IgSF11 homophilic cell adhesion molecules regulate the synaptic laminar specificity of cortical chandelier cells.
Abstract
The most prominent structural hallmark of the mammalian neocortical circuitry is the layer-based organization of specific cell types and synaptic inputs. Accordingly, cortical inhibitory interneurons (INs), which shape local network activity, exhibit subtype-specific laminar specificity of synaptic outputs. However, the underlying molecular mechanisms remain unknown. Here, we demonstrate that Immunoglobulin Superfamily member 11 (IgSF11) homophilic adhesion proteins are preferentially expressed in one of the most distinctive IN subtypes, namely, chandelier cells (ChCs) that specifically innervate axon initial segments of pyramidal neurons (PNs), and their synaptic laminar target. Loss-of-function experiments in either ChCs or postsynaptic cells revealed that IgSF11 is required for ChC synaptic development in the target layer. While overexpression of IgSF11 in ChCs enlarges ChC presynaptic boutons, expressing IgSF11 in nontarget layers induces ectopic ChC synapses. These findings provide evidence that synapse-promoting adhesion proteins, highly localized to synaptic partners, determine the layer-specific synaptic connectivity of the cortical IN subtype.
INTRODUCTION
The mammalian neocortex is organized into six cytoarchitecturally distinct layers. Each layer contains specific types of excitatory pyramidal neurons (PNs) that are defined by gene expression, morphology, electrophysiological property, and remote projection targets (1, 2). PNs in distinct layers receive inputs from a different set of intracortical and subcortical neurons, generating specific output signals (3–8). Among these multiple sources of inputs to PNs, cortical inhibitory interneurons (INs) locally innervate PNs and play a key role in shaping their activity at synaptic, cellular, and network levels (9–12). They display a high degree of diversity in cellular properties such as morphology, physiology, gene expression, and connectivity (9–11). Recent studies showed that distinct IN subtypes are functionally specialized (13–15). Thus, assembling inhibitory inputs from a specific combination of IN subtypes is likely one of the crucial steps to building layer-specific computational modules. Individual IN subtypes exhibit one or a few preferential synaptic target layers (6, 14, 16–19), suggesting that they are assigned to participate in layer-specific information processing. For example, somatostatin-positive (SOM+) Martinotti cells in deeper layers predominantly innervate the apical dendrites of PNs in layer 1 (L1), where information from intracortical and subcortical neurons are integrated (6, 14). One type of L4 fast-spiking parvalbumin-positive (PV+) INs forms reciprocal connections with excitatory spiny neurons in the same layer (19).
The assembly of layer-specific local networks comprising INs and PNs involves different developmental events including cell migration, laminar positioning, axonal morphogenesis, and synapse formation (11, 20–22). Accumulating evidence has indicated that cell type–specific interactions between INs and PNs contribute to at least some of these developmental processes. Fate-converting corticofugal PNs to callosal PNs in L5 by manipulating a master transcriptional regulator reduces the number of L5 PV+ and SOM+ INs, which are normally enriched in deep cortical layers (21). Genetic reprogramming that fate-switches callosal PNs into corticofugal PNs in L2/3 induces more perisomatic boutons from PV+ INs onto the fate-converted PNs at the level equivalent to endogenous L5 corticofugal PNs (20). These findings suggest the cell-extrinsic mechanisms that mediate cell type–specific communications between INs and PNs during the assembly of layer-specific circuit modules. However, little is known about the molecular mechanisms underlying the synaptic matching between INs and their target layer PNs.
A recent study using RNA sequencing (RNA-seq) techniques has highlighted subtype-specific molecular programs emerging in cortical INs during early postnatal development and identified a few cell surface proteins that mediate synapse formation of IN subtypes (23). These molecules appear to be expressed in multiple more specific subtypes within a broad subgroup (i.e., PV+ INs and SOM+ INs) throughout cortical layers (23). In addition, their binding partners are widely distributed in the cortex (24–26). Therefore, they are unlikely involved in the laminar specificity of IN synaptic outputs. Besides these newly identified molecules, several studies have found synapse organizers that specifically promote the formation and/or maturation of inhibitory synapses (27, 28). However, these synaptogenic molecules are ubiquitously distributed in the cortex (29, 30), and layer-specific synapse-promoting factors have not been reported.
Addressing this issue requires a proper experimental system that enables genetic targeting of a bona fide IN subtype that reproducibly exhibit layer-specific synapse assembly. The chandelier cell (ChC) is a distinct IN subtype that specifically innervates axon initial segments (AISs) of PNs and powerfully controls their spike generation (17, 31–33). Consistent with their functional significance in the normal brain indicated by prior studies, deficits in ChC synapses have been implicated in brain disorders such as schizophrenia and epilepsy (34, 35). In the medial prefrontal cortex (mPFC) and the anterior cingulate cortex (aCC), upper L2/3 (uL2/3) ChCs establish their synaptic terminals on AISs of uL2/3 PNs (17, 18, 36). The stereotypy of the laminar and subcellular distribution of synaptic outputs makes ChCs an ideal model to study the molecular mechanisms underlying the wiring of IN subtypes. Our group has developed genetic strategies that target ChC progenitors and make ChCs genetically tractable (36, 37).
Here, combining RNA-seq and fluorescent in situ hybridization (FISH), we identify IgSF11 (38) as the homophilic cell adhesion molecule (CAM) that is preferentially expressed in ChCs and uL2/3 PNs in the mPFC. With a multifaceted approach, we demonstrate that IgSF11 is necessary for morphological, structural, and functional differentiation of ChC presynaptic boutons in the target layer. Furthermore, ectopically overexpressing IgSF11 in deep layers induces ectopic synapses from ChCs. Our work thus indicates that the homophilic CAM that is specifically expressed in a synaptic pair shapes the laminar specificity of ChC synaptic outputs through its synapse-promoting activity.
RESULTS
IgSF11 mRNAs are preferentially enriched in ChCs compared to SOM+ INs and VIP+ INs
A recent study using RNA-seq–based gene expression profiling indicated that each developing IN subtype expresses a specific set of genes that are involved in its synaptic assembly (23). We hypothesized that such subtype-specific genes contain those encoding cell surface proteins determining the layer-specific synaptic connectivity of IN subtypes. To identify genes that are differentially expressed in developing IN subtypes, we independently performed RNA-seq–based gene expression comparisons among ChCs, SOM+ INs, and vasoactive intestinal peptide–positive INs (VIP+ INs) at postnatal day five (P5) when INs begin to form synapses (Fig. 1A) (23, 36, 39, 40). SOM+ INs and VIP+ INs originate from the medial ganglionic eminence (MGE) and the caudal ganglionic eminence (CGE), respectively, and are largely nonoverlapping heterogenous populations (10, 11). We specifically labeled these IN subtypes with green fluorescent proteins (GFPs) using genetically engineered mice (ChC-GFP, SOM-GFP, and VIP-GFP mice) (36, 41, 42), manually collected cells of interest from upper layers of the mPFC where ChCs are more efficiently labeled in ChC-GFP mice compared to other cortical areas (36), and then carried out Illumina bulk RNA-seq (Fig. 1B). Representative marker genes that are known to exhibit expression specificity in these subtypes displayed different numbers of reads among them as expected (Fig. 1C). For example, the Lhx6 gene encoding a homeodomain protein that is highly expressed in MGE-derived INs but not CGE-derived INs (11, 43) showed significantly more reads in ChCs and SOM+ INs than VIP+ INs. In contrast, the Prox1 gene encoding a homeobox protein that is specifically expressed in CGE-derived INs (43, 44) showed a significantly larger number of reads in VIP+ INs compared to ChCs and SOM+ INs. In addition, the SOM gene and the VIP gene showed significantly more reads in SOM+ INs and VIP+ INs, respectively, compared to the other two IN subtypes. These results suggest that our RNA-seq datasets are reliable for comparing gene expression profiles among these IN subtypes.
Fig. 1. IgSF11 mRNAs are preferentially expressed in both ChCs and their laminar target.
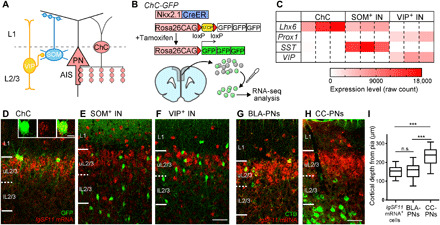
(A) Schematic illustration of cortical inhibitory circuits. ChCs specifically innervate AISs of uL2/3 PNs, whereas SOM+ INs and VIP+ INs form synapses on dendrites of PNs and other types of INs, respectively. (B) Schematic of genetic labeling and manual sorting of ChCs for RNA-seq. (C) Heatmap showing expression of the representative marker genes in ChCs, SOM+ INs, and VIP+ INs. n = 3 biological replicates for each IN subtype. (D to F) IgSF11 mRNA expression in the mPFC of P16 ChC-GFP (D), SOM-GFP (E), and VIP-GFP mice (F). Insets in (D) are confocal single optical section images of the ChC soma expressing IgSF11 mRNA. IgSF11 mRNA FISH signals and GFP signals are shown in red and green, respectively. Scale bars, 50 and 10 μm (inset). (G and H) FISH signals for IgSF11 mRNAs (red) in the mPFC containing BLA-projecting PNs (G) and CC-projecting PNs (H) retrogradely labeled with cholera toxin B (CTB) (green). 84.3% of BLA-PNs (n = 64 cells from three mice) and 11.7% of CC-PNs (n = 111 cells from three mice) in L2/3 express IgSF11 mRNAs. Scale bars, 50 μm. (I) Cortical depth of IgSF11 mRNA+ cells, BLA-projecting PNs, and CC-projecting PNs in the mPFC. One-way analysis of variance (ANOVA); ***P < 0.001. n = 80 cells from three mice for each condition. In the box plots, center line, whiskers, and box limits represent median, minimum to maximum, and the 25th to 75th percentile, respectively. n.s., not significant; pia, The pia matterSP.
On the basis of our RNA-seq data, we then searched for cell surface molecules that are predominantly expressed in ChCs. Screening with our arbitrary criteria (more than twofold difference; P < 0.05) identified 7 of 927 genes encoding cell surface proteins. Since homophilic cell adhesion is known to mediate synaptic partner matching in several neuronal systems (45–48), we reasoned that homophilic CAMs may be ideal candidates for the molecules controlling the laminar specificity of ChC synaptic outputs. Accordingly, we selected three CAMs for further analysis: Cadherin-6 (Cdh6), Cdh12, and IgSF11 (fig. S1A). Cdh6 and Cdh12 belong to the cadherin superfamily that mediates a variety of neurobiological processes (49, 50). Our previous polymerase chain reaction (PCR)–based screening also identified Cdh6 as a cadherin that is preferentially expressed in developing ChCs (51), verifying the specificity of our RNA-seq data again. IgSF11, a member of the immunoglobulin superfamily, is known to mediate AMPA receptor trafficking in dendritic spines of hippocampal PNs (16, 38), but its roles in INs are completely unknown.
