Abstract
Multiple myeloma (MM), a terminally differentiated B cell malignancy, remains difficult to cure. Understanding the molecular mechanisms underlying the progression of MM may identify therapeutic targets and lead to a fundamental shift in treatment of the disease. Deubiquitination, like ubiquitination, is a highly regulated process, implicated in almost every cellular process. Multiple deubiquitinating enzymes (DUBs) have been identified, but their regulation is poorly defined. Here, we determined that TRIP13 increases cellular deubiquitination. Overexpression of TRIP13 in mice and cultured cells resulted in excess cellular deubiquitination by enhancing the association of the DUB USP7 with its substrates. We show that TRIP13 is an oncogenic protein because it accelerates B cell tumor development in transgenic mice. TRIP13-induced resistance to proteasome inhibition can be overcome by a USP7 inhibitor in vitro and in vivo. These findings suggest that TRIP13 expression plays a critical role in B cell lymphoma and MM by regulating deubiquitination of critical oncogenic (NEK2) and tumor suppressor (PTEN, p53) proteins. High TRIP13 identifies a high-risk patient group amenable to adjuvant anti-USP7 therapy.
Keywords: Hematology
Keywords: Drug therapy, Oncogenes, Ubiquitin-proteosome system
Introduction
Multiple myeloma (MM), a malignancy of terminally differentiated antibody-secreting plasma cells, can be separated into high and low risk based on the differential expression of 70 genes on gene expression profiling (GEP) (1). High-risk GEP, present in approximately 20% of newly diagnosed disease, is characterized by the increased expression of 52 genes, the majority of which map to chromosome 1q, combined with reduced expression of 18 genes, the majority of which map to chromosome 1p. While the combination of autologous stem cell transplantation (ASCT) with novel agents, such as proteasome inhibitors, immunomodulatory agents, and monoclonal antibodies (2, 3), has significantly improved progression-free and overall survival (OS) in low-risk disease, the dismal outcome of high-risk disease remains a significant clinical problem (4). Also, the acquisition of a high-risk signature at disease relapse is linked to shorter post-relapse survival, strongly suggesting that high-risk cells represent only a small fraction of tumor bulk at diagnosis in low-risk disease but survive chemotherapeutic interventions (1). Consistent with this concept, the copy number of chromosome 1q and the percentage of cells with 1q gains both increase at relapse in a significant fraction of cases of MM (5). These data suggest that chromosome 1q copy number gains are a likely driver of aggressive tumor behavior. The GEP70 signature not only represents a highly robust collection of risk biomarkers; its composition also reveals key biological features of disease progression, and potential targets for therapeutic intervention.
The gene thyroid hormone receptor–interacting protein 13 (TRIP13) represents one of the genes within the 70-gene signature overexpressed in high-risk MM (1) and is also one of 10 genes constituting a chromosome instability (CIN) signature in MM (6). In addition to MM (7, 8), high TRIP13 expression is associated with aggressive disease in chronic lymphocytic leukemia (9), prostate cancer (10, 11), breast cancer (12, 13), colorectal cancer (14), hepatocellular carcinoma (15), lung cancer (16, 17), and neck and head cancer (18).
TRIP13 is a member of the AAA (ATPases associated with diverse cellular activities) ATPase family of proteins that alter the conformation of client macromolecules and thereby affect cellular signaling. This enzyme is involved in various cellular biological processes and is classically considered a regulator of chromosomal events including chromosome synapsis, checkpoint signaling, and DNA break formation and recombination (19). Premature exit from the spindle assembly checkpoint blocks DNA repair and leads to mis-segregation of chromosomes, causing both structural and numerical aneuploidy events. Recent studies have shown that high levels of TRIP13 promote both the nonhomologous end joining and the homology-directed repair of double-strand breaks (18, 20), potentially leading to CIN, cancer cell survival, metastasis, and enhanced drug resistance.
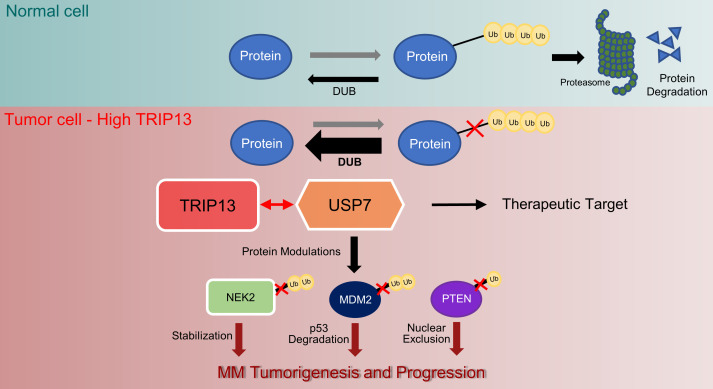
The ubiquitin-proteasome system (UPS) enables a dynamic homeostasis of proteins through a highly regulated process of protein ubiquitination and proteasomal degradation. The UPS is deregulated in MM and represents a potential target of intervention for MM therapy. By comparing gene expression profiles of MM cells prior to and after administration of a test dose of the proteasome inhibitor bortezomib (BTZ), we were able to show that upregulation of the proteasome within 48 hours of drug administration is associated with poor outcome in MM patients treated with immunomodulatory drugs (IMiDs), proteasome inhibitors, and ASCT (21). The centrosomal kinase never in mitosis A–related (NIMA-related) kinase 2A (NEK2), mapping to chromosome 1q, is another copy number–sensitive gene in MM and is expressed at elevated levels in high-risk MM and other cancers (6, 22, 23). NEK2 increases proteasome activity in MM cells (24), and binds to and is stabilized by the deubiquitinase ubiquitin-specific protase-7 (USP7; also known as HAUSP), leading to the activation of the canonical NF-κB signaling pathway, resulting in increased drug resistance and bone destruction (25, 26). TRIP13 expression causes spindle assembly checkpoint errors through enhanced ubiquitin-mediated, proteasomal degradation of MAD2 (7).
Here, we provide further evidence that TRIP13 enhances cellular protein deubiquitination by binding the deubiquitinase USP7. Moreover, we show that TRIP13 plays a critical role in stabilizing NEK2 via a TRIP13/USP7-dependent mechanism. Using a small-molecule inhibitor of USP7, we have targeted the TRIP13/USP7/NEK2 complex and shown that this intervention abrogates B cell tumorigenesis and overcomes drug resistance.
Results
Generation of Trip13 tissue-specific transgenic murine model.
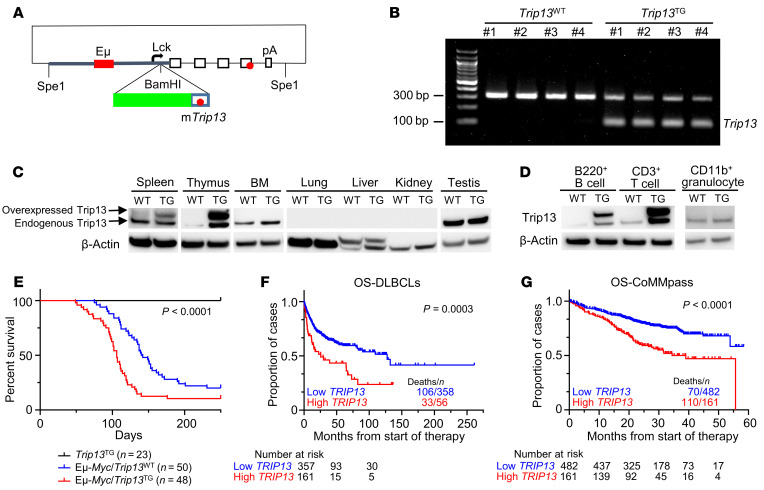
To determine whether Trip13 can drive lymphoid-derived tumorigenesis, we inserted the complete coding region of mouse Trip13 into the p1026x-HA3P vector (27). The p1026x vector includes a 3.2 kb fragment of the proximal promoter of the mouse Lck gene, a 0.92 kb fragment of the Eμ enhancer, and a 2.1 kb mutated (non-translatable) version of the human growth hormone gene (hGH) driving expression in T and B cells (Figure 1A). Three lines were selected and backcrossed to a C57BL/6 background for at least 10 generations (Figure 1B). Western blotting was used to confirm the tissue-specific expression of Trip13 protein in transgenic (Trip13TG) mice relative to wild-type (Trip13WT) littermate mice. Figure 1C shows that Trip13 protein was endogenously expressed in mouse spleen, thymus, and bone marrow, and was significantly increased in Trip13TG mice. As expected, testis as a positive control expressed high Trip13 protein in both Trip13TG and Trip13WT mice, while lung, liver, and kidney had no detectable expression. To further determine the cell specificity of Trip13 expression, we enriched B cells from spleen and T cells from thymus of Trip13TG mice using magnetic beads conjugated to B (anti-B220) and T (anti-CD3) lymphoid lineage–specific antibodies. Western blots of protein isolated from enriched cells showed that Trip13 protein was dramatically increased in Trip13TG mice compared with Trip13WT mice in B220+ B cells and CD3+ T cells but not in CD11b+ granulocytes (Figure 1D). There was no significant difference in the survival of Trip13TG mice relative to Trip13WT mice, nor in lymphoid tissue sizes (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI146893DS1). To further investigate whether lymphoid lineage subsets changed with Trip13 transgene, we performed flow cytometry on mouse spleen, lymph nodes, thymus, and bone marrow, and found that percentages of both B- and T-lineage subsets were similar in Trip13TG mice compared with Trip13WT littermate controls (data not shown). These data strongly suggest that Trip13 overexpression by itself does not initiate tumorigenesis, as is the case with most oncogenes.
Figure 1. TRIP13 accelerates tumorigenesis in Eμ-Myc mouse model and is linked to poor prognosis in patients with B cell malignancies.
