. 2A). Similarly, KD of LEDGF or HDGF2 alone exhibited a partial effect on reducing tumor size and extending mice survival, while their co-KD manifested a much more robust effect (Fig. 2B). Of note, we have observed a rapid selection advantage of clones that escaped from shRNA-mediated KD in cultured H3K27M-DIPG cells, likely due to the strong inhibition of cell proliferation by shRNAs. We further confirmed that the engrafted tumors formed in NSG mice also escaped from shRNA KD by examining the end-stage tumor lysates from control and double KD (dKD) groups (fig. S2, A and B).
Fig. 2. KD of NSD1/2 or LEDGF/HDGF2 delays tumor onset and extends survival in a xenograft mouse model.
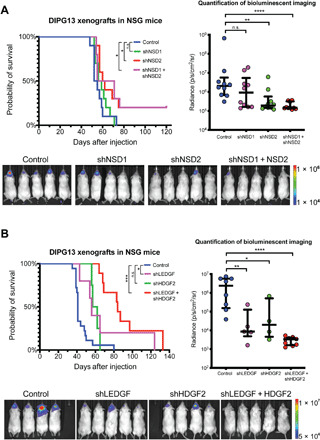
(A) Left: A Kaplan-Meier survival curve plot of mice bearing xenograft tumors with endpoints defined by a sign of distress. DIPG13 cells stably expressing a firefly luciferase and control or shRNAs against NSD1 and/or NSD2 were implanted in the cortex of NSG mice by intracranial injection. A total of 250,000 cells were implanted in each mouse (n = 10 mice for each condition). Right: A quantification of firefly luciferase signals in mice described in the left panel. Bioluminescent imaging (BLI) data were presented at day 35 after injection before the first mouse exhibited a sign of distress. Bottom: Representative BLI images from indicated conditions. (B) Left: A Kaplan-Meier survival curve of mice implanted with DIPG13 cells stably expressing control or shRNAs against LEDGF and/or HDGF2 in the same experimental conditions described in (A) (n = 10 mice for control; n = 5 for shLEDGF; n = 4 for shHDGF2; and n = 9 for shLEDGF + shHDGF2). Right: A quantification of BLI signals in mice describe in the left panel. Bottom: Representative BLI images from indicated conditions. *P < 0.05 and ***P < 0.001 by log-rank test; n.s., not significant (P > 0.05) for Kaplan-Meier survival analysis. *P < 0.05, **P < 0.01 and ***P < 0.001 by a nonparametric Mann-Whitney test for BLI quantifications.
LEDGF/HDGF2 spread with H3K36me2, correlating with mRNA expression
As loss-of-function of LEDGF/HDGF2 phenocopied that of NSD1/2, we speculated that LEDGF/HDGF2 function as the main readers that mediate the downstream effect of NSD1/2 in regulating gene expression profiles and pathways in the context of DIPG. Chromatin immunoprecipitation sequencing (ChIP-seq) of H3K36me2, LEDGF, and HDGF2 gave evidence of a general positive correlation among their chromatin occupancies (Fig. 3A) (25). In a comparison between H3-WT and H3K27M DIPG cells, the enriched occupancy of LEDGF/HDGF2 correlated highly with the spreading of H3K36me2 at derepressed loci elicited by H3K27M, in stark contrast to those genes decorated by H3K27me3 shown by heatmaps and metaprofiles of ChIP-seq (Fig. 3A and fig. S3, A and B). Genes with higher messenger RNA (mRNA) expression levels by RNA sequencing (RNA-seq) exhibited substantially higher enrichments in H3K36me2, LEDGF, and HDGF2 occupancies by ChIP-seq (Fig. 3B). Furthermore, in an isogenic HEK293T-based system with inducible H3 histone, either WT or H3K27M, we consistently observed increased LEDGF/HDGF2 occupancy at genes that had lost H3K27me3 and gained H3K36me2 upon induction of the H3K27M oncohistone (Fig. 3C). Next, we generated NSD1 and NSD2 knockout (KO) lines in DIPG13 cells by using CRISPR-Cas9 to disrupt the respective catalytic SET domain. Consistent with RNA interference–based results, NSD2-KO cells exhibited extremely poor proliferation and very low H3K36me2 levels, whereas changes in these criteria were almost inconsequential in the NSD1-KO cells (fig. S4A). NSD2-KO cells also exhibited largely reduced chromatin occupancies of LEDGF and HDGF2 (Fig. 3D). Together, these data ascertain the hierarchical relationship of NSD1/2-LEDGF/HDGF2 in regulating gene expression in DIPG cells. Of note, we were unable to obtain a NSD1/2 double KO (dKO) line in DIPG13, and the NSD2-KO lines were barely maintainable, becoming very sensitive to lentiviral infection and the selection process, thereby preventing a successful rescue using a WT NSD2 complementary DNA. In parallel, we also generated an NSD1 and NSD2 dKO by CRISPR-Cas9 in HEK293T cells and corroborated that loss of NSD1/2 and H3K36me2 only slightly affected its proliferation (figs. S1D and S4B).
Fig. 3. LEDGF/HDGF2 occupancy on chromatin is dependent on NSD1/2-H3K36me2 and positively correlates with gene expression.