IgSF11 mRNAs are specifically expressed in both ChCs and PNs in their laminar target
To further screen these homophilic CAM candidates, we investigated whether they are specifically expressed in the synaptic target layer (uL2/3) of ChCs in the developing neocortex using FISH. We found that Cdh6 and Cdh12 mRNAs are broadly expressed across neocortical layers at P16 (fig S1, B and C), excluding them from strong candidates. Similar results were confirmed in the Allen Brain Atlas (Cdh6: http://developingmouse.brain-map.org/experiment/show/100056565; Cdh12: http://developingmouse.brain-map.org/experiment/show/100053582). We then carried out IgSF11 mRNA FISH using P16 brains. To confirm that IgSF11 mRNAs are preferentially expressed in ChCs among the IN subtypes while testing the laminar specificity of IgSF11 mRNA expression, we combined IgSF11 mRNA FISH with GFP immunohistochemistry in brain sections from ChC-GFP, SOM-GFP, and VIP-GFP mice. IgSF11 mRNA FISH signals were highly localized to uL2/3 cells in the mPFC (Fig. 1, D to F). In addition, consistent with the results from RNA-seq–based gene expression comparisons, a great majority of ChCs expressed IgSF11 mRNAs (88.5%, n = 35 cells) (Fig. 1D), whereas only a small fraction of SOM+ INs (4.6%, n = 44 cells) and VIP+ INs (5.4%, n = 56 cells) tested positive for IgSF11 mRNA expression (Fig. 1, E and F). We also found that this expression pattern is established as early as P7 when ChCs begin to form synapses (fig. S2A) and that the expression level obviously increases between P7 and P12 when ChCs actively add synapses (fig. S2B) and then persists until P16 (fig. S2C). This spatial and temporal expression pattern of IgSF11 mRNAs is consistent with the view that IgSF11 plays a role in establishing the synaptic laminar specificity of ChCs.
To further characterize the identity of IgSF11+ cells, we performed a series of experiments combining IgSF11 mRNA FISH with different cell identification approaches. IgSF11 mRNAs exhibited dense expression in uL2/3 and sparse expression in L1 (Fig. 1, D to F). Since excitatory PNs constitute 80% of cortical neurons in uL2/3 and inhibitory INs are one of the major cell types and the only neuronal cells in L1, we reasoned that a major fraction of IgSF11+ cells in uL2/3 and L1 may represent PNs and INs, respectively. Consistent with this idea, double FISH against IgSF11 mRNAs and Gad1 (a pan-IN marker) mRNAs showed that a great majority of IgSF11+ cells in L1 are Gad1 positive (88.1%, n = 67 cells), whereas a vast majority of IgSF11+ cells in uL2/3 are Gad1 negative (92.5%, n = 265 cells) (fig. S3, A to C). To directly show that uL2/3 PNs express IgSF11 mRNAs, we randomly labeled a fraction of uL2/3 PNs with GFP using in utero electroporation (IUE) and examined what percentage of GFP+ cells express IgSF11 mRNAs. We found that nearly all GFP+ uL2/3 PNs express IgSF11 mRNAs (96.0%, n = 75 cells) (fig. S3, D and E). A recent study showed that ChCs preferentially innervate PNs projecting axons to the basolateral amygdala (BLA) but not the contralateral cortex (CC) (17). To test whether BLA-projecting PNs but not CC-projecting PNs predominantly express IgSF11, we combined cholera toxin B (CTB)–mediated retrograde labeling of PNs and IgSF11 mRNA FISH. The cortical depth of CC-projecting PNs was significantly greater than those of BLA-projecting PNs and IgSF11+ cells in L2/3 (Fig. 1, G to I), suggesting that the laminar position of BLA-projecting PNs overlaps with that of IgSF11+ cells. A larger fraction of BLA-projecting PNs (84.3%) expressed IgSF11 mRNAs compared to CC-projecting PNs (11.7%). Together, these results support the idea that IgSF11 is a strong candidate for the homophilic CAM that mediates synaptic partner matching between ChCs and PNs in their synaptic target layer.
IgSF11 proteins distribute all over the cell in both ChCs and uL2/3 PNs
To determine the cellular and subcellular localization of IgSF11 proteins, we tagged endogenous IgSF11 genes with hemagglutinin (HA) sequences using a homology-independent targeted integration (HITI) technique (52). Correct insertions of HA tags into the IgSF11 gene loci were confirmed with single-cell genotyping (Fig. 2A) (53). To implement HITI in ChCs, we cotransfected the embryonic day 15 (E15) MGE that contains ChC progenitors with HA donor plasmids, CRISPR-Cas9 plasmids, and GFP plasmids using exo utero electroporation and then transplanted the electroporated MGE explants into the brains of P1 host pups (Fig. 2B). GFP+ neurons migrated out of the explant and settled in several cortical areas including the mPFC and the aCC. Most of them were ChCs whose morphology is indistinguishable from endogenous ones in adults. The transplanted ChC precursors normally developed following their internal developmental schedule: For example, the degree of axonal arborization of the transplanted ChCs in P21 host animals approximately corresponds to that of P16 endogenous ChCs (Fig. 2C). Hereafter, we use the age of 5 days younger than host animals as the age of transplanted ChCs [equivalent postnatal day (EP) to endogenous ChCs]. This genetic strategy was also used to manipulate ChCs in other experiments of the present study. Consistent with our finding that IgSF11 mRNAs are preferentially expressed in ChCs among the IN subtypes, HA signals were only detected in ChCs among GFP+ INs at EP16 (four GFP+/HA+ ChCs out of 10 GFP+ ChCs among 20 GFP+ INs) (Fig. 2D). We also found that HA-tagged IgSF11 proteins are distributed throughout the subcellular compartments including ChC synaptic cartridges, arrays of presynaptic boutons aligned with AISs (AIS boutons) (Fig. 2D). To apply HITI to PNs, we then transfected L2/3 PN progenitors with HA donor plasmids, CRISPR-Cas9 plasmids, and GFP plasmids using IUE at E15 (Fig. 2E). At P16, although GFP+ PNs were broadly found in L2/3, GFP+/HA+ PNs were restricted to uL2/3 in accordance with the layer-specific IgSF11 mRNA expression (Fig. 2F). All subcellular compartments including the AIS displayed HA signals (Fig. 2G). Thus, these results suggest that IgSF11 proteins are present both on ChC axonal terminals and their subcellular synaptic targets in the specific layer during ChC synaptogenesis.
Fig. 2. IgSF11 proteins distribute all over the cell in both ChCs and uL2/3 PNs.
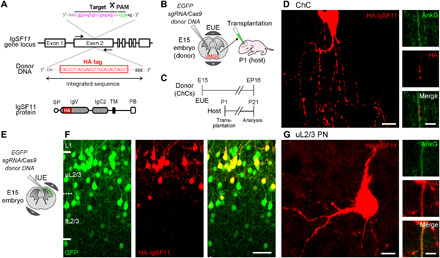
(A) Schematic of HITI-based gene tagging of IgSF11 genomic loci. The HA sequence is inserted into the exon 2 of the IgSF11 gene locus (top). HA tag is designed to be inserted at the extracellular immunoglobulin V (IgV) domain (bottom). SP, signal peptide; IgC2, immunoglobulin C2 like; TM, transmembrane; PB, PDZ binding domain. (B) Schematic of HITI experiments to target the IgSF11 gene locus with HA in ChCs. (C) Experimental timeline of HITI experiments in ChCs. (D) Expression of HA-tagged endogenous IgSF11 proteins (red) in ChC. AnkG is shown in green. Scale bars, 10 μm (left panels) and 5 μm (right panels). (E) Schematic of HITI experiments to target the IgSF11 gene locus with HA in uL2/3 PNs. (F) Expression of HA-tagged endogenous IgSF11 proteins (red) in L2/3 GFP+ PNs (green). Scale bar, 50 μm. (G) Expression of HA-tagged endogenous IgSF11 proteins (red) in uL2/3 PN. AnkG is shown in green. AnkG, Ankyrin G. Scale bars, 10 μm (left panels) and 5 μm (right panels).
IgSF11 in ChCs is necessary for the formation and morphological differentiation of their presynaptic boutons on AISs
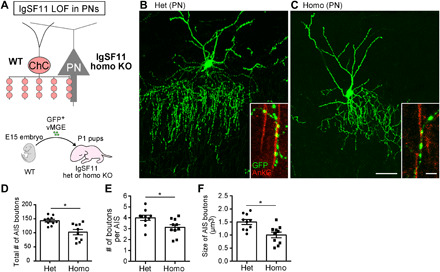
To dissect the functional role of IgSF11 in ChC synaptic development, we first performed loss-of-function (LOF) experiments using CRISPR-Cas9 genome editing technology (fig. S4A). Homozygous (homo) mutant cells were identified by a single-cell genotyping method after morphological analysis (fig. S4, D and E). At EP21, the total number of AIS boutons and the number of boutons per AIS in homo mutant ChCs were significantly lower than those in control ChCs transfected with LacZ-sgRNAs (fig. S4, B to G). In addition, the size of AIS boutons in homo mutant ChCs significantly was reduced compared to control ChCs (fig. S4, B to E and H). To further validate these findings, we used germline knockout (KO) mice (16) as donors of ChCs. ChC progenitors from heterozygous (het) or homo KO mice were electroporated with GFP plasmids and transplanted into wild-type (WT) host animals (Fig. 3A). Similar to the results from CRISPR-Cas9–based LOF experiments, homo KO ChCs displayed significant reductions in the number and size of AIS boutons compared to control het KO ChCs (Fig. 3, B to F). Homo KO ChCs showed normal expression of their genetic markers as demonstrated by FISH against FGF13 (control het KO: 100%, n = 12 cells; homo KO: 100%, n = 9 cells) and Trps1 (control het KO: 100%, n = 12 cells; homo KO: 100%, n = 10 cells) mRNAs (23, 54), suggesting that basic fate specification of ChCs properly occurs in the absence of IgSF11 (fig. S5). These results indicate that IgSF11 in ChCs is necessary for the formation and morphological differentiation of their AIS boutons.
Fig. 3. IgSF11 in ChCs is necessary for morphological, structural, and functional development of ChC presynaptic boutons on AISs of uL2/3 PNs.