(A) Schematic diagram of the modified p1026x vector. Briefly, a 10.4 kb Lck-(Eμ)-mTrip13-hGH fragment was inserted into the p1026x vector by Spe1. The modified vector contains a 3.2 kb fragment of the mouse Lck proximal promoter to +37 with respect to the transcription start site (thick gray line), a 0.92 kb fragment of the immunoglobulin heavy chain intronic enhancer (Eμ) (solid red box) inserted into the Lck fragment, a 2.1 kb mutated (non-translatable) version of the human growth hormone gene (hGH) (open boxes), and the Trip13 cDNA (green box) inserted into the BamHI site of the Lck promoter fragment. The polyadenylation site of the hGH gene is indicated. Stop codons in the Trip13 cDNA and hGH genes are indicated by red circles. (B) Genotyping of Trip13-transgenic (Trip13TG) mice performed on ear tissue DNA by PCR. The 324 bp band for Trip13WT and the 100 bp band for Trip13TG are indicated. (C) Western blot analysis of Trip13 protein in different tissues of Trip13TG and Trip13WT mice. (D) Western blot analysis of Trip13 protein in B220+ B cells, CD3+ T cells, and CD11b+ granulocytes of Trip13TG and Trip13WT mice. (E) Kaplan-Meier analysis of Trip13TG (n = 23), Eμ-Myc/Trip13WT (n = 50), and Eμ-Myc/Trip13TG (n = 48) mice (P < 0.0001 by log-rank test). The number of evaluated mice is indicated in parentheses. (F) Kaplan-Meier analysis of DLBCL patients with high versus low TRIP13 (best cutoff, P = 0.0003 by log-rank test). (G) Kaplan-Meier analysis of MM patients with high (quartile 4) versus low (quartile 1–3) TRIP13 (P < 0.0001 by log-rank test).
Eμ-Myc/Trip13TG mice develop an aggressive B cell malignancy.
The MYC oncogene is widely dysregulated in a variety of human cancers, including MM, lymphomas, and solid tumors (28–30), and the Eμ-Myc mouse is a well-established preclinical model for B cell malignancies including diffuse high-grade lymphomas, diffuse large B cell lymphomas (DLBCLs), and plasmacytomas (31, 32). To determine whether Trip13 does cooperate with Myc in tumorigenesis, we crossed the Trip13TG with the Eμ-Myc mice. Mouse survival was compared between Eμ-Myc mice and Eμ-Myc/Trip13TG mice. The Eμ-Myc/Trip13TG mice developed tumors at a younger age than the Eμ-Myc mice, resulting in a significantly shorter survival (median survival: Eμ-Myc/Trip13TG mice 106 days vs. Eμ-Myc/Trip13WT mice 141 days; P = 0.0002; Figure 1E). With lymphoma progression, mice developed pronounced cervical, inguinal, axillary, and intestinal lymphadenopathy, splenomegaly, and an enlarged thymus (Supplemental Figure 2, A and B), respiratory distress, hunched posture, limited mobility, and paralysis. Flow cytometry (Supplemental Figure 2, C and D) further confirmed that the spontaneous tumors were lymphomas of B cell origin. Thus, Trip13 cooperates with Myc to accelerate the development of B cell malignancies.
Elevated TRIP13 expression is linked to poor prognosis.
We examined whether elevated expression of TRIP13 is associated with inferior clinical outcome in DLBCLs and MM. Kaplan-Meier analysis based on TRIP13 expression levels in DLBCL demonstrated a significantly inferior OS associated with high TRIP13 (using best cutoff) (Figure 1F; P = 0.0003). Interestingly, this association was specific to activated B cell DLBCLs (ABC DLBCLs) (Supplemental Figure 2E; P = 0.01573), but was not observed in germinal center or unclassified B cell DLBCLs. ABC DLBCLs have been associated with a significantly worse OS compared with germinal center B cell DLBCLs (33, 34). Our data indicate that TRIP13 expression is a significant prognostic factor in ABC DLBCL. In an independent MM data set, the CoMMpass data set, Kaplan-Meier analysis of MM patients also validated a significantly inferior OS associated with high TRIP13 (quartile 4) (Figure 1G; P < 0.0001), confirming our earlier findings in MM patients treated with tandem transplants (7).
TRIP13 enhances cellular protein deubiquitination.
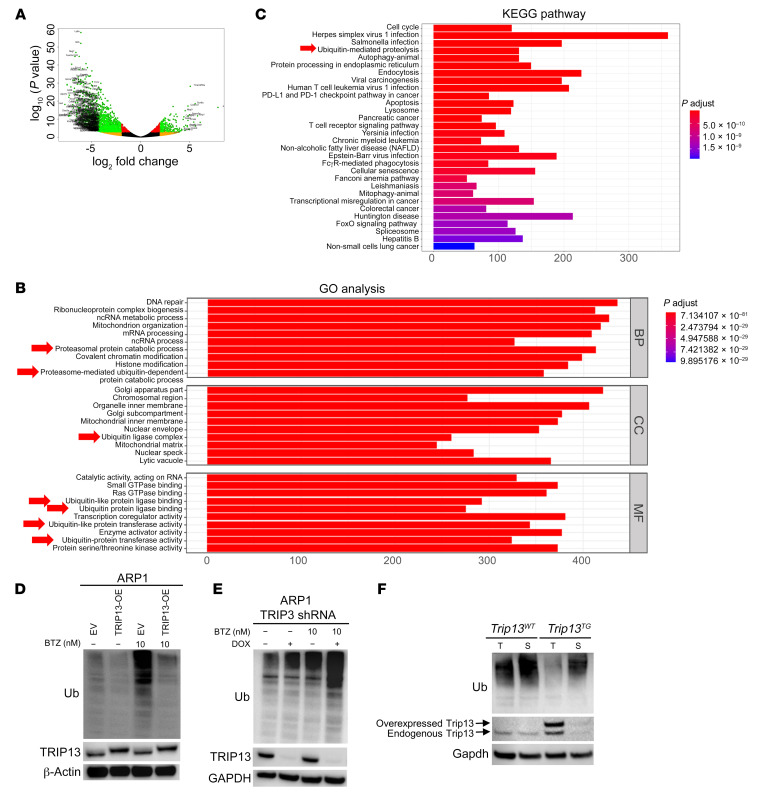
To further characterize the role of Trip13 in Eμ-Myc–induced B cell malignancies, we collected premalignant B cells (B220+) from both Eμ-Myc/Trip13TG and Eμ-Myc/Trip13WT mice at 6 weeks of age and performed RNA sequencing analyses. As shown in Figure 2A, more than 1900 genes were differentially expressed in B cells from these 2 strains (P < 0.001). We then analyzed signaling pathways using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The GO analysis revealed that proteasome-mediated and ubiquitination-dependent GO genes were significantly enriched for biological processes (BP), cellular components (CC), and molecular functions (MF) in Eμ-Myc/Trip13TG mice (Figure 2B; P < 10–7). In the MF group, 4 of the top 10 upregulated GO terms were associated with ubiquitin-related pathways. KEGG analysis confirmed that the ubiquitin-mediated proteolysis pathway was significantly enriched (Figure 2C). To determine the relationship between high TRIP13 and sensitivity to chemotherapeutic drugs in MM, the MM cell lines ARP1 and H929, with or without TRIP13 overexpression, were treated with the alkylating agent melphalan, the DNA-damaging topoisomerase inhibitor etoposide, or the proteasome inhibitor BTZ (Supplemental Figure 3). TRIP13 overexpression only prevented BTZ-induced cell death but did not alter sensitivity to melphalan or etoposide. Our clinical experience, however, indicates that a combination of BTZ, IMiDs, and ASCTs does not markedly improve patient outcome in MM with a high-risk GEP70 score or high TRIP13. These findings suggest that TRIP13 may play a significant role in regulating drug response through its ability to modulate the UPS within tumor cells.
Figure 2. TRIP13 enhances cellular protein deubiquitination.
(A–C) Representational and pathway analysis of approximately 1900 differentially expressed genes in B cells from Eμ-Myc/Trip13TG versus Eμ-Myc/Trip13WT mice (P < 0.001) displayed in volcano plot (A) and bar graphs of GO terms (B) and KEGG pathways (C). Ubiquitin-related (Ub-related) and proteasome-related terms or pathways are indicated by red arrows. (D) Western blot analysis of Ub, TRIP13, and β-actin in EV and TRIP13-OE ARP1 cells treated overnight with or without 10 nM BTZ. (E) Western blot analysis of Ub, TRIP13, and GAPDH in ARP1 cells transfected with a doxycycline-inducible (DOX-inducible) TRIP13 shRNA, treated with or without DOX for 72 hours, followed by treatment with or without 10 nM BTZ. (F) Western blot analysis of Ub, Trip13, and Gapdh in thymus (T) and spleen (S) from Trip13TG and Trip13WT mice.
To address this possibility further, we examined the effect of TRIP13 expression on cellular ubiquitination. In TRIP13-overexpressed ARP1 and H929 MM cell lines, the total cellular ubiquitinated protein levels were significantly decreased compared with those in cells transfected with empty vector (EV) (Figure 2D, lane 1 vs. lane 2, and Supplemental Figure 4). Inhibition of protein degradation with the proteasome inhibitor BTZ highlighted the decreased cellular ubiquitination seen in TRIP13-overexpressing (TRIP13-OE) cells compared with EV cells (Figure 2D, lane 3 vs. lane 4). We further examined the relationship between TRIP13 and cellular ubiquitin levels by using ARP1 cells stably transfected with a doxycycline-inducible vector expressing TRIP13 shRNA. Knockdown of TRIP13 resulted in elevated ubiquitinated proteins, and this was even more pronounced upon treatment with BTZ (Figure 2E). To confirm this observation in vivo, we used the newly developed Trip13TG mice. Immunoblotting for ubiquitinated proteins in Trip13TG mice showed a decreased signal in both thymus (Figure 2F, lanes 1 and 3) and spleen tissue (Figure 2F, lanes 2 and 4) relative to Trip13WT littermate controls.
