Abstract
Objective
The primary objective of this study was to evaluate the correlation of large mitochondrial DNA (mtDNA) deletions in skin samples of people with HIV (PWH) with measures of neuropathy and prior exposure to therapy. We hypothesized that deletions would be associated with neuropathy. As secondary objectives, we determined the correlation of deletion burden with demographic data and neuropathy measures.
Methods
In this retrospective cohort study, we measured the accumulation of large mtDNA deletions in skin biopsies from PWH recruited as part of the AIDS Clinical Trials Group (ACTG). Our cohort includes individuals with and without sensory neuropathy, as well as individuals with normal or abnormal skin biopsies. Skin biopsies, sural and peroneal nerve conduction studies, total neuropathy score, and deletion burden scores were measured, along with baseline demographic data such as age, CD4+ cell count, viral counts, and prior nucleoside reverse transcriptase inhibitor exposures.
Results
Sixty-seven PWH were enrolled in the study. The mean age of the cohort (n = 67) was 44 years (SD 6.8, range 32–65 years), and 9 participants were female. The mean CD4+ T-cell count was 168 cells/mm3 (SD 97 cells/mm3, range 1–416 cells/mm3) and mean viral load was 51,129 copies/mL (SD 114,586 copies/mL, range 147–657,775 copies/mL). We determined that there was a correlation between the total mtDNA deletion and intraepidermal nerve fiber density (IENFD) (r = −0.344, p = 0.04) and sural nerve amplitude (r = −0.359, p = 0.004).
Conclusions
Both IENFD and sural nerve amplitude statistically correlate with mitochondrial mutation burden in PWH, specifically in those with HIV-associated sensory neuropathy as assessed by skin biopsy.
Trial Registration Information
HIV-associated sensory neuropathy (HIV-SN) is one of the most common complications limiting the quality of life of people with HIV (PWH). The pathogenesis is likely to be multifactorial, including indirect neurotoxicity of HIV envelope protein gp120 and direct toxicity of antiretroviral drugs.1-4 Within neuronal cells, mitochondria are assembled in the cell body, transported via microtubule-based machinery, and docked in the axon based on energy requirements. Thus, susceptibility of the distal axon to degeneration in HIV-SN can be hypothesized to be related to both its unique temporal and spatial distribution with distal segments containing older mitochondria.5 This allows accumulation of mitochondrial DNA (mtDNA) deletion mutations in distal nerves.
Large deletion mutations in mtDNA mutations have been observed in muscle, peripheral blood mononuclear cells, and the sural nerves of autopsied individuals with HIV-SN.6-8 Neurotoxic antiretroviral drugs, especially nucleoside reverse transcriptase inhibitors (dNRTIs), further compound the damage by decreasing the processivity of mtDNA polymerase gamma or by allowing the expansion of deleted genomes.9 Genetic polymorphisms, e.g., in CAMKK2 and TNF, have been associated with the development of HIV-SN after treatment with staduvine.10,11
In this study we analyzed the effect of HIV infection on neuropathy and mtDNA integrity in skin biopsies in a cohort of PWH. Using an assay that allows measurement of mutation burden across the mitochondrial genome, we have analyzed the frequency of mutations in this cohort and have correlated the degree of mutation deletion with demographic factors and objective measurements of neuropathy.
Methods
Standard Protocol Approvals, Registrations, and Patient Consents
This study was approved by the Johns Hopkins Institutional Review Board (study NA_00077011). Written informed consent was obtained from all patients (or guardians of patients) participating in the study (consent for research) as part of enrollment in the AIDS Clinical Trials Group (ACTG) network. This study has been registered as a public trial at ClinicalTrials.gov (NCT00017771).
Characteristics of the Study Participant Samples
Clinical data and specimens for this study were obtained through the ACTG network. The ACTG is the world's largest and longest-running HIV clinical trials network.12 This study was approved by the Johns Hopkins Institutional Review Board (study NA_00077011). PWH (n = 67) had undergone skin biopsies and evaluation for the presence of HIV-SN at baseline and then were enrolled in 3 different dNRTI treatment arms. We were granted access to prior dNRTI exposure, including didanosine, d4T, and dideoxycytidine. The clinical criteria used for the diagnosis of neuropathy included 2 of 3 examination findings of reduced pinprick and vibration sensation distally in the feet and reduced ankle deep-tendon reflexes. Further assessment of limb strength and sensory and motor symptoms and electrophysiologic assessment of sural and peroneal amplitudes were carried out by a board-certified neurologist to calculate the total neuropathy score (TNS).13 Blinded pathologic evaluation of intraepidermal nerve fiber density (IENFD) was carried out by a board-certified neurologist using skin biopsy samples obtained from the proximal thigh and distal leg of each participant. Patients with other potential causes of neuropathy such as chronic alcohol use or diabetes were excluded from the study.
