Abstract
Androgen receptor–positive prostate cancer (PCa) and estrogen receptor–positive luminal breast cancer (BCa) are generally less responsive to immunotherapy compared with certain tumor types such as melanoma. However, the underlying mechanisms are not fully elucidated. In this study, we found that FOXA1 overexpression inversely correlated with interferon (IFN) signature and antigen presentation gene expression in PCa and BCa patients. FOXA1 bound the STAT2 DNA-binding domain and suppressed STAT2 DNA-binding activity, IFN signaling gene expression, and cancer immune response independently of the transactivation activity of FOXA1 and its mutations detected in PCa and BCa. Increased FOXA1 expression promoted cancer immuno- and chemotherapy resistance in mice and PCa and BCa patients. These findings were also validated in bladder cancer expressing high levels of FOXA1. FOXA1 overexpression could be a prognostic factor to predict therapy resistance and a viable target to sensitize luminal PCa, BCa, and bladder cancer to immuno- and chemotherapy.
Keywords: Cell Biology, Therapeutics
Keywords: Cancer, Cancer immunotherapy, Molecular biology
Introduction
Patients with certain cancer types such as melanoma and non–small cell lung cancer have significantly benefited from immunotherapies such as immune checkpoint inhibitors (ICIs) (1). However, ICIs including the anti–cytotoxic T lymphocyte antigen 4 (anti-CTLA4) antibody ipilimumab and anti–PD-1 antibody pembrolizumab exhibit limited anticancer activity in prostate cancer (PCa), most of which are androgen receptor positive (AR+) (2). Similarly, breast cancer (BCa), especially the estrogen receptor–positive (ER+) subtype, also exhibits very limited response to immunotherapy (3). Thus, AR+ PCa and ER+ BCa are generally considered immunologically “cold” cancer types and it is of paramount importance to decipher the underlying mechanisms that drive resistance to immunotherapy. One factor underlying immunologically cold PCa and BCa relates to low levels of tumor mutation burden (TMB). Surprisingly, some studies suggest that the ICI therapy response is not significantly associated with TMB in BCa and melanoma (4, 5), while others show that upregulation of interferon γ (IFN-γ) response pathway genes are associated with improved clinical responses to ICI in PCa with low TMB (6).
Activation of signaling pathways in tumors in response to IFNs including type I (such as IFN-α and IFN-β), type II (IFN-γ), and type III IFN (such as IFN-λs) is essential for cytotoxic T lymphocyte–mediated (CTL-mediated) killing of cancer cells (7–12). High expression of IFN signaling genes is associated with greater response to ICIs in melanoma and neck squamous cell carcinoma (13). In cancer cells, IFN induces expression of IFN-stimulated genes (ISGs) including antigen presentation machinery (APM) genes such as major histocompatibility complex class I (MHC I) (14). Presentation of cancer-specific neoantigens, which is regulated by IFN signaling, is a key factor affecting CTL activity and ICI therapy efficacy (15–17). This critical step is governed by MHC or human leukocyte antigen (HLA) that presents intracellular peptides on the cell surface that are recognized by the T cell receptor (TCR). As a result, this immune-stimulating signaling cascade triggers infiltration of cytotoxic lymphocytes, mainly CTLs and natural killer (NK) cells, ultimately leading to granule exocytosis as a common mechanism to destroy cancer cells by expressing and releasing pore-forming proteins including perforin 1 (PRF1), granule-associated enzymes (or granzymes, GZMs), and NK cell granule protein 7 (NKG7).
Improved clinical outcomes have been achieved for certain cancer types such as melanoma after treatment with immunotherapies including adoptive transfer of tumor-infiltrating lymphocytes (TILs) and application of ICIs (e.g., CTLA4, PD-1, and PD-L1 blockade antibodies; refs. 18–21). However, disease relapse often occurs after initial tumor regression and immunotherapy resistance can develop. Notably, as demonstrated in melanoma, such resistance has been linked to genetic alterations such as mutations and somatic copy number alterations in IFN/receptor signaling and APM genes (7, 18, 22, 23).
The transcription factor FOXA1 is a well-studied pioneer factor required for AR and ER activities in PCa and BCa cells (24–26). The FOXA1 gene is also implicated in these two cancer types, owing to its frequent mutations (27–30). The frequency of somatic point mutations of FOXA1 is approximately 4% to 8% and FOXA1 mutations promote cancer progression by reprogramming the functions of AR and other factors in these cancer types (27–31). In the present study, we identified a role of FOXA1 in suppressing IFN signaling and the cancer immune response, which drives cancer immune evasion and therapy resistance. Importantly, this function is independent of the well-known pioneer-factor function of FOXA1 and its mutations detected in PCa and BCa.
Results
FOXA1 inversely correlates with IFN signaling activity in PCa and BCa.
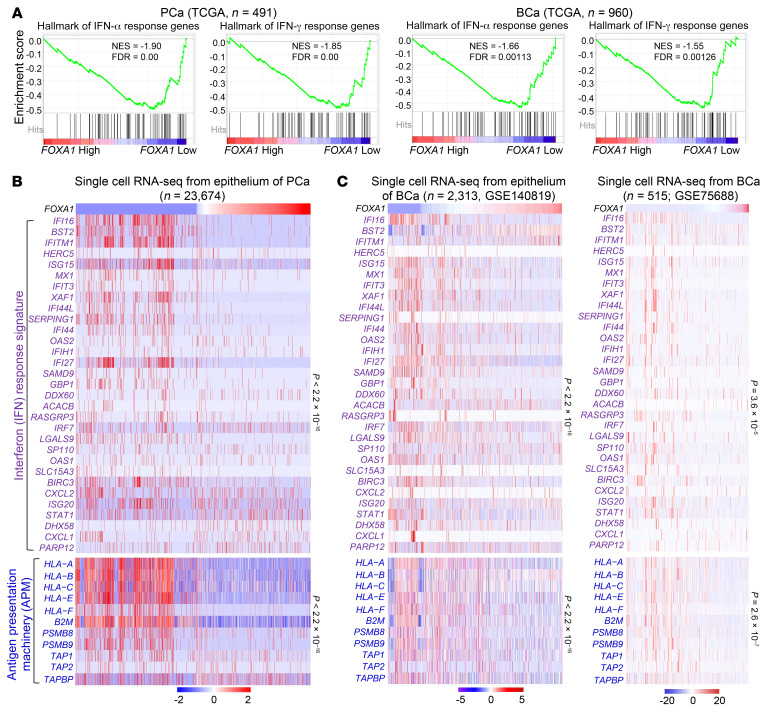
The so-called immune coldness commonly observed in a few cancer types such as AR+ PCa and ER+ luminal BCa prompted us to hypothesize that there might be a common immune evasion mechanism shared by different cancers. As demonstrated in melanoma, low IFN activation predicts unfavorable prognosis of ICI immunotherapy (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI147025DS1) and deletion of IFN pathway genes links to ICI resistance (5, 7, 13), suggesting that inactivation of the IFN pathway may confer immune evasiveness on cancers. We demonstrated that the expression signature of IFN response genes (termed IFN activity or score) in The Cancer Genome Atlas (TCGA) cohort of PCa and luminal BCa was significantly lower than that in melanoma, lung, and kidney cancers, which are generally responsive to ICIs (refs. 32–34 and Supplemental Figure 1B). Therefore, we performed meta-analysis of TCGA PCa and BCa RNA sequencing (RNA-seq) data sets to search for genes that are negatively correlated with IFN activity. We demonstrated that FOXA1 is the only common gene among the top 5 hits, the expression of which negatively correlated with IFN activity in both PCa and BCa cohorts (Supplemental Figure 1C). Gene set enrichment analysis (GSEA) also revealed a negative correlation between expression of FOXA1 and IFN response genes in TCGA cohorts of PCa and BCa patients (Figure 1A). To corroborate the meta-analysis data, we performed single-cell RNA-seq (scRNA-seq) in PCa patient samples and demonstrated that FOXA1 level was inversely associated with expression of IFN response signature genes and APM genes in the luminal cell population (Figure 1B, Supplemental Figure 1D, and Supplemental Tables 1 and 2). Similar results were obtained from BCa cells through meta-analysis of scRNA-seq data from 2 cohorts of BCa patient specimens (refs. 35, 36, and Figure 1C). These findings are consistent with the results obtained from the analysis of RNA-seq data from bulk tissues in different cohorts with primary and metastatic PCa and BCa, respectively (Supplemental Figure 2). Aberrant activation of IFN signaling genes in FOXA1-low tumors in both TCGA PCa and BCa cohorts was unlikely caused by either genomic alterations in IFN response genes or overall TMB because there was no obvious correlation between FOXA1 expression and genomic alterations in IFN response genes or overall TMB (Supplemental Figure 3). FOXA1 mRNA expression was highly upregulated but intriguingly, CD274 (PD-L1) mRNA expression was downregulated in PCa and BCa tissues compared with normal tissues in various cohorts examined (Supplemental Figure 4). Together, our data indicate that overexpression of FOXA1 may play a pivotal role in regulating immune coldness by modulating IFN response gene expression in PCa and BCa.
Figure 1. FOXA1 levels inversely correlate with IFN signaling gene expression in prostate (PCa) and breast cancer (BCa).
(A) GSEA enrichment plots show the inverse correlation between FOXA1 expression and IFN response hallmark genes in PCa and BCa from TCGA database. The FOXA1-high and -low groups were defined using the median as the cutoff. (B and C) Heatmaps show the inverse correlation of FOXA1 expression with IFN response signature genes and APM genes in single-cell RNA-seq data of PCa (B) and BCa (C) samples. Statistical significance was determined by Pearson’s correlation test.
FOXA1 binds the DNA-binding domain and inhibits DNA-binding ability of STAT2.