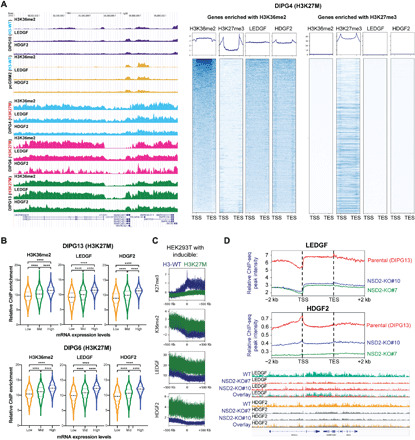
(A) Left: Representative ChIP-seq tracks of H3K36me2, LEDGF, and HDGF2 chromatin co-occupancy in H3-WT (DIPG10 and pcGBM2) and H3K27M (DIPG4, DIPG6, and DIPG13) cells. Right: Representative heatmaps of ChIP-seq for DIPG4 cells centered to H3K36me2- or H3K27me3-enriched genes. (B) Violin plots showing enrichment of H3K36me2, LEDGF, or HDGF2, respectively, for genes categorized as low, mid, or high based on their mRNA expression. The central thick dash line indicates the mean value of each plot; the upper thin dash line indicates the top 25% percentile and the lower one for the bottom 25% percentile. (C) Metaprofile plots of H3K27me3, H3K36me2, LEDGF, and HDGF2 ChIP-seq data in HEK293T cells ectopically expressing H3-WT or H3K27M for 24 hours. Data were presented within a 500-kb window upstream or downstream from TSS. (D) Top: Metaprofile plots of LEDGF and HDGF2 occupancy in WT or NSD2-KO DIPG13 cells (clone #7 and clone #10). Bottom: Representative ChIP-seq tracks for the top panel. Overlaid panels were presented at the bottom to better illustrate the differences. TSS, transcription start site; TES, transcription end site. ****P < 0.0001 by Student’s t test.
NSD1/2 and LEDGF/HDGF2 control protumorigenic gene expression signatures
We then investigated the functional consequence to gene expression profiles and the pathways affected upon loss of NSD1/2 or LEDGF/HDGF2 in DIPG13 cells. We performed RNA-seq analysis using NSD2-KO cells in which we transiently knocked down NSD1 using siRNAs (NSD2-KO + siNSD1), as well as LEDGF/HDGF2 dKD cells. By gene set enrichment analysis (GSEA), we detected the down-regulation of a total of 1208 previously established gene signatures/pathways in NSD2-KO + siNSD1 cells (clone #7), two-thirds (788 of 1208, 65%) of which overlapped with those down-regulated in LEDGF/HDGF2 dKD cells (Fig. 4A, left). A similar result (752 of 1129, 66%) was observed in a second independent NSD2-KO clone (clone #10) (Fig. 4A, right). The high-ranked overlapping signatures/pathways included ESC stemness gene signature, CHEK2 pathway, epidermal growth factor receptor (EGFR) signaling, MYC targets, pre-B lymphocyte developmental genes, and hypoxia–down-regulated genes (Fig. 4B and fig. S5A), indicating that the NSD1/2-H3K36me2-LEDGF/HDGF2 writer-reader module is necessary for maintaining the distinctive tumorigenic gene expression pattern in H3K27M DIPG cells (Fig. 4B). Of note, the down-regulated pathways in NSD2-KO cells are highly overlapped with those in NSD2-KO + siNSD1 cells since DIPG13 cells are mainly dependent on NSD2 for its endogenous H3K36me2 levels (fig. S5B). On the other hand, co-KD of DNMT3A and DNMT3B exhibited some, but less, correlation with loss of NSD1/2 as gauged by RNA-seq and GSEA analyses (fig. S5C). Moreover, 42.7% (516 of 1208) of down-regulated pathways in NSD2-KO + siNSD1 and 41.4% (754 of 1822) of those in LEDGF/HDGF2-dKD cells are overlapped with those in H3K27M (DIPG13) when being compared to H3-WT (DIPG10) cells (fig. S5D), corroborating a relevance of H3K36me2-dysregulated pathways being adapted in H3K27M cells.
Fig. 4. NSD1/2 and LEDGF/HDGF2 coregulate protumorigenic pathways and can be disrupted by a H3K36me2-mimicking peptide.
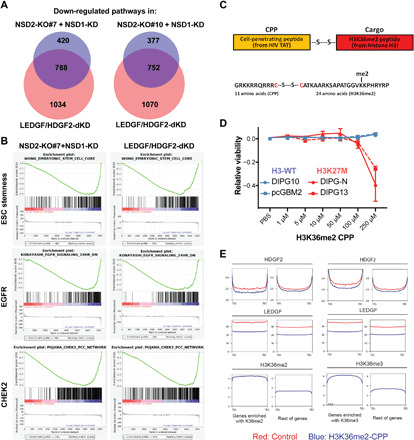
(A) Venn diagrams showing overlaps of gene signatures/pathways down-regulated in LEDGF/HDGF2 dKD DIPG13 cells and NSD2-KO#7 + siNSD1 or NSD2-KO#10 + siNSD1 DIPG13 cells (P = 4.2 × 10−9 and 1.8 × 10−4, respectively). The alteration of gene signatures/pathways was detected by GSEA. P values were obtained by the hypergeometric distribution to compute the significance of the overlap of two pathway sets. (B) Representative images of highly ranked GSEA signatures/pathways detected in (A), including a CHEK2 pathway, an embryonic stem cell (ESC) steamness signature, and a set of EGFR signaling target genes. (C) Top: A schematic illustration of the design of CPP. A HIV-based cell entry peptide was linked to a H3K36me2 peptide (histone H3 21 to 43 amino acids, cargo peptide) by a disulfide bond. Bottom: Amino acid sequence of H3K36me2-linked CPP. (D) A CellTiter-Glo cell survival assay for H3-WT (DIPG10 and pcGBM2) and H3K27M (DIPG-N and DIPG13) cells treated with control (vehicle only) or H3K36me2-CPP. Cells were assayed at 72 hours after dosing with a titration of control or H3K36me2-CPP and data were presented by ratios of CellTiter-Glo signals in control versus H3K36me2-CPP treated cells. PBS, phosphate-buffered saline. (E) Metaprofiles of ChIP-seq analysis for changes in LEDGF and HDGF2 occupancy of genes enriched with H3K36me2 or H3K36me3 and the rest of genes in DIPG13 cells treated with a control vehicle (red) or a H3K36me2-CPP (blue).