PCR Detection of Mitochondrial Deletion Mutations
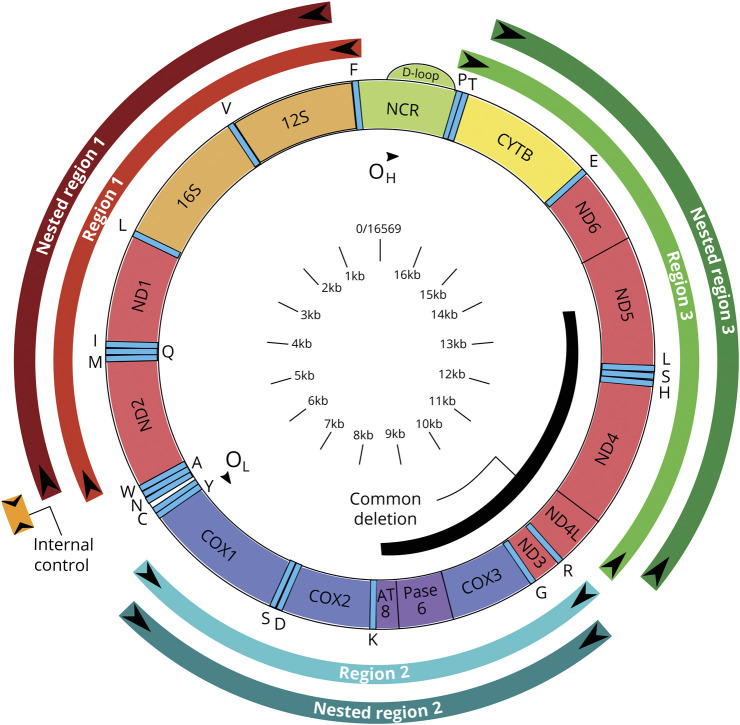
A nested PCR was used to determine the total mitochondrial genomic deletion mutation burden in epidermal tissue. Each sample was incubated with 20 μL NaOH (50 mM) at 95°C for 25 minutes, and reaction was stopped by adding 2.2 μL Tris-HCl (pH 8.0). PCR amplifications were performed with the One puReTaq Ready-To-Go PCR bead (GE Healthcare, Piscataway, NJ) in 25 µL final volume reactions. Forward and reverse inner and outer primers were obtained from Integrated DNA Technologies (Coralville, IA) (figure 1 and eTable 1 give all primers sequences, doi.org/10.5061/dryad.8pk0p2nmh) and added to a final concentration of 0.5 μM. The amplification was carried out in an MJ DNA Engine Opticon 2 (Bio-Rad, Hercules, CA). First-round cycling conditions consisted of 95°C for 2 minutes followed by 30 cycles at 95°C × 20 seconds, 57°C × 20 seconds, 72°C × 2 minutes, and, by final extension, 72°C for 10 minutes. Two microliters of PCR product from the first round was used in the second-round amplification, and the cycling conditions were as follows: 95°C for 2 minutes for 30 cycles (95°C × 20 seconds, 58°C × 20 seconds, 72°C × 2 minutes), followed by final extension at 72°C for 10 minutes. For the internal control, 2 μL PCR product from the first PCR reaction was amplified with a pair of primers resulting in a short (300 bp) PCR product. The PCR product was then cleaned to remove excess primers with a PCR Purification Kit (Qiagen, Valencia, CA), according to the protocol provided by the manufacturer. After 3 washes, the PCR product was eluted in 200 μL 90°C dH2O. The amplicon fluorescence value was divided for the value obtained for the control region in that reaction. The total deletion score (TDS) was calculated by adding the values of the 3 regions examined. First-round amplification was as follows: region 1: forward primer, 5′-CCCTAACACCAGCCTAACAA(nt369-388); reverse primer, 5′- GTTGATGCAGAGTGGGGTTT(nt5598-5617); region 2: forward primer, 5′- CTCCTACTCCTGCTCGCATC(nt6228-6247); reverse primer, 5′- GGAGTGGGTGTTGAGGGTTA(nt10968-10987); and region 3: forward primer, 5′- CCTGACTCCTACCCCTCACA(nt10968-10987); reverse primer, 5′- CTTGGCTATCATCACCCGAT(nt11146-11165); second-round nested PCR–fluorescent tag+ **FAM: region 1: forward primer, 5′-TTATTTTCCCCTCCCACTCC(nt449-468); reverse primer, 5’-*Fam-GTGGGGTTTTGCAGTCCTTA(nt5586-5606); region 2: forward primer, 5′- CTTAGGGGCCATCAATTTCA(6377–6396); reverse primer, 5’-**Fam/TGTAAATGAGGGGCATTTGG(nt10466-10485); and region 3: forward primer, 5′- CTTGGCTATCATCACCCGAT(nt11146-11165); reverse primer: 5’-**Fam/AGGACAGGCCCATTTGAGTA(nt15873-15892) (internal control PCR).
Figure 1. Mitochondrial DNA Map Showing Locations of Markers Used During This Study.
Short Fragment Amplification in Region 1
The forward primer was 5′- CACCATCATAGCCACCATCA(nt5306-5325), and the reverse primer was same as that in the second round: FAM/GTGGGGTTTTGCAGTCCTTA(nt.5586–5606).