TRIP13 enhances cellular deubiquitination by binding USP7.
To evaluate how TRIP13 enhances cellular deubiquitination, we took advantage of the fact that overexpression of TRIP13 induces a BTZ-resistant phenotype in MM and other cancer cells. Using ARP1 and H929 MM cell lines, we tested a series of widely used ubiquitination and deubiquitination inhibitors, such as PYR-41 (E1 inhibitor), PR-619 (nonselective USP inhibitor), spautin-1 (USP10 inhibitor), or P5091 (USP7 inhibitor). Cell viability analysis showed that only the USP7 inhibitor P5091 reduced TRIP13-mediated drug resistance (Supplemental Figure 5).
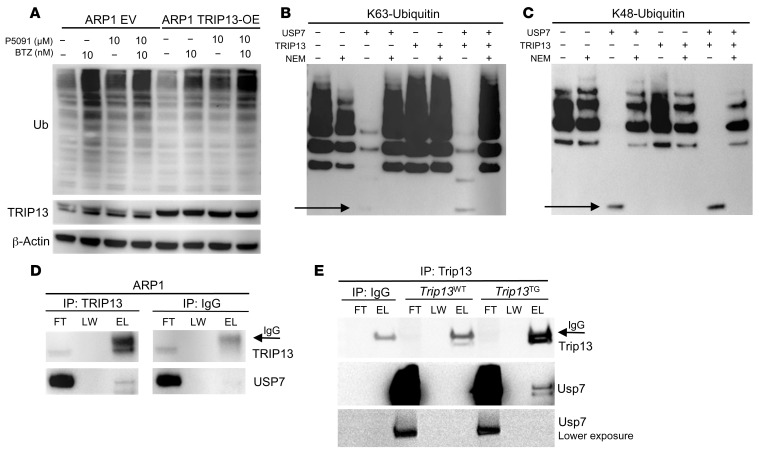
We analyzed the ubiquitin levels in ARP1 EV and ARP1 TRIP13-OE cells treated with the USP7 inhibitor P5091 overnight, with or without BTZ. TRIP13-OE cells showed lower ubiquitination when compared with EV cells (Figure 3A, lane 1 vs. lane 5 and lane 2 vs. lane 6). However, this decrease was reversed by the P5091 treatment (Figure 3A, lane 3 vs. lane 7 and lane 4 vs. lane 8). Since there is a high correlation of TRIP13, NEK2, and USP7 (25), USP7 might interact with TRIP13 to mediate cellular deubiquitination.
Figure 3. TRIP13 enhances cellular deubiquitination by binding USP7.
(A) Western blot analysis of Ub, TRIP13, and β-actin in EV and TRIP13-OE ARP1 cells treated overnight with or without 10 nM BTZ or 10 μM P5091 alone or in combination. (B and C) Western blot analysis of Ub following USP7 deubiquitination of recombinant K63-Ub or K48-Ub in the presence or absence of purified TRIP13 and N-ethylmaleimide (NEM). (D) TRIP13 immunoprecipitation (IP) in ARP1 cells. Western blots of flow-through (FT), last wash (LW), and elution (E) of the IP were probed with TRIP13 and USP7 antibodies. Arrow indicates nonspecific IgG band. (E) Trip13 IP from thymus of Trip13TG and Trip13WT mice. Western blots were probed for Trip13 and Usp7 antibodies.
To evaluate whether TRIP13 acts as a deubiquitinating enzyme (DUB) or regulates USP7 activity, we performed an in vitro deubiquitination assay using 2 different ubiquitin substrates: K63 and K48. Western blot analysis showed that purified TRIP13 did not change the ubiquitination of K63 (Figure 3B, lane 5 vs. lane 1) or K48 (Figure 3C, lane 5 vs. lane 1), suggesting that TRIP13 does not function as a DUB. Commercially available recombinant USP7 proteins were able to degrade ubiquitin (Figure 3, B and C, lane 3 vs. lane 1), and this degradation was inhibited when the deubiquitinase inhibitor N-ethylmaleimide was added to the in vitro deubiquitination assays (Figure 3, B and C, lane 4 vs. lane 3). As shown in Figure 3, B and C (lane 3 vs. lane 1), the DUB USP7 released the ubiquitin monomeric form (indicated by arrows), while polyubiquitin chains were degraded in vitro. We observed that the USP7-mediated ubiquitin degradation was further enhanced by the addition of TRIP13 as evidenced by a stronger signal of ubiquitin monomers released (Figure 3, B and C, lane 5 vs. lane 1). These data indicate that TRIP13 enhances USP7-mediated ubiquitin degradation.
To determine whether TRIP13 binds to USP7, coimmunoprecipitation (co-IP) of endogenous TRIP13 in ARP1 cells was performed using TRIP13 antibodies. USP7 was found to bind to endogenous TRIP13 (Figure 3D, TRIP13 IP). This result was also confirmed by TRIP13-FLAG overexpression and IP in H1299 cells using anti-FLAG antibodies (Supplemental Figure 6). To further confirm that Trip13 binds to Usp7 in vivo, a Trip13 IP was performed on thymus protein lysates isolated from Trip13TG mice compared with Trip13WT littermate controls (Figure 3E). Western blots detected that Usp7 was bound to Trip13 in Trip13TG mice. This association was not detected in Trip13WT littermate controls, most likely because endogenous Trip13 levels were too low to detect USP7 binding by Western blotting.
TRIP13 plays a critical role in stabilizing the USP7 target NEK2.
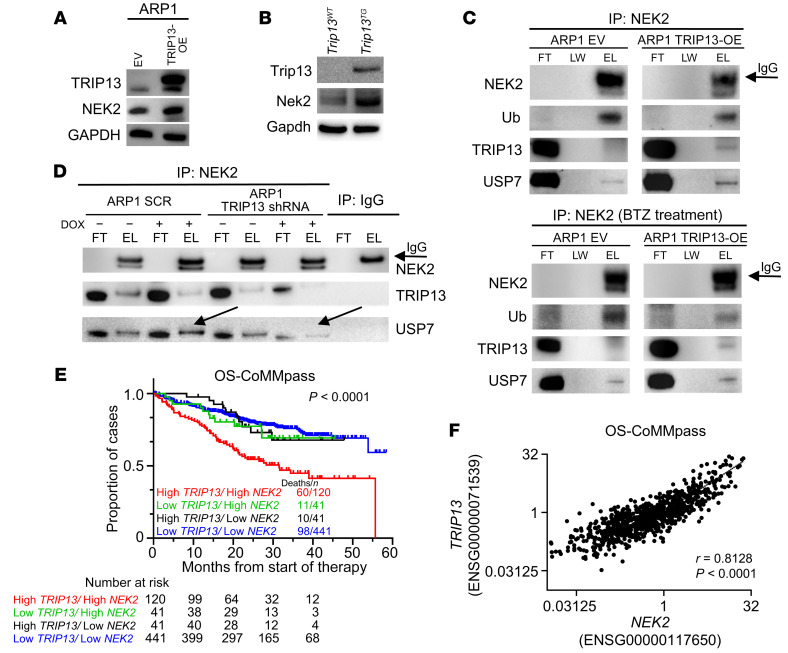
NEK2 protein binds to USP7, and USP7 stabilizes NEK2, resulting in increased drug resistance to proteasome inhibition (25). To explore whether TRIP13 and NEK2 interact with each other, we performed Western blot analysis for NEK2 in cell lysates from ARP1 EV and TRIP13-OE MM cells. Our results showed that TRIP13 overexpression led to an increase in total NEK2 expression (Figure 4A). Similar results were observed in 3T3 cells (Supplemental Figure 7A) and in cell lysates from Trip13TG mice (Figure 4B). Transfection of cells with a TRIP13 shRNA resulted in decreased NEK2 protein (Supplemental Figure 7, B and C). To address whether the observed increase in NEK2 is mediated by USP7 deubiquitination, ubiquitinated NEK2 levels were assessed. Western blot analysis revealed that the NEK2 ubiquitination signal was reduced when TRIP13 was overexpressed, while USP7 protein was increased (Figure 4C). As an additional control, we treated those cells with BTZ prior to cell lysis and IP. Similar levels of NEK2 were pulled down in both EV and TRIP13-OE cells treated with BTZ. However, there was less ubiquitinated NEK2 in the presence of elevated TRIP13 (Figure 4C, lower panel). Additionally, when NEK2 was immunoprecipitated, USP7 and TRIP13 protein were found to be bound to NEK2. Similar results were observed in H1299 cells when TRIP13 was immunoprecipitated using HA antibodies (Supplemental Figure 7D). These results strongly suggest that TRIP13 physically interacts with USP7 and NEK2 to promote NEK2 deubiquitination.
Figure 4. TRIP13 plays a critical role in stabilizing the USP7 target NEK2.
(A and B) Western blot analysis of TRIP13, NEK2, and GAPDH in ARP1 EV and TRIP13-OE cells (A) and tissues from Trip13TG and Trip13WT mice (B). (C) NEK2 IP from ARP1 EV and TRIP13-OE cells treated with or without 10 nM BTZ for 1 hour followed by Western blot analysis of NEK2, Ub, TRIP13, and USP7. (D) NEK2 IP from ARP1 cells stably expressing scramble (SCR) or TRIP13 shRNA treated with or without DOX for 48 hours followed by Western blot analysis of NEK2, TRIP13, and USP7. Arrows indicate the reduction in USP7 signal after TRIP13 shRNA induction. (E) Kaplan-Meier analysis of MM patients with high TRIP13 and high NEK2 versus MM with one or both genes low (P < 0.0001 by log-rank test). (F) Scatterplot of NEK2 and TRIP13 gene expression intensities from the CoMMpass data set. Pearson’s correlation (r) value and corresponding P value are indicated.