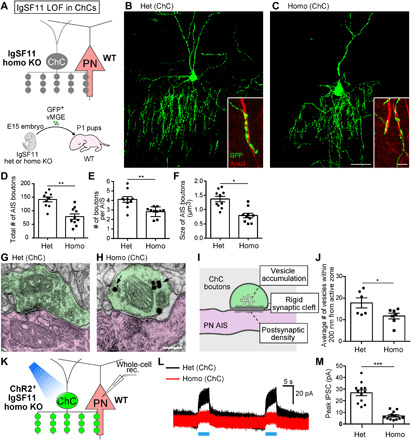
(A) Schematic illustration of IgSF11 LOF experiments in ChCs. (B and C) Representative images of IgSF11 het (B) and homo (C) KO ChCs at EP21. Scale bars, 50 and 5 μm (insets). (D to F) Quantification of the total number of AIS boutons (D), the number of boutons per AIS (E), and the size of AIS boutons (F) in IgSF11 het and homo KO ChCs. n = 10 cells from five mice per condition. Student’s t test, *P < 0.05 and **P < 0.01. (G to J) Ultrastructural analysis of AIS boutons in IgSF11 het and homo KO ChCs. (G and H) Representative images of ultrastructure of AIS boutons in IgSF11 het (G) and homo (H) KO ChCs. The synapse represented in (H) is an incomplete synapse. AIS boutons and AISs are represented in green and magenta, respectively. Arrows in (H) indicate gold nanoparticles conjugated to GFP antibodies. Although the bouton in (G) displays no gold nanoparticles, some other serial sections containing the same bouton show them. Scale bar, 200 nm. (I) Schematic of the ChC bouton that meets three criteria for the complete synapse. (J) Quantification of the average number of vesicles near the active zone in IgSF11 het and homo KO ChCs. n = 6 boutons from four mice (het) and n = 8 boutons from three mice (homo). Student’s t test, *P < 0.05. (K to M) IPSCs in uL2/3 PNs elicited by activation of IgSF11 het or homo KO ChCs. (K) Schematic of the optogenetics-assisted inhibitory postsynaptic currents (IPSC) recording. (L) Representative IPSC traces from uL2/3 PNs evoked by blue light illumination to ChR2-YFP–expressing IgSF11 het (black) and homo (red) KO ChCs. (M) Quantification of the amplitude of IPSCs in PNs evoked by activation of IgSF11 het and homo KO ChCs. n = 12 cells from six mice for each condition. Student’s t test, ***P < 0.001. Data are means ± SEM.
IgSF11 in ChCs is necessary for the structural and functional differentiation of their presynaptic boutons on AISs
We next addressed whether the remaining AIS boutons in homo KO ChCs normally differentiate by examining their ultrastructure with pre-embedding immunoelectron microscopy (Fig. 3, G to J). The AIS was structurally identified as the portion of the axon exhibiting a characteristic electron-dense region. AIS boutons of transplanted ChCs were distinguished by the presence of gold particles conjugated to anti-GFP antibodies, and profiles were analyzed using successive ultrathin sections. We defined “complete synaptic boutons” as those meeting three criteria: rigid synaptic cleft, vesicle accumulation, and thin postsynaptic density (Fig. 3I). All AIS boutons of control het KO ChCs were complete synaptic boutons (n = 9 boutons) (Fig. 3G), while approximately 30% of AIS boutons in homo KO ChCs displayed incomplete synaptic boutons (four incomplete synapses out of 14 boutons) (Fig. 3H). The average number of synaptic vesicles near the active zone per bouton in homo KO ChCs was significantly lower than those in control ChCs despite only analyzing complete synaptic boutons from homo KO ChCs (Fig. 3J). Thus, IgSF11 in ChCs is necessary for the structural differentiation of AIS synaptic boutons.
To further determine whether homo KO ChCs have functional deficits in synaptic transmission, we performed optogenetics-assisted electrophysiological recording. We expressed channelrhodopsin 2 in control het KO ChCs or homo KO ChCs and recorded inhibitory postsynaptic currents (IPSCs) from PNs that are innervated by the ChC under exposure to blue light at EP21 (Fig. 3K). The amplitude of IPSCs induced by stimulation of homo KO ChCs was significantly lower than that in controls (Fig. 3, L and M). Thus, IgSF11 is essential for establishing synaptic transmission from ChCs to PNs.
IgSF11 in ChCs is sufficient to promote morphological differentiation of their presynaptic boutons on AISs
The above results indicate that IgSF11 in ChCs is necessary for morphological, structural, and functional differentiation of their synaptic boutons in the target layer during development. We then asked whether IgSF11 in ChCs is sufficient to promote morphological maturation of their AIS boutons by examining ChCs overexpressing IgSF11. Since a plasmid-based approach was not able to achieve reliable expression of IgSF11 in ChCs, we used an adeno-associated virus (AAV)–mediated expression system. Flp-dependent (FlpD-) AAV-HA-IgSF11 and FlpD-AAV-GFP were co-injected into the brains of P1 ChC-Flp mice (Fig. 4A), and ChCs that express HA-IgSF11 were analyzed at P16. Overexpressing HA-IgSF11 in ChCs severely disrupted the overall organization of AIS boutons: The total number of AIS boutons and the number of boutons per AIS in ChCs overexpressing HA-IgSF11 were significantly reduced compared to those in control ChCs expressing only GFP (Fig. 4, B, C, F, and G). This observation suggests that ChCs overexpressing HA-IgSF11 fail to continuously form AIS boutons, likely due to enhanced adhesion to AISs. However, notably, ChCs overexpressing HA-IgSF11 developed significantly larger AIS boutons than control ChCs (Fig. 4, D, E, and H). These enlarged boutons were not found on subcellular compartments other than AISs, suggesting that subcellular synapse specificity of ChCs overexpressing HA-IgSF11 is maintained. These results indicate that IgSF11 in ChCs is sufficient to promote morphological differentiation of AIS boutons.
Fig. 4. IgSF11 in ChCs is sufficient to promote morphological differentiation of ChC presynaptic boutons on AISs of uL2/3 PNs.
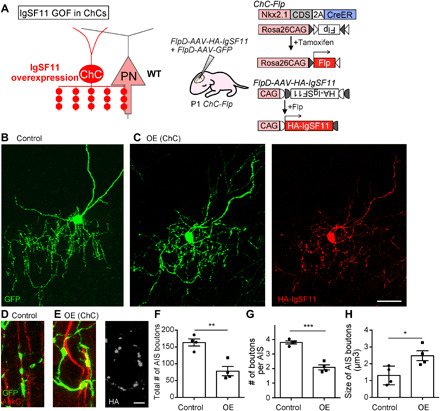
(A) Schematic of IgSF11 gain-of-function (GOF) experiments in ChCs. (B and C) Representative images of P16 control GFP+ ChC (B) and GFP+/HA-IgSF11+ ChC (C). GFP and HA-IgSF11 are shown in green and red, respectively. Scale bar, 50 μm. (D and E) Enlarged view of AIS boutons from P16 control GFP+ ChC (D) and GFP+/HA-IgSF11+ ChC (E). GFP, AnkG, and HA-IgSF11 are shown in green, red, and light gray, respectively. Scale bar, 5 μm. (F to H) Quantification of the total number of AIS boutons (F), the number of boutons per AIS (G), and the size of AIS boutons (H) in GFP+ ChCs and GFP+/HA-IgSF11+ ChCs. n = 5 cells from three mice per condition. OE, overexpression. Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001. Data are means ± SEM.
To further test whether overexpressing IgSF11 in presynaptic INs is generally sufficient to promote morphological development of presynaptic boutons, we overexpressed IgSF11 in different IN subgroups/subtypes. Overexpressing IgSF11 in SOM+ INs and VIP+ INs appears to cause neither changes in their bouton morphology nor formation of obvious synaptic cartridges on AISs in uL2/3 (fig. S6). We also found that overexpressing IgSF11 in L5 PV+ INs does not redirect their innervation to uL2/3 (fig. S7). Thus, presynaptic IgSF11 may require additional ChC-specific molecules for transmitting synapse-promoting signals.
IgSF11 in target layer PNs is necessary for the formation and morphological differentiation of ChC presynaptic boutons on AISs
Given that IgSF11 proteins have homophilic adhesion activity (38), it is plausible to assume that IgSF11 in uL2/3 PNs serves as a ligand for IgSF11 in ChCs. If this is the case, then removal of IgSF11 from uL2/3 PNs must cause phenotypes of ChC presynaptic boutons similar to those observed in IgSF11 homo KO ChCs. To test this possibility, we created the condition where IgSF11 is deleted in postsynaptic cells but not ChCs by transplanting WT ChC progenitors into homo KO pups (Fig. 5A). Homo KO cortices appeared to exhibit the normal distribution of layer-specific PN markers such as Cux1 (a supragranular/granular PN marker) and Ctip2 (an infragranular PN marker) (55) at a gross level, validating that basic fate specification of PNs properly occurs in the absence of IgSF11 (fig. S8, A and B). As expected, IgSF11 LOF in postsynaptic cells significantly reduced the total number of AIS boutons, the number of boutons per AIS, and the size of AIS boutons in EP21 ChCs compared to the control condition where het KO pups were used as host animals (Fig. 5, B to F). These results suggest that IgSF11 in target layer PNs is necessary for the formation and morphological differentiation of AIS boutons of ChCs.
Fig. 5. IgSF11 in PNs is necessary for morphological development of ChC presynaptic boutons on AISs of uL2/3 PNs.
(A) Schematic illustration for IgSF11 LOF experiments in PNs. (B and C) Representative images of EP21 ChCs transplanted into IgSF11 het (B) and homo (C) KO host brains. Scale bars, 50 and 5 μm (insets). (D to F) Quantification of the total number of AIS boutons (D), the number of boutons per AIS (E), and the size of AIS boutons (F) in ChCs transplanted into IgSF11 het and homo KO host brains. n = 10 cells from five mice per condition. Student’s t test, *P < 0.05.