Fluorescence Measurement of PCR Products
Short DNA can be amplified much more effectively under the designed amplification conditions; thus, fluorescence measurement from the amplified PCR product will mostly represent mtDNA with large deletions. A plate with 100 μL of the PCR product was used for fluorescence reading in a FlexStation 3 Fluorescence Microplate Reader (Molecular Devices, Eugene, OR). The ratio between the amplified PCR product fluorescence measurement and the internal control PCR fluorescence measurement was used to compare the amount of mitochondrial genomic DNA deletions to original mitochondrial genomic DNA template.
Statistical Analysis
Statistical analysis was carried out with JASP 0.12.2 software (University of Amsterdam, Netherlands) and Microsoft Excel (Redmond, WA). To analyze correlations for continuous variables, the Pearson correlation coefficient and its r and p values were used; to determine statistical significance between 2 groups, the Student independent-samples t test was used. Missing data points occurred when not all the clinical information was available for all patients. For 1 patient, the deletion score occurred beyond the 99th percentile distribution, likely due to experimental error. The rest of the data were still used in the study. For PCR analysis, a single point occurred beyond the 99% distribution and was removed.
Data Availability
Individual participant data and a data dictionary defining each field in the set will be made available to investigators for work that was not initially proposed as part of the parent protocol on a case-by-case basis via an online request to the ACTG. Completion of an ACTG data use agreement might be required.
Results
Demographics
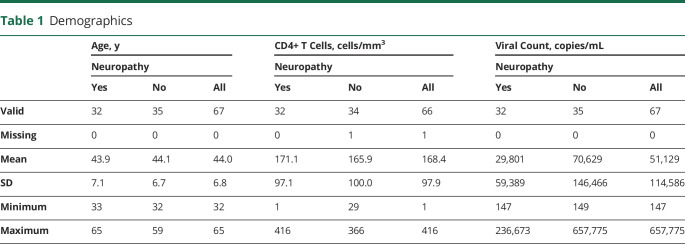
The mean age of the cohort (n = 67) was 44 years (SD 6.8 years, range 32–65 years), and 9 participants were female (table 1). HIV disease severity was assessed with CD4+ T-cell count (available for 66 of 67 patients) and plasma HIV RNA copy number (available for all patients). The mean CD4+ T-cell count was 168 cells/mm3 (SD 97 cells/mm3, range 1–416 cells/mm3) and mean viral load was 51,129 copies/mL (SD 114,586 copies/mL, range 147–657,775 copies/mL). Data for demographics are available from Dryad: datadryad.org/stash/share/jAwLdjvcmou7YvmfPl_uNhwsFKK7HsQKdorQ34Wwffw.
Table 1.
Demographics
Assessment of Neuropathy
Neuropathy Signs
The clinical criteria used for the diagnosis of neuropathy included 2 of 3 examination findings of reduced pinprick and vibration sensation distally in the feet and reduced ankle deep-tendon reflexes. Further assessment of limb strength and sensory and motor symptoms and electrophysiologic assessment of sural and peroneal amplitudes were carried out by a board-certified neurologist to calculate the TNS. On the basis of this assessment, we stratified the cohort into 2 groups: those without and those with neuropathy (table 1).
Neuropathy Measures
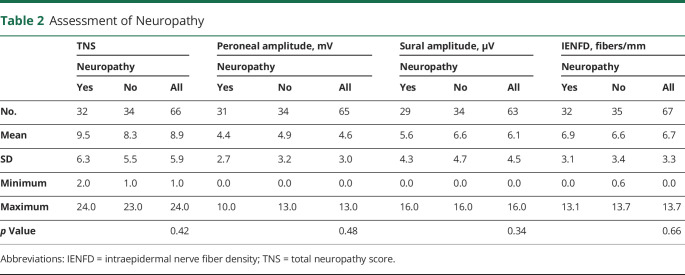
Two objective measures of neuropathy were also evaluated: (1) sural and peroneal conduction amplitudes and (2) IENFD and fiber morphology as determined from skin biopsy samples (table 2). Biopsies in which axonal swelling or other abnormalities were noted were scored as abnormal, despite a normal fiber density, because they can represent early signs of neuropathic changes. The presence of axonal swelling at any age was considered abnormal. For the cohort sural nerve mean action potential average amplitude was 6.1 μV (SD 4.5 μV, range 0–16.1 μV, n = 63), while the peroneal mean compound motor action potential amplitude was 4.6 mV (SD 2.9 mV, range 0–12.5 mV, n = 65). In terms of the other objective measures for the cohort, the TNS was 8.9 (SD 5.9, range 1–24) and the IENFD was 6.7 (SD 3.3, range 0–13.7) (table 2). With the use of the Student independent t test, there were no significant differences in the values between the neuropathy and the no neuropathy group. Analysis of prior exposure to dNRTIs did not reveal any association between demographic data and prior exposure.
Table 2.