To determine whether TRIP13 binding to USP7 and NEK2 was critical for NEK2 deubiquitination, we analyzed the binding between USP7 and NEK2 in the absence of TRIP13. Using NEK2 antibody to pull down binding proteins, USP7 levels were significantly lower in ARP1 cells lacking TRIP13 (Figure 4D, TRIP13 shRNA) compared with controls. These data indicate that TRIP13 mediates the interaction of USP7 with NEK2.
As in Figure 1G, we have previously shown that high TRIP13 is linked to a shorter OS of MM patients. Using the CoMMpass data set, we further found that MM patients with high TRIP13 and high NEK2 (both quartile 4) had an inferior OS (Figure 4E; P < 0.0001) in comparison with disease with low TRIP13/low NEK2, low TRIP13/high NEK2 (P = 0.0228), or high TRIP13/low NEK2 (P = 0.0093). Kaplan-Meier analysis of a University of Arkansas for Medical Sciences (UAMS) microarray data set also showed that MM with high TRIP13/high NEK2 (both quartile 4) was associated with an inferior outcome compared with the disease lacking elevated expression of both genes (Supplemental Figure 7E; P < 0.0001). A strong correlation between expression of TRIP13 and expression of NEK2 was found in both the CoMMpass (Figure 4F; r = 0.8128, P < 0.0001) and the UAMS data sets (Supplemental Figure 7F; r = 0.7091, P < 0.0001). These data suggest that elevated expression of both TRIP13 and NEK2 is linked to a more aggressive clinical phenotype in MM. Gene set enrichment analysis (GSEA) from CoMMpass shows that ubiquitin-mediated proteolysis pathways were significantly enriched in MM patients with high TRIP13 or with both high TRIP13 and high NEK2, but not in those with high NEK2 but low TRIP13 (Supplemental Figure 8).
TRIP13 dysregulates other USP7 targets.
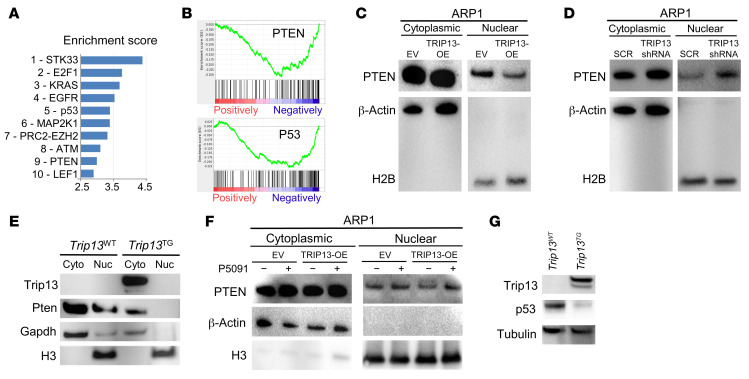
USP7 stabilizes multiple oncogenes (35), resulting in PTEN nuclear exclusion (36) and p53 degradation (37). To test whether TRIP13 was involved in USP7-mediated oncogene stabilization, we performed GSEA on the RNA sequencing data of Eμ-Myc/Trip13TG and Eμ-Myc/Trip13WT mouse B cells (Figure 5A). Ten significant pathways distinguished B cells of the Eμ-Myc/Trip13TG mice from those of the Eμ-Myc/Trip13WT mice. PTEN signaling and p53 signaling were significantly suppressed by Trip13 (Figure 5B). We then analyzed PTEN localization in MM cells overexpressing TRIP13. Immunoblotting on nuclear and cytoplasmic fractions was performed in EV and TRIP13-OE ARP1 cells. The data confirmed that nuclear PTEN was significantly reduced in TRIP13-OE ARP1 cells (Figure 5C) compared with EV ARP1 cells. We then analyzed nuclear levels of PTEN when TRIP13 was knocked down (Figure 5D, TRIP13 shRNA) and found that in the absence of TRIP13, there was an enrichment of nuclear PTEN. Consistent with these in vitro data, nuclear Pten was undetectable in Trip13TG mice, but it was detectable in Trip13WT littermate controls (Figure 5E). We further investigated whether USP7 inhibition could increase nuclear PTEN in TRIP13-OE MM cells. Indeed, similar levels of nuclear PTEN were observed in P5091-treated EV and TRIP13-OE ARP1 cells (Figure 5F). The p53 protein was barely detectable in Trip13TG mice thymus relative to Trip13WT littermate controls (Figure 5G). These results indicate that TRIP13 regulates levels of Pten and p53.
Figure 5. TRIP13 dysregulates other USP7 targets.
(A and B) Gene set enrichment analysis (GSEA) of the 1900 most significantly differentially expressed genes in comparison of Eμ-Myc/Trip13TG and Eμ-Myc/Trip13WT B cells, showing a bar view of the top 10 pathways (A) and an enrichment plot for PTEN (top plot) and p53 pathways (bottom plot) indicating suppression of these pathways when Trip13 is overexpressed (B). (C and D) Western blot analysis of PTEN, β-actin, and histone 2B (H2B) from nuclear and cytosolic fractions of EV and TRIP13-OE ARP1 cells (C) and ARP1 cells stably transduced with SCR and TRIP13 shRNA after 72 hours of induction with DOX (D). (E) Western blot analysis of Trip13, Pten, Gapdh, and histone 3 (H3) from nuclear and cytosolic fractions of thymus from Trip13TG and Trip13WT mice. (F) Western blot analysis of PTEN, β-actin, and H3 in nuclear and cytosolic fractions from EV and TRIP13-OE ARP1 MM cells treated with 16 μM P5091 for 5 hours. (G) Western blot analysis of Trip13, p53, and tubulin in thymus tissues from Trip13TG and Trip13WT mice.
The AAA ATPase of TRIP13 is required for USP7 deubiquitinase function.
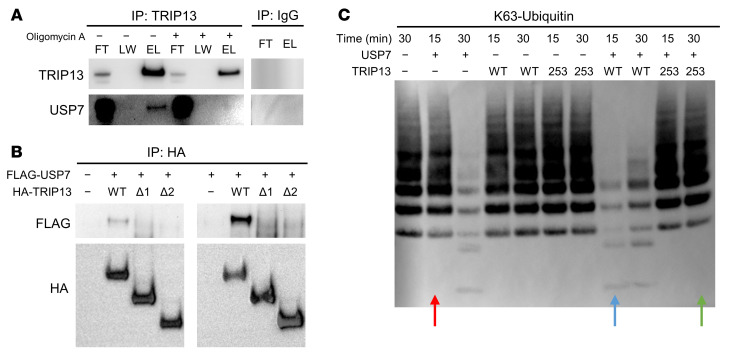
To evaluate how TRIP13 regulates USP7 activity, we first analyzed the level of USP7 phosphorylation. No difference was detected in USP7 phosphorylation between EV and TRIP13-OE ARP1 cells (Supplemental Figure 9). We next tested whether stabilization might be an ATP-dependent process driven by TRIP13 as reported for the TRIP13-p31comet/C-MAD2/CDC20 complex (38). Treatment of TRIP13-OE ARP1 cells with oligomycin, an inhibitor of ATPase synthase (39), followed by TRIP13 IP revealed that TRIP13 was not bound to USP7 (Figure 6A). We also generated two C-terminal deletion TRIP13 constructs lacking the ATPase domain, one lacking the first 100 amino acids (Δ1) and a second missing the first 200 amino acids (Δ2). HEK293T cells were transduced with TRIP13-WT or truncated constructs (Δ1 and Δ2 in combination with WT FLAG-USP7). IP using HA antibody followed by Western blot analysis showed that TRIP13-WT was binding USP7 while TRIP13-Δ1 and -Δ2 failed to do so (Figure 6B). We used the TRIP13 ATP hydrolysis–defective mutant TRIP13-E253Q (40) in an in vitro deubiquitination time course. As shown in Figure 6C (red arrow), USP7 alone could not degrade ubiquitin within 15 minutes. In the presence of TRIP13-WT, ubiquitin degradation was detected at 15 minutes (Figure 6C, blue arrow), whereas TRIP13-E253Q did not enhance USP7-mediated ubiquitin degradation. Remarkably, not only did we observe that TRIP13-E253Q did not enhance USP7 DUB activity, but no ubiquitin degradation was detected at all within the 30-minute time frame of the assay (Figure 6C, green arrow). These data indicate that (a) TRIP13 enhances and accelerates USP7-mediated ubiquitin degradation, (b) TRIP13 ATP hydrolysis is required for TRIP13/USP7 DUB activity, and (c) TRIP13 ATPase is required for TRIP13-USP7 binding as well as for the subsequent DUB activity of USP7.
Figure 6. The AAA ATPase of TRIP13 is required for USP7 deubiquitinase function.
(A) TRIP13 IP of ARP1 TRIP13-OE cells treated with oligomycin A for 6 hours followed by Western blot analysis of FT, LW, and E for TRIP13 and USP7. (B) TRIP13 IP from HEK293T cells transduced with FLAG-USP7 or FLAG-USP7 and WT HA-TRIP13, truncation 1 (Δ1) HA-TRIP13, or truncation 2 (Δ2) HA-TRIP13 followed by Western blot analysis of FLAG or HA. (C) Deubiquitination time course and Western blot analysis of Ub derived from recombinant K63-Ub in the presence of recombinant USP7 with or without purified TRIP13-WT or TRIP13-E253Q mutant (“253”).