H3K36me2-mimicking peptide inhibits H3K27M-DIPG proliferation by displacing chromatin-bound LEDGF/HDGF2
Last, we adopted a chemical approach to tackle the H3K36me2 writer-reader pathway. We engineered a transportable H3K36me2 peptide comprising a cell-penetrating peptide (CPP) of 11 amino acids derived from an HIV TAT protein–based cell membrane entry signal, in a disulfate bond fusion with an H3 peptide (amino acids 21 to 43) having a dimethyl chemical modification at lysine-36 (H3K36me2-CPP) (Fig. 4C). The CPPs enter the nucleus as evidenced by the literature (28). Upon cell entry, the disulfate bond is reduced in the intracellular redox environment, and the released H3K36me2 peptide acts as an endogenous competitive inhibitor against the PWWP domains of LEDGF and HDGF. Notably, incubation with this peptide largely reduced the proliferation of H3K27M DIPG cells (DIPG6 and DIPG13) but had little impact on that of H3-WT cells (DIPG10 and pcGBM2) (Fig. 4D). A treatment including extended titrations of H3K36me2-CPP further demonstrated that the median inhibitory concentration of H3K36me2-CPP for DIPG13 is ~173 μM (fig. S5E). Treatment with H3K36me2-CPP in DIPG13 cells competitively dislodged endogenous LEDGF and HDGF2 from chromatin, pointing to its functional efficacy, and such effects appear to be general and genome-wide as evident by examining genes with high or low enrichment of H3K36m2 or H3K36me3, respectively (Fig. 4E). Of note, H3K36me2-CPP was more efficient at dislodging LEDGF relative to HDGF2, possibly due to HDGF2 having a stronger preferential binding to H3K36me3 peptides and nucleosomes than LEDGF (24, 25).
DISCUSSION
Similar to the case of PRC2 and H3K27me3, NSD1/2 and H3K36me2 are essential for normal development but also play pleiotropic and context-dependent roles in human cancer (29). For example, inactivating mutations in NSD1 were frequently found in head and neck cancers (30), whereas an activating NUP98-NSD1 fusion protein that arises from a chromosomal translocation drives leukemogenesis in human acute myeloid leukemia (31, 32), and further, a gain-of-function mutation in NSD2 is present in acute lymphoblastic leukemia (33, 34). Here, we established that LEDGF/HDGF2 are functional readers mediating protumorigenic effects downstream of NSD1/2 in the context of H3K27M-DIPG. H3K27M-DIPG acquire a previously unidentified dependency on this H3K36me2 writer-reader axis through its maintenance of a tumor-promoting gene expression profile. However, the tumor evolution process that leads to this acquired dependency following the initial H3K27M-mediated epigenome remodeling remains to be defined. While NSD1 and NSD2 functionally converge, H3K27M-DIPG cell lines exhibited a spectrum of dependency on either of them, similar to the case with LEDGF and HDGF2. Additional layers of regulation may foster such individual differences, including the ratio of expression levels and involvement of potential cofactors. In addition, as H3K36me2 is antagonistic to H3K27me2/me3, depletion of NSD1/2 could partially restore H3K27me3 at some normally repressed loci, which might also contribute to a tumor-inhibiting effect even with the presence of H3K27M in DIPG cells, as implied by a recent study reporting the generation of an isogenic DIPG system (16).
Importantly, these H3K36me2 writers and readers can be potentially targeted by pharmacological approaches through their functional domains, such as the PWWP domains and the SET domains of NSD1/2. However, extensive efforts to target the SET domains of NSD proteins with potent inhibitors have proven unsuccessful at the nanomolar scale. In this regard, a recent cryo–electron microscopy study revealed that a series of residues within and flanking the SET domain are crucial for unwrapping the linker and nucleosomal DNA such that an autoinhibitory state inherent to NSD proteins is converted to an active conformation (35, 36). Understanding these unique features of NSDs could further facilitate the structurally assisted design of new inhibitors. Other pharmacological targeting strategies that might function in a more specific manner involve the PWWP domains of LEDGF/HDGF2. Fortunately, two selective PWWP domain binding ligands have been reported recently by the Structural Genome Consortium (www.thesgc.org). Such prototype compounds will provide insights into future pharmacological development. In addition to H3K27M-DIPG, an impaired PRC2 activity and loss of H3K27me2/me3 also drive tumorigenesis of several other types of cancer, including malignant peripheral nerve sheath tumors having frequent genetic deletions in multiple PRC2 core subunits (37) and posterior fossa type A ependymoma having aberrant expression of EZHIP, an endogenous protein that inhibits PRC2 (38–40). The results here expand our understanding of epigenetic dysregulations in H3K27M-DIPG and suggest that disruption of the H3K36me2 pathway should be taken into consideration for developing and designing therapeutic interventions for a broad spectrum of tumors that exhibit impaired PRC2 activity. Recently, several studies have also explored the transcriptional and epigenetic vulnerabilities of H3K27M-DIPG and suggested a number of potential targets, including CDK7 of TFIIH, bromodomain proteins binding to acetylated histones, and the residual activity of PRC2 (27, 41, 42). Together with our results, these approaches may provide a foundation for combinatory therapies.