Assessment of Neuropathy
Further stratifying the neuropathy group between those with normal or abnormal biopsy, we see that although the TNS remains about the same (8.3 vs 8.4), there is a clear trend toward worse sural (8.1 μV vs 4.8 μV) and peroneal (5.7 mV vs 3.9 mV) and IENFD (8.2 fibers/mm vs 4.3 fibers/mm). Only the latter reached statistical significance (p < 0.001) (table 3). The latter value indicates to us that criteria for abnormal skin biopsies lead to a statistical meaningful separation of the 2 groups.
Table 3.
Neuropathy Subgroup
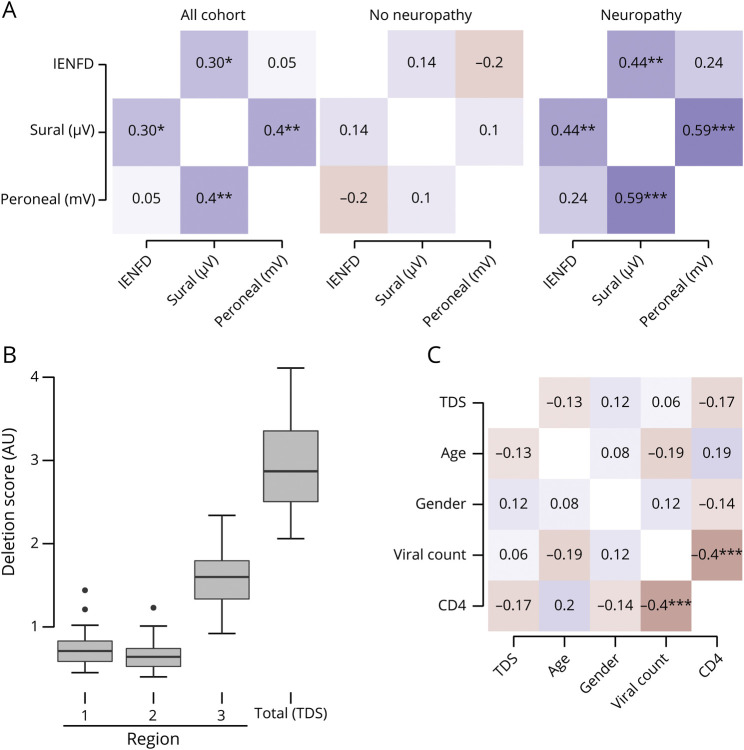
We then performed an internal validation of the neuropathy measures with the IENFD measurements for the whole cohort. We found that sural nerve amplitudes had a positive statistically significant correlation with IENFD (r = 0.304, p = 0.016), while peroneal nerve amplitudes (r = 0.05, p = 0.691) did not (figure 2A). Within the group of patients with neuropathy, the association between IENFD and sural amplitudes is stronger (r = 0.44, p = 0.009). In both groups, the sural and peroneal amplitudes are correlated with each other as well. These results are in agreement with previous studies that have shown the diagnostic utility of IENFD measurements to determine the presence of HIV neuropathy.14,15
Figure 2. Neuropathy Measurements and Demographics.
(A) Heat map showing Pearson correlation scores and statistical significance between sural nerve amplitudes and intraepidermal nerve fiber density (IENFD) for the cohort and the neuropathy and no neuropathy subgroups. (B) Deletion scores for each region were measured, and the total deletion score (TDS) for the sample was calculated. (C) There was no correlation between any of the demographic data and TDS. There was an expected strong negative correlation between CD4 count and viral load. AU = arbitrary units.
Deletion Burden in the mtDNA
Using our assay, we were able to develop an mtDNA deletion burden score that is based on fluorescence for each of the 3 regions. The individual region scores were summed to give the TDS (figure 2B). The fluorescence for each region is normalized to the same small control region and is scored as arbitrary units (AU). We found mean values of 0.718 AU (SD 0.19 AU, range 0.450–1.440 AU) for region 1, 0.652 AU (SD 0.16 AU, range 0.40–1.23 AU) for region 2, and 1.59 AU (SD 0.545 AU, range 0.9–2.3 AU) for region 3, indicating that region 3, which spans the majority of the common mitochondrial deletion, contained the largest number of deletions. The TDS was 2.96 AU (SD 0.545 AU, range 2.1–4.11 AU). The differences between the different regions are statistically significant. When a 1-tailed t test is used, region 1 vs 2 has p = 0.015, while regions 1 or 2 vs region 3 has p < 0.0001. Region 3 appears to be the region that accumulates the most deletion mutations. This region contains a section of the common mitochondrial deletion, but it does not span the whole area.
Total mtDNA Deletion Mutation Burden Does Not Correlate With Age, CD4+ Cell Count, or Viral Load
Given the deletion rates, we then determined whether mitochondrial deletion mutation burden was associated with established risk factors of HIV neuropathy, namely age and advanced HIV disease. Only CD4+ T-cell counts at the time of biopsy were available. There was no correlation between the cohort demographics, including age (r = −0.129, p = 0.300), sex (r = 0.116, p = 0.349), and viral count (r = 0.056, p = 0.651), and the TDS (figure 2C). As expected, we did find a correlation between CD4 count and viral load (r = −0.416, p < 0.001).