The USP7 inhibitor reduces TRIP13-induced BTZ drug resistance in MM.
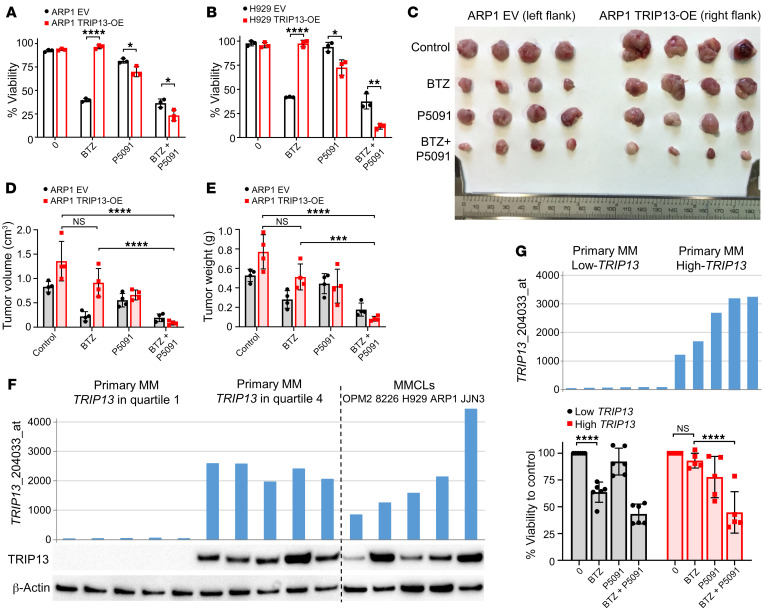
As shown in Figure 7, A and B, TRIP13-OE ARP1 and H929 MM cells were markedly resistant to BTZ, while TRIP13 EV MM cell lines were sensitive. However, TRIP13-OE MM cells were rendered BTZ sensitive by addition of P5091 to BTZ, demonstrating that USP7 inhibition reduces TRIP13-induced BTZ drug resistance in MM cells. We tested the effects of P5091 on TRIP13-OE and EV ARP1 MM cells grown in the NSG subcutaneous xenograft mouse model. Mice injected with EV cells in the left flank and TRIP13-OE cells in the right flank were grouped and treated with vehicle control, P5091 (10 mg/kg, i.v., twice a week from day 3), BTZ (1 mg/kg, i.p., twice a week from day 7), or a combination of P5091 and BTZ. As shown in the pictures of dissected tumors (Figure 7C), TRIP13 significantly enhanced MM tumor growth, and BTZ had little effect in reducing the TRIP13-OE MM tumor burden. However, P5091 alone and in combination with BTZ significantly reduced MM tumor size (Figure 7, D and E). We also confirmed that TRIP13 mRNA expression and protein expression are strongly correlated in primary MM samples (Figure 7F). To further validate the clinical relevance of USP7 inhibition reducing TRIP13-induced BTZ drug resistance, we performed ex vivo treatment on 6 low-TRIP13 and 6 high-TRIP13 primary CD138-positive MM samples as determined by TRIP13 gene expression signal. TRIP13 gene expression signals of these individual primary CD138-positive MM samples were analyzed. In agreement with in vitro MM cell line results, primary MM cells with high TRIP13 exhibited drug resistance to BTZ; however, combination treatment with P5091 and BTZ significantly reduced MM cell viability, while primary MM cells with low TRIP13 were sensitive to both BTZ and combination treatment (Figure 7G). The additive/synergistic effect of BTZ and P5091 suggests that combined USP7 and proteasome inhibition overcomes TRIP13- and NEK2-induced MM drug resistance and tumor growth.
Figure 7. The USP7 inhibitor P5091 reduces TRIP13-induced BTZ drug resistance in MM.
(A and B) ARP1 (A) and H929 (B) EV and TRIP13-OE cells treated with or without 10 nM BTZ or 10 μM P5091 alone or in combination for 48 hours followed by cell viability determination by trypan blue staining (n = 3 per condition). (C) ARP1 EV and TRIP13-OE cells (~0.5 × 106 cells per injection) were injected into the left and right flanks, respectively, of NSG mice. Mice were treated with vehicle control, BTZ (1 mg/kg, i.p., twice a week from day 7), P5091 (10 mg/kg, i.v., twice a week from day 3), and a combination. Mice were sacrificed and tumors were dissected and photographed by week 3 (n = 4 per group). (D and E) Tumor volume (D) and tumor weight (E) were measured and quantified from C. (F) TRIP13 gene expression signal is plotted on the y axis. Primary MM with TRIP13 expression in quartile 1 (n = 5) and quartile 4 (n = 5) and MM cell lines (MMCLs, n = 5) are grouped and plotted along the x axis. Corresponding TRIP13 and β-actin protein levels were analyzed from cell lysates of aliquot CD138-positive cells and MMCLs by Western blot. (G) TRIP13 gene expression signal of primary MM with low TRIP13 expression in quartile 1 (n = 6) and high TRIP13 expression in quartile 4 (n = 5) is plotted on the y axis. Corresponding CD138-positive cells were treated with or without 5 nM BTZ or 2.5 μM P5091 alone or in combination for 24 hours followed by cell viability determination by trypan blue staining. One high-TRIP13 sample was excluded because of low cell viability after thawing. Data are represented as mean ± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 by Student’s t test in A and B and by Tukey’s test for multiplicity-adjusted P values in D, E, and G.
Usp7 inhibition abrogates B cell lymphomas in vivo.
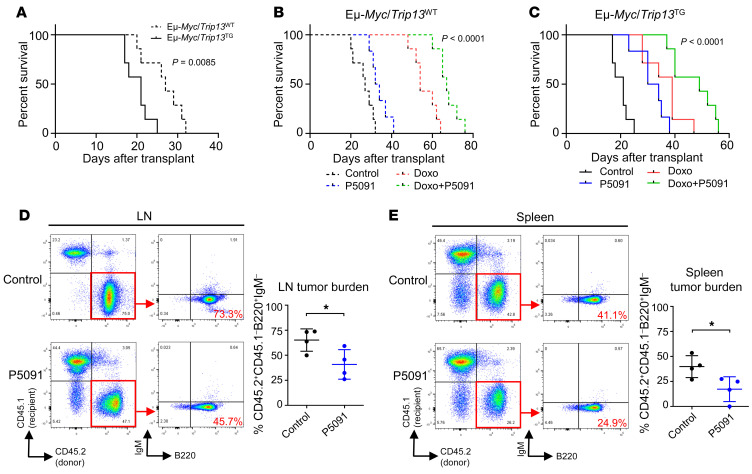
To investigate the effects of Usp7 inhibition on B cell lymphomas in vivo, we generated B cell tumors in recipient WT syngeneic C57BL/6 mice by transplantation of spontaneously arising Eμ-Myc/Trip13WT and Eμ-Myc/Trip13TG lymphoma cells. Mice with transplanted Eμ-Myc/Trip13TG cells exhibited a shorter survival compared with mice with transplanted Eμ-Myc/Trip13WT cells (Figure 8A; P = 0.0085). This result indicates that overexpression of Trip13 in Eμ-Myc produces a more aggressive B cell malignancy, consistent with results of double-transgenic Eμ-Myc/Trip13TG mice in Figure 1E and the MM xenograft model in Figure 7C. Compared with vehicle controls, treatment with P5091 significantly improved the survival of mice that underwent transplantation of both Eμ-Myc/Trip13WT lymphoma cells (Figure 8B, blue dashed line vs. black dashed line, median survival 33 days vs. 27 days) and the more aggressive Eμ-Myc/Trip13TG lymphoma cells (Figure 8C, blue solid line vs. black solid line, median survival 32 days vs. 21 days). Single-agent treatment with doxorubicin, a cytotoxic agent used to treat lymphoma, was highly efficacious in recipient mice bearing Eμ-Myc/Trip13WT lymphoma (Figure 8B, red dashed line). The combination of doxorubicin and P5091 further extended mouse survival (combination treatment: median survival 67 days; doxorubicin only: 54 days). The median survival of mice bearing Eμ-Myc/Trip13TG lymphoma treated with single-agent doxorubicin was only 39 days compared with 21 days in vehicle control–treated animals, indicating decreased efficacy of doxorubicin in Trip13-overexpressing B cell lymphomas. However, mice bearing Eμ-Myc/Trip13TG lymphomas survived a median of 49 days when treated with the combination of doxorubicin and P5091.
Figure 8. Pharmacological inhibition of Usp7 improves survival and abrogates lymphoma growth in transplanted Myc-driven B cell lymphomas in vivo.
(A) Kaplan-Meier analysis of C57BL/6 mice with transplanted Eμ-Myc lymphoma cells (clone 1 [Trip13WT, dashed lines] and clone 2 [Trip13TG, solid lines]). (B and C) Mice with transplanted Eμ-Myc/Trip13WT (B) or Eμ-Myc/Trip13TG (C) lymphoma, treated with vehicle control (black lines), P5091 (10 mg/kg, i.v., twice a week from day 3 after transplant; blue lines), doxorubicin (Doxo; 10 mg/kg, i.p., once on day 7 after transplant; red lines), and combination (green lines) (P values between all groups of each cohort by log-rank test are indicated; n = 6–7 per group). (D and E) Representative flow cytometry plots demonstrating the loss of Eμ-Myc/Trip13TG donor lymphoma cells (CD45.2+CD45.1–B220+IgM–) after P5091 treatment in CD45.1+ recipient mouse lymph node (D) and spleen (E) tumor tissues. Data are represented as mean ± SD. *P < 0.05 by Student’s t test; n = 4 per group.