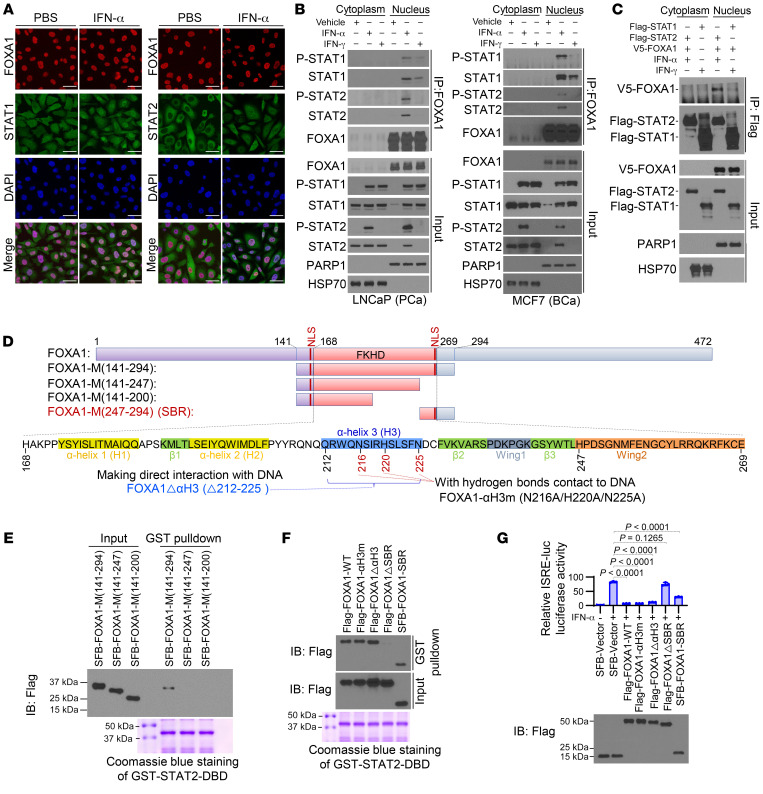
To explore possible molecular mechanisms underlying FOXA1 inhibition of IFN signaling, we first examined the colocalization of FOXA1 with STAT1 and STAT2, two major effectors of IFN signaling. FOXA1 protein was primarily detected in the nucleus in LNCaP cells regardless of IFN-α treatment (Figure 2A). STAT1 protein was localized in both the cytoplasm and nucleus, while STAT2 was mainly localized in the cytoplasm of LNCaP cells grown in regular medium without additional IFN-α stimulation (Figure 2A). IFN-α treatment increased localization of both STAT1 and STAT2 proteins in the nucleus where they were colocalized with FOXA1 (Figure 2A). Because phosphorylation of STAT1 and STAT2 is important for their nuclear localization, we sought to determine whether FOXA1 affects STAT1 and STAT2 phosphorylation. We found that neither ectopic expression of FOXA1 in FOXA1-negative cell lines such as DU145 and MDA-MB-231 nor knockdown (KD) of endogenous FOXA1 in LNCaP and MCF7 cell lines had any effect on STAT1 and STAT2 protein phosphorylation (Supplemental Figure 5, A–D). Consistent with the nuclear localization status of FOXA1 (Figure 2A), coimmunoprecipitation (co-IP) assay showed that FOXA1 interacted with STAT1 and STAT2 at endogenous levels in the nuclei of LNCaP and MCF7 cells treated with IFN-α (Figure 2B). Similarly, we observed that FOXA1 binds to STAT1 in the nucleus in both cell lines stimulated with IFN-γ, although STAT1 homodimer association with FOXA1 was much weaker than that of the STAT1-STAT2 heterodimer (Figure 2B), and this result was confirmed by reciprocal co-IP experiments (Figure 2C). FOXA1 overexpression also had no effect on STAT2-STAT1-IRF9 and STAT1-STAT1 complex formation following the stimulation with IFN-α and IFN-γ, respectively (Supplemental Figure 5E). These data suggest that FOXA1 expression does not affect the formation of STAT1 and/or STAT2 protein-containing complexes in cells stimulated with type I or II IFNs.
Figure 2. FOXA1 binds the STAT2 DBD and impedes STAT2 DNA-binding ability.
(A) Immunofluorescent cytochemical analysis of STAT1, STAT2, and FOXA1 in LNCaP cells treated with vehicle (PBS) or IFN-α. (B) Co-IP shows the interaction of FOXA1 with STAT1 and STAT2 at endogenous levels in LNCaP and MCF7 cells treated with IFN-α or IFN-γ. See complete unedited blots in the supplemental material. (C) Co-IP analysis of interaction among ectopically expressed FOXA1, STAT1, and STAT2 proteins in 293T cells. (D) Diagram shows the domain structure of the FOXA1 forkhead (FKHD) DNA-binding domain (DBD) and FOXA1 truncation and missense mutation expression constructs. NLS, nuclear localization signal; SBR, STAT2-binding region. (E and F) GST pulldown assay shows the interaction of GST-tagged STAT2 DBD with the indicated FOXA1 mutants expressed in 293T cells. ΔαH3, deletion of α-helix 3; ΔSBR, deletion of STAT2-binding region. (G) Luciferase reporter assay shows the inhibitory effect of the indicated WT FOXA1 or mutants on ISRE-luc reporter gene activity in DU145 cells. Data shown as mean ± SD (n = 3). Statistical significance was determined by 1-way ANOVA with Bonferroni’s correction for multiple tests.
Because binding of FOXA1 with STAT2-STAT1 in IFN-α–stimulated cells was much stronger than that with STAT1-STAT1 in IFN-γ–treated cells (Figure 2, B and C), we chose to further characterize how FOXA1 binds to STAT2. Co-IP assays using 2 STAT2 N-terminal truncation mutants indicated that FOXA1 binds the STAT2 DNA-binding domain (DBD) (Supplemental Figure 5F). Glutathione S-transferase (GST) pulldown assay confirmed that FOXA1 directly bound the STAT2 DBD in vitro (Supplemental Figure 5G). We further performed in vitro protein binding assays using different GST-STAT2 DBD truncation mutants and demonstrated that FOXA1 bound more than two-thirds (aa 366–486) of the STAT2 DBD (aa 312–486; Supplemental Figure 5H).
We also determined which region in FOXA1 interacts with STAT2. Mutagenesis and GST pulldown assays showed that the middle portion of FOXA1 (FOXA1-M, aa 141–294), which contains the forkhead domain (FKHD, aa 168–269), bound the STAT2 DBD (Supplemental Figure 5I). We further generated 3 additional C-terminal truncation mutants from FOXA1-M (Figure 2D) and utilized them for GST pulldown and co-IP assays. We found that deletion of the C-terminal portion of FOXA1-M (aa 247–294) abolished FOXA1’s interaction with the STAT2 DBD and this fragment is referred to as the STAT2-binding region (SBR) of FOXA1 (Figure 2, D and E, and Supplemental Figure 5J). The protein binding results were fully supported by the data obtained from IFN-sensitive response element–based (ISRE-based) luciferase reporter gene assay (Supplemental Figure 5K). These findings suggest that the SBR is required for FOXA1 binding of STAT2 and suppression of IFN activity.
Since the SBR contains the Wing2 motif (aa 247–269), a key region involved in regulating the DNA-binding activity of the FOXA1 DBD (refs. 29, 30, 37, and Figure 2D), we sought to determine whether the DNA-binding ability of FOXA1 is required for inhibition of IFN signaling. The third α-helix (αH3, aa 212–225), especially residues N216, H220, and N225, in the FKHD domain of FOXA1 have direct contact with DNA (37). We generated 2 DNA binding–deficient mutants, FOXA1-N216A/H220A/N225A (FOXA1-αH3m) and FOXA1Δ212–225 (FOXA1ΔαH3), by mutating these 3 residues to alanine and deleting the entire αH3. Results from electrophoretic mobility shift assay (EMSA) and the luciferase reporter assay confirmed that these mutants lost their ability to bind to the cognate FOXA1-response DNA element (FRE) in the KLK3 (prostate-specific antigen) gene enhancer and to initiate FOXA1 gene transactivation activity (refs. 29, 30, and Supplemental Figure 5, L and M). However, apart from the FOXA1ΔSBR mutant, these 2 DNA binding–deficient mutants of FOXA1 were still able to bind the STAT2 DBD and inhibit IFN activity (Figure 2, F and G), suggesting that FOXA1 suppresses IFN signaling independently of its DNA-binding function. We further performed a biotin pulldown assay using a biotin-labeled ISRE DNA oligonucleotide (biotin-ISRE; Supplemental Figure 5N, top). We demonstrated that addition of in vitro–generated WT FOXA1 and FOXA1ΔαH3 mutant (deletion of DNA-contacting region) proteins prohibited STAT2 binding of its targeting DNA sequence ISRE in the nuclear extract of IFN-α–stimulated FOXA1-negative DU145 cells (Supplemental Figure 5N, bottom). However, this role of FOXA1 was abolished by deletion of the SBR (FOXA1ΔSBR; Supplemental Figure 5N, bottom). Moreover, no FOXA1 protein was pulled down by the biotin-labeled ISRE DNA oligonucleotide, but rather FOXA1 bound STAT2 in a dose-dependent manner in the chromatin-free fraction of the nuclear extract (Supplemental Figure 5N, bottom). These data suggest that high-level FOXA1 inhibits the ability of STAT2 to bind to its target DNA sequence (ISRE). This notion is further supported by the results from EMSA showing that STAT2 binding of the ISRE probe was inhibited in lysates of DU145 cells transfected with WT FOXA1 and the FOXA1ΔαH3 mutant, but no such effect was observed for the FOXA1ΔSBR mutant or AR (Supplemental Figure 5O). Together, these data indicate that FOXA1 inhibits STAT2 DNA-binding ability and this function of FOXA1 does not rely on its DNA-binding ability.
FOXA1 impedes IFN-α–induced STAT2 chromatin occupancy and the effect is independent of its mutations in cancer.