Total mtDNA Deletion Mutation Burden Correlates With IENFD and Sural Nerve Amplitudes
We then measured the correlation between TDS and both subjective and objective measures of neuropathy (figure 3). We determined that there was a modest correlation between the TDS and IENFD (r = −0.344, p = 0.04) and sural nerve amplitude (r = −0.359, p = 0.004) but not with the TNS, which incorporates neuropathy symptoms into the score, or the peroneal motor amplitude.
Figure 3. Correlation Between TDS and Objective Measures of Neuropathy.
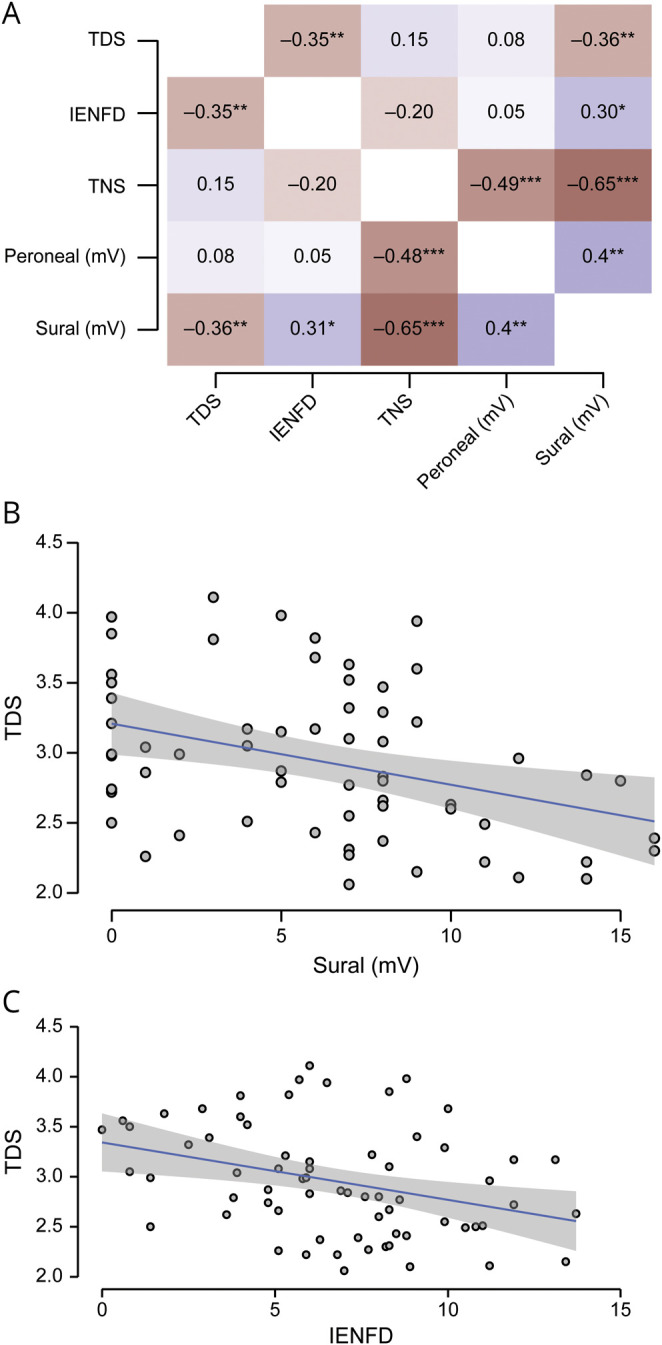
(A) The total neuropathy score (TNS) and peroneal nerve amplitude did not correlate with total deletion score (TDS) for the whole cohort. Intraepidermal nerve fiber density (IENFD) and sural nerve amplitude did. Linear regression analysis of the (B) sural amplitude and (C) IENFD demonstrated that a small fraction of the data were explained by the association. Gray area represents 95% confidence interval.