METHODS
Cell culture
SU-DIPG-4, SU-DIPG-6, SU-DIPG-13, and SU-DIPG-38 cells are gifts from the laboratory of M. Monje (Stanford). DIPG-N is generated by D.G.P. at the New York University (NYU). All DIPG cell lines were cultured and maintained in tumor stem medium, which contains 1:1 mixture of Neurobasal-A and Dulbecco’s Modified Eagle Medium (DMEM)/F12 medium (Life Technologies), supplemented with 1% antibiotic/antimycotic solution (Life Technologies), 2 mM GlutaMAX (Life Technologies), 10 mM Hepes buffer (Life Technologies), 1 mM sodium pyruvate (Sigma-Aldrich), 1% MEM nonessential amino acid solution (Sigma-Aldrich), B-27 supplement minus vitamin A (Gibco), human EGF (20 ng/ml) (Shenandoah Biotech), and human fibroblast growth factor (20 ng/ml) (Shenandoah Biotech). HEK293T and HEK293-FT cells were maintained in DMEM supplemented with 10% fetal bovine serum,1 mM sodium pyruvate (Sigma-Aldrich), 2 mM l-glutamine (Sigma-Aldrich), and 1% penicillin/streptomycin solution (Sigma-Aldrich).
Antibodies
The antibodies used in this study are as follows: LEDGF (Proteintech) rabbit polyclonal, catalog no. 25504-1-AP; HDGF2 (Proteintech) rabbit polyclonal, catalog no. 15134-1-AP; H3K27me3 (Cell Signaling Technology) rabbit monoclonal C36B11, catalog no. 9733; H3K36me2 (Cell Signaling Technology) rabbit monoclonal C75H12, catalog no. 2901; H3K36me3 (Cell Signaling Technology) rabbit monoclonal D5A7, catalog no. 4909; NSD1 (Bethyl Laboratories) rabbit monoclonal, catalog no. A300-BL715 (discontinued); NSD1 (University of California, Davis/National Institutes of Health NeuroMab Facility) mouse monoclonal, N312/10; NSD2 (Millipore) mouse monoclonal 29D1, catalog no. MABE191; NSD3 (Cell Signaling Technology) rabbit monoclonal D4N9N, catalog no. 92056; ASH1L (Bethyl Laboratories) rabbit polyclonal, catalog no. A301-748A; SETD2 (Bio-Rad) mouse monoclonal OTI1E1, catalog no. VMA00449; glyceraldehyde-3-phosphate dehydrogenase (Cell Signaling Technology) rabbit monoclonal D16H11, catalog no. 5174; histone H3 (Abcam) rabbit monoclonal EPR16987, catalog no. ab176842; anti-Flag (Sigma-Aldrich) mouse monoclonal M2, catalog no. F1804; and H2Av (Active Motif) rabbit polyclonal, catalog no. 39715.
shRNA constructs and lentivirus production
pLKO.1-based shRNAs against NSD1, NSD2, LEDGF, and HDGF2 were purchased from Sigma-Aldrich for lentiviral production and delivery. A lentiviral vector expressing firefly luciferase and mCherry was used for bioluminescence imaging experiments (43). For the production of viral particles, 10 μg of lentiviral vectors were cotransfected with 2.5 μg of pcREV, 3 μg of BH-10, and 5 μg of pVSV-G packaging vectors into 293-FT cells. The virus-containing medium was collected 48 hours after transfection, and the target cells were spin infected. Polybrene was added to the viral medium at a concentration of 8 μg/ml. Infected cells were selected by puromycin (1 ng/ml) for 2 days, G-418 (400 μg/ml) for 5 days, or fluorescence-activated cell sorting (FACS)–sorted for mCherry. The target sequence of each shRNA used is as follows: NSD1, AGGAGTGGATGGGACATATAA (TRCN0000238370); NSD2, CGGAAAGCCAAGTTCACCTTT (TRCN0000274182); LEDGF, GCAGCTACAGAAGTCAAGATT (TRCN0000286344); and HDGF2, GCAGGAGAGCAGAGCAGAGAA (TRCN0000107975).
siRNA transfection
The siRNAs used in this study were purchased from Dharmacon (ON-TARGETplus SMARTpool), including the ones against NSD1, NSD2, NSD3, ASH1L, SETD2, DNMT3A, and DNMT3B. Five nanomoles of each pooled siRNA was transfected into DIPG cells at ~50% confluency in six-well plates using Lipofectamine RNAiMAX according to the manufacturer’s instructions.
CRISPR-Cas9 genome editing
Single-guide RNAs (sgRNAs) were designed using CRISPR design tool in https://benchling.com. All sgRNAs used were cloned in pSpCas9(BB)-2A–green fluorescent protein (GFP) (plasmid 48138, Addgene). The sgRNAs were transfected into DIPG cells using Lipofectamine 2000 (Life Technologies). Single clones from GFP-positive cells were isolated individually into each well of 96-well plates by FACS. The sequence of sgRNA that targets the SET domain of NSD1 is GTAGCTTTACAGTTGCAACG and that of NSD2 is CCCACAGATGAGAATCCTTG.