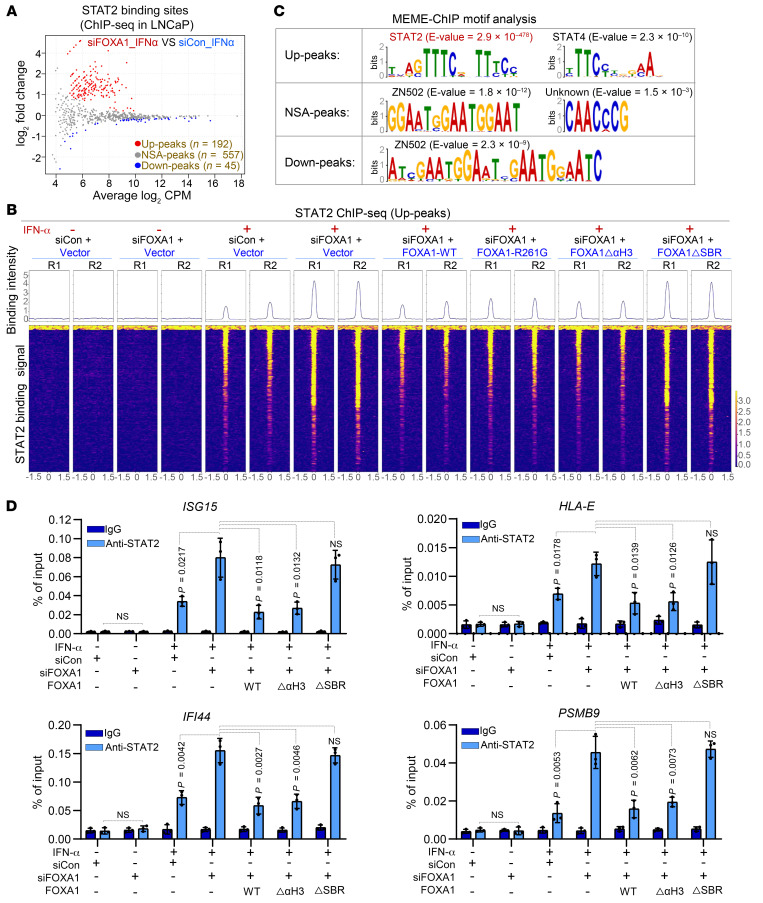
Next, we sought to determine whether FOXA1 affects STAT2 genome-wide binding on chromatin. We first knocked down the endogenous FOXA1 in FOXA1-high LNCaP cells with small interfering RNA (siRNA) specifically targeting the 3′ untranslated region (3′UTR) and then rescued with non–siRNA-targeted WT FOXA1, DNA binding–deficient mutant FOXA1ΔαH3, or STAT2 binding–deficient mutant FOXA1ΔSBR (Supplemental Figure 6A). These FOXA1 expression–manipulated cell lines were utilized for STAT2 chromatin immunoprecipitation and sequencing (ChIP-seq) analysis. The ChIP-seq replicates in each cellular condition were well correlated (Supplemental Figure 6B). We identified 794 STAT2 binding peaks, including 192 significantly upregulated peaks (termed Up-peaks), 45 downregulated peaks (termed Down-peaks), and 557 peaks with no significant alterations (termed NSA-peaks) following FOXA1 KD (Figure 3, A and B and Supplemental Figure 6, C and D). The Up-peaks tended to distribute toward gene promoters, whereas the NSA-peaks and Down-peaks appeared to go in opposite directions (Supplemental Figure 6E). MEME-ChIP motif analysis revealed that the STAT2 motif was the top hit among the Up-peaks, but no such enrichment was observed in the NSA-peaks and Down-peaks (Figure 3C). Gene ontology biological process (GO-BP) analysis revealed that the genes associated with FOXA1 KD–induced Up-peaks were highly related to STAT2-relevant pathways (such as IFN and immune responses), but no such enrichment was observed in the NSA-peaks and Down-peaks (Supplemental Figure 6F).
Figure 3. The effect of FOXA1 on STAT2 cistrome in PCa cells.
(A) MA plot of STAT2 ChIP-seq data in LNCaP cells transfected with control (siCon) or FOXA1-specific siRNAs in the presence or absence of IFN-α treatment. Red dots (Up-peaks) and blue dots (Down-peaks) represent increased and decreased peaks (FDR < 0.05), respectively, and gray dots indicate peaks with no significant alterations (NSAs) (FDR > 0.05) after FOXA1 KD. (B) Heatmaps show the signaling intensity of 192 STAT2 ChIP-seq Up-peaks in LNCaP cells under the indicated cellular conditions. (C) MEME-ChIP DNA motif analysis in 192 STAT2 ChIP-seq Up-peaks, 557 NSA-peaks, and 45 Down-peaks caused by FOXA1 KD in LNCaP cells. (D) ChIP-qPCR analysis of STAT2 occupancy at genomic loci of STAT2 target genes ISG15, IFI44, HLA-E, and PSMB9 under the indicated cellular conditions. Data shown as mean ± SD (n = 3). Statistical significance was determined by 1-way ANOVA with Bonferroni’s correction for multiple tests.
IFN-α–stimulated and FOXA1 KD–enhanced STAT2 binding on chromatin was confirmed at the canonical IFN signaling gene loci such as ISG15, IFI44, HLA-E, and PSMB9 (Supplemental Figure 7A). These results were further validated by ChIP-qPCR data in both LNCaP and MCF7 cell lines (Figure 3D and Supplemental Figure 7B). Restored expression of both WT FOXA1 and FOXA1ΔαH3 but not FOXA1ΔSBR reversed FOXA1 KD–enhanced STAT2 binding at target loci (Figure 3D and Supplemental Figure 7A). These results support the notion that FOXA1 suppresses STAT2 chromatin binding and IFN signaling in a manner dependent on FOXA1 binding of STAT2, but this effect does not require the DNA-binding activity of FOXA1.
In contrast with the FOXA1 impact on STAT2 occupancy on chromatin, IFN-α treatment had little or no effect on genome-wide chromatin engagement of FOXA1 in LNCaP cells (Supplemental Figure 8A). Less than 0.5% of total FOXA1 peaks overlapped with STAT2 binding peaks identified in LNCaP cells (Supplemental Figure 7A and Supplemental Figure 8B). FOXA1 impact on STAT2 chromatin occupancy but not vice versa could be explained, at least in part, by the observation that the abundance of STAT2 mRNA was significantly lower compared with FOXA1 mRNA in LNCaP and PCa patient samples (Supplemental Figure 8C).
These results support a model wherein FOXA1 forms a protein complex with STAT2 and inhibits STAT2 occupancy at its target gene loci containing a STAT2 DNA-binding motif, but has no obvious effect on loci without such motif. This model is consistent with the finding from the biotin-labeled-ISRE pulldown assay showing that FOXA1 inhibits STAT2 DNA-binding ability and has no association with the STAT2 DNA-binding motif (Supplemental Figure 5N). This model is also supported by the observation that STAT2 binding peaks containing a STAT2 DNA-binding motif (Up-peaks) had very minimal (≤5%) overlap with FOXA1 binding peaks identified in LNCaP and PCa patient samples (ref. 38 and Supplemental Figure 8D). These results support the working model in which FOXA1 binds to the DBD of STAT2, thereby blocking STAT2 binding of the canonical STAT2 DNA-binding motif, but not other motifs indirectly associated with STAT2 (Supplemental Figure 8E).
FOXA1 suppression of STAT2 occupancy on chromatin is independent of its mutations in cancer.
The FOXA1 gene is frequently mutated in hormone-receptor-driven cancers such as PCa and BCa (27–30). FOXA1 mutations reprogram its pioneer-factor activity and enhance PCa aggressiveness (29, 30). We first investigated whether cancer-associated FOXA1 missense mutations affect STAT2 function and IFN activity. We found that PCa-derived hotspot missense mutants of FOXA1, including FOXA1-H247Q, FOXA1-R261G, and FOXA1-F266L, were able to bind STAT2 to an extent similar to that of WT FOXA1 (Supplemental Figure 9A). Using the hotspot mutant FOXA1-R261G as a working model, STAT2 ChIP-seq data revealed that restored expression of FOXA1-R261G completely reversed FOXA1 depletion–enhanced STAT2 occupancy on chromatin in IFN-α–treated LNCaP cells, an effect similar to that of WT FOXA1 (Figure 3B and Supplemental Figure 7A). We also confirmed that, similar to WT FOXA1, restored expression of PCa-derived mutants FOXA1-H247Q and FOXA1-R261G reversed FOXA1 KD–enhanced expression of IFN signaling pathway proteins such as ISG15 and MHC I in both LNCaP and VCaP PCa cell lines (Supplemental Figure 9, B and C). We further verified the STAT2-inhibitory effect of these cancer-associated FOXA1 mutants using ISRE-based luciferase reporter assays (Supplemental Figure 9D). Moreover, there was no significant difference in expression of IFN response and APM genes between FOXA1 WT and mutated prostate and breast tumors in TCGA cohorts (Supplemental Figure 9E). Collectively, these data suggest that the PCa-derived FOXA1 missense mutants, at least those hotspot mutants we examined, retain the ability to inhibit STAT2 chromatin occupancy, IFN activity, and cancer immune response gene expression. Although a significant fraction of FOXA1 mutations in cancer are C-terminal truncation mutations (29, 30), we demonstrated that different from the SBR-deficient mutant FOXA1 (aa 1–247), the 2 PCa-associated C-terminal truncation mutants FOXA1 (aa 1–290) and FOXA1 (aa 1–268) (29, 30) retained the ability to bind to STAT2 and suppress IFN-induced STAT2 transcriptional activity (Supplemental Figure 9, F–H), suggesting that the STAT2-inhibiting effect of FOXA1 is also not affected by FOXA1 C-terminal truncation mutations.
Identification of FOXA1-affected STAT2 target genes via transcriptome analysis.