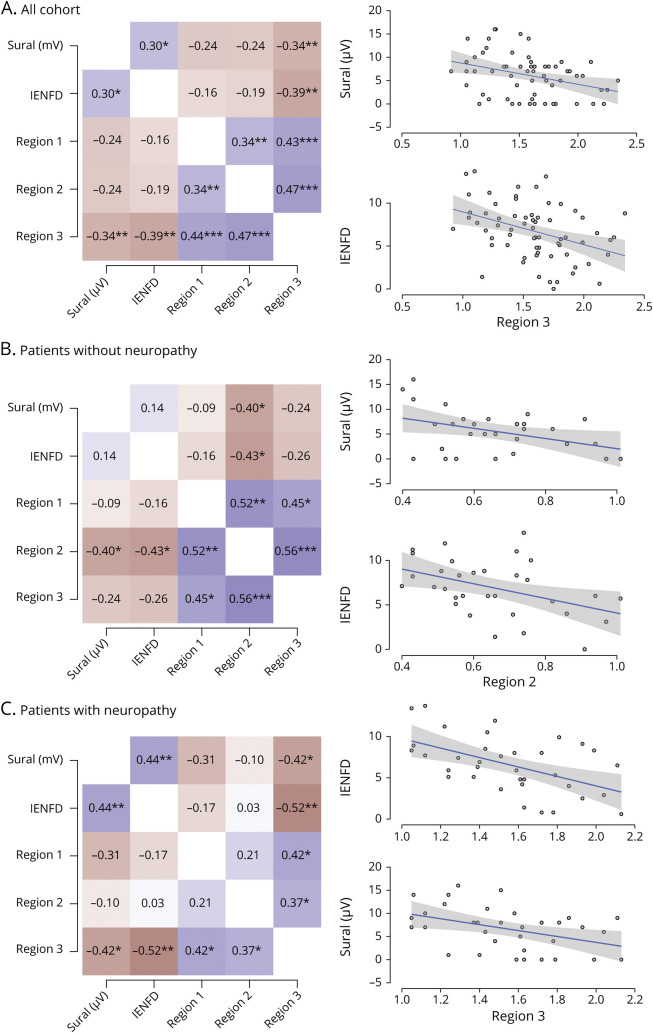
Because we used 3 sets of primers to measure deletion load throughout the mitochondrial genome, we were able to determine which deleted section of the genome correlates best with IENFD and sural amplitudes (figure 4). We observed that region 3, which encompasses nucleotides 10,968 through 16,054 and spans the majority of the common deletion, is responsible for the correlation between both IENFD (r = −0.387, p = 0.001) and sural amplitude (r = −0.341, p = 0.001) and the TDS. When assessed independently, neither region 1 nor 2 was statistically correlated.
Figure 4. Subregion Analysis of Deletions.
(A) For the total cohort, region 3 is responsible for most of the correlation with neuropathy measures. When those (B) without neuropathy and (C) with neuropathy are separated, regions 2 and 3, respectively, are correlated. Gray area represents 95% confidence interval. IENFD = intraepidermal nerve fiber density.
Discussion
In this study, we examined the association between mtDNA deletions and indices of peripheral neuropathy in patients with HIV-SN. Large mtDNA deletions have been associated with aging and are thought to result from inefficiencies in mtDNA replication and DNA repair. The common deletion mutation has been used as a marker of mtDNA aging and has previously been shown to correlate with neuropathy in human postmortem sural nerve samples.16 It has also been associated with other neurologic disorders such as Kearns-Sayre syndrome.17
HIV replication has been found to be limited in the peripheral nerve and is restricted mainly to the proximal segment, within dorsal root ganglia macrophages and neurons.18-20 Degeneration is thought to occur through indirect neurotoxicity from exposure to Tat and a gp120-dependent process, as well as via production of proinflammatory cytokines.19,21-25 This may constitute an independent mechanism that is unrelated to one driving distal small fiber degeneration and may indeed contribute less significantly to neuropathy symptoms and signs. For this reason, only CD4+ nadir and viral load >5,000 copies, representing advanced infection, have been shown to correlate with neuropathy, as opposed to CD4+ T-cell count and viral load in antiretroviral therapy–exposed PWH.26,27 A limitation of our study is that only CD4+ T-cell count at the time of enrollment was available.
We observed that in those patients assessed not to have neuropathy, region 2 correlated with sural amplitude and IENFD with r = − 0.402 (p = 0.031) and r = −0.433 (p = 0.013), respectively. In those noted to have neuropathy, region 3 again correlated with sural amplitude and IENFD with r = − 0.422 (p = 0.013) and r = −0.524 (p = 0.001), respectively. The common mtDNA4977 spans regions 2 and 3. This information suggests that in the total cohort and in those with neuropathy, region 3 is responsible for most the correlation. However, in those without neuropathy, there is a better correlation with mutations in region 2. Our assay does not allow measurement of total mtDNA quantity. However, the difference in deletion site preference may reflect differences in the mechanism responsible for them.
Our data demonstrate that both IENFD and sural nerve amplitude statistically correlate with mitochondrial mutation burden in PWH with and without neuropathy. The preferred regions of deletions are different. Previous studies have shown IENFD reduction as a reliable marker of symptoms, correlating with pain in patients with HIV-SN and as an early marker to predict neuropathy with abnormalities detected in asymptomatic HIV-seropositive individuals before the development of clinical neuropathy.28,29 In particular, we saw an increased incidence of deletions in region 3 of the mtDNA, which includes the 3′ end of the well-known common deletion. The association of deletions within region 2 in those without neuropathy suggests that perhaps mutations in that patient subset accumulate via a different mechanism that is not related to the development of neuropathy. Only region 3 mutations were statistically correlated with IENFD in those patients with HIV-SN. This suggests that a different hot spot for deletions is seen in patients in HIV-SN compared to aging individuals. It would be interesting to see whether this observation also extends to patients without HIV. An additional compounding factor is that all patients had some prior exposure to dNRTIs, and the role of this exposure cannot be ruled out as contributing to deletions. Another limitation is that specific information about other causes of neuropathy (e.g., other medications and alcohol use) was not part of the study.