To further confirm that Usp7 inhibition decreased lymphoma tumor burden, we took advantage of the C57BL/6 congenic strain expressing the CD45.1 allele to distinguish host cells from donor tumor cells expressing the CD45.2 allele. Both vehicle control– and P5091-treated mice were sacrificed once vehicle control mice achieved the preset endpoint; flow cytometry was performed to evaluate tumor burden. The Eμ-Myc/Trip13TG donor lymphoma cell population (CD45.2+CD45.1–B220+IgM–) significantly decreased after P5091 treatment in both lymph nodes (Figure 8D) and spleen (Figure 8E), demonstrating that P5091 inhibits Eμ-Myc/Trip13TG–driven lymphoma growth.
Discussion
The dramatic difference in survival of MM patients with high- and low-risk disease, reflected in tumor cell GEP, can be traced to specific chromosomal lesions, namely gains of chromosome 1q and losses of chromosome 1p. Through sequential GEPs, we have demonstrated that TRIP13 expression is correlated with disease progression and drug resistance in MM (6). The important role of TRIP13 in cancer extends beyond MM, with elevated TRIP13 expression correlated to poor prognosis in multiple malignancies (9–16, 18). These observations highlight TRIP13 as a potential therapeutic target.
Here, we present evidence of a role for TRIP13 in the modulation of ubiquitin homeostasis of B cell malignancies. We demonstrate the clinical importance of high TRIP13 expression associated with inferior patient outcomes. Using a transgenic murine model, RNA sequencing, and systemic in vitro and in vivo biochemical approaches, we have shown that TRIP13 enhances cellular deubiquitination by binding the deubiquitinase USP7, resulting in a decrease in cellular ubiquitination levels as observed upon stable TRIP13 overexpression in MM cells and in transgenic mice. Applying in vitro kinase assays of polyubiquitin chains K48 and K63, we demonstrated that the decreased ubiquitination was not the result of endogenous catalytic activity of TRIP13 but occurs through TRIP13 interaction with the deubiquitinase USP7. The type of ubiquitination modification determines the cellular fate and function of the protein. Monoubiquitin is involved in endocytosis, protein transport, histone regulation, and DNA repair, while multiubiquitin is mostly involved in endocytosis and polyubiquitin is involved in various cellular processes (41). Polyubiquitin chains formed on lysine-48 (K48) of ubiquitin have a well-established role in targeted protein degradation by the 26S proteasome (42), whereas polyubiquitin chains formed on lysine-63 (K63) are involved in DNA repair and endocytosis. However, a recent study showed that K63 ubiquitylation also triggers proteasomal degradation (43). Our in vitro ubiquitinase studies on both K48 and K63 polyubiquitin chains highlighted the potential cellular function of the TRIP13/USP7 complex in the modulation of substrate proteasomal degradation.
Our findings of TRIP13 involvement in deubiquitination pathways are in agreement with findings of other investigators demonstrating that AAA ATPases are able to bind DUBs and promote their activity. For example, p97 has been shown to recruit and promote deubiquitination activity of several DUBs, such as vcip135, Otu1, and YOD1 (44, 45). We hypothesized that TRIP13 AAA ATPase activity would provide the chemical energy for the ATP-dependent ubiquitin-protease activity of USP7. Indeed, the AAA ATPase–defective TRIP13-E253Q mutant abrogated the in vitro deubiquitinase activity of USP7. USP7 maintained deubiquitination even in the absence of TRIP13, indicating that TRIP13 does not provide ATP for USP7 deubiquitination. The presence of TRIP13, however, leads to a more rapid and more thorough deubiquitination by USP7. These results indicate that TRIP13 binding promotes USP7 deubiquitination activity and that a mutation of TRIP13 at glutamate-253 disrupts this binding. While the crystallization of TRIP13 with USP7 has not been performed, we would predict that TRIP13 binding to USP7 occurs in the ubiquitin-like (UBL) 1–3 region and that a mutation at E253Q would disrupt this interaction sufficiently to interfere with the stabilization of the USP7 switching loop. Early evidence including point mutation and C-terminal deletion in TRIP13 suggests that this binding of USP7 is dependent on the carboxy terminus of TRIP13.
Ubiquitination can mediate protein function through changes in localization, stability, and interactions. PTEN (36, 46) and p53 (37, 47–49) each exert a critical function in regulating genomic stability, and are USP7 targets. In this study, we found that TRIP13 dysregulates these well-known USP7 targets, resulting in tumor progression. Specifically, we found evidence that TRIP13 causes the nuclear exclusion of the tumor suppressor PTEN and leads to decreased stability of p53. Application of the USP7 small-molecule inhibitor P5091 demonstrated that these activities of TRIP13 occur through the deubiquitinase USP7. Data presented in this work indicate that increased TRIP13 activity suppresses the TP53 pathway and thus indicate that this might be an alternative pathway to disable TP53 activity in MM and other malignancies, not requiring inactivation of TP53 by deletion or mutations. Future work will be aimed at studying the interplay between TRIP13 and TP53.
Elevated expression of the NEK2 gene is highly correlated with CIN, drug resistance, and poor prognosis in MM (6, 26). We recently discovered that NEK2 protein binds to the DUB USP7. USP7 stabilizes NEK2 by preventing proteasomal degradation of NEK2 in MM cells, resulting in increased drug resistance, which is a prominent feature of the high-risk phenotype (25). We also showed that TRIP13 plays a critical role in stabilizing NEK2 through its interaction with USP7. GSEA from CoMMpass indicates that in high-risk MM, TRIP13 and not NEK2 primarily exerts a ubiquitin-mediated proteolysis function; this ubiquitination process appears to have an additive effect in high-risk MM patients. Importantly, using a small-molecule inhibitor of USP7 in vivo in both the MM xenograft and B cell lymphoma mouse models, we have shown that abrogation of USP7 function leads to reduced tumor growth and decreased drug resistance. This study reveals a function of TRIP13 in cancer biology and identifies the TRIP13/USP7/NEK2 complex as a promising target of therapeutic intervention for high-risk MM.
Our findings provide 3 focal points for therapeutic intervention in high-risk MM: (a) direct targeting of TRIP13, which has already shown good efficacy (50); (b) targeting of the TRIP13 effector deubiquitinase USP7; and (c) targeting of the USP7 target NEK2. Our efforts presented here were primarily focused on MM. The fact that elevated expression of TRIP13 is observed in many cancers, combined with our data in the H1299 lung cancer cell line and the Eμ-Myc/Trip13TG mouse model, suggests that targeting of TRIP13-mediated ubiquitination may have much broader applications in cancer.
Methods
Establishment of Trip13-transgenic mice.
Trip13TG mice were generated at the Genome Editing Core Facility of the University of Iowa. We used the p1026x vector containing the Lck proximal promoter and the immunoglobulin heavy chain intronic enhancer (Eμ). The mouse Trip13 complete sequenced coding region was inserted into the p1026x vector (a gift from Brian M. Iritani, University of Washington, Seattle, Washington, USA). Briefly, a 10.4 kb Lck-(Eμ)-mTrip13-hGH fragment was inserted into the p1026x vector by Spe1. The modified vector contained a 3.2 kb fragment of the mouse Lck proximal promoter to +37 with respect to the transcription start site, a 0.92 kb fragment of the Eμ inserted into the Lck fragment, a 2.1 kb mutated (nontranslatable) version of the hGH gene, and the Trip13 cDNA inserted into the BamHI site of the Lck promoter fragment. The Lck-(Eμ)-mTrip13-hGH fragment is cut out from the p1026x vector by Spe1 and injected into C57BL/6 CBA hybrid embryos. Founders were selected by genotyping by PCR from ear tissue DNA with the following Trip13 primers: forward, 5′-ACCATGTACCCATACGATGTTCCAG-3′, and reverse, 5′-GCCCTTGTCATCGTCATCCTTGTAG-3′. Internal control was used with the following primers: forward, 5′-CTAGGCCACAGAATTGAAAGATCT-3′, and reverse, 5′-GTAGGTGGAAATTCTAGCATCATCC-3′. Three lines were selected and backcrossed to a C57BL/6 background for at least 10 generations.
Cell culture.
Human myeloma cell lines (ARP1, H929, OCI-MY5, and their derivative cell lines) were cultured in RPMI 1640 medium (Gibco) supplemented with 10% heat-inactivated FBS (Gibco), penicillin (100 IU/mL), and streptomycin (100 μg/mL) in a humidified incubator at 37°C and 5% CO2/95% air. HEK293T, H1299, and 3T3 cell lines were cultured with DMEM (Gibco) with the same supplements. Primary MM cells were obtained from MM patient bone marrow aspirates by density gradient centrifugation using Ficoll-Paque and purification with CD138 immunomagnetic bead selection with the autoMACs automated separation system (Miltenyi Biotec). Purity of the plasma cells was determined by flow cytometry, and aliquots were cryopreserved in liquid nitrogen routinely. Purified MM cells were thawed and cultured in the same conditions with the myeloma cell lines previously mentioned.
Vectors and transfections.
Human TRIP13 was cloned into the pCDH vector under the promoter of CAG and tagged with 3× FLAG and HA. The shRNA sequences for TRIP13 were 5′-CGATTATGTGATGACAACTTTCTCGAGAAAGTTGTCATCACATAATCGT-3′ and 5′-GCTAATACACTACTGTTGAAAGAGCTCTTTCAACAGTAGTGTATTAGC-3′ and were inserted into AgeI and EcoRI sites of doxycycline-inducible expression pLKO-Tet-On shRNA. NEK2 was cloned into the pCMV-AIG vector. TRIP13 deletion constructs (Δ1: 100-aa deletion from the C-terminus; Δ2: 200-aa deletion from the C-terminus) were generated by PCR amplification and cloned using XbaI and BamHI enzymes. USP7-FLAG vector was obtained from Addgene (plasmid 16655). Lentiviruses were packaged in HEK293T cells using VSV-G and psPAX2 helper vectors (Addgene) using calcium phosphate transfection (Promega). Transient transfections were done with Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s guidelines.