Expressing IgSF11 in nontarget layer is sufficient to induce ectopic presynaptic boutons from ChCs
Last, we addressed whether localized expression of IgSF11 in uL2/3 is critical to layer-restricted formation of ChC synapses. To this end, we ectopically overexpressed IgSF11 in nontarget deeper layers, lower L2/3 (lL2/3) and L5, by injecting AAV-HA-IgSF11 into P1 host pups that simultaneously had transplantation of GFP-labeled ChC progenitors (Fig. 6A). The distribution of Cux1 (RFP+ cells: 6.7 ± 0.7%, n = 173 cells; HA-IgSF11+ cells: 6.4 ± 0.7%, n = 194 cells; three brains for each) and Ctip2 (RFP+ cells: 41.2 ± 3.3%, n = 173 cells; HA-IgSF11+ cells: 40.7 ± 4.1%, n = 287 cells; three brains for each) in L5 was similar between control brains infected with AAV-RFP and those infected with AAV-HA-IgSF11, suggesting that overexpressing IgSF11 does not disrupt normal fate specification of PNs (fig. S8, C to F). In control samples, EP16 ChCs developed few, if any, axonal boutons in lL2/3 and L5 as observed in samples without infection (Fig. 6B). In contrast, when HA-IgSF11 was expressed in deeper layers, ChCs notably developed ectopic axonal boutons (Fig. 6C). These ectopic boutons included those containing vesicular gamma-aminobutyric acid transporter (VGAT), a presynaptic protein of inhibitory synapses, suggesting that HA-IgSF11 expressed in deeper layers induces presynaptic boutons from ChCs (Fig. 6, D and E). The number of VGAT+ ChC boutons in deeper layers expressing HA-IgSF11 was significantly higher than that in control (Fig. 6F). Note that ectopic expression of HA-IgSF11 induces ChC presynaptic boutons both on AISs and somata. However, intriguingly, the ectopic boutons formed on AISs exhibited a greater ratio of VGAT+ boutons compared to those formed on somata (Fig. 6, D, E, and G), suggesting that IgSF11 synapse-promoting activity in PNs is differentially regulated at distinct subcellular compartments. These results suggest that when expressed in deeper layer PNs, IgSF11 is sufficient to induce ectopic presynaptic boutons from ChCs. Therefore, layer-specific expression of IgSF11 is essential for ensuring the layer-restricted organization of ChC synapses. The phenotype caused by expressing IgSF11 in ectopic layers could be explained by the secondary outcome following increased axonal ingrowth. However, our finding that overexpression of HA-IgSF11 in deeper layer PNs induce perisomatic presynaptic boutons from ChCs (Fig. 6E) suggests direct synapse-promoting activity of IgSF11 since control ChCs fail to form them. Furthermore, when HA-IgSF11 was overexpressed in uL2/3, ChCs developed perisomatic boutons surrounding IgSF11-expressing PNs without changing the overall axonal density (figs. S9 and S10), negating the indirect effect of IgSF11 on ChC synapse formation.
Fig. 6. IgSF11 in AISs of PNs is sufficient for development of ChC presynaptic boutons in the laminar target.
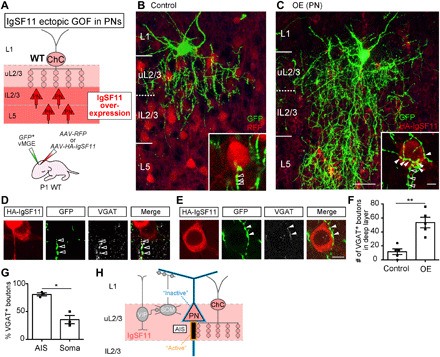
(A) Schematic of IgSF11 GOF experiments in PNs. (B and C) Representative images of EP16 ChCs (green) transplanted into brains expressing RFP [red (B)] or HA-IgSF11 [red (C)]. Scale bars, 50 and 5 μm (insets). Filled and empty arrowheads in insets represent putative perisomatic and AIS boutons, respectively. (D and E) Representative images of ectopic ChC synapses on the AIS [(D) empty arrowheads] and the cell body [(E) filled arrowheads] of IgSF11-overexpressing L5 PNs. HA-IgSF11, GFP, and VGAT are shown in red, green, and light gray, respectively. Scale bar, 10 μm. (F) Quantification of the number of VGAT+ ChC presynaptic boutons in lL2/3 and L5 of brains expressing RFP or HA-IgSF11. n = 5 cells from three mice per condition. Student’s t test, **P < 0.01. (G) Quantification of the percentage of VGAT+ boutons in putative AIS and perisomatic boutons in lL2/3 and L5 of brains expressing HA-IgSF11. n = 60 boutons (AIS) and n = 57 boutons (perisomatic) from three ChCs. Three mice were analyzed. Student’s t test, *P < 0.05. (H) Diagram representing a model of IgSF11 functions in regulating formation and differentiation of layer-specific ChC synapses on AISs. Data are means ± SEM.
DISCUSSION
Our results demonstrate that IgSF11 homophilic CAMs are specifically expressed in synaptic partners, constituting the IN-PN circuit motif that plays a key role in shaping PN spiking patterns, pre- and postsynaptically in ChCs and uL2/3 PNs, respectively. Notably, our findings also reveal that IgSF11 proteins positively regulate ChC synapse formation and differentiation per se in the synaptic target layer in vivo. There are a couple of examples in excitatory synapses of the mammalian brain where binding pairs of CAMs as well as ligands and receptors are selectively expressed in presynaptic cells and their postsynaptic cellular/subcellular targets and directly promote synapse formation and synaptic partner matching (48, 56, 57). However, such cell surface molecules that dictate inhibitory synapse specificity have not been found. Thus, to our knowledge, our study provides the first insight into synapse-promoting CAMs that exhibit the highly restricted expression in local IN-PN synaptic partners and determine synaptic specificity of the IN subtype, particularly the laminar specificity of its synapses (Fig. 6H). In the mPFC, ChCs innervate PNs projecting axons to the BLA, which is the nuclei essential for fear learning, and both neurons express IgSF11 (Fig. 1, D and G to I). A previous study showed that aversive signals that are used as unconditioned stimuli in fear conditioning activate ChCs in vivo (58). Therefore, IgSF11 may contribute to the assembly of functionally relevant IN-PN pairs. Our results showed that L1 INs and a small fraction of SOM+ INs and VIP+ INs in uL2/3 also express IgSF11. It would be intriguing to test whether these IgSF11+ INs are integrated into microcircuits including BLA-projecting PNs.
Deleting endogenous IgSF11 reduced ChC synapses and axonal branches in the target layer, while expressing IgSF11 in nontarget layers induced ectopic innervation by ChCs. These results suggest that IgSF11-mediated synapse formation stabilizes ChC axonal arbors. Nevertheless, IgSF11 LOF did not lead to overshooting of ChC axons to nontarget layers, suggesting that IgSF11 itself and IgSF11-mediated synapse formation is not necessary for confining ChC axons to the target layer. Unknown attractants and repellents in the target and nontarget layers, respectively, may work in concert to largely prevent ChC axons from entering the nontarget layers. In addition, ChC axons that may incidentally overshoot the target layer could likely be destabilized because of failure to establish synaptic contacts. Thus, IgSF11-dependent synapse formation and axonal stabilization in the target layer may cooperate with IgSF11-independent laminar restriction of axonal growth to certainly establish layer-specific innervation by ChCs.
We showed that although IgSF11 gain of function (GOF) in PNs induces ChC axonal boutons surrounding the PN somata (Fig. 6E and fig. S10), most of them are incomplete synapses (Fig. 6G). On the other hand, ectopically expressed IgSF11 in nontarget PNs reliably induced ChC presynaptic boutons on AISs (Fig. 6, D and G). These results indicate differential regulations of IgSF11 synapse-promoting activity against ChCs in distinct subcellular compartments. It is conceivable that AIS-specific molecules gate IgSF11 signaling to promote ChC synaptic development. In addition, IgSF11 activity in subcellular domains other than AISs may be inactivated by unknown molecular interactions to preclude ChCs from initiating synapse formation. These subcellular regulations of IgSF11 activity could explain how ChCs form synaptic boutons only at AISs despite ubiquitous subcellular expression of IgSF11 in PNs (Fig. 6H). We propose that IgSF11 serve as an indispensable component to induce ChC synapses at AISs while determining their laminar specificity. It should be noted that recently identified transmembrane molecules in postsynaptic cells that are involved in subcellular synapse formation of cortical INs also exhibit ubiquitous subcellular distribution (59, 60). This implies that modifying activity of ubiquitous synapse-promoting molecules depending on subcellular locations may be common mechanisms by which INs establish subcellular synapse specificity.
A recent study suggested that more distinct IN subtypes within a broadly defined IN subgroup such as PV+ INs, SOM+ INs, and VIP+ INs play different roles in cortical processing (13–15). Therefore, the ultimate understanding of functional and wiring principles of cortical inhibitory circuits may require investigations on bona fide IN subtypes such as ChCs. Our present study elucidating molecular mechanisms by which highly specific IN-PN circuit motifs are established corresponds with this direction. ChCs have been implicated in the network oscillations (12, 58) and brain disorders such as schizophrenia and epilepsy (34, 35). Thus, our work may also provide an entry point into understanding the etiology of neurodevelopmental disorders caused by circuit deficits in different IN subtypes.
MATERIALS AND METHODS
Study design
The major findings of this manuscript were reproduced in all samples observed for this study. To ensure reproducibility, we used both male and female animals and repeated experiments at least three times per condition. No animals or data points were excluded from the analysis in this study.
Animals
All experimental procedures using live animals were approved by the Institutional Animal Care and Use Committee of the Max Planck Florida Institute for Neuroscience and performed in accordance with institutional and federal guidelines. The mice were kept under a 12-hour light/12-hour dark cycle and housed in standard cages with water and food ad libitum. E0 and P0 are defined by the day of plug and the day of birth, respectively. Both males and females were used for this study.
Mouse strains
Heterozygous Nkx2.1-CreER knock-in mice were bred to homo Cre-responsive loxP-STOP-loxP-triple GFP (Ai47) reporter mice to obtain Nkx2.1CreER/+;Ai47GFP/+. To get Nkx2.1CreER/+;Ai47GFP/GFP mice, Nkx2.1CreER/+;Ai47GFP/+mice were further bred to Ai47GFP/GFP mice. To obtain Nkx2.1CreER/+;Ai47GFP/+mice (ChC-GFP mice) for experiments, Nkx2.1CreER/+;Ai47GFP/GFP male mice were crossed with Swiss Webster (SW) female mice.
Heterozygous Nkx2.1-2A-CreER mice were bred to homo FLEX-Flp mice to obtain Nkx2.12A-CreER/+;FLEX-FlpFlp/+. To get Nkx2.12A-CreER/+; FLEX-FlpFlp/Flp mice, Nkx2.12A-CreER/+;FLEX-FlpFlp/+mice were further bred to homo FLEX-FlpFlp/Flp mice. To obtain Nkx2.12A-CreER/+; FLEX-FlpFlp/+ mice (ChC-Flp mice) for experiments, Nkx2.12A-CreER/+; FLEX-FlpFlp/Flp male mice were crossed with SW female mice.
SOM-ires-Cre and VIP-ires-Cre knock-in mice were maintained as homozygotes. To obtain SOMires-Cre/+;Ai47GFP/+ (SOM-GFP mice) or VIPires-Cre/+;Ai47GFP/+ mice (VIP-GFP mice), SOMires-Cre/ires-Cre or VIPires-Cre/ires-Cre homo male mice were crossed with Ai47GFP/GFP female mice. To obtain VIPires-Cre/+ or SOMires-Cre/+ het mice, VIPires-Cre/ires-Cre or SOMires-Cre/ires-Cre homo male mice were crossed with SW female mice.
IgSF11 het (Igsf11+/−) KO male mice and IgSF11 homo (Igsf11−/−) KO female mice were maintained. To obtain Igsf11+/− and Igsf11−/− mice for experiments, Igsf11−/− females were crossed with Igsf11+/− males.