Mouse intracranial injections and bioluminescence imaging
Mice were housed within NYU Langone Medical Center’s Animal Facilities. All procedures were performed according to our IACUC (Institutional Animal Care and Use Committee)-approved protocol as previously described (44). Briefly, 6- to 8-week old NSG-null mice were anesthetized by intraperitoneal injection of ketamine/xylazine (10 and 100 mg/kg, respectively), mounted on a stereotactic frame (Harvard Apparatus), in which a high-speed drill was used to drill a hole in the calvaria (2 mm off the midline and 2 mm anterior to the coronal suture), and stereotactically injected with 5 μl of a suspension of human DIPG cells (50,000 cells/μl) at a depth of 3 mm. Animals were imaged for luciferase expression at the time points indicated. Mice were injected with luciferin (Gold Biotechnology 115144-35-9) at a dose of 200 mg/kg 15 min before imaging on the PerkinElmer IVIS Spectrum instrument. The resulting images were analyzed using PerkinElmer’s Living Image software package.
ChIP-seq and RNA-seq
ChIP-seq experiments were performed as previously described (11). Briefly, cells were cross-linked with 1% formaldehyde for 10 min. Following nuclei isolation, the chromatin was extracted and fragmented to ~250 base pairs (bp) using a Diagenode Bioruptor. ChIP was performed with the specific antibodies listed above. For quantification, (spike-in) chromatin from Drosophila (1:100 ratio to the experimental chromatin) with Drosophila-specific H2Av antibody was added to each sample as a spike-in control, allowing ChIPs to be compared to one another. Libraries were prepared using 1 to 30 ng of immunoprecipitated DNA as previously described (45). For RNA-seq experiments, RNA was isolated using RNeasy mini spin columns (Qiagen) according to the manufacturer’s instruction. Total RNA (1 to 5 μg) was then processed by oligo(dT) selection and library preparation using the Automated KAPA Library Prep Kit.
Bioinformatics
ChIP-seq and RNA-seq analyses were processed as previously described (11, 14, 25). Briefly, the ChIP-seq data were first mapped to human genome (hg38) using the Bowtie2 software package (version 2.3.0). All reads that failed to align to the human genome were mapped to the fly genome (dm6). The total library size was then adjusted to the reference genome (fly). MACS2 software package (version 2.1.1) was used for calling significantly enriched peaks at a false discovery rate less than 5% relative to the input samples. For RNA-seq data, STAR (version 2.6.1) (46) and RSEM (version 1.3.2) (47) indices were created on the basis of the mouse 10 ensemble genome and gene annotations downloaded from UCSC genome browser. Paired-end 79-bp reads were directly mapped to this STAR index with command line options “--outFilterMismatchNmax 3 outFilterMultimapNmax 20 --winAnchorMultimapNmax 50 --quantMode TranscriptomeSAM GeneCounts.” RSEM software package was then used to estimate relative gene expressions with parameter settings “--paired-end --strandedness reverse” on the alignments generated by STAR. For all genes with at least one of the libraries above zero transcript per million, the average expression values across biological replicates were compared between samples for detecting differentially expressed genes, using DESeq2 (48). Gene Ontology (GO) term enrichment analysis was performed using DAVID Functional Annotation Tool (49). The complete list of GO term categories with statistically significant enrichment was extracted. GSEA was conducted using GSEAPreranked (50) software package on differential analysis results generated by DESeq2.
Metagene profile analyses
Metagene profiles for Figs. 3C and 4E and fig. S3 were generated with deepTools v2.3.3. Genes were divided in three equal categories (low, mid, and high) by their RNA-seq counts. Only genes containing peaks for H3K36me2, LEDGF, and HDGF2, respectively, were selected and then plotted (Fig. 3B). Genes with peaks in the control condition for H3K36me2 and H3K36me3 were selected and plotted for HDGF2 and LEDGF enrichment, respectively, for control and H3K36me2-CPP conditions (Fig. 4E). Genes with peaks for K27me3, H3K36me2, and H3K36me3, respectively, were selected and then plotted for K27me3, H3K36me2, H3K36me3, HDGF2, and LEDGF and compared to multiple WT and K27M DIPG cell lines (fig. S3).
CPP treatment
CPPs were purchased from LifeTein. The CPP was reconstituted and diluted in phosphate-buffered saline as vehicle. For CellTiter-Glo cell survival assays, cells were treated for 72 hours with a titration of cell penetrating peptide as indicated. For ChIP-seq, cells were treated with 250 μM CPP or vehicle only (control) for 16 hours and then cross-linked with 1% formaldehyde for 10 min and processed for ChIP-seq.