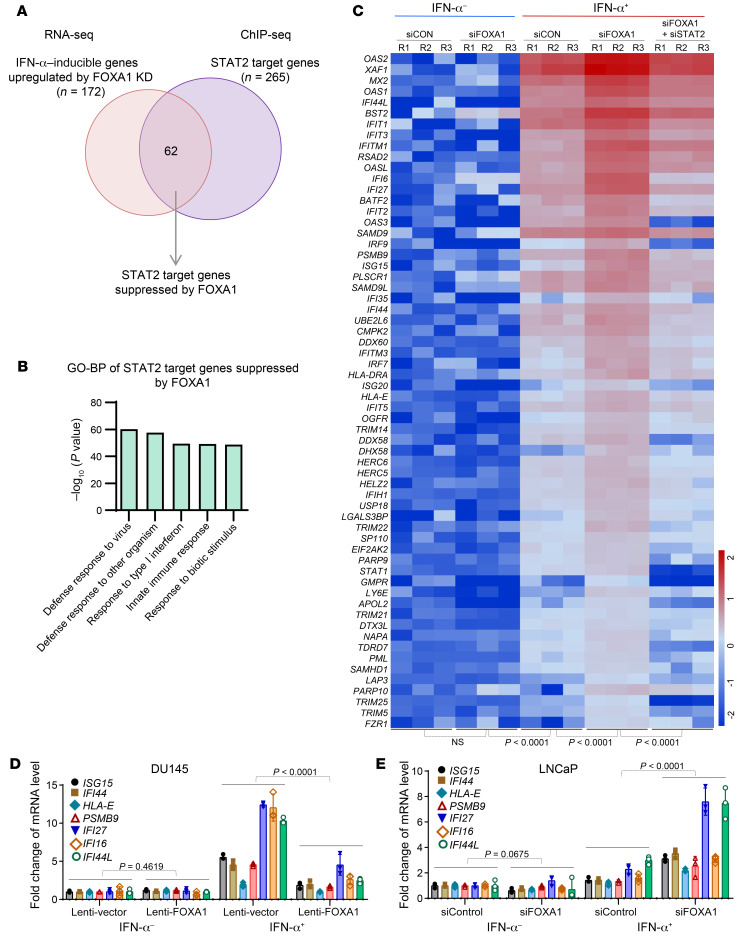
We globally assessed the impact of FOXA1 on STAT2 target gene expression at the transcriptional level. We knocked down FOXA1 in LNCaP cells in the presence or absence of IFN-α treatment and harvested these cells for RNA-seq analysis. The replicates of RNA-seq data correlated very well (Supplemental Figure 10A). Through unsupervised cluster analysis of differentially expressed genes, we identified a subset of IFN-α–stimulated genes (n = 172), the expression of which was significantly increased after FOXA1 KD (Supplemental Figure 10B). We further performed integrated analysis of STAT2 ChIP-seq and RNA-seq data and identified 62 FOXA1-suppressed STAT2 target genes (Figure 4A). GO-BP analysis revealed that this set of genes was strongly relevant to IFN or immune responses (Figure 4B). Importantly, we found that FOXA1 KD induced upregulation of this set of genes in LNCaP cells treated with IFN-α; however, this effect was reversed by STAT2 co-KD (Figure 4C and Supplemental Figure 10C). By performing RT-qPCR analysis, we confirmed that FOXA1 overexpression decreased the expression of a subset of STAT2 target genes in FOXA1-negative DU145 cells and that FOXA1 KD in FOXA1-high LNCaP cells increased expression of this subset of genes under IFN-α stimulation (Figure 4, D and E). Similar to the impact on the FOXA1 cistrome due to lower abundance of STAT2 compared with FOXA1 (Supplemental Figure 8), IFN-α treatment or STAT2 KD had little or no effect on expression of FOXA1 target genes (both up- and downregulated) in LNCaP cells (Supplemental Figure 11). These results indicate that FOXA1 inhibits expression of a set of IFN-α–stimulated STAT2 target genes that are highly related to IFN signaling and immune response.
Figure 4. Identification of IFN-α response genes suppressed by FOXA1.
(A) Venn diagram showing the overlap of IFN-α–stimulated genes upregulated by FOXA1 KD with the STAT2 target genes identified by STAT2 ChIP-seq. (B) GO-BP pathway analysis of the 62 STAT2 target genes suppressed by FOXA1. (C) Heatmap shows the differential expression of the 62 STAT2 target genes suppressed by FOXA1 in LNCaP cells under the indicated cellular conditions. (D and E) RT-qPCR analysis of expression of STAT2 target genes ISG15, IFI44, HLA-E, PSMB9, IFI27, IFI16, and IFI44L in FOXA1-negative DU145 cells infected with lentivirus expressing vector or FOXA1 (D) and in FOXA1-high LNCaP cells transfected control or FOXA1-specific siRNAs (E). Data shown as mean ± SD (n = 3). Statistical significance was determined by 1-way ANOVA.
FOXA1 overexpression suppresses anticancer immune responses in mice and patients.
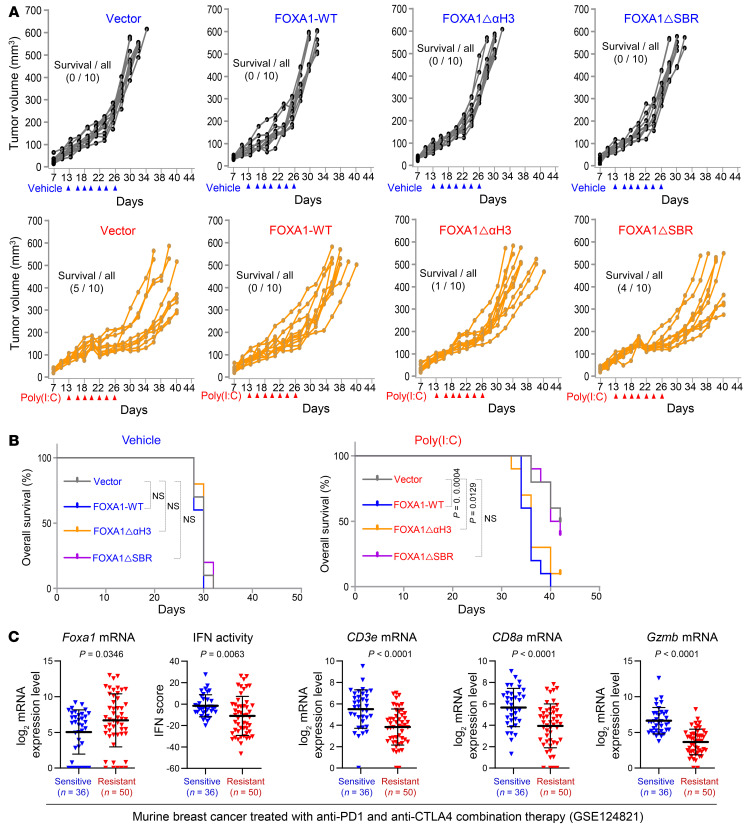
To determine whether FOXA1 inhibits anticancer immune responses in vivo, we generated murine PCa TRAMP-C2 (lacking endogenous Foxa1 expression) stable cell lines expressing control vector, WT FOXA1, DNA binding–deficient mutant FOXA1ΔαH3, or STAT2 binding–deficient mutant FOXA1ΔSBR for animal studies. Stable FOXA1 expression in these cell lines and their response to IFN-α were confirmed by Western blot analysis (Supplemental Figure 12A) before they were injected s.c. into syngeneic C57BL/6 male mice. Poly(I:C) was intratumorally injected to trigger a type I IFN immune response (39). Poly(I:C) administration decreased the growth of control (TRAMP-C2-Vector) tumors in the majority of mice and prolonged the overall mouse survival (Figure 5, A and B, and Supplemental Figure 12B). In contrast, the growth-inhibiting effect of poly(I:C) was largely diminished in tumors with overexpression of WT FOXA1 and FOXA1ΔαH3 but not FOXA1ΔSBR (Figure 5, A and B, and Supplemental Figure 12B). Furthermore, TILs, especially CD8+ T cells (including cytotoxic [GZMB+] CD8+ T cells), were discernibly increased in TRAMP-C2-Vector tumors treated with poly(I:C); however, such an effect was diminished in tumors with overexpression of WT FOXA1 and FOXA1ΔαH3 but not FOXA1ΔSBR (Supplemental Figure 12, C and D, and Supplemental Figure 13A). The tumor response to poly(I:C) treatment and FOXA1 expression was further reflected in expression of murine IFN signaling genes including Isg15, Ifi44, H2-k1, and Psmb9 (Supplemental Figure 13B). It is worth noting that FOXA1 and FOXA subfamily members FOXA2 and FOXA3 share similar FKHDs (DNA binding) and the same SBR motifs in both human and mouse (Supplemental Figure 14A). Like FOXA1, both FOXA2 and FOXA3 also interacted with and suppressed IFN-induced transcriptional activity of STAT2 (Supplemental Figure 14, B and C). It has been reported that FOXA2 is expressed in neuroendocrine PCa (NEPC) (40). Compared with the typical NEPC cell line NCI-H660 and NE-like cell line PC-3, TRAMP-C2 cells express relatively low levels of Foxa2 (Supplemental Figure 14D). The limited Foxa2 expression along with the observation that little or no Foxa1 is expressed in TRAMP-C2 cells is consistent with the finding that TRAMP-C2 tumors elicited measurable anticancer immune responses in mice. Consistent with these observations in the murine PCa model, meta-analysis of data reported previously (41) showed that Foxa1 mRNA levels were much higher in resistant tumors compared with sensitive tumors, while the IFN activity and expression of effector T (Teff) cell markers such as CD3e, CD8a, and Gzmb positively correlated with the response of murine breast tumors to anti–PD-1 and anti-CTLA4 combined therapy (Figure 5C). These data support a role of Foxa1 in negatively regulating IFN signaling and immune response in PCa and BCa in mice.
Figure 5. FOXA1 overexpression suppresses PCa immune response in mice.
(A) Growth of TRAMP-C2 prostate tumors stably expressing vector, WT FOXA1, FOXA1ΔαH3, or FOXA1ΔSBR treated with vehicle or poly(I:C) at the indicated time points (arrowheads) in C57BL/6 mice. (B) Tumor-free survival of syngeneic mice bearing TRAMP-C2 tumors stably expressing vector, WT FOXA1, FOXA1ΔαH3, or FOXA1ΔSBR treated with vehicle or poly(I:C). Statistical significance was determined by log-rank (Mantel-Cox) test. (C) Analysis of RNA-seq data from a cohort of murine BCa (GSE124821) showing the association of high expression of Foxa1 and low expression of IFN response genes (IFN activity) and CD3e, CD8a, and Gzmb T cell marker genes with the responsiveness to anti–PD-1 and anti-CTLA4 combination therapy (resistant versus sensitive). Statistical comparison was done using Mann-Whitney U test.