Generation of Nkx2.1-2A-CreER and FLEX-Flp knock-in mice
For Nkx2.1-2A-CreER knock-in mice in which a 2A-CreER cassette was inserted in frame immediately after an open reading frame of an Nkx2.1 gene, the targeting vector containing 5′ and 3′ homology arms, a 2A-CreER cassette, an frt-Neo-frt cassette, and an HSV-TK gene was constructed. For FLEX-Flp mice, the targeting vector containing Rosa26 homology arms, a CAG promoter, and a FLEX-Flp cassette was constructed. Both targeting vectors were generated using a PCR-based cloning strategy. 129SVj/B6 F1 hybrid embryonic stem (ES) cells (V6.5) were electroporated with the targeting vectors and subjected to drug resistance tests. Neomycin-resistant ES clones for Nkx2.1-2A-CreER and FLEX-Flp were screened by mini Southern blotting and PCR, respectively, for correct targeting. Positive ES clones were used for tetraploid complementation to obtain male het mice following standard procedures.
Genotyping
The following primers were used to determine genotypes: Cre (forward, 5′-cggtcgatgcaacgagtgatg-3′; reverse, 5′-agcctgttttgcacgttcacc-3′) for Nkx2.1-CreER, Nkx2.1-2A-CreER, SOM-ires-Cre, and VIP-ires-Cre mice; ROSA (forward, 5′-cccaaagtcgctctgagttg ttatc-3′), ROSA mutant (reverse, 5′-gaaggagcgggagaatggatatg-3′), and ROSA WT (reverse, 5′-ccaggcgggccatttaccgtaag-3′) for FLEX-Flp and Ai47 mice; and IgSF11-P1 (5′-acatgcacaggaaggtcctcatt-3′), IgSF11-P3 (5′-gcgccgtcaaacatttgtccac-3′), and IgSF11-P4 (5′-aggatgtccttaagagtacacagga g-3′) for IgSF11 KO mice.
Tamoxifen induction
Tamoxifen was administered to timed pregnant SW females that were bred to Nkx2.1CreER/+;Ai47GFP/GFP males or Nkx2.12A-CreER/+; FLEX-FlpFlp/Flp males by gavaging at E17 to induce CreER activity in the offspring. To achieve dense labeling of ChCs for RNA-seq and FISH experiments, the dose was adjusted to 3.0 mg/30 g of body weight. Nkx2.1CreER/+;Ai47GFP/+ pups were identified by GFP expression in the lung at P0 or P1 under a fluorescence dissecting stereomicroscope. To achieve sparse expression of Flp in ChCs for IgSF11 GOF experiments, the dose was adjusted to 0.15 mg/30 g of body weight. Nkx2.12A-CreER/+;FLEX-FlpFlp/+ pups were identified by genotyping PCR before surgery.
Vector construction
The plasmids for AAV-HA-IgF11 (pAAV-CAG-HA-IgSF11), FlpD-AAV-GFP (pAAV-CAG-dfrt-GFP), and FlpD-AAV-HA-IgSF11 (pAAV-CAG-dfrt-HA-IgSF11) were generated by infusion of the HA-IgSF11, dfrt-GFP, and dfrt-HA-IgSF11 sequences, respectively, into pAAV-CAG vectors. To generate the plasmid for CreD-AAV-HA-IgSF11 (pAAV-CAG-FLEX-HA-IgSF11), the egfp sequence of pAAV-CAG-FLEX-egfp was replaced with that of HA-IgSF11.
The construction of single guide RNA (sgRNA) vectors was performed as previously described (53). Briefly, to generate plasmids harboring LacZ- and IgSF11-sgRNA, the annealed oligonucleotides containing the target sequence were inserted into pU6-sgRNA (backbone)-pCBh-Cas9. The sense sequences of sgRNA oligonucleotides are as follows: 5′-tgcgaatacgcccacgcgat-3′ (LacZ-sgRNA), 5′-aatgacattgaggttcagga-3′ (IgSF11-sgRNA for LOF), and 5′-aatgacattgaggttcagga-3′ (IgSF11-sgRNA for HITI). To create the donor DNA for HITI experiments (HA-IgSF11 donorDNA-2PAM), the promoter region of PX330 (#58778, Addgene) was removed and the donor sequence including HA tag was inserted. The donor sequence used for IgSF11 is as follows: 5′-cctgggtctc ggacacttccaggatacccttacgacgttccagactacgctggcctgggtctcgg acact tccag-3′.
Single-cell genotyping
A genotypic analysis of IgSF11-sgRNA–transfected cells was performed as previously described (53) with slight modifications. Briefly, after confocal scanning, sections (60 μm) that contained the morphologically analyzed ChCs or PNs were incubated in 30% sucrose in phosphate buffered saline (PBS) overnight at 4°C and flash-frozen on a flat optimal cutting temperature compound block (Sakura), recut into 20-μm sections using a cryostat (Leica), and placed on polyethylene naphthalate (PEN) membrane coverslips (Leica). The soma of the morphologically reidentified ChCs or PNs was cut out by laser microdissection (Leica), collected in PCR tubes, and digested in 5 μl of DirectPCR Lysis Reagent (Cell) (Viagen Biotech) with proteinase K (0.1 mg/ml final concentration; Ambion). After two rounds of PCR amplifying an approximately 200–base pair (bp) fragment around the sgRNA binding site, this fragment was cloned into pBlueskript vectors using infusion cloning (Clontech). Five plasmid clones were randomly selected and subjected to DNA sequencing with M13 forward primer. Primers used in the first PCR are the following: forward 5′-tgtgatgctcaaagagctgc-3′ and reverse 5′-atgatgcttccctccctatctc-3′. Primers used in the second PCR are the following: forward 5′-atgggaccaaattgctccttc-3′ and reverse 5′-acctgttcgggctggtttgc-3′.
Adeno-associated viruses
AAV-RFP (AAV-hSyn-mCherry, #114472) and CreD-AAV-GFP (AAV-CAG-FLEX-EGFP, #51502) were purchased from Addgene. AAV-HA-IgF11, FlpD-AAV-GFP, FlpD-AAV-HA-IgSF11, and CreD-AAV-HA-IgSF11 were packaged by Vigene Biosciences and aliquoted and stored at −80°C until use.
Exo utero electroporation of the MGE and transplantation of MGE grafts.
Timed pregnant SW mice were deeply anesthetized at gestational day 15 using isofluorane. After cervical dislocation, the uterus was dissected and kept in ice-cold PBS. Brains were removed from embryonic heads, and plasmid solution was injected into each ventricle. Electroporation was conducted dorsoventrally using a forceps electrode (5 mm in diameter; NEPA GENE) with the plus pole positioned ventrally (70 V, 50-ms duration, 950-ms interval, five pulses, and 10% decay; NEPA GENE). The following combination of plasmids were used: pCAG-GFP (1.5 μg/μl) for labeling ChCs, pCAG-GFP (1.0 μg/μl)/pU6-IgSF11-sgRNA-pCBh-Cas9 or pU6-LacZ-sgRNA-pCBh-Cas9 (3.0 μg/μl) for CRISPR-Cas9–mediated IgSF11 LOF experiments, pCAG-DsRed (1.0 μg/μl)/pCAG-ChR2-YFP (3.0 μg/μl) for electrophysiological recording, and HA-IgSF11 donorDNA-2PAM (2.0 μg/μl)/pU6-IgSF11-sgRNA (2.0 μg/μl)/pCAG-GFP (1.5 μg/μl) for HITI-mediated gene tagging. Afterward, the brains were cut into 400-μm sections using a tissue chopper (Intracell Technology). Brain slices were kept in Hibernate buffer (Hibernate-E, 100× GlutaMAX, and 50× B27 supplement; Gibco). The ventral MGE was dissected and collected in 1 ml of Hibernate buffer and manually cut approximately into 200 μm by 200 μm grafts.
P1 SW mice were anesthetized on ice for 2 min, and the absence of pain perception was assured. Embryonic tissue from one embryo was injected at 0.2 mm anterior and 0.2 mm lateral of the bregma at a depth of 150 μm using pulled glass pipets (model no. G150F-4, Warner Instruments) in combination with a stereotactic apparatus (Kopf) and a picospritzer (Parker). The incision was closed with a vet bond (Patterson Veterinary), and the pups were placed on a heat plate at 37°C until full recovery and subsequently returned to the mother.
In IgSF11 LOF experiments in ChCs, E15 Igsf11+/− and Igsf11−/− embryos were obtained from the same litter. Brains from embryos were individually collected, and their genotypes were determined by PCR. In IgSF11 LOF experiments in postsynaptic cells, P1 Igsf11+/− and Igsf11−/− pups from the same litter were used as host animals. After birth, their genotypes were determined by PCR before surgery. Individual host animals were differentially marked by the tattooing to the paws.
IgSF11 GOF experiments
To overexpress IgSF11 in ChCs, the mixture of FlpD-AAV-HA-IgSF11 and FlpD-AAV-GFP was injected into P1 ChC-Flp mice. To ectopically overexpress IgSF11 in SOM+ INs or VIP+ INs, the mixture of CreD-AAV-HA-IgSF11 and CreD-AAV-GFP was injected into P1 VIP-ires-Cre or SOM-ires-Cre mice. For ectopic overexpression of IgSF11 in deep-layer PNs, AAV-HA-IgSF11 or AAV-RFP was injected into P1 SW mice. After anesthetizing the pups on ice for 2 min, 500 nl of AAV solution was injected at 0.2 mm anterior and 0.1 mm lateral of the bregma at a depth of 0.15 mm using pulled glass pipets in combination with a stereotactic apparatus and a picospritzer. To visualize ChCs in IgSF11 GOF experiments in PNs, GFP-electroporated MGE tissues of E15 SW embryo were transplanted into the same hemisphere of the neocortex as described above. To overexpress IgSF11 in INs, Cre-electroporated MGE tissues of E15 SW embryo were transplanted into the hemisphere of the neocortex before injecting the mixture of CreD-AAV-HA-IgSF11 and CreD-AAV-GFP as described above. The incision was closed with the vet bond, and the pups were placed on a heat plate at 37°C until full recovery and subsequently returned to the mother.
In utero electroporation
To target L2/3 PNs for HITI experiments, IUE targeting cortical progenitors was performed at E15. Plasmid DNAs diluted in PBS, which contain pCAG-GFP (1.0 μg/μl)/HA-IgSF11 donorDNA-2PAM (1.5 μg/μl)/pU6-IgSF11 sgRNA-pCBh-Cas9 (1.5 μg/μl), were injected into the cerebral ventricles of embryos using sharp pulled glass pipettes (~2 μl per embryo). To examine IgSF11 mRNA FISH signals in uL2/3 PNs, E15 PN progenitors were electroporated with pCAG-GFP plasmids (1.0 μg/μl) using IUE. The following conditions were used for electroporation: two poring pulses of 50 V followed by five pulses of 35 V with 10% decay rate (NEPA21 super electroporator, NEPA GENE).