Acknowledgments
We thank L. D. Vales for constructive comments and proofreading of the manuscript and S. Baker and N. Jabado for scientific discussion. We also thank the past and present members of D.R.’s laboratory for discussion and L. Popoca, D. Hernandez, and H. Yang for technical assistance. Funding: The New York University Flow Cytometry Core, Proteomics Laboratory, and Genome Technology Center were partially supported by the New York University Grossman School of Medicine and the Laura and Isaac Perlmutter Cancer Center support grant, National Cancer Institute (P30CA016087). The work in D.R.’s laboratory is supported by National Institutes of Health (NIH) grant R01CA199652, the Howard Hughes Medical Institute (HHMI), and the Making Headway Foundation St. Baldrick’s Research Grant (189290). J.-R.Y. is supported by the American Cancer Society (DMC-PF-17-035-01). G.L. is supported by the Hyundai Hope on Wheels Research Grant, the Making Headway Foundation St. Baldrick’s Research Grant (189290), and the Alex’s Lemonade Stand Foundation. J.M.S. was supported as a Simons Foundation’s junior fellow and by NIH grant K99AA024837. J.D.F. was supported by a NYSTEM Stem Cell Biology training grant to NYU Grossman School of Medicine (no. C322560GG). D.G.P. was supported by NIH/NINDS R01NS102665, NYSTEM (NY State Stem Cell Science) IIRP C32595GG, NIH/NIBIB R01EB028774 (to S. Baete at NYU Grossman School of Medicine), NYU Grossman School of Medicine, and DFG (German Research Foundation) FOR2149 as Mercator fellow. Author contributions: J.-R.Y., G.L., J.M.S., and D.R. conceptualized and designed the study. J.-R.Y., G.L., and J.M.S. conducted the experiments. D.B. and J.D.F. conducted the mouse work under the supervision of D.G.P. R.S.-M., Y.J., and N.D. performed bioinformatics analyses. J.-R.Y. and G.L. wrote the manuscript under the guidance of D.R. Competing interests: D.R. is a cofounder of Constellation Pharmaceuticals and Fulcrum Therapeutics. J.M.S. is a co-founder of Nzumbe Inc. D.G.P. is an inventor on a European Union patent related to this work filed by New York University (“Method for treating high-grade gliomas,” no. 3289104, filed 29 April 2016, published 16 November 2020). D.G.P. has received consultant fees from Tocagen, Synaptive Medical, Monteris, and Robeaute. The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The sequencing data have been uploaded to NCBI-GEO under reference no. GSE174006. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/29/eabg7444/DC1
REFERENCES AND NOTES
- 1.Schwartzentruber J., Korshunov A., Liu X. Y., Jones D. T. W., Pfaff E., Jacob K., Sturm D., Fontebasso A. M., Quang D. A. K., Tönjes M., Hovestadt V., Albrecht S., Kool M., Nantel A., Konermann C., Lindroth A., Jäger N., Rausch T., Ryzhova M., Korbel J. O., Hielscher T., Hauser P., Garami M., Klekner A., Bognar L., Ebinger M., Schuhmann M. U., Scheurlen W., Pekrun A., Frühwald M. C., Roggendorf W., Kramm C., Dürken M., Atkinson J., Lepage P., Montpetit A., Zakrzewska M., Zakrzewski K., Liberski P. P., Dong Z., Siegel P., Kulozik A. E., Zapatka M., Guha A., Malkin D., Felsberg J., Reifenberger G., von Deimling A., Ichimura K., Collins V. P., Witt H., Milde T., Witt O., Zhang C., Castelo-Branco P., Lichter P., Faury D., Tabori U., Plass C., Majewski J., Pfister S. M., Jabado N., Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Wu G., Broniscer A., McEachron T. A., Lu C., Paugh B. S., Becksfort J., Qu C., Ding L., Huether R., Parker M., Zhang J., Gajjar A., Dyer M. A., Mullighan C. G., Gilbertson R. J., Mardis E. R., Wilson R. K., Downing J. R., Ellison D. W., Zhang J., Baker S. J.; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project , Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sturm D., Witt H., Hovestadt V., Khuong-Quang D. A., Jones D. T. W., Konermann C., Pfaff E., Tönjes M., Sill M., Bender S., Kool M., Zapatka M., Becker N., Zucknick M., Hielscher T., Liu X. Y., Fontebasso A. M., Ryzhova M., Albrecht S., Jacob K., Wolter M., Ebinger M., Schuhmann M. U., van Meter T., Frühwald M. C., Hauch H., Pekrun A., Radlwimmer B., Niehues T., von Komorowski G., Dürken M., Kulozik A. E., Madden J., Donson A., Foreman N. K., Drissi R., Fouladi M., Scheurlen W., von Deimling A., Monoranu C., Roggendorf W., Herold-Mende C., Unterberg A., Kramm C. M., Felsberg J., Hartmann C., Wiestler B., Wick W., Milde T., Witt O., Lindroth A. M., Schwartzentruber J., Faury D., Fleming A., Zakrzewska M., Liberski P. P., Zakrzewski K., Hauser P., Garami M., Klekner A., Bognar L., Morrissy S., Cavalli F., Taylor M. D., van Sluis P., Koster J., Versteeg R., Volckmann R., Mikkelsen T., Aldape K., Reifenberger G., Collins V. P., Majewski J., Korshunov A., Lichter P., Plass C., Jabado N., Pfister S. M., Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Jansen M. H., Veldhuijzen van Zanten S. E., Sanchez Aliaga E., Heymans M. W., Warmuth-Metz M., Hargrave D., van der Hoeven E. J., Gidding C. E., de Bont E. S., Eshghi O. S., Reddingius R., Peeters C. M., Schouten-van Meeteren A. Y. N., Gooskens R. H. J., Granzen B., Paardekooper G. M., Janssens G. O., Noske D. P., Barkhof F., Kramm C. M., Vandertop W. P., Kaspers G. J., van Vuurden D. G., Survival prediction model of children with diffuse intrinsic pontine glioma based on clinical and radiological criteria. Neuro Oncol. 