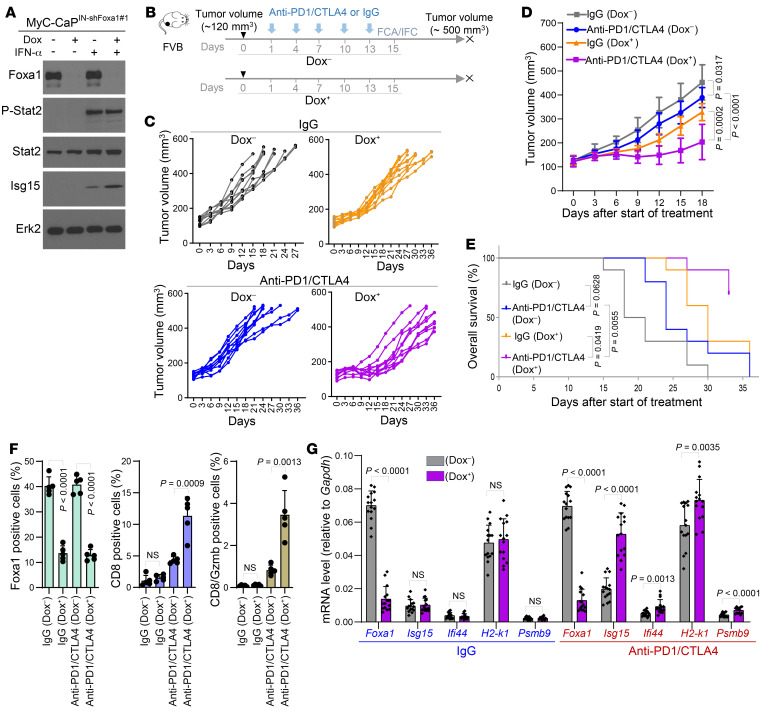
Next, we examined whether FOXA1 overexpression confers resistance to ICI immunotherapy. We effectively knocked down endogenous Foxa1 in the murine MyC-CaP PCa cell line using doxycycline-inducible shRNAs and demonstrated that Foxa1 KD enhanced IFN signaling in MyC-CaP cells following IFN-α treatment (Figure 6A and Supplemental Figure 15A). We generated a syngeneic PCa mouse model using these stable MyC-CaP cell lines and treated mice with anti–PD-1 and anti-CTLA4 antibodies in combination with or without doxycycline. We demonstrated that KD of Foxa1 by doxycycline treatment enhanced the therapeutic efficacy of anti–PD-1 and anti-CTLA4 in MyC-CaP tumors (Figure 6, B–E, and Supplemental Figure 15, B and C). KD of Foxa1 also increased IFN response gene expression and the accumulation of cytotoxic CD8+ cells in MyC-CaP tumors (Figure 6, F and G, and Supplemental Figure 15, B and C). These data not only indicate that FOXA1 mediates resistance to ICI immunotherapy, but also imply that FOXA1 is a viable target to sensitize PCa to ICIs.
Figure 6. Foxa1 knockdown sensitizes murine PCa to anti–PD-1 and anti-CTLA4 combination therapy.
(A) Western blot analysis of indicated proteins in MyC-CaP murine PCa cells stably expressing doxycycline-inducible lentiviral shFoxa1#1 (MyC-CaPIN-shFoxa1#1) and treated with or without doxycycline (Dox) or/and IFN-α. Erk2 served as a loading control. (B) Schematic diagram of generation and anti–PD-1/anti-CTLA4 treatment of MyC-CaPIN-shFoxa1#1 prostate tumors in syngeneic mice. Dox–, without doxycycline treatment; Dox+, with doxycycline treatment; FCA, flow cytometric analysis; IFC, immunofluorescent cytochemistry. (C and D) Growth of MyC-CaPIN-shFoxa1#1 prostate tumors treated with IgG or combination of anti–PD-1/anti-CTLA4 at the indicated time points (arrowheads) in FVB mice (n = 10 mice/group). Statistical significance was determined by 2-way ANOVA. (E) Tumor-free survival of syngeneic mice bearing MyC-CaPIN-shFoxa1#1 prostate tumors treated with IgG or anti–PD-1/anti-CTLA4 (n = 10 mice/group). Statistical significance was determined by log-rank (Mantel-Cox) test. (F) Flow cytometric analysis of Foxa1-, CD8-, and Gzmb-positive cells in MyC-CaPIN-shFoxa1#1 tumors from mice 2 days after the last administration of IgG or anti–PD-1/anti-CTLA4. Data are shown in the bar graphs as mean ± SD (n = 5 mice/group). Statistical significance was determined by 1-way ANOVA with Bonferroni’s correction for multiple tests. (G) RT-qPCR analysis of Foxa1 and murine Stat2 target genes Isg15, Ifi44, H2-k1, and Psmb9 in MyC-CaPIN-shFoxa1#1 tumors from mice 2 days after the last administration of IgG or anti–PD-1/anti-CTLA4. The data are presented as the mean ± SD (n = 5 mice/group). Statistical significance was determined by 1-way ANOVA with Bonferroni’s correction for multiple tests.
To validate our findings from animal studies in clinical settings, we performed meta-analysis of the clinical data from castration-resistant prostate cancer (CRPC) patients treated with personalized peptide vaccines (PPVs) (42). PPVs constitute an immunotherapy that uses multiple cancer peptides knowingly associated with preexisting host immunity. A previous phase III study suggests that the median overall survival in PPV-treated patients with high lymphocyte counts was significantly longer than placebo-treated patients (43). By analyzing the gene expression and clinical data in the patients pretreated with PPV, we demonstrated that patients with high expression of FOXA1 mRNA had lower overall survival (Supplemental Figure 15D). We further showed that expression of IFN response and CTL marker genes such as CD3E, PRF1, and GZMB were significantly lower in FOXA1-high tumors compared with FOXA1-low counterparts (Supplemental Figure 15D), supporting a possible IFN signal–inhibiting and immune-suppressing function of FOXA1 in PCa patients.
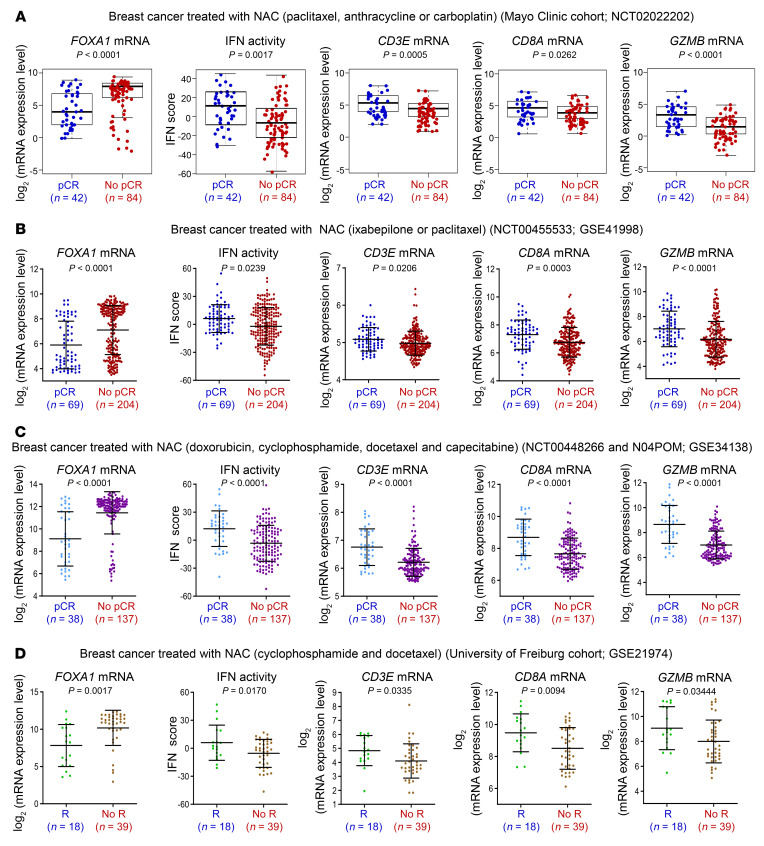
Activation of IFNs is required for anticancer immune responses elicited by chemotherapy and the IFN-related genetic signature correlates with clinical responses to this chemotherapeutic regimen in BCa (44). Immune responses are also implicated in chemotherapy of BCa because higher levels of TILs prior to neoadjuvant chemotherapy (NAC) are associated with higher rates of pathological complete response (pCR) (45, 46). To explore whether FOXA1 associates with the NAC-associated immune response, we analyzed RNA-seq data from the prospective BEAUTY study of BCa patients treated with NAC at the Mayo Clinic (ref. 47 and Supplemental Figure 16A). We observed that FOXA1 mRNA levels were significantly higher in tumors without pCR (No-pCR) compared with tumors with pCR, whereas the IFN activity was opposite (Figure 7A). In comparison with pCR tumors, No-pCR tumors expressed much lower levels of Teff cell markers such as CD3E, CD8A, and GZMB while having much higher expression of FOXA1 (Figure 7A), supporting a role of FOXA1 in negatively regulating IFN signaling and immune response in BCa patients. The association of high FOXA1 expression with low IFN activity and NAC resistance was also observed in 3 independent BCa cohorts (Figure 7, B–D). Almost all luminal tumors in the Mayo Clinic cohort expressed FOXA1 at high levels, but had a very low rate of pCR (no single pCR case in luminal-A tumors and only 10.8% pCR rate in luminal-B tumors; Supplemental Figure 16, A and B). FOXA1 is generally considered a luminal gene. However, we did find that it was expressed in triple-negative breast cancer (TNBC), although the expression level was lower compared with that in luminal subtypes (Supplemental Figure 16B). Although the overall pCR rate was very high (64%) in TNBC (Supplemental Figure 16A), there was no statistically significant difference in FOXA1 expression between pCR and No-pCR in the TNBC cohort (Supplemental Figure 16B). One possible reason could be the small sample size of TNBC cases (n = 42) in the Mayo Clinic cohort. This notion is supported by the finding from a larger cohort (n = 119) of NAC-treated TNBC where FOXA1 mRNA level was significantly higher in No-pCR TNBC than those with pCR (Supplemental Figure 16C). Intriguingly, neither NAC responsiveness nor FOXA1 expression was associated with TMB in the Mayo Clinic cohort (Supplemental Figure 16, D and E). These results suggest that FOXA1 overexpression confers resistance to NAC.
Figure 7. FOXA1 overexpression associates with resistance of neoadjuvant chemotherapy (NAC) in cancer.
(A–C) Analysis of RNA-seq data from a cohort of 126 BCa patients at the Mayo Clinic (A) and microarray data from 2 independent cohorts of BCa (GSE41998 and GSE34138) (B and C) showing the association of high expression of FOXA1 and low expression of IFN response genes (IFN activity) and CD3E, CD8A, and GZMB T cell marker genes with the responsiveness to NAC (No-pCR versus pCR). pCR, pathological complete response; No-pCR, no pathological complete response. Statistical comparison was done using Mann-Whitney U test. (D) Analysis of microarray data from a cohort of BCa (GSE21974) showing the association of high expression of FOXA1 and low expression of IFN response genes (IFN activity) and CD3E, CD8A, and GZMB T cell marker genes with the responsiveness to NAC (No-R versus R). R, response; No-R, no response. Statistical comparison was done using Mann-Whitney U test.