Although this study demonstrates an association between the extent of mtDNA deletions in the skin and sensory neuropathy as assessed by both EINFD and sural amplitude among PWH, it does not establish a causal link, especially because the deletion burden is assessed from whole skin and not individual fibers. An additional limitation of our work is that normal biopsies were not available to serve as controls for deletion distribution. We speculate that the indirect toxic milieu in the dorsal root ganglia of PWH might exacerbate the proliferation of mitochondrial deletions in already vulnerable sensory axons. Further studies analyzing the effect of HIV toxic products on mtDNA replication in peripheral nerve neurons are needed to address this issue.
Acknowledgment
R.H.R. is supported in part, through philanthropic support from Dr. Peter Buck and additional anonymous contributors. A.R.K., under grant R01 MH124530-01, has NIH support from the National Institute of Mental Health. M.F.O. is supported through R01AG061066 and the CNPq-Brazil (245954/2012). M.M.D. is supported by the Fogarty International Center at the NIH under grant D43TW009343. J.C.M. is supported through grants JCM 5P30MH075673-13, 5P01MH105280-05, and 5R01NS049465-04. A.H. received support from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Glossary
- ACTG
AIDS Clinical Trials Group
- AU
arbitrary units
- dNRTI
nucleoside reverse transcriptase inhibitor
- IENFD
intraepidermal nerve fiber density
- mtDNA
mitochondrial DNA
- PWH
people with HIV
- TDS
total deletion score
- TNS
total neuropathy score
Appendix. Authors
Study Funding
The authors report no targeted funding.
Disclosure
R.H. Roda has served as a consultant to Argenx. D. Bargiela, W. Chen, K. Perry, and R.J. Ellis have no conflicts of interest to report. D.B. Clifford has served as a consultant to Biogen, Takeda, Millennium, Genzyme, Amgen, Roche/Genentech, Glaxo Smith Kline, Serono, Inhibikase, Dr. Reddy's Lab, Bristol Myers Squibb, Atara, Mitsubishi Tanabe, Excision BioTherapeutics, Up to Date, and Wolters Kluwer; has served on Data Safety Monitoring Boards for Genentech/Roche, Wave, EMD Serono, Shire/Takeda, Pfizer, and Sanofi; and has provided legal consulting for Cook County, State Farm, Wilke & Wilke PC, Shevlin Smith, and Sal Indomenico PC. He has NIH support from National Institute of Mental Health, National Institute of Neurological Disorders and Stroke, National Institute of Allergy and Infectious Diseases, National Institute on Aging, and National Center for Advancing Translational Sciences. A. Bharti, A.R. Kallianpur, M.F. Oliveira, M. Diaz , and L.H. Rubin have no conflicts of interest to report. C. Gavegnano serves as unpaid consultant for Eli Lilly. J.C. McArthur has no conflict of interest to report. A. Hoke served on the Safety Advisory Board of Disarm Therapeutics and AxoProtego Therapeutics and has stock options. He has consulted with Pfizer, Galapagos, and TEVA. He has research funding from the NIH, Department of Defense, and Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. M. Polydefkis has served as a consultant to Biogen, Alnylam, Akcea, Vertex, and Pfizer. Go to Neurology.org/N for full disclosures.
References
- 1.Keswani SC, Jack C, Zhou C, Höke A. Establishment of a rodent model of HIV-associated sensory neuropathy. J Neurosci. 2006;26(40):10299-10304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keswani SC, Polley M, Pardo CA, Griffin JW, McArthur JC, Hoke A. Schwann cell chemokine receptors mediate HIV-1 gp120 toxicity to sensory neurons. Ann Neurol. 2003;54(3):287-296. [DOI] [PubMed] [Google Scholar]
- 3.Keswani SC, Hoke A, Schifitto G, McDermott MP, Sacktor N. Incidence of and risk factors for HIV-associated distal sensory polyneuropathy. Neurology. 2003;61(2):279-280. [DOI] [PubMed] [Google Scholar]
- 4.Hahn K, Triolo A, Hauer P, McArthur JC, Polydefkis M. Impaired reinnervation in HIV infection following experimental denervation. Neurology. 2007;68(16):1251-1256. [DOI] [PubMed] [Google Scholar]
- 5.Mandal A, Drerup CM. Axonal transport and mitochondrial function in neurons. Front Cell Neurosci. 2019:13:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M, Foli Y, Liu Z, et al. High frequency of mitochondrial DNA mutations in HIV-infected treatment-experienced individuals. HIV Med. 2017;18(1):45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maagaard A, Holberg-Petersen M, Kollberg G, Oldfors A, Sandvik L, Bruun J. Mitochondrial (mt)DNA changes in tissue may not be reflected by depletion of mtDNA in peripheral blood mononuclear cells in HIV-infected patients. Antivir Ther. 2006;11(5):601-608. [PubMed] [Google Scholar]