Immunohistochemistry
Following procedures previously described in (37), we performed immunohistochemistry. Mice were deeply anesthetized with an intraperitoneal injection of ketamine and xylazine [ketamine (50 mg/kg) and xylazine (5 mg/kg)] and transcardially perfused with 15 ml of cold 0.9% saline solution followed by 20 ml of 4% paraformaldehyde (PFA) in PBS. Brains were dissected and postfixed in 2% PFA overnight at 4°C and afterward stored in PBS until further use. Next, coronal brain sections (60 μm) were prepared using a vibratome (Leica), permeabilized in 0.3% Triton X-100 in PBS for 10 min, followed by blocking in 0.3% Triton X-100 and 10% donkey serum (Jackson ImmunoResearch Laboratories) in PBS for 1 hour. Subsequently, slices were incubated with primary antibodies in blocking solution overnight at 4°C. The following primary antibodies were used: rat anti-HA (1:500; Roche, catalog no. 11-867-423-001), rabbit anti-HA (1:500; Cell Signaling Technologies, catalog no. 3724), chicken anti-GFP (1:2000; Abcam, catalog no. ab13970), mouse anti-AnkG (1:500; UC Davis/NIH Neuromab, catalog no. clone N106/36 75-146), rabbit anti-VGAT (1:3000; Synaptic Systems, catalog no. 131003), rat anti-Ctip2 (1:1000; Abcam, catalog no. ab18465), rabbit anti-Cux1 (1:500; Proteintech, catalog no. 117331), goat anti-PV (Swant, catalog no. PVG-214), and rabbit anti-RFP (1:800; Rockland, catalog no. 600-401-379). After three washing steps, each for 10 min in PBS, the slices were incubated in appropriate fluorescently labeled secondary antibodies in blocking reagent for visualization at room temperature for 2 hours. All donkey secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. The following secondary antibodies were used at a concentration of 1:1000: anti-chicken Alexa Fluor 488 (catalog no. 703-545-155), anti-mouse Alexa Fluor 488 (catalog no. 715-545-151), anti-rabbit Alexa Fluor 488 (catalog no. 711-545-152), anti-rabbit Cy3 (catalog no. 711-165-152), anti-mouse Cy3 (catalog no. 715-165-151), anti-mouse Cy5 (catalog no. 715-175-150), and anti-rat Alexa Fluor 594 (catalog no. 712-585-150). After four washing steps in PBS, slices were mounted in DAKO fluorescent mounting medium (Agilent, S3023) and stored at 4°C.
Imaging of fixed brain samples
All images were acquired using a confocal microscope [Zeiss CLSM 880; 20× PLAN ApoChromat, numerical aperture (NA): 0.8 m; 63× PLAN ApoChromat, NA: 1.4 m].
Morphological analysis of ChC axonal boutons
For morphological analysis of ChC axonal boutons, confocal images were taken from the soma-containing slice. The area of 100 μm by 100 μm square with the soma of the cell located in the upper middle of the square was selected. A swelling structure, whose width is twice more than neighbor axons, was defined as “axonal bouton.” Furthermore, axonal boutons contacting or overlapping with AnkG+ AISs in a single optical section were defined as “AIS boutons.” The total number of AIS boutons and the number of boutons per AIS were manually counted in the Z-stack using the “multipoint” function of the Fiji image analysis software. The axonal arbor was semiautomatically traced and measured using ImageJ software and NeuronJ plug-in. The size of AIS boutons was measured using marching cube-type surface generation of Imaris software (Bitplane).
Ultrastructual analysis by electron microscopy
Following procedures previously described in (37), we carried out ultrastructural analysis by electron microscopy. Mice were anesthetized with ketamine xylazine mixture and perfused transcardially with 0.9% NaCl in 0.025 M Sorensen’s phosphate buffer (PB; pH 7.4) followed by 4% PFA and 0.25% glutaraldehyde in 0.1 M PB for 12 min. After postfixation in 4% PFA in PB overnight and washing in 0.1 M PB, 50-μm-thick coronal sections of the cortex were obtained using a vibratome (Leica). GFP expressing single isolated ChCs were identified using an epifluorescence microscope [Olympus BX51; 10× UPlanSApo, NA: 0.4 (Olympus) or 20× UPlanFl, NA: 0.5 (Olympus)]. To find the overlapping of ChC axonal boutons and HA-IgSF11–overexpressing PNs, confocal microscopy (Zeiss CLSM 780; 20× PLAN ApoChromat, NA: 0.8) was used. Sections were treated with 15 and 30% sucrose in 0.1 M PB. Sections were permeabilized by submersion in liquid nitrogen followed by incubation in a blocking solution of 10% normal goat serum (NGS), 1% fish skin gelatin (FSG) in 50 mM tris buffered saline (TBS) for 1 hour. Sections were incubated with rabbit anti-GFP antibodies (0.125 μg/ml; Abcam, ab#6556) diluted in TBS with 1% NGS and 0.1% FSG for 2 days, washed with TBS, and then incubated with nanogold-conjugated anti-rabbit antibodies (1:50; Nanoprobes, catalog 2003) diluted in TBS with 1% NGS and 0.1% FSG. For labeling HA-IgSF11–overexpressing PNs, rabbit anti-HA antibodies (0.067 μg/ml; Cell Signaling Technologies, #3724) were added together with anti-GFP antibodies. Immunogold-labeled sections were washed and a silver-enhanced using HQ SILVER intensification kit (Nanoprobes, catalog 2012), then osmificated with 0.5% OsO4, en bloc stained with 1% uranyl acetate, and dehydrated with a series of ethanol and acetone. Dehydrated sections were embedded in Fluka Durcupan resin (Sigma-Aldrich) and polymerized at 60°C for 2 days. Immunogold-labeled ChCs were trimmed out under a dissecting scope and serially sectioned at a thicknes of 45 nm with an ultramicrotome (UC7, Leica). Serial ultrathin sections were counterstained with uranyl acetate and lead citrate. Samples were examined in a Tecnai G2 Spirit electron microscope (Thermo Fisher Scientific) at 100 kV. Images were acquired with a Veleta charge-coupled device camera (Olympus) operated by TIA software (Thermo Fisher Scientific).
Transmission electron microscopy data collection and analysis
GFP-immunogold–positive profiles with a diameter of more than 200 nm were identified, and those profiles were serially imaged to cover whole varicosities from neck to neck. A typical varicosity spans 13 to 19 serial sections. Transmission electron microscopy image analysis was performed with Photoshop (Adobe) and Fiji image analysis software [National Institutes of Health (NIH)]. For synaptic analysis, we defined a “complete synapse” as having (i) a pre- and postsynaptic membrane forming a rigid synaptic cleft, (ii) a thin postsynaptic density, and (iii) presynaptic accumulation of vesicles. We selected three successive images showing close to the middle of the active zone and showing vesicles clearly and manually counted the number of vesicles within 200 nm from each active zone (i.e., the presynaptic membrane opposing the postsynaptic density) using Fiji. The boutons in which the active zone was not observed were excluded from the quantification of vesicles near the active zone. Immunogold particles in the profiles were thresholded and subtracted from the total area.
Slice preparation for electrophysiological recordings
Following procedures previously described in (61), we prepared brain slices for electrophysiological recordings. Acute neocortical slices were obtained from 4-week-old mice, into which ChR2-YFP–electropolated MGE explants of IgSF11 het or homo KO mice were transplanted. The mice were anesthetized with isoflurane and decapitated. Brains were quickly removed and chilled in ice-cold high-magnesium cutting solution containing the following: 100 mM choline chloride, 25 mM NaHCO3, 2.5 mM KCl, 0.5 mM CaCl2, 7 mM MgCl2, 1.25 mM NaH2PO4, 25 mM glucose, 20 mM Hepes, 3.1 mM Na-pyruvate, and 5 mM Na-ascorbate. pH and osmolarity were adjusted to 7.4 and ~300 mOsm, respectively. The isolated brain was glued onto the stage of a vibratome (Leica VT1000, Leica Biosystems, Buffalo Grove, IL, USA), and coronal slices (270 μm thick) were cut. The slices were transferred and incubated at 34°C for 30 min in a slice container superfused with artificial cerebrospinal fluid (ACSF) solution containing the following: 124 mM NaCl, 26 mM NaHCO3, 3.2 mM KCl, 2.5 mM CaCl2, 1.3 mM MgCl2, 1.25 mM NaH2PO4, and 10 mM glucose, saturated with 95% O2 and 5% CO2 gas. Thereafter, slices were maintained at room temperature for the experiments.
Electrophysiological recordings
Whole-cell voltage-clamp recordings from uL2/3 PNs in the frontal cortex were carried out at room temperature while the recording chamber was perfused with ACSF at 1 to 1.5 ml/min. The recordings were made using a MultiClamp 700B amplifier controlled by Clampex 10.2 via Digidata 1440A data acquisition system (Molecular Devices, Sunnyvale, CA, USA). The pipette solution contained the following: 120 mM CsCl, 10 mM Hepes, 10 mM Na2-phosphocreatine, 8 mM NaCl, 5 mM QX-314, 4 mM Mg–adenosine triphosphate, 0.4 mM Na2–guanosine triphosphate, and 2 mM EGTA (pH = 7.3 with CsOH, 295 mOsm). After forming a whole-cell patch on the soma of a PN through the pipette filled with Alexa Fluor 594 for post hoc morphological reconstruction, the membrane potential was held at 0 mV under the voltage clamp conditions. Under this condition, we monitored IPSC in the absence or presence of 470-nm wavelength of blue light stimulation through water-immersion 40× objective coupled to a light-emitting diode (pE-100, CoolLED) to activate adjacent ChR2-expressing ChCs. The peak amplitude of IPSCs from each genotype was measured upon 5 s of blue light.
RNA sequencing
RNAs were purified from manually collected ChCs, SOM+ INs, or VIP+ INs as described previously (51). Briefly, fresh brains were taken from P5 ChC-GFP, SOM-GFP, and VIP-GFP pups, and 300- to 400-μm-thick slices were prepared using a tissue chopper (Intracell Technology). Cortical strips containing L2/3 cells were dissected from the mPFC and the aCC and dissociated into individual cells in Hibernate E medium (Invitrogen) containing pronase E (1 mg/ml; Sigma-Aldrich) and deoxyribonuclease I (5 U/ml; Thermo Fisher Scientific) at 37°C for 10 min. After washing with Hibernate E medium once, cells were spread in 35-mm dishes. One hundred fifty GFP+ cells were picked up using a glass micropipette (Warner Instruments, #203-776-064) pulled with a micropipette puller (Sutter Instrument, #P-1000) under a fluorescence dissecting stereomicroscope. RNA was extracted by an RNeasy Micro kit (QIAGEN). The samples were then kept on ice and processed for library preparation using Illumina Seq according to the manufacturer’s protocol. Samples were sequenced in the NextSeq 500 with paired-end 75-bp reads. Demultiplexed and quality filtered raw reads (fastq) generated from the NextSeq 500 were trimmed (Illumina adaptor sequences) using Flexbar 2.4 and aligned to the reference genome (Mus musculus, mm10) using TopHat version 2.0.9. htseq-count version 0.6.1 was used to generate gene counts, and differential gene expression analysis was performed using DESeq2.