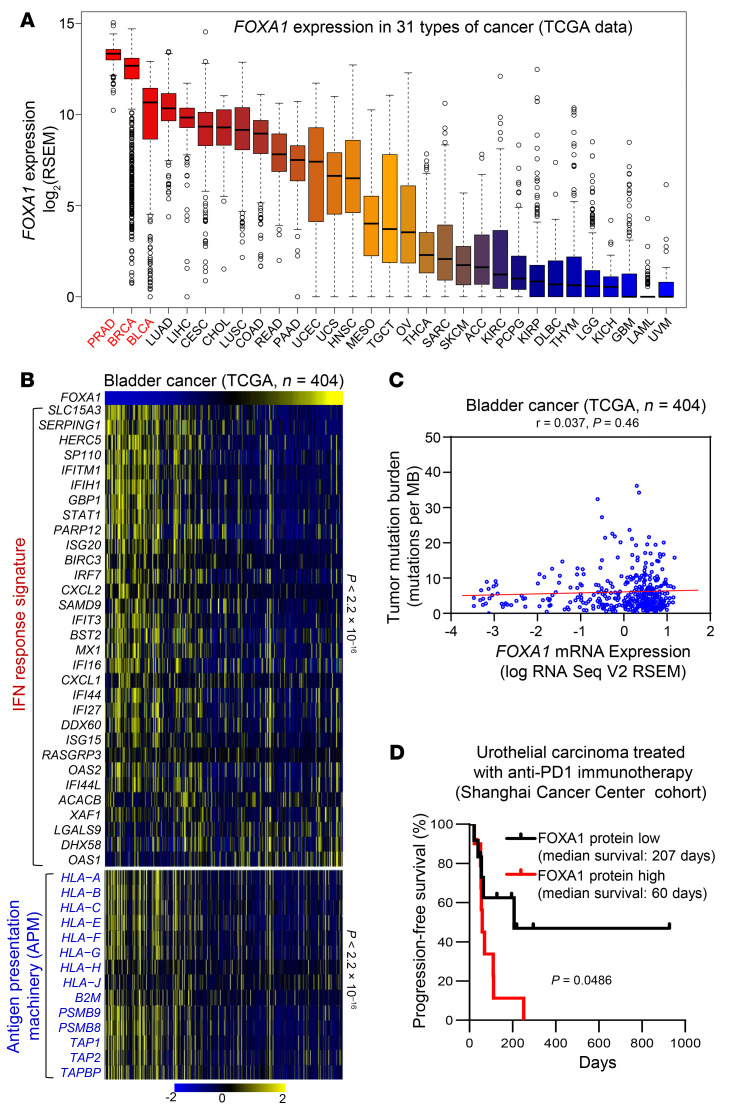
In addition to PCa and luminal BCa, 2 steroid hormone–driven cancers, bladder cancer is the third cancer type in TCGA database that has the highest expression of FOXA1 (Figure 8A). Therefore, we were also interested in evaluating the association of FOXA1 with IFN activation and ICI immunotherapy response in bladder cancer, a malignancy not necessarily driven by AR or ER. Similar to the results in PCa and BCa, FOXA1 expression inversely correlated with the levels of IFN response signature and APM genes, but not TMB (Figure 8, B and C). FOXA1 can also bind to STAT proteins and suppress IFN signaling in bladder cancer cells (Supplemental Figure 17). Furthermore, we examined FOXA1 protein expression using immunohistochemistry (IHC) in a set of 22 urothelial carcinomas of the bladder, ureter, and renal pelvis collected prior to anti–PD-1 therapy. We found that patients with tumors expressing higher FOXA1 protein levels had much lower rates of progression-free survival (Figure 8D, Supplemental Figure 18, and Supplemental Table 3). Similar results were also obtained from bladder cancer patients treated with Bacillus Calmette-Guerin (BCG) immunotherapy (ref. 48 and Supplemental Figure 19A) or chemotherapy consisting of methotrexate, vinblastine, doxorubicin, and cisplatin (MVAC; ref. 49 and Supplemental Figure 19B). Together, the data from PCa, BCa, and bladder cancer patients suggest that FOXA1 overexpression contributes to immune evasion and immuno- and chemotherapy resistance in the clinic.
Figure 8. FOXA1 confers immunotherapy resistance in bladder cancer.
(A) Comparison of FOXA1 mRNA level among 31 cancer types in TCGA database, including PRAD (prostate adenocarcinoma), BRCA (breast invasive carcinoma), BLCA (bladder urothelial carcinoma), LUAD (lung adenocarcinoma), LIHC (liver hepatocellular carcinoma), CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma), CHOL (cholangiocarcinoma), LUSC (lung squamous cell carcinoma), COAD (colon adenocarcinoma), READ (rectum adenocarcinoma), PAAD (pancreatic adenocarcinoma), UCEC (uterine corpus endometrial carcinoma), UCS (uterine carcinosarcoma), HNSC (head and neck squamous cell carcinoma), MESO (mesothelioma), TGCT (testicular germ cell tumors), OV (ovarian serous cystadenocarcinoma), THCA (thyroid carcinoma), SARC (sarcoma), SKCM (skin cutaneous melanoma), ACC (adrenocortical carcinoma), KIRC (kidney renal clear cell carcinoma), PCPG (pheochromocytoma and paraganglioma), KIRP (kidney renal papillary cell carcinoma), DLBC (lymphoid neoplasm diffuse large B-cell lymphoma), THYM (thymoma), LGG (brain lower grade glioma), KICH (kidney chromophobe), GBM (glioblastoma multiforme), LAML (acute myeloid leukemia), and UVM (uveal melanoma). (B) Heatmaps show the negative correlation of FOXA1 expression with the expression levels of IFN response signature genes and APM genes in TCGA cohort of bladder cancers. Samples are ranked based on FOXA1 transcript levels. Statistical significance was determined by Pearson’s correlation test. (C) The correlation between FOXA1 level and TMB in TCGA cohort of bladder cancers. Statistical significance was determined by Pearson’s correlation test. (D) Progression-free survival of patients with FOXA1-low or -high urothelial carcinomas treated with anti–PD-1 immunotherapy. Statistical significance was determined by log-rank (Mantel-Cox) test. See also Supplemental Figure 18 and Supplemental Table 3 for FOXA1 IHC staining and patient clinical information, respectively.
Discussion
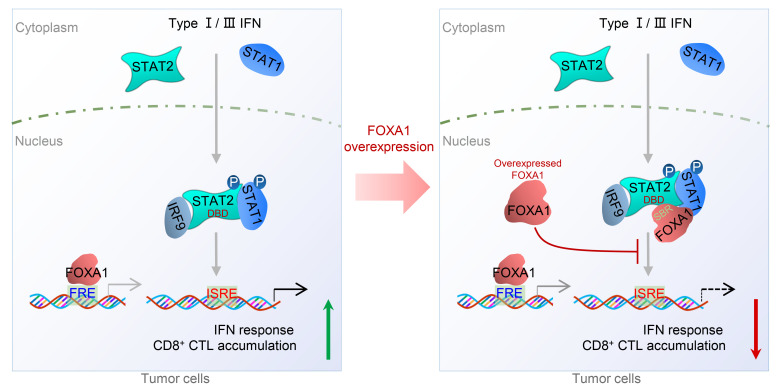
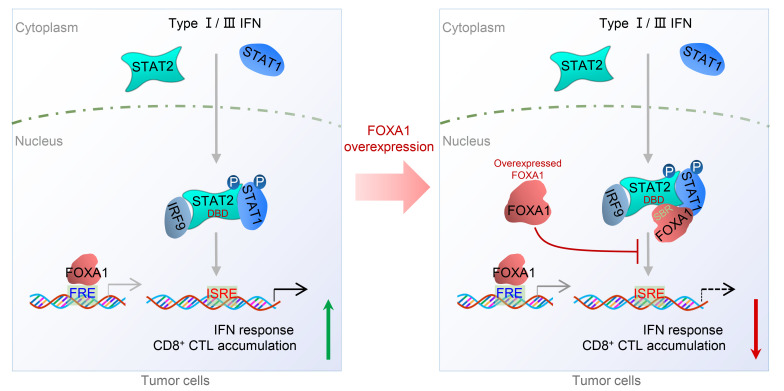
Genetic inactivation (deletions or inactivation mutations) of genes encoding IFN response/receptor and antigen presentation pathway proteins has been linked to immune evasion of human cancers such as melanoma (7, 18, 22, 23). In the present study, we demonstrate that FOXA1 directly binds a large portion (more than two-thirds) of the STAT2 DBD and inhibits STAT2 binding of its DNA-binding sequence (ISRE). Through genome-wide ChIP-seq and RNA-seq analyses, we further show that overexpressed FOXA1 prohibits STAT2 occupancy on gene loci harboring a typical STAT2 DNA-binding motif and impedes expression of STAT2 targets including IFN signaling and APM genes. We also provide evidence that FOXA1 overexpression not only inhibits anticancer immune responses in mice, but also associates with decreased expression of IFN response and CTL marker genes in patient samples. Our findings therefore reveal FOXA1 overexpression–mediated inhibition of STAT2 DNA-binding activity and transcriptional suppression of IFN signaling and antigen presentation gene expression as a mechanism of cancer cell escape from immune surveillance (Figure 9).
Figure 9. A hypothetical model deciphering FOXA1 overexpression–mediated suppression of IFN signaling and cancer immune response.
Upon stimulation of cells with type I/III IFNs, STAT1 and STAT2 proteins are phosphorylated, dimerize (STAT2-STAT1 heterodimer), and translocate into the nucleus to initiate the transcription of IFN-stimulated genes (ISGs) by binding to the ISRE and ultimately promote cancer immune response (left). However, in FOXA1-overexpressing tumor cells, the overexpressed FOXA1 protein, in a manner independent of its binding to chromatin DNA, reduces the accessibility of the STAT protein complex and impairs ISG expression, thereby suppressing cancer immune response (right). FRE, forkhead response element; ISRE, IFN stimulation response element; CTL, cytotoxic T lymphocyte; DBD, DNA-binding domain; SBR, STAT2-binding region; P, phosphorylation.
