Abstract
Glycosaminoglycans (GAGs) are a family of complex carbohydrates vital to all mammalian organisms and involved in numerous biological processes. Chondroitin and dermatan sulfate, an important class of GAGs, are linear macromolecules consisting of disaccharide building blocks of N-acetylgalactosamine and two different uronic acids. The varying degree and the site of sulfation render their characterization challenging. Here, we combine mass spectrometry with cryogenic infrared spectroscopy in the wavenumber range from 1000 to 1800 cm–1. Fingerprint spectra were recorded for a comprehensive set of disaccharides bearing all known motifs of sulfation. In addition, state-of-the-art quantum chemical calculations were performed to aid the understanding of the differences in the experimental fingerprint spectra. The results show that the degree and position of charged sulfate groups define the size of the conformational landscape in the gas phase. The detailed understanding of cryogenic infrared spectroscopy for acidic and often highly sulfated glycans may pave the way to utilize the technique in fragment-based sequencing approaches.
Introduction
Carbohydrates or glycans are biological macromolecules that make up a significant amount of organic matter on earth and fulfill essential functional and structural roles in living organisms.1 Glycosaminoglycans (GAGs) are a major family of glycans, which participate in biological processes, such as hemostasis, inflammation, cell migration, proliferation, and differentiation.2−4 GAGs are long, acidic, and often sulfated, and within this family, an important class is chondroitin sulfate and dermatan sulfate (CS/DS). CS/DS are usually a part of larger glycoproteins, so-called proteoglycans, and often have chain lengths of over 50 disaccharide units.5−7 A characteristic for the glycan backbone of CS/DS are repeating disaccharide units of two different uronic acids (UA) linked to N-acetylgalactosamine (GalNAc). The UA in CS is glucuronic acid (GlcA), whereas the backbone of DS contains iduronic acid (IdoA) monosaccharides. Disaccharide fragments from larger chains are the product of bacterial chondroitinase digestion, which introduces a double bond between C4 and C5 in the UA (then abbreviated ΔUA). Varying O-sulfations at C2 of the uronic acid and at C4 and C6 of the GalNAc further increase the complexity of CS/DS chains significantly.8 The most informative level of GAG analysis resides in its disaccharide building blocks; therefore, disaccharide analysis is essential for any oligosaccharide or full-length GAG sequence.
Recent developments in mass spectrometry-based techniques, such as electron-based dissociation9−11 and ion mobility-mass spectrometry,12,13 have improved the sensitivity and integrity in GAG analysis toward varying uronic acid stereochemistry and in sulfation.14,15 Nevertheless, the distinction of certain sulfation motifs, especially O-sulfations at C4 and C6 in CS/DS, is still exceptionally challenging by mass spectrometry-based techniques.13,16−20
A novel approach for GAG structural characterization is infrared (IR) spectroscopy of ions in the gas phase.21 GAGs and their fragments ranging from monosaccharides to pentasaccharides have shown to exhibit a wealth of characteristic spectral features.22−25 Cryogenic temperatures have enhanced the spectral quality significantly in the midinfrared26 and the OH-stretching region.24 Here, we assess the capability of cryogenic IR spectroscopy for the differentiation and structural characterization of uronic acids of CS/DS disaccharides with all known motifs of sulfation.
Methods
Sample Preparation
Glycosaminoglycans were purchased from Iduron (Alderley Edge, U.K.) and used without further purification. For each disaccharide, a stock solution of 1 mM in H2O was further diluted prior to use to yield a 50 μM analyte solution in H2O/MeOH (v/v, 1/1).
Ion Mobility-Mass Spectrometry
Drift tube (DT) ion mobility-mass spectrometry measurements were performed on a modified Synapt G2-S HDMS instrument (Waters, Manchester, U.K.) containing a drift tube instead of the commercial traveling wave cell27 and equipped with a nanoelectrospray ionization source. Ion mobilities were determined employing the stepped-field approach, and collision cross sections (CCS) were derived from the respective mobilities using the Mason–Schamp equation.28
Cryogenic Infrared Spectroscopy
The home-built experimental setup has been described in detail recently.29−31 In brief, anions are produced in a nanoelectrospray ionization source, mass-to-charge selected in a quadrupole mass filter, and stored in an ion trap at ca. 90 K. Generated in a pulsed Even-Lavie valve, superfluid helium nanodroplets traverse the ion trap and pick up single ions. It has been shown that in the event of the pick-up, the ions are, with the rate of cooling, kinetically trapped in their present conformation. In a detection region of the instrument, the doped nanodroplets overlap with the laser beam of the Fritz-Haber-Institut free-electron laser (FHI FEL). At a resonant wavelength, the ion is vibrationally excited and immediately cooled to its ground state by evaporative cooling of the helium bath. After several iterations, the ion is eventually released from the nanodroplet and detected in a time-of-flight mass analyzer. The ion signal is plotted as a function of the incident photon energy and divided by the corresponding laser power as a first-order approximation. Relative intensities are, therefore, considered as a general reference. Infrared spectra are the result of an average of two individual scans.
Computational Methods
The conformational space of the disaccharides was sampled using the evolutionary algorithm FAFOOM32 utilizing external software FHI-aims33 for local DFT optimization of each sampled structure at the PBE + vdWTS34 level of theory and light basis set settings. Mutation of all rotatable bonds and ring puckers was allowed during the conformational search. The algorithm allows functional groups to interact. In total ca. 250 structures were sampled for each disaccharide using the described approach. The methodology has previously yielded excellent accuracy for small glycans.35,36
Results and Discussion
Chondroitin sulfate disaccharides ranging from nonsulfated species up to triply sulfated species were chosen to cover all known sulfate combinations. In Figure 1a, the chemical structure of the triply sulfated disaccharide 1 is depicted. To assure molecular stability in the gas phase and limit charge migration, the disaccharides were investigated as deprotonated anionic species with a charge state equal to the number of sulfates at which the charges are localized.23,26 The nonsulfated disaccharide is investigated with a charge state equal to the number of carboxyl groups, i.e., as singly charged species.
Figure 1.