Cell lysis, IP, and co-IP.
Cell pellets were resuspended in lysis buffer (150 mM NaCl, 10 mM EDTA, 10 mM Tris [pH 7.4], and 1% Triton X-100 supplemented with protease inhibitor cocktail, DTT, and 25 mM N-ethylmaleimide), vortexed vigorously for 10 seconds, incubated on ice for 10 minutes, vortexed again, and centrifuged at 16,200g for 10 minutes at 4°C. The cell pellet was discarded, and supernatant was analyzed for Western blot or frozen at –80°C. For cell fractionations, the Nuclear/Cytosol Fractionation kit (BioVision Inc.) was used according to the manufacturer’s instructions. For IPs, lysates were collected, and the same amounts of protein were incubated with 25 μL of either Protein A or Protein G Dynabeads (Invitrogen) and 3 μL of primary antibody overnight. Samples were washed 3 times with IP buffer and eluted by rotating for 20 minutes at room temperature with sample buffer supplemented with 350 mM DTT. Co-IPs were performed using IP buffer (Thermo Fisher Scientific) supplemented with the protease inhibitor cocktail. Samples were vortexed for 5 seconds, incubated on ice for 20 minutes, vortexed again, and centrifuged at 16,200g for 10 minutes at 4°C, and supernatants were collected.
Western blot.
Protein concentration was calculated by NanoDrop (Thermo Fisher Scientific). Proteins were separated with NuPAGE 4%–12% Bis-Tris Gel (NOVEX) at 180 V, and then transferred to a nitrocellulose membrane for 1 hour at 400 mA at 4°C. Membrane was blocked for 30 minutes with 10% milk at room temperature. Antibodies for anti-USP7 (D17C6 XP, Cell Signaling Technology), anti-TRIP13 (C-4: sc-514314, Santa Cruz Biotechnology), anti-NEK2 (D-8: sc-55601, Santa Cruz Biotechnology), anti-ubiquitin (3933S, Cell Signaling Technology), anti-HA (C29F4, Cell Signaling Technology), anti-PTEN (D.4.3 XP, Cell Signaling Technology), anti-p53 (1C12, Cell Signaling Technology), anti-GAPDH (14C10, Cell Signaling Technology), anti–β-actin (D6A8, Cell Signaling Technology), and anti-H2B (D2H6, Cell Signaling Technology) were incubated overnight at 4°C with a dilution of 1:500 or 1:1000. Secondary rabbit antibody (goat anti-rabbit IgG [H+L], HRP-conjugated, Cell Signaling Technology) and secondary mouse antibody (goat anti-mouse IgG-HRP: sc-2005, Santa Cruz Biotechnology) were incubated for 1 hour at a dilution of 1:10,000 at room temperature. For exposure, Immobilon Western HRP Substrate Peroxide Solution (Millipore) was used. Imaging was done with a ChemiDoc XRS+ System with Image Lab Software (Bio-Rad).
In vitro and ex vivo drug treatment conditions.
For shRNA induction, doxycycline was used at 100 ng/mL for 48 or 72 hours. Bortezomib (BTZ; Selleckchem), P5091 (Sigma-Aldrich), etoposide (Sigma-Aldrich), melphalan (Sigma-Aldrich), PYR-41 (Selleckchem), PR-619 (Selleckchem), and spautin-1 (Selleckchem) were used at indicated concentrations for different time periods. N-ethylmaleimide was solubilized in ethanol and added to lysis buffer immediately before use at a concentration of 10 mM.
Cell viability.
For trypan blue staining, 10 μL of cell culture was mixed with 10 μL of trypan blue. Samples were counted in a hemocytometer; translucent cells were counted as alive and viable, and blue-stained cells were counted as dead. Cell viability was calculated by division of viable cell number by total cell number; each sample was done in triplicate. For MTT assay, briefly, cells were seeded in 96-well plates and assay performed on triplicated wells for every condition. MTT was then added for 2 hours at a final concentration of 2.5 mg/mL. Plates were centrifuged and the soluble fraction decanted. Crystal pellets were suspended in DMSO and absorbance read at 590 nm, and cell viability was presented as percentage normalized to untreated cells.
In vitro deubiquitination.
Recombinant USP7 His6 tag (DUB), USP10 His6 tag (DUB), and K48- and K63-specific ubiquitins (pUb) were purchased from Boston Biochem. TRIP13 was not commercially available and therefore was isolated from ARP1 TRIP13-OE cells by IP using HA antibodies. For the in vitro deubiquitination, 4 reactions were prepared: (a) deUb buffer (18 μL) + pUb (1 μg) + deUb buffer (2 μL); (b) deUb buffer (18 μL) + pUb (1 μg) + DUB (0.1 μg) + deUb buffer (1 μL); (c) deUb buffer (18 μL) + pUb (1 μg) + TRIP13 (1 μg) + deUb buffer (1 μL); and (d) deUb buffer (18 μL) + pUb (1 μg) + DUB (0.1 μg) + TRIP13 (1 μg). All these reactions were repeated in the presence of 25 mM N-ethylmaleimide. Reactions were mixed by gentle pipetting and incubated at 35°C. After 60 minutes, samples were analyzed by Western blot. Ponceau S stain was used to determine the efficiency of transfer. DeUb assay buffer was made from 50 mM Tris-HCl (pH 7.4), 50 mM NaCl, 2 mM MgCl2, 1 mM DTT, and 5% glycerol.
Mouse xenograft models and experiments.
For Figure 7, C–E, 8- to 10-week-old NSG mice (bought from The Jackson Laboratory, bred and housed at the University of Iowa Animal Facilities) were subcutaneously injected with 0.5 × 106 ARP1 EV and TRIP13-OE cells into the left and right flank, respectively. Mice were treated with vehicle control, BTZ (1 mg/kg, i.p., twice a week from day 7), P5091 (10 mg/kg, i.v., twice a week from day 3), and combination. Mice were sacrificed and tumors were dissected and assessed by week 3.
For Figure 8, A–C, 8- to 10-week-old C57BL/6 mice (bought from The Jackson Laboratory, bred and housed at the University of Iowa Animal Facilities) were injected i.v. with 2 × 106 Eμ-Myc/Trip13WT or 2 × 106 Eμ-Myc/Trip13TG lymphoma cells via the tail vein on day 0 and treated with the Usp7 inhibitor P5091 from 3 days after lymphoma cell injection, doxorubicin once on day 7, and a combination of P5091 and doxorubicin, comparing with vehicle control group. P5091 (10 mg/kg; Selleckchem) was injected i.v. twice a week. Doxorubicin (10 mg/kg; Selleckchem) was injected i.p. once on day 7. For survival analyses, treatment of mice continued until an ethical endpoint was reached. Mice were sacrificed at the ethical endpoint: enlarged lymph nodes (palpable diameter of approximately 1 cm), loss of weight (more than 20%), labored breathing, hunched posture, ruffled fur, limited mobility or paralysis. For the P5091 and combination groups, P5091 treatment ended when the first mouse in the group reached the ethical endpoint.
For Figure 8, D and E, to investigate tumor burden after P5091 treatment, 8- to 10-week-old CD45.1+ C57BL/6 mice were injected i.v. with 2 × 106 CD45.2+ Eμ-Myc/Trip13TG lymphoma cells via the tail vein, and P5091 treatment was administered as described above. All mice were sacrificed once the first vehicle control mice reached the ethical endpoint.
Flow cytometry.
Mouse lymph nodes and spleens were dissected and crushed into single-cell suspension in FACS buffer (1× PBS with 2% FBS) and filtered through 70 μm cell strainers. Single-cell suspension was collected by rinsing with FACS buffer. Cells from spleens were lysed with red blood cell lysis buffer (K-D Medical) to remove red blood cells. After washing with 1× PBS, 0.5 × 106 to 1 × 106 cells were resuspended in 100 μL FACS buffer and preincubated with anti–mouse CD16/32 antibody (BioLegend) for blocking prior to labeling with antibodies of CD45.2-PerCP-Cy5.5, CD45.1-PE, B220-APC-Cy7, and IgM-FITC (BioLegend) for 20 minutes at 4°C. Cells were then washed with FACS buffer and analyzed on a BD LSR II UV flow cytometer at the University of Iowa flow cytometry facility. All flow results were analyzed using FlowJo software version 9 (BD Biosciences).
Microarray data sets.
Gene expression profiling data were obtained from previous studies and are available in the NCBI’s Gene Expression Omnibus under accession numbers GSE2658 (1) and GSE10846 (51).
Statistics.
Mouse survival analysis and patient survival analysis were carried out using GraphPad Prism software (version 8.0), and significance was determined using log-rank (Mantel-Cox) test. All other statistical comparisons were analyzed by unpaired, 2-tailed, independent Student’s t test, unless otherwise described in the figure legends. A P value less than 0.05 was considered significant. Results are presented as mean ± SD, and n is indicated in figure legends.
Study approval.
NSG mice (The Jackson Laboratory) were bred and maintained in compliance with the guidelines for institutional animal care at the University of Iowa. The animal studies were performed according to the guidelines of the Institutional Animal Care and Use Committee and the local veterinary office and ethics committee of the University of Iowa under IACUC-approved protocol 5081482. Deidentified primary samples were obtained from MM patients during UAMS clinic visits. Signed institutional review board–approved informed consent forms are kept on record at the UAMS Tissue Biorepository and Procurement Service under approved protocol IRB 260731.