17, 160–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassan H., Pinches A., Picton S. V., Phillips R. S., Survival rates and prognostic predictors of high grade brain stem gliomas in childhood: A systematic review and meta-analysis. J. Neurooncol 135, 13–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis P. W., Muller M. M., Koletsky M. S., Cordero F., Lin S., Banaszynski L. A., Garcia B. A., Muir T. W., Becher O. J., Allis C. D., Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan K.-M., Fang D., Gan H., Hashizume R., Yu C., Schroeder M., Gupta N., Mueller S., James C. D., Jenkins R., Sarkaria J., Zhang Z., The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margueron R., Reinberg D., The polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J.-R., Lee C.-H., Oksuz O., Stafford J. M., Reinberg D., PRC2 is high maintenance. Genes Dev. 33, 903–935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury III W. J., Voigt P., Martin S. R., Taylor W. R., de Marco V., Pirrotta V., Reinberg D., Gamblin S. J., Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C.-H., Yu J.-R., Kumar S., Jin Y., Roy G. L., Bhanu N., Kaneko S., Garcia B. A., Hamilton A. D., Reinberg D., Allosteric activation dictates PRC2 activity independent of its recruitment to chromatin. Mol. Cell 70, 422–434.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oksuz O., Narendra V., Lee C.-H., Descostes N., Roy G. L., Raviram R., Blumenberg L., Karch K., Rocha P. P., Garcia B. A., Skok J. A., Reinberg D., Capturing the onset of PRC2-mediated repressive domain formation. Mol. Cell 70, 1149–1162.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escobar T. M., Oksuz O., Saldaña-Meyer R., Descostes N., Bonasio R., Reinberg D., Active and repressed chromatin domains exhibit distinct nucleosome segregation during DNA replication. Cell 179, 953–963.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stafford J. M., Lee C.-H., Voigt P., Descostes N., Saldaña-Meyer R., Yu J.-R., Leroy G., Oksuz O., Chapman J. R., Suarez F., Modrek A. S., Bayin N. S., Placantonakis D. G., Karajannis M. A., Snuderl M., Ueberheide B., Reinberg D., Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci. Adv. 4, eaau5935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An S., Camarillo J. M., Huang T. Y. T., Li D., Morris J. A., Zoltek M. A., Qi J., Behbahani M., Kambhampati M., Kelleher N. L., Nazarian J., Thomas P. M., Saratsis A. M., Histone tail analysis reveals H3K36me2 and H4K16ac as epigenetic signatures of diffuse intrinsic pontine glioma. J. Exp. Clin. Cancer Res. 39, 261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harutyunyan A. S., Chen H., Lu T., Horth C., Nikbakht H., Krug B., Russo C., Bareke E., Marchione D. M., Coradin M., Garcia B. A., Jabado N., Majewski J., H3K27M in gliomas causes a one-step decrease in H3K27 methylation and reduced spreading within the constraints of H3K36 methylation. Cell Rep. 33, 108390 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitges F. W., Prusty A. B., Faty M., Stützer A., Lingaraju G. M., Aiwazian J., Sack R., Hess D., Li L., Zhou S., Bunker R. D., Wirth U., Bouwmeester T., Bauer A., Ly-Hartig N., Zhao K., Chan H., Gu J., Gut H., Fischle W., Müller J., Thomä N. H., Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Yuan W., Xu M., Huang C., Liu N., Chen S., Zhu B., H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka Y., Katagiri Z., Kawahashi K., Kioussis D., Kitajima S., Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene 397, 161–168 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Li Y., Trojer P., Xu C. F., Cheung P., Kuo A., Drury W. J. III, Qiao Q., Neubert T. A., Xu R. M., Gozani O., Reinberg D., The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J. Biol. Chem. 284, 34283–34295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan W., Xie J., Long C., Erdjument-Bromage H., Ding X., Zheng Y., Tempst P., Chen S., Zhu B., Reinberg D., Heterogeneous nuclear ribonucleoprotein L Is A Subunit Of Human KMT3a/Set2 complex required for H3 Lys-36 trimethylation activity in vivo. J. Biol. Chem. 284, 15701–15707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharya S., Levy M. J., Zhang N., Li H., Florens L., Washburn M. P., Workman J. L., The histone methyltransferase SETD2 couples transcription and splicing by engaging pre-mRNA processing factors through its SHI domain. bioRxiv 2020.06.06.138156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C., Zhu B., Roles of H3K36-specific histone methyltransferases in transcription: Antagonizing silencing and safeguarding transcription fidelity. Biophys. Rep. 4, 170–177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sankaran S. M., Wilkinson A. W., Elias J. E., Gozani O. A., A PWWP domain of histone-Lysine N-methyltransferase NSD2 binds to dimethylated Lys-36 of histone H3 and regulates NSD2 function at chromatin. J. Biol. Chem. 291, 8465–8474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LeRoy G., Oksuz O., Descostes N., Aoi Y., Ganai R. A., Kara H. O., Yu J.-R., Lee C.