Retrograde tracing of PC subtypes
P10 mice were deeply anesthetized using isofluorane. Mice were mounted in a stereotaxic head frame. Bregma coordinates were identified for brain areas: CC [anteroposterior (A/P): 2.0 mm; mediolateral (M/L): 0.1 mm; dorsoventral (D/V): 1.2 mm depth from the pial surface] and BLA (A/P: −1.0 mm, M/L: 2.5 mm; D/V: 3.0 mm). An incision was made over the scalp; a small hole was made into the skull by a 32-gauge needle. Five hundred nanoliters of CTB subunit conjugated with Alexa Fluor 488 (0.3 μl, 2% in PBS; Life Technologies) was injected into the BLA and CC of the brain using pulled glass pipets in combination with a stereotactic apparatus and a picospritzer. The incision was closed with the vet bond, and mice were placed on a heat plate at 37°C until full recovery and subsequently returned to the mother. At P16, mice were perfused, and the tissue was processed by the protocols for FISH.
Fluorescent in situ hybridization
Following procedures previously described in (51), we performed FISH to detect IgSF11, Cdh6, Cdh12, FGF13, Trps1, and Gad1 mRNAs. Briefly, the fragment of each gene was amplified by reverse transcription PCR with the following primers: forward 5′-tcagtgccctctcttccg-3′ and reverse 5′-caggccacttcacacacg-3′ (IgSF11), forward 5′-acattaaggaaggaaggagg-3′ and reverse 5′-gttgggtcgtcagcatcag-3′ (Cdh6), forward 5′-agaccttgatgtgggc-3′ and reverse 5′-ccatctgagtcacacc-3′ (Cdh12), forward 5′-tgtgcccaaactggtcct-3′ and reverse 5′-tggccgatgattctggtt-3′ (Gad1), forward 5′-tcgctcatccggcaaaag-3′ and reverse 5′-ggttctgttatagagcc-3′ (FGF13), and forward 5′-atcaagcccctcgattcc-3′ and reverse 5′-cagcccgtaacagcgact-3′ (Trps1). The amplified fragment was cloned into pBluescript vectors using an In-Fusion kit (TaKaRa). Digoxigenin (DIG)–labeled single-strand riboprobes were synthesized using T7 or T3 RNA polymerase and DIG RNA-labeling mix (Roche).
Mice were perfused, and the tissue was processed as described above. Sections (50 μm thick) prepared from whole brains were treated with proteinase K (40 μg/ml for 30 min at room temperature) and hybridized at 63°C with DIG-labeled antisense riboprobes in a hybridization buffer (Enzo Life Sciences). The sections were washed twice in 1× SSC (Invitrogen) containing 50% formamide and once in 0.1× SSC at 63°C, followed by two washes with 0.1 M maleic buffer (pH 7.5) containing 0.1% Tween 20 and 150 mM NaCl. Then, these sections were incubated with anti-DIG antibodies conjugated with alkaline phosphatase overnight at 4°C, followed by three washes in PBT solution (PBS containing 0.1% Triton X-100). Sections were incubated with chick polyclonal anti-GFP antibodies overnight at 4°C. After washing three times in PBT, sections were incubated with biotinylated anti-chick immunoglobulin G antibodies overnight at 4°C followed by three washes in PBT. The sections were then incubated with Alexa Fluor 488–streptavidin (1:1000; Jackson ImmunoResearch Laboratories, #016-540-084) to visualize GFP+ cells for 2 hours at room temperature. For FISH, the color development for mRNA expression was performed in the presence of 2-hydroxy-3-naphthoic acid-2’-phenylanilide phosphate (HNPP)/FastRed solution [HNPP (100 μg/ml) and FastRed (250 μg/ml); Roche) for 20 min at room temperature. The sections were washed 1 min in PBS and mounted with CC/Mount tissue mounting medium (Sigma-Aldrich, #C9368). Confocal images were taken immediately after color development.
For double FISH of IgSF11 and Gad1 mRNAs, in addition to DIG-labeled IgSF11 RNA probes, Gad1 RNA probes were synthesized by using a fluorescein isothiocyanate (FITC) labeling kit (Roche, catalog no. 11685619910). After hybridization, FITC signals were converted to biotin by using HRP-conjugated FITC antibodies (1:1000; Roche, catalog no. 11426346910) and biotin-conjugated HRP antibodies (1:500; Jackson ImmunoResearch Laboratories, catalog no. 325-065-021). Biotin signals were amplified by using an ABC kit (Vector Laboratories, catalog no. PK-6100), and the sections were incubated with Alexa Fluor 488–streptavidin to visualize Gad1 mRNA signals before the color development of IgSF11 mRNA signals by HNPP/FastRed reaction.
Statistical analyses
Except for the analysis of layer distribution of retrogradely labeled L2/3 PNs by CTB (Fig. 1I), graphs and results are presented as the means ± SEM throughout experiments. In Fig. 1I, data are box plots. Except for RNA-seq, all statistical analyses were performed using Prism 6.0 (GraphPad). Comparisons were done by unpaired two-tailed Student’s t test for two groups or one-way analysis of variance (ANOVA) for more than two groups. After ANOVA, a Bonferroni post hoc test was performed to analyze statistical significance between groups. Differential gene expression analysis of RNA-seq dataset was performed using DESeq2. P values of less than 0.05 were considered significant. Statistical significance is presented in figures in the following manner: *P < 0.05, **P < 0.01, and ***P < 0.001.
Acknowledgments
We would like to thank G. Miyoshi, G. Di Cristo, R. Yasuda, and Y. Yoshida for comments on the manuscript; the Genomics Core Facility at Scripps Florida for performing RNA-seq experiments; and Taniguchi lab members for the helpful discussion. Funding: This work was supported by funding from National Institutes of Health grants MH107460 (to H.-B.K.), DP1MH119428 (to H.-B.K.), and NS119488 (to H.T.); Citizens United for Research in Epilepsy (CURE) (to H.T.); and the Max Planck Society (to H.-B.K., N.K., and H.T.). Author contributions: H.T. conceived and supervised the project. Y.H. and H.T. designed experiments, interpreted results, and wrote the manuscript. Y.I. collected cells, prepared mRNA samples for RNA-seq, and identified ChC-specific cell surface molecules. C.G.O. (LOF experiments), E.P. (GOF experiments), C.I.T., D.G.-G., and N.K. performed EM experiments and data analyses. J.H.H. performed all electrophysiology experiments and data analyses. A.S. performed CRISPR-Cas9–based KO experiments. H.T. designed knock-in constructs. A.S. and Y.O. generated knock-in constructs for FLEX-Flp and Nkx2.1-2A-CreER, respectively, and established mouse lines. E.K. provided IgSF11 germ line KO mice. H.-B.K. provided comments and edited the manuscript. Y.H. conducted all the other experiments and analyses. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All materials except IgSF11 germ line KO mice can be provided by H.T.’s pending scientific review and a completed material transfer agreement. Requests for these materials should be submitted to H.T. IgSF11 germline KO mice can be provided by E.K. under the condition of collaboration with E.K. Requests for IgSF11 germline KO mice should be submitted to E.K. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, H.T. (hiroki.taniguchi@mpfi.org). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/29/eabf1600/DC1
REFERENCES AND NOTES
- 1.Tasic B., Yao Z., Graybuck L. T., Smith K. A., Nguyen T. N., Bertagnolli D., Goldy J., Garren E., Economo M. N., Viswanathan S., Penn O., Bakken T., Menon V., Miller J., Fong O., Hirokawa K. E., Lathia K., Rimorin C., Tieu M., Larsen R., Casper T., Barkan E., Kroll M., Parry S., Shapovalova N. V., Hirschstein D., Pendergraft J., Sullivan H. A., Kim T. K., Szafer A., Dee N., Groblewski P., Wickersham I., Cetin A., Harris J. A., Levi B. P., Sunkin S. M., Madisen L., Daigle T. L., Looger L., Bernard A., Phillips J., Lein E., Hawrylycz M., Svoboda K., Jones A. R., Koch C., Zeng H., Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greig L. C., Woodworth M. B., Galazo M. J., Padmanabhan H., Macklis J. D., Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adesnik H., Naka A., Cracking the function of layers in the sensory cortex. Neuron 100, 1028–1043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronoff R., Petersen C. C., Layer, column and cell-type specific genetic manipulation in mouse barrel cortex. Front. Neurosci. 2, 64–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Souza R. D., Burkhalter A., A laminar organization for selective cortico-cortical communication. Front. Neuroanat. 11, 71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yavorska I., Wehr M., Somatostatin-expressing inhibitory interneurons in cortical circuits. Front. Neural Circuits 10, 76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X., Shen S., Cadwell C. R., Berens P., Sinz F., Ecker A. S., Patel S., Tolias A. S., Principles of connectivity among morphologically defined cell types in adult neocortex. Science 350, aac9462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris K. D., Shepherd G. M., The neocortical circuit: Themes and variations. Nat. Neurosci. 18, 170–181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tremblay R., Lee S., Rudy B., GABAergic interneurons in the neocortex: From cellular properties to circuits. Neuron 91, 260–292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kepecs A., Fishell G., Interneuron cell types are fit to function. Nature 505, 318–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim L., Mi D., Llorca A., Marin O., Development and functional diversification of cortical interneurons. Neuron 100, 294–313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klausberger T., Magill P. J., Márton L. F., Roberts J. D., Cobden P. M., Buzsáki G., Somogyi P., Brain-state- and cell-type-specific firing of hippocampal interneurons in vivo. Nature 421, 844–848 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Junco-Clemente P., Ikrar T., Tring E., Xu X., Ringach D. L., Trachtenberg J. T., An inhibitory pull-push circuit in frontal cortex. Nat. Neurosci. 20, 389–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munoz W., Tremblay R., Levenstein D., Rudy B., Layer-specific modulation of neocortical dendritic inhibition during active wakefulness. Science 355, 954–959 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Bortone D. S., Olsen S. R., Scanziani M., Translaminar inhibitory cells recruited by layer 6 corticothalamic neurons suppress visual cortex. Neuron 82, 474–485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang S., Oh D., Lee Y., Hosy E., Shin H., van Riesen C., Whitcomb D., Warburton J. M., Jo J., Kim D., Kim S. G., Um S. M., Kwon S. K., Kim M. H., Roh J. D., Woo J., Jun H., Lee D., Mah W., Kim H., Kaang B. K., Cho K., Rhee J. S., Choquet D., Kim E., Synaptic adhesion molecule IgSF11 regulates synaptic transmission and plasticity. Nat. Neurosci. 19, 84–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J., Tucciarone J., Padilla-Coreano N., He M., Gordon J. A., Huang Z. J., Selective inhibitory control of pyramidal neuron ensembles and cortical subnetworks by chandelier cells. Nat. Neurosci. 20, 1377–1383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X., Tucciarone J., Jiang S., Yin F., Wang B.-S., Wang D., Jia Y., Jia X., Li Y., Yang T., Xu Z., Akram M. A., Wang Y., Zeng S., Ascoli G. A., Mitra P., Gong H., Luo Q., Huang Z. J., Genetic single neuron anatomy reveals fine granularity of cortical axo-axonic cells. Cell Rep. 26, 3145–3159.