TMB has been linked to neoantigen availability and immune response to ICIs in various human cancers, especially those cancer types with high TMB such as melanoma and non–small cell lung cancer (50–52). It has been confirmed by multiple studies that PCa possesses a lower TMB rate than many other epithelial tumor types (27, 50, 53). Low TMB appears to be a possible reason for the coldness of PCa to immunotherapy. However, response to ICIs is not significantly associated with TMB in some cancers such as BCa (4). Increasing evidence indicates that high IFN-γ–response gene expression is necessarily required for responses to ICIs in PCa with low TMB (6). Of note, a recent clinical study showed that responders do not have significantly higher TMB than nonresponders when stratified by melanoma subtypes (5). In our study, we found that there was little or no association of TMB with FOXA1 expression, infiltration of CD8+ Teff cells, or NAC efficacy in different BCa cohorts. These observations suggest that TMB is not the only major player affecting the therapeutic efficiency of ICIs, but other factors such as IFN activation in cancer are also critical for ICI responses. This notion is further supported by our finding that FOXA1 overexpression causes IFN suppression, immune evasion, and immunotherapy resistance in cancer. However, a limitation in our study in bladder cancer is that we do not have high-throughput sequencing data of TMB in tumor samples treated with anti–PD-1 immunotherapy and therefore future investigation is warranted to determine whether or not FOXA1 overexpression correlates with anti–PD-1 resistance in bladder cancer patients with high TMB.
FOXA1 is a well-known pioneer factor, owing to its function in activation of steroid hormone receptors such as AR and ER, and its expression is essential for maintenance of the luminal phenotype of PCa and BCa (24–26). AR+ PCa and ER+ BCa are generally less responsive to immunotherapies (2, 3, 54). It is highly relevant to explore whether the immune nonresponsiveness in these 2 cancer types is related to luminal epithelial origins, hormone dependence, or other factors commonly present in PCa and BCa such as the pioneer factor FOXA1. We demonstrated that FOXA1 expression inversely correlates with expression of IFN response and APM genes in both PCa and BCa; however, the association is unrelated to tumor stages such as primary PCa versus CRPC, suggesting that the correlation appears to be not entirely hormone related. A high level of TILs has been shown to be associated with favorable response to NAC in BCa (45, 55). Similar to the response to immunotherapy in PCa, we found that a high level of FOXA1 also associates with reduced IFN activity and T cell infiltration in BCa treated with NAC. Although BCa can be divided into multiple molecular subtypes with distinct traits such as luminal versus TNBC, similar to the outcome in luminal BCa, a significant association of high-level FOXA1 expression with NAC response was also detected in TNBC even when the luminal AR+ TNBC was excluded. Thus, our data show that FOXA1 overexpression rather than luminal epithelial origins or hormone dependence is one of the key factors contributing to the immunotherapy nonresponsiveness in both PCa and BCa.
The importance of the pioneer-factor function of FOXA1 in PCa and BCa is further accentuated by the finding that the FOXA1 gene is frequently mutated in human cancers such as PCa and BCa (27–30). FOXA1 mutants are reported to drive proliferation of cancer cells or organoids cultured in vitro and tumor growth in immune-deficient mice in a manner dependent on its pioneer-factor activities such as DNA binding and chromatin regulation (29, 30). However, the role of FOXA1 mutations in regulation of cancer immune response has not been explored to our knowledge. Our findings from cancer cell lines, mouse tumors, and patient samples indicate that FOXA1 inhibition of IFN and APM signaling is independent of the missense and C-terminal truncation mutations of FOXA1 detected in human cancers. Moreover, we provide evidence that FOXA1 suppresses IFN signaling gene expression and cancer immune response and that this effect does not depend on its DNA-binding function. Our results indicate that besides via its gene mutation–mediated cell proliferation and invasion, FOXA1 can also drive cancer progression through overexpression-mediated suppression of IFN signaling and inhibition of cancer immune response in a manner independent of its pioneer-factor function (Figure 9).
In summary, we identify FOXA1 overexpression–mediated suppression of STAT2 DNA binding and IFN-signaling gene expression as a mechanism of immune evasion in different cancer types including PCa, BCa, and bladder cancer. This role of FOXA1 is neither dependent on its pioneer-factor function nor its mutations detected in human cancers. Our findings stress that prospective studies are warranted to investigate whether FOXA1 overexpression could be a potential prognostic factor to predict immunotherapy response in a manner independent of luminal cell origins or TMB of tumors. Our results also imply that further exploration is needed to determine whether eliminating FOXA1 protein could be a viable strategy to turn FOXA1-overexpressed “immune-cold” tumors into “immune-hot” tumors. Pioneer factors such as FOXA1 are notoriously difficult to target and currently there are no clinical-grade agents that target FOXA1. The emerging technology proteolysis targeting chimeras (PROTACs), especially the olionucleotide-based PROTACs (56), could be a promising option to develop novel anticancer agents to target FOXA1.
Methods
scRNA-seq data analysis.
Detailed clinical information of PCa samples used for scRNA-seq is provided in Supplemental Table 1. For data generated with the 10× Genomics platform, raw data were processed using Cell Ranger v2.1.0 (10× Genomics) into the unique molecular identifier (UMI) matrix. The maximum number of cells in one sample was estimated by Cell Ranger across 13 samples and was used as the estimation of cell number. For each cell, we calculated 3 quality measures: the percentage of total genes that were mitochondrial genes, number of total genes expressed, and number of housekeeping genes expressed (57). We removed cells that had higher than 20% expression of mitochondrial genes, fewer than 200 or greater than 5218 total genes expressed, or fewer than 56 housekeeping genes expressed (57). The resultant gene expression matrix was imported into the R v3.5.1 (https://www.r-project.org/) statistical environment for further analysis. Data normalization and annotation were performed using R package scran (v1.10.2). To calculate the pool-based normalization factor, cells for each sample were first split into sensible clusters using the quickCluster function (maximum size = 3000). The fastMNN function (k = 5, d = 50, approximate = TRUE, auto.order = TRUE) was used to apply a mutual nearest-neighbor method to correct for batch effect among samples. Similarly, the scRNA-seq data from BCa (GSE140819 including GSM4186971, GSM4186973, GSM4186975, GSM4186977, GSM4186972, and GSM4186980; ref. 35) were analyzed accordingly. To investigate the expression correlation between FOXA1 level and the expression of IFN response signature genes as well as APM genes, these genes were ranked according to the increased levels of FOXA1 transcript revealed by the scRNA-seq data from PCa or BCa as described above, and heatmaps were generated accordingly.
IFN activity score and gene expression correlation analysis.
To calculate the IFN activity score and other gene expression activity scores, gene expression values (log2[FPKM + 1]) of each sample were converted to z scores using z = (x – μ)/σ, where μ is the average gene expression value across all samples of a gene and σ is the standard deviation of the gene expression value across all samples of a gene. The z scores were then summed across all genes for each sample to represent the gene expression activity score. The summed z scores were converted to a percentile and normalized between 0 and 1, with 0 being the lowest and 1 being the highest (Supplemental Figure 1, D and E). The IFN response signature for activity genes (ACACB, BIRC3, BST2, CXCL1, CXCL2, DDX60, DHX58, GBP1, HERC5, IFI16, IFI27, IFI44, IFI44L, IFIH1, IFIT3, IFITM1, IRF7, ISG15, ISG20, LGALS9, MX1, OAS1, OAS2, PARP12, RASGRP3, SAMD9, SERPING1, SLC15A3, SP110, STAT1, and XAF1), which has been shown to be associated with favorable prognosis in melanoma (39), and APM genes (B2M, HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, HLA-G, HLA-H, HLA-J, PSMB8, PSMB9, TAP1, TAP2, and TAPBP) were included in the analyses unless the expression data of specific gene(s) were not available in the data set.
To identify which factors contribute to the suppression of IFN activity in immunologically cold tumors, we analyzed RNA-seq expression data from TCGA cohorts of PCa (n = 490; ref. 53) and BCa (n = 960; ref. 58) and generated a list of genes whose expression negatively correlated with IFN activity by performing Spearman’s ρ rank analysis. Genes commonly present in the list of the top hits in both PCa and BCa cohorts were considered further as the potential candidate(s) that may be able to suppress IFN activity in immunologically cold cancers (such as PCa and BCa). All data were derived from cBioPortal (http://www.cbioportal.org/) (59).
To investigate the expression correlation between FOXA1 level and IFN response signature genes or APM genes, these genes were ranked according to the increased levels of FOXA1 transcript revealed by bulk tissue RNA-seq data from PCa (TCGA, n = 490; ref. 53), metastatic PCa (SU2C/PCF Dream Team, source of file: data_mRNA_seq_fpkm_polya.txt, n = 270; ref. 60), bone metastatic PCa from the Mayo Clinic (dbGaP: phs001141.v1.p1, n = 54; ref. 61), BCa from TCGA (n = 960; ref. 58), BCa from the METABRIC database (n = 1904; ref. 62), and bladder cancer from TCGA (n = 404; ref. 63), and heatmaps were generated accordingly. The expression levels of IFN response signature genes and APM genes were scored. Their correlations with FOXA1 expression were determined based on the Pearson’s r values and P values.
ChIP-seq and bioinformatics analyses.
For ChIP experiments, cells were cross-linked for 15 minutes at room temperature with 1% formaldehyde/PBS solution. Cross-linked chromatin was sonicated, diluted, and immunoprecipitated with Protein A/G–agarose prebound with antibodies at 4°C overnight. Antibodies used for ChIP were anti-STAT2 (2 μg/sample; 72604S, Cell Signaling Technology) and anti-FOXA1 (2 μg/sample; ab23738, Abcam). Precipitated protein-DNA complexes were eluted and cross-linking was reversed at 65°C for 12 hours. High-throughput sequencing (51 nt, paired-end) was performed using the Illumina HiSeq 4000 platform at the Mayo Clinic Genome Core Facility. All short reads were mapped to the human reference genome (GRCh38/hg38) using bowtie2 v2.1.0 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) with default configurations. Reads mapped to multiple positions (greater than 2) were discarded, and the remained reads were used for peak calling using MACS2 (version 2.0.10) with a P-value cutoff of 1 × 10–5 (macs2 call peak -bdg -SPTMR -f BAM -p 1e-5; ref. 64). Peaks located in blacklists such as centromere regions were removed (https://sites.google.com/site/anshulkundaje/projects/blacklists). ChIP-seq tag intensity tracks (bedGraph files) were generated by MACS2, and converted into bigWig files using the UCSC wigToBigWig tool (https://genome.ucsc.edu/goldenPath/help/bigWig.html). Heatmaps were drawn by deepTools 2.0 (https://deeptools.readthedocs.io/en/develop/). A set of peaks identified by ChIP-seq were validated by qPCR using Power SYBR Green (Thermo Fisher Scientific, 4368708). Primer sequences used for ChIP-qPCR are listed in Supplemental Table 4.
Statistics.