- 8.Lehmann HC, Chen W, Borzan J, Mankowski J, Höke A. Mitochondrial dysfunction in distal axons contribute to HIV sensory neuropathy. Ann Neurol. 2011;69(1):100-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherry CL, Nolan D, James IR, et al. Tissue-specific associations between mitochondrial DNA levels and current treatment status in HIV-infected individuals. J Acquir Immune Defic Syndr. 2006;42(4):435-440. [DOI] [PubMed] [Google Scholar]
- 10.Gaff J, Pillay P, Cherry C, Laws SM, Price P, Kamerman P. The role of CAMKK2 polymorphisms in HIV-associated sensory neuropathy in South Africans. J Neurol Sci. 2020;416:116987. [DOI] [PubMed] [Google Scholar]
- 11.Gaff J, Octaviana F, Pillay P, et al. TNF-block genotypes influence susceptibility to HIV-associated sensory neuropathy in Indonesians and South Africans. Int J Mol Sci. 2020:21(2):380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirsch MS. Antiviral drug development for the treatment of human immunodeficiency virus infections: an overview. Am J Med. 1988;85(2A):182-185. [PubMed] [Google Scholar]
- 13.Cornblath DR, Chaudhry V, Carter K, et al. Total neuropathy score: validation and reliability study. Neurology. 1999;53(8):1660-1664. [DOI] [PubMed] [Google Scholar]
- 14.Polydefkis M, Yiannoutsos CT, Cohen BA, et al. Reduced intraepidermal nerve fiber density in HIV-associated sensory neuropathy. Neurology. 2002;58(1):115-119. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann DN, McDermott MP, Sowden JE, et al. Is skin biopsy a predictor of transition to symptomatic HIV neuropathy? A longitudinal study. Neurology. 2006;66(6):857-861. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann HC, Chen W, Borzan J, Mankowski JL, Höke A. Mitochondrial dysfunction in distal axons contributes to human immunodeficiency virus sensory neuropathy. Ann Neurol. 2011;69(1):100-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein A, Falk MJ. Mitochondrial DNA deletion syndromes. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews® [online]. University of Washington, Seattle; 1993. [PubMed] [Google Scholar]
- 18.Nagano I, Shapshak P, Yoshioka M, Xin K, Nakamura S, Bradley WG. Increased NADPH-diaphorase reactivity and cytokine expression in dorsal root ganglia in acquired immunodeficiency syndrome. J Neurol Sci. 1996;136(1-2):117-128. [DOI] [PubMed] [Google Scholar]
- 19.Brannagan TH, Nuovo GJ, Hays AP, Latov N. Human immunodeficiency virus infection of dorsal root ganglion neurons detected by polymerase chain reaction in situ hybridization. Ann Neurol. 1997;42(3):368-372. [DOI] [PubMed] [Google Scholar]
- 20.Yoshioka M, Shapshak P, Srivastava AK, et al. Expression of HIV-1 and interleukin-6 in lumbosacral dorsal root ganglia of patients with AIDS. Neurology. 1994;44(6):1120-1130. [DOI] [PubMed] [Google Scholar]
- 21.Tyor WR, Wesselingh SL, Griffin JW, McArthur JC, Griffin DE. Unifying hypothesis for the pathogenesis of HIV-associated dementia complex, vacuolar myelopathy, and sensory neuropathy. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;9(4):379-388. [PubMed] [Google Scholar]
- 22.Milligan ED, Mehmert KK, Hinde JL, et al. Thermal hyperalgesia and mechanical allodynia produced by intrathecal administration of the human immunodeficiency virus-1 (HIV-1) envelope glycoprotein, gp120. Brain Res. 2000;861(1):105-116. [DOI] [PubMed] [Google Scholar]
- 23.Herzberg U, Sagen J. Peripheral nerve exposure to HIV viral envelope protein gp120 induces neuropathic pain and spinal gliosis. J Neuroimmunol. 2001;116(1):29-39. [DOI] [PubMed] [Google Scholar]
- 24.Oh SB, Tran PB, Gillard SE, Hurley RW, Hammond DL, Miller RJ. Chemokines and glycoprotein120 produce pain hypersensitivity by directly exciting primary nociceptive neurons. J Neurosci. 2001;21(14):5027-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Li G, Steiner J, Nath A. Role of Tat protein in HIV neuropathogenesis. Neurotox Res. 2009;16(3):205-220. [DOI] [PubMed] [Google Scholar]
- 26.Tagliati M, Grinnell J, Godbold J, Simpson DM. Peripheral nerve function in HIV infection: clinical, electrophysiologic, and laboratory findings. Arch Neurol. 1999;56(1):84-89. [DOI] [PubMed] [Google Scholar]
- 27.Childs EA, Lyles RH, Selnes OA, et al. Plasma viral load and CD4 lymphocytes predict HIV-associated dementia and sensory neuropathy. Neurology. 1999;52(3):607-613. [DOI] [PubMed] [Google Scholar]
- 28.Winer JB, Bang B, Clarke JR, et al. A study of neuropathy in HIV infection. Q J Med. 1992;83(302):473-488. [PubMed] [Google Scholar]
- 29.McCarthy BG, Hsieh ST, Stocks A, et al. Cutaneous innervation in sensory neuropathies: evaluation by skin biopsy. Neurology. 1995;45(10):1848-1855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Individual participant data and a data dictionary defining each field in the set will be made available to investigators for work that was not initially proposed as part of the parent protocol on a case-by-case basis via an online request to the ACTG. Completion of an ACTG data use agreement might be required.