Author contributions
CL performed the experiments, collected and analyzed the data, generated the figures, and wrote and edited the manuscript. JX and RFM performed experiments and analyzed the data. FC and YH reviewed the data and edited the manuscript. TCA and FT analyzed the RNA sequencing data. HX maintained the mouse strains and analyzed the data. DL, DG, SKJ, and FVR reviewed the data. SJ, JDS, and GT reviewed the data and edited the manuscript. IF performed experiments, designed and supervised experiments, and analyzed data. FZ designed and supervised this study, collected and analyzed data, and wrote and edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute (NCI) grant R01CA236814 (to FZ), US Department of Defense grant CA180190 (to FZ), Leukemia and Lymphoma Society grant TRP 6549-18 (to FZ), a Myeloma Crowd Research Initiative Award (to FZ), the Riney Family Multiple Myeloma Research Program Fund (to FZ), NCI grant R01CA151354 (to SJ), and UAMS Winthrop P. Rockefeller Cancer Institute start-up funds (to FZ). We would like to thank Hasem Habelhah (University of Iowa) for providing us with the protocol of the in vitro ubiquitination assay, Zhimin Gu for making the Lck-(Eμ)-mTrip13-hGH/p1026x construct to generate transgenic mice, and the UAMS Tissue Biorepository and Procurement Core and UAMS Proteomics Core for their support.
Version 1. 06/01/2021
In-Press Preview
Version 2. 07/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(14):e146893.https://doi.org/10.1172/JCI146893.
Contributor Information
Can Li, Email: cli2@uams.edu.
Jiliang Xia, Email: jiliangxia2009@163.com.
Fangping Chen, Email: xychenfp@qq.com.
Yanjuan He, Email: 15973130688@163.com.
Timothy Cody Ashby, Email: tcashby@uams.edu.
Feixiang Teng, Email: fxteng@126.com.
Hongwei Xu, Email: HXu@uams.edu.
Dingxiao Liu, Email: dingxiao-liu@uiowa.edu.
Dongzheng Gai, Email: DGai@uams.edu.
Sarah K. Johnson, Email: skjohnson@uams.edu.
Frits van Rhee, Email: vanrheefrits@uams.edu.
Siegfried Janz, Email: sjanz@mcw.edu.
Guido Tricot, Email: GJTricot@uams.edu.
Ivana Frech, Email: ivana.frech@gmail.com.
Fenghuang Zhan, Email: FZhan@uams.edu.
References
- 1.Shaughnessy JD, Jr , et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 2.Kumar SK, et al. Natural history of relapsed myeloma, refractory to immunomodulatory drugs and proteasome inhibitors: a multicenter IMWG study. Leukemia. 2017;31(11):2443–2448. doi: 10.1038/leu.2017.138. [DOI] [PubMed] [Google Scholar]
- 3.Rajkumar SV. Multiple myeloma: 2012 update on diagnosis, risk-stratification, and management. Am J Hematol. 2012;87(1):78–88. doi: 10.1002/ajh.22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlogie B, et al. Curing myeloma at last: defining criteria and providing the evidence. Blood. 2014;124(20):3043–3051. doi: 10.1182/blood-2014-07-552059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanamura I, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108(5):1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou W, et al. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell. 2013;23(1):48–62. doi: 10.1016/j.ccr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao Y, et al. TRIP13 impairs mitotic checkpoint surveillance and is associated with poor prognosis in multiple myeloma. Oncotarget. 2017;8(16):26718–26731. doi: 10.18632/oncotarget.14957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heuck CJ, et al. Five gene probes carry most of the discriminatory power of the 70-gene risk model in multiple myeloma. Leukemia. 2014;28(12):2410–2413. doi: 10.1038/leu.2014.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou K, et al. Loss of thyroid hormone receptor interactor 13 inhibits cell proliferation and survival in human chronic lymphocytic leukemia. Oncotarget. 2017;8(15):25469–25481. doi: 10.18632/oncotarget.16038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larkin SE, et al. Identification of markers of prostate cancer progression using candidate gene expression. Br J Cancer. 2012;106(1):157–165. doi: 10.1038/bjc.2011.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong L, et al. TRIP13 is a predictor for poor prognosis and regulates cell proliferation, migration and invasion in prostate cancer. Int J Biol Macromol. 2019;121:200–206. doi: 10.1016/j.ijbiomac.2018.09.168. [DOI] [PubMed] [Google Scholar]
- 12.Rhodes DR, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin KJ, et al. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One. 2008;3(8):e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdul Aziz NA, et al. A 19-gene expression signature as a predictor of survival in colorectal cancer. BMC Med Genomics. 2016;9(1):58. doi: 10.1186/s12920-016-0218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao J, et al. Silencing TRIP13 inhibits cell growth and metastasis of hepatocellular carcinoma by activating of TGF-β1/smad3. Cancer Cell Int. 2018;18:208. doi: 10.1186/s12935-018-0704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang JU, et al. Gain at chromosomal region 5p15.33, containing TERT, is the most frequent genetic event in early stages of non-small cell lung cancer. Cancer Genet Cytogenet. 2008;182(1):1–11. doi: 10.1016/j.cancergencyto.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, et al. The oncogenic role of TRIP13 in regulating proliferation, invasion, and cell cycle checkpoint in NSCLC cells. Int J Clin Exp Pathol. 2019;12(9):3357–3366. [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee R, et al. TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat Commun. 2014;5:4527. doi: 10.1038/ncomms5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vader G. Pch2(TRIP13): controlling cell division through regulation of HORMA domains. Chromosoma. 2015;124(3):333–339. doi: 10.1007/s00412-015-0516-y. [DOI] [PubMed] [Google Scholar]
- 20.Clairmont CS, et al. TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nat Cell Biol. 2020;22(1):87–96. doi: 10.1038/s41556-019-0442-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaughnessy JD, Jr, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood. 2011;118(13):3512–3524. doi: 10.1182/blood-2010-12-328252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbagallo F, et al. Increased expression and nuclear localization of the centrosomal kinase Nek2 in human testicular seminomas. J Pathol. 2009;217(3):431–441. doi: 10.1002/path.2471. [DOI] [PubMed] [Google Scholar]
- 23.Stricker TP, et al. Expression profiling of 519 kinase genes in matched malignant peripheral nerve sheath tumor/plexiform neurofibroma samples is discriminatory and identifies mitotic regulators BUB1B, PBK and NEK2 as overexpressed with transformation. Mod Pathol. 2013;26(7):930–943. doi: 10.1038/modpathol.2012.242. [DOI] [PubMed] [Google Scholar]
- 24.Meng L, et al. Inhibition of Nek2 by small molecules affects proteasome activity. Biomed Res Int. 2014;2014:273180. doi: 10.1155/2014/273180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franqui-Machin R, et al. Destabilizing NEK2 overcomes resistance to proteasome inhibition in multiple myeloma. J Clin Invest. 2018;128(7):2877–2893. doi: 10.1172/JCI98765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao M, et al. NEK2 induces osteoclast differentiation and bone destruction via heparanase in multiple myeloma. Leukemia. 2017;31(7):1648–1650. doi: 10.1038/leu.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iritani BM, et al. Control of B cell development by Ras-mediated activation of Raf. EMBO J. 1997;16(23):7019–7031. doi: 10.1093/emboj/16.23.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misund K, et al. MYC dysregulation in the progression of multiple myeloma. Leukemia. 2020;34(1):322–326. doi: 10.1038/s41375-019-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horn H, et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121(12):2253–2263. doi: 10.1182/blood-2012-06-435842. [DOI] [PubMed] [Google Scholar]
- 31.Harris AW, et al. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167(2):353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rempel RE, et al. Utilization of the Eμ-Myc mouse to model heterogeneity of therapeutic response. Mol Cancer Ther. 2014;13(12):3219–3229. doi: 10.1158/1535-7163.MCT-13-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz R, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, et al. USP7: target validation and drug discovery for cancer therapy. Med Chem. 2018;14(1):3–18. doi: 10.2174/1573406413666171020115539. [DOI] [PubMed] [Google Scholar]
- 36.Song MS, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455(7214):813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416(6881):648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 38.Alfieri C, et al. Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature. 2018;559(7713):274–278. doi: 10.1038/s41586-018-0281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Symersky J, et al. Oligomycin frames a common drug-binding site in the ATP synthase. Proc Natl Acad Sci U S A. 2012;109(35):13961–13965. doi: 10.1073/pnas.1207912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye Q, et al. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. Elife. 2015;4:e07367. doi: 10.7554/eLife.07367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popovic D, et al. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20(11):1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 42.Mallette FA, Richard S. K48-linked ubiquitination and protein degradation regulate 53BP1 recruitment at DNA damage sites. Cell Res. 2012;22(8):1221–1223. doi: 10.1038/cr.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohtake F, et al. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc Natl Acad Sci U S A. 2018;115(7):E1401–E1408. doi: 10.1073/pnas.1716673115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ernst R, et al. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell. 2009;36(1):28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, et al. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164(7):973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trotman LC, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128(1):141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004;3(6):689–692. [PubMed] [Google Scholar]
- 48.Li M, et al. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13(6):879–886. doi: 10.1016/S1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 49.Shen Y, et al. TSPY1 suppresses USP7-mediated p53 function and promotes spermatogonial proliferation. Cell Death Dis. 2018;9(5):542. doi: 10.1038/s41419-018-0589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, et al. A small-molecule inhibitor targeting TRIP13 suppresses multiple myeloma progression. Cancer Res. 2020;80(3):536–548. doi: 10.1158/0008-5472.CAN-18-3987. [DOI] [PubMed] [Google Scholar]
- 51.Lenz G, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.