GraphPad Prism version 8.0 or SPSS version 17.0 (IBM) was used for statistical analyses of results from RT-qPCR, luciferase reporter, and cell proliferation assays. Other statistical analyses were conducted in the statistical computing environment R (v. 3.3.1). Statistical comparison was done using the Mann-Whitney U test or 1-way analysis of variance (ANOVA) with Bonferroni’s correction for multiple tests. Because treatment and time course were investigated, 2-way ANOVA was also applied. Spearman’s ρ correlation test and Pearson’s correlation test were used for evaluating the statistical significance of correlations. Statistical analysis is specifically described in the figure legends. A P value of less than 0.05 was considered significant.
Study approval.
The protocol for the animal experiments including the TRAMP-C2 murine PCa model and MyC-CaP murine PCa model was approved by the Mayo Clinic Institutional Animal Cure and Use Committee (IACUC). The protocol for study of urothelial carcinoma treated with anti–PD-1 immunotherapy was approved by the Ethical and Scientific Committee (equivalent to Institutional Review Board [IRB]) at Fudan University Shanghai Cancer Center. All patients provided written informed consent before enrolling in the study.
Author contributions
HH and YH conceived of and designed the study. YH, SR, MPG, DPH, and SH acquired data. YH, Liguo Wang, TW, YTX, DPH, KRK, ZY, H Sheng, and XT analyzed and interpreted the data. HH, SR, YH, MPG, CMP, HD, VJS, Liguo Wang, TW, and YY prepared the manuscript. JMC, DY, HD, H Su, JM, SW, HX, YP, Liewei Wang, RW, JCB, SJW, XH, XZ, AHB, and CMP provided administrative, technical, or material support. HH and SR supervised the study.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health grants R01 CA130908 and CA203849 (to HH); the Mayo Clinic Foundation (to HH); Mayo Clinic Breast SPORE grant P50CA116201-9 (to MPG, Liewei Wang, KRK, and VJS); Mayo Clinic Cancer Center Support grant P50CA015083; Mayo Clinic Center for Individualized Medicine; Nadia’s Gift Foundation; John P. Guider; The Eveleigh Family; George M. Eisenberg Foundation for Charities; Prospect Creek Foundation; and the Randy Shaver Cancer Research and Community Fund. YH is supported by a Mayo Clinic Edward C. Kendall Fellowship.
Version 1. 06/08/2021
In-Press Preview
Version 2. 07/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(14):e147025.https://doi.org/10.1172/JCI147025.
Contributor Information
Yundong He, Email: He.yundong@mayo.edu.
Liguo Wang, Email: wang.liguo@mayo.edu.
Ting Wei, Email: wei.ting@mayo.edu.
Yu-Tian Xiao, Email: xiaoyutian212@gmail.com.
Haoyue Sheng, Email: sheng.haoyue@mayo.edu.
Hengchuan Su, Email: suhengchuan@163.com.
Daniel P. Hollern, Email: hollernd@email.unc.edu.
Xiaoling Zhang, Email: xiaolingzhang@jlu.edu.cn.
Jian Ma, Email: majian0922@gmail.com.
Simeng Wen, Email: simeng_wen@tmu.edu.cn.
Hongyan Xie, Email: hxie6@mgh.harvard.edu.
Yuqian Yan, Email: Yan.Yuqian@mayo.edu.
Yunqian Pan, Email: pan.yunqian@mayo.edu.
Xiaonan Hou, Email: Hou.Xiaonan@mayo.edu.
Xiaojia Tang, Email: Tang.Xiaojia@mayo.edu.
Vera J. Suman, Email: suman.vera@mayo.edu.
Jodi M. Carter, Email: carter.jodi@mayo.edu.
Richard Weinshilboum, Email: weinshilboum.richard@mayo.edu.
Liewei Wang, Email: wang.liewei@mayo.edu.
Krishna R. Kalari, Email: Kalari.Krishna@mayo.edu.
Saravut J. Weroha, Email: weroha.saravut@mayo.edu.
Alan H. Bryce, Email: bryce.alan@mayo.edu.
Judy C. Boughey, Email: boughey.judy@mayo.edu.
Haidong Dong, Email: Dong.Haidong@mayo.edu.
Charles M. Perou, Email: cperou@med.unc.edu.
Dingwei Ye, Email: dwyeli@163.com.
Matthew P. Goetz, Email: goetz.matthew@mayo.edu.
Shancheng Ren, Email: renshancheng@gmail.com.
Haojie Huang, Email: huang.haojie@mayo.edu.
References
- 1.Wei SC, et al. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8(9):1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 2.Rescigno P, de Bono JS. Immunotherapy for lethal prostate cancer. Nat Rev Urol. 2019;16(2):69–70. doi: 10.1038/s41585-018-0121-y. [DOI] [PubMed] [Google Scholar]
- 3.Esteva FJ, et al. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019;20(3):e175–e186. doi: 10.1016/S1470-2045(19)30026-9. [DOI] [PubMed] [Google Scholar]
- 4.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu D, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med. 2019;25(12):1916–1927. doi: 10.1038/s41591-019-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subudhi SK, et al. Neoantigen responses, immune correlates, and favorable outcomes after ipilimumab treatment of patients with prostate cancer. Sci Transl Med. 2020;12(537):eaaz3577. doi: 10.1126/scitranslmed.aaz3577. [DOI] [PubMed] [Google Scholar]
- 7.Gao J, et al. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(2):397–404. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishizuka JJ, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565(7737):43–48. doi: 10.1038/s41586-018-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kranz LM, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534(7607):396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 10.Borden EC. Interferons α and β in cancer: therapeutic opportunities from new insights. Nat Rev Drug Discov. 2019;18(3):219–234. doi: 10.1038/s41573-018-0011-2. [DOI] [PubMed] [Google Scholar]
- 11.Aznar MA, et al. Immunotherapeutic effects of intratumoral nanoplexed poly I:C. J Immunother Cancer. 2019;7(1):116. doi: 10.1186/s40425-019-0568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162(5):974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayers M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borden EC, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6(12):975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rooney MS, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Restifo NP, et al. Loss of functional beta 2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. J Natl Cancer Inst. 1996;88(2):100–108. doi: 10.1093/jnci/88.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leach DR, et al. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 20.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 22.Zaretsky JM, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davoli T, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355(6322):eaaf8399. doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132(6):958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer. 2012;12(6):381–385. doi: 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- 27.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1, and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams EJ, et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature. 2019;571(7765):408–412. doi: 10.1038/s41586-019-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parolia A, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature. 2019;571(7765):413–418. doi: 10.1038/s41586-019-1347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arruabarrena-Aristorena A, et al. FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell. 2020;38(4):534–550. doi: 10.1016/j.ccell.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eggermont AMM, et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N Engl J Med. 2018;378(19):1789–1801. doi: 10.1056/NEJMoa1802357. [DOI] [PubMed] [Google Scholar]
- 33.Gandhi L, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 34.Rini BI, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2019;380(12):1116–1127. doi: 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- 35.Slyper M, et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med. 2020;26(5):792–802. doi: 10.1038/s41591-020-0844-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark KL, et al. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 38.Pomerantz MM, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat Genet. 2020;52(8):790–799. doi: 10.1038/s41588-020-0664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bald T, et al. Immune cell-poor melanomas benefit from PD-1 blockade after targeted type I IFN activation. Cancer Discov. 2014;4(6):674–687. doi: 10.1158/2159-8290.CD-13-0458. [DOI] [PubMed] [Google Scholar]
- 40.Park JW, et al. FOXA2 is a sensitive and specific marker for small cell neuroendocrine carcinoma of the prostate. Mod Pathol. 2017;30(9):1262–1272. doi: 10.1038/modpathol.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollern DP, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179(5):1191–1206. doi: 10.1016/j.cell.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Araki H, et al. Haptoglobin promoter polymorphism rs5472 as a prognostic biomarker for peptide vaccine efficacy in castration-resistant prostate cancer patients. Cancer Immunol Immunother. 2015;64(12):1565–1573. doi: 10.1007/s00262-015-1756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noguchi M, et al. A randomized phase III trial of personalized peptide vaccination for castration‑resistant prostate cancer progressing after docetaxel. Oncol Rep. 2021;45(1):159–168. doi: 10.3892/or.2020.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sistigu A, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20(11):1301–1309. doi: 10.1038/nm.3708. [DOI] [PubMed] [Google Scholar]
- 45.Denkert C, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol. 2015;33(9):983–991. doi: 10.1200/JCO.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 46.Savas P, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13(4):228–241. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 47.Goetz MP, et al. Tumor sequencing and patient-derived xenografts in the neoadjuvant treatment of breast cancer. J Natl Cancer Inst. 2017;109(7):djw306. doi: 10.1093/jnci/djw306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim YJ, et al. Gene signatures for the prediction of response to Bacillus Calmette-Guerin immunotherapy in primary pT1 bladder cancers. Clin Cancer Res. 2010;16(7):2131–2137. doi: 10.1158/1078-0432.CCR-09-3323. [DOI] [PubMed] [Google Scholar]
- 49.McConkey DJ, et al. A prognostic gene expression signature in the molecular classification of chemotherapy-naïve urothelial cancer is predictive of clinical outcomes from neoadjuvant chemotherapy: a phase 2 trial of dose-dense methotrexate, vinblastine, doxorubicin, and cisplatin with bevacizumab in urothelial cancer. Eur Urol. 2016;69(5):855–862. doi: 10.1016/j.eururo.2015.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hellmann MD, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(5):843–852. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venturini NJ, Drake CG. Immunotherapy for prostate cancer. Cold Spring Harb Perspect Med. 2019;9(5):a030627. doi: 10.1101/cshperspect.a030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salgado R, et al. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with Lapatinib and Trastuzumab: a secondary analysis of the NeoALTTO Trial. JAMA Oncol. 2015;1(4):448–454. doi: 10.1001/jamaoncol.2015.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paiva SL, Crews CM. Targeted protein degradation: elements of PROTAC design. Curr Opin Chem Biol. 2019;50:111–119. doi: 10.1016/j.cbpa.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abida W, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A. 2019;116(23):11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, et al. A prospective genome-wide study of prostate cancer metastases reveals association of wnt pathway activation and increased cell cycle proliferation with primary resistance to abiraterone acetate-prednisone. Ann Oncol. 2018;29(2):352–360. doi: 10.1093/annonc/mdx689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robertson AG, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(3):540–556. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.