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koelbl C., Helmstaedter M., Lubke J., Feldmeyer D., A barrel-related interneuron in layer 4 of rat somatosensory cortex with a high intrabarrel connectivity. Cereb. Cortex 25, 713–725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye Z., Mostajo-Radji M. A., Brown J. R., Rouaux C., Tomassy G. S., Hensch T. K., Arlotta P., Instructing perisomatic inhibition by direct lineage reprogramming of neocortical projection neurons. Neuron 88, 475–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodato S., Rouaux C., Quast K. B., Jantrachotechatchawan C., Studer M., Hensch T. K., Arlotta P., Excitatory projection neuron subtypes control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron 69, 763–779 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wester J. C., Mahadevan V., Rhodes C. T., Calvigioni D., Venkatesh S., Maric D., Hunt S., Yuan X., Zhang Y., Petros T. J., McBain C. J., Neocortical projection neurons instruct inhibitory interneuron circuit development in a lineage-dependent manner. Neuron 102, 960–975.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favuzzi E., Deogracias R., Marques-Smith A., Maeso P., Jezequel J., Exposito-Alonso D., Balia M., Kroon T., Hinojosa A. J., Maraver E. F., Rico B., Distinct molecular programs regulate synapse specificity in cortical inhibitory circuits. Science 363, 413–417 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Konno K., Matsuda K., Nakamoto C., Uchigashima M., Miyazaki T., Yamasaki M., Sakimura K., Yuzaki M., Watanabe M., Enriched expression of GluD1 in higher brain regions and its involvement in parallel fiber-interneuron synapse formation in the cerebellum. J. Neurosci. 34, 7412–7424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hepp R., Hay Y. A., Aguado C., Lujan R., Dauphinot L., Potier M. C., Nomura S., Poirel O., El Mestikawy S., Lambolez B., Tricoire L., Glutamate receptors of the delta family are widely expressed in the adult brain. Brain Struct. Funct. 220, 2797–2815 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Sagane K., Hayakawa K., Kai J., Hirohashi T., Takahashi E., Miyamoto N., Ino M., Oki T., Yamazaki K., Nagasu T., Ataxia and peripheral nerve hypomyelination in ADAM22-deficient mice. BMC Neurosci. 6, 33 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graf E. R., Zhang X., Jin S. X., Linhoff M. W., Craig A. M., Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119, 1013–1026 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi H., Katayama K., Sohya K., Miyamoto H., Prasad T., Matsumoto Y., Ota M., Yasuda H., Tsumoto T., Aruga J., Craig A. M., Selective control of inhibitory synapse development by Slitrk3-PTPdelta trans-synaptic interaction. Nat. Neurosci. 15, 389–398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai J. K., Doering L. C., Foster J. A., Developmental expression of the neuroligins and neurexins in fragile X mice. J. Comp. Neurol. 524, 807–828 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Beaubien F., Cloutier J.-F., Differential expression of Slitrk family members in the mouse nervous system. Dev. Dyn. 238, 3285–3296 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Szabadics J., Varga C., Molnar G., Olah S., Barzo P., Tamas G., Excitatory effect of GABAergic axo-axonic cells in cortical microcircuits. Science 311, 233–235 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Szentágothai J., Arbib M., Conceptual models of neural organization. Neurosci. Res. Program Bull. 12, 305–510 (1974). [PubMed] [Google Scholar]
- 33.Somogyi P., A specific ‘axo-axonal’ interneuron in the visual cortex of the rat. Brain Res. 136, 345–350 (1977). [DOI] [PubMed] [Google Scholar]
- 34.Lewis D. A., The chandelier neuron in schizophrenia. Dev. Neurobiol. 71, 118–127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeFelipe J., Chandelier cells and epilepsy. Brain 122 (Pt. 10), 1807–1822 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi H., Lu J., Huang Z. J., The spatial and temporal origin of chandelier cells in mouse neocortex. Science 339, 70–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinecke A., Hozhabri E., Tapanes S., Ishino Y., Zeng H., Kamasawa N., Taniguchi H., Neocortical chandelier cells developmentally shape axonal arbors through reorganization but establish subcellular synapse specificity without refinement. eNeuro 4, ENEURO.0057-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada H., Suzu S., Hayashi Y., Okada S., BT-IgSF, a novel immunoglobulin superfamily protein, functions as a cell adhesion molecule. J. Cell. Physiol. 204, 919–926 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Marques-Smith A., Lyngholm D., Kaufmann A.-K., Stacey J. A., Hoerder-Suabedissen A., Becker E. B., Wilson M. C., Molnár Z., Butt S. J., A transient translaminar GABAergic interneuron circuit connects thalamocortical recipient layers in neonatal somatosensory cortex. Neuron 89, 536–549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuncdemir S. N., Wamsley B., Stam F. J., Osakada F., Goulding M., Callaway E. M., Rudy B., Fishell G., Early somatostatin interneuron connectivity mediates the maturation of deep layer cortical circuits. Neuron 89, 521–535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taniguchi H., He M., Wu P., Kim S., Paik R., Sugino K., Kvitsiani D., Fu Y., Lu J., Lin Y., Miyoshi G., Shima Y., Fishell G., Nelson S. B., Huang Z. J., A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daigle T. L., Madisen L., Hage T. A., Valley M. T., Knoblich U., Larsen R. S., Takeno M. M., Huang L., Gu H., Larsen R., Mills M., Bosma-Moody A., Siverts L. A., Walker M., Graybuck L. T., Yao Z., Fong O., Nguyen T. N., Garren E., Lenz G. H., Chavarha M., Pendergraft J., Harrington J., Hirokawa K. E., Harris J. A., Nicovich P. R., McGraw M. J., Ollerenshaw D. R., Smith K. A., Baker C. A., Ting J. T., Sunkin S. M., Lecoq J., Lin M. Z., Boyden E. S., Murphy G. J., da Costa N. M., Waters J., Li L., Tasic B., Zeng H., A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174, 465–480.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liodis P., Denaxa M., Grigoriou M., Akufo-Addo C., Yanagawa Y., Pachnis V., Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. 27, 3078–3089 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyoshi G., Young A., Petros T., Karayannis T., McKenzie Chang M., Lavado A., Iwano T., Nakajima M., Taniguchi H., Huang Z. J., Heintz N., Oliver G., Matsuzaki F., Machold R. P., Fishell G., Prox1 regulates the subtype-specific development of caudal ganglionic eminence-derived GABAergic cortical interneurons. J. Neurosci. 35, 12869–12889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berns D. S., DeNardo L. A., Pederick D. T., Luo L., Teneurin-3 controls topographic circuit assembly in the hippocampus. Nature 554, 328–333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osterhout J. A., Josten N., Yamada J., Pan F., Wu S. W., Nguyen P. L., Panagiotakos G., Inoue Y. U., Egusa S. F., Volgyi B., Inoue T., Bloomfield S. A., Barres B. A., Berson D. M., Feldheim D. A., Huberman A. D., Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron 71, 632–639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams M. E., Wilke S. A., Daggett A., Davis E., Otto S., Ravi D., Ripley B., Bushong E. A., Ellisman M. H., Klein G., Ghosh A., Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron 71, 640–655 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuwako K. I., Nishimoto Y., Kawase S., Okano H. J., Okano H., Cadherin-7 regulates mossy fiber connectivity in the cerebellum. Cell Rep. 9, 311–323 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Takeichi M., The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 8, 11–20 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Sanes J. R., Zipursky S. L., Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell 181, 536–556 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Ishino Y., Yetman M. J., Sossi S. M., Steinecke A., Hayano Y., Taniguchi H., Regional cellular environment shapes phenotypic variations of hippocampal and neocortical chandelier cells. J. Neurosci. 37, 9901–9916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E. J., Hatanaka F., Yamamoto M., Araoka T., Li Z., Kurita M., Hishida T., Li M., Aizawa E., Guo S., Chen S., Goebl A., Soligalla R. D., Qu J., Jiang T., Fu X., Jafari M., Esteban C. R., Berggren W. T., Lajara J., Nuñez-Delicado E., Guillen P., Campistol J. M., Matsuzaki F., Liu G.-H., Magistretti P., Zhang K., Callaway E. M., Zhang K., Belmonte J. C., In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinecke A., Kurabayashi N., Hayano Y., Ishino Y., Taniguchi H., In vivo single-cell genotyping of mouse cortical neurons transfected with CRISPR/Cas9. Cell Rep. 28, 325–331.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paul A., Crow M., Raudales R., He M., Gillis J., Huang Z. J., Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell 171, 522–539.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costa M. R., Muller U., Specification of excitatory neurons in the developing cerebral cortex: Progenitor diversity and environmental influences. Front. Cell. Neurosci. 8, 449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuda K., Miura E., Miyazaki T., Kakegawa W., Emi K., Narumi S., Fukazawa Y., Ito-Ishida A., Kondo T., Shigemoto R., Watanabe M., Yuzaki M., Cbln1 is a ligand for an orphan glutamate receptor δ2, a bidirectional synapse organizer. Science 328, 363–368 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Sando R., Jiang X., Sudhof T. C., Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science 363, eaav7969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massi L., Lagler M., Hartwich K., Borhegyi Z., Somogyi P., Klausberger T., Temporal dynamics of parvalbumin-expressing axo-axonic and basket cells in the rat medial prefrontal cortex in vivo. J. Neurosci. 32, 16496–16502 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tai Y., Gallo N. B., Wang M., Yu J.-R., Van Aelst L., Axo-axonic innervation of neocortical pyramidal neurons by GABAergic chandelier cells requires Ankyring-associated L1CAM. Neuron 102, 358–372.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fossati M., Assendorp N., Gemin O., Colasse S., Dingli F., Arras G., Loew D., Charrier C., Trans-synaptic signaling through the glutamate receptor Delta-1 mediates inhibitory synapse formation in cortical pyramidal neurons. Neuron 104, 1081–1094.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yetman M. J., Washburn E., Hyun J. H., Osakada F., Hayano Y., Zeng H., Callaway E. M., Kwon H. B., Taniguchi H., Intersectional monosynaptic tracing for dissecting subtype-specific organization of GABAergic interneuron inputs. Nat. Neurosci. 22, 492–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/29/eabf1600/DC1