-H., Stafford J., Shilatifard A., Reinberg D., LEDGF and HDGF2 relieve the nucleosome-induced barrier to transcription in differentiated cells. Sci. Adv. 5, eaay3068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinberg D. N., Papillon-Cavanagh S., Chen H., Yue Y., Chen X., Rajagopalan K. N., Horth C., McGuire J. T., Xu X., Nikbakht H., Lemiesz A. E., Marchione D. M., Marunde M. R., Meiners M. J., Cheek M. A., Keogh M. C., Bareke E., Djedid A., Harutyunyan A. S., Jabado N., Garcia B. A., Li H., Allis C. D., Majewski J., Lu C., The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piunti A., Hashizume R., Morgan M. A., Bartom E. T., Horbinski C. M., Marshall S. A., Rendleman E. J., Ma Q., Takahashi Y. H., Woodfin A. R., Misharin A. V., Abshiru N. A., Lulla R. R., Saratsis A. M., Kelleher N. L., James C. D., Shilatifard A., Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Med. 23, 493–500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaro J. L., Vekich J. E., Tran T., Shen W.-C., Nuclear localization of cell-penetrating peptides is dependent on endocytosis rather than cytosolic delivery in CHO cells. Mol. Pharm. 6, 337–344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Husmann D., Gozani O., Histone lysine methyltransferases in biology and disease. Nat. Struct. Mol. Biol. 26, 880–889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Network , Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaju R. J., Fidler C., Haas O. A., Strickson A. J., Watkins F., Clark K., Cross N. C., Cheng J. F., Aplan P. D., Kearney L., Boultwood J., Wainscoat J. S., A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 98, 1264–1267 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Wang G. G., Cai L., Pasillas M. P., Kamps M. P., NUP98–NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat. Cell Biol. 9, 804–812 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., Morais P., Meltzer J., Korejwa A., Jané-Valbuena J., Mapa F. A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I. H., Cheng J., Yu G. K., Yu J., Aspesi P., de Silva M., Jagtap K., Jones M. D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R. C., Liefeld T., MacConaill L., Winckler W., Reich M., Li N., Mesirov J. P., Gabriel S. B., Getz G., Ardlie K., Chan V., Myer V. E., Weber B. L., Porter J., Warmuth M., Finan P., Harris J. L., Meyerson M., Golub T. R., Morrissey M. P., Sellers W. R., Schlegel R., Garraway L. A., The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., Troche C., Swaroop A., Kulis M., Oyer J., Will C., Dupere-Richer D., Riva A., Pipe C., Norton S., Bennett R. L., Licht J. D., A gain of function mutation in the NSD2 histone methyltransferase drives glucocorticoid resistance of acute lymphoblastic leukemia. Blood 132, 653–653 (2018). [Google Scholar]
- 35.Qiao Q., Li Y., Chen Z., Wang M., Reinberg D., Xu R. M., The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J. Biol. Chem. 286, 8361–8368 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W., Tian W., Yuan G., Deng P., Sengupta D., Cheng Z., Cao Y., Ren J., Qin Y., Zhou Y., Jia Y., Gozani O., Patel D. J., Wang Z., Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases. Nature , 498–503 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee W., Teckie S., Wiesner T., Ran L., Prieto Granada C. N., Lin M., Zhu S., Cao Z., Liang Y., Sboner A., Tap W. D., Fletcher J. A., Huberman K. H., Qin L. X., Viale A., Singer S., Zheng D., Berger M. F., Chen Y., Antonescu C. R., Chi P., PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 46, 1227–1232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hübner J.-M., Müller T., Papageorgiou D. N., Mauermann M., Krijgsveld J., Russell R. B., Ellison D. W., Pfister S. M., Pajtler K. W., Kool M., EZHIP/CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol. 21, 878–889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain S. U., Do T. J., Lund P. J., Rashoff A. Q., Diehl K. L., Cieslik M., Bajic A., Juretic N., Deshmukh S., Venneti S., Muir T. W., Garcia B. A., Jabado N., Lewis P. W., PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat. Commun. 10, 2146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piunti A., Smith E. R., Morgan M. A. J., Ugarenko M., Khaltyan N., Helmin K. A., Ryan C. A., Murray D. C., Rickels R. A., Yilmaz B. D., Rendleman E. J., Savas J. N., Singer B. D., Bulun S. E., Shilatifard A., CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci. Adv. 5, eaax2887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagaraja S., Vitanza N. A., Woo P. J., Taylor K. R., Liu F., Zhang L., Li M., Meng W., Ponnuswami A., Sun W., Ma J., Hulleman E., Swigut T., Wysocka J., Tang Y., Monje M., Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell 31, 635–652.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohammad F., Weissmann S., Leblanc B., Pandey D. P., Højfeldt J. W., Comet I., Zheng C., Johansen J. V., Rapin N., Porse B. T., Tvardovskiy A., Jensen O. N., Olaciregui N. G., Lavarino C., Suñol M., de Torres C., Mora J., Carcaboso A. M., Helin K., EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 23, 483–492 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Yu J.-R., Tai Y., Jin Y., Hammell M. C., Wilkinson J. E., Roe J. S., Vakoc C. R., van Aelst L., TGF-β/Smad signaling through DOCK4 facilitates lung adenocarcinoma metastasis. Genes Dev. 29, 250–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bayin N. S., Modrek A. S., Dietrich A., Lebowitz J., Abel T., Song H. R., Schober M., Zagzag D., Buchholz C. J., Chao M. V., Placantonakis D. G., Selective lentiviral gene delivery to CD133-expressing human glioblastoma stem cells. PLOS ONE 9, e116114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narendra V., Rocha P. P., An D., Raviram R., Skok J. A., Mazzoni E. O., Reinberg D., CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/29/eabg7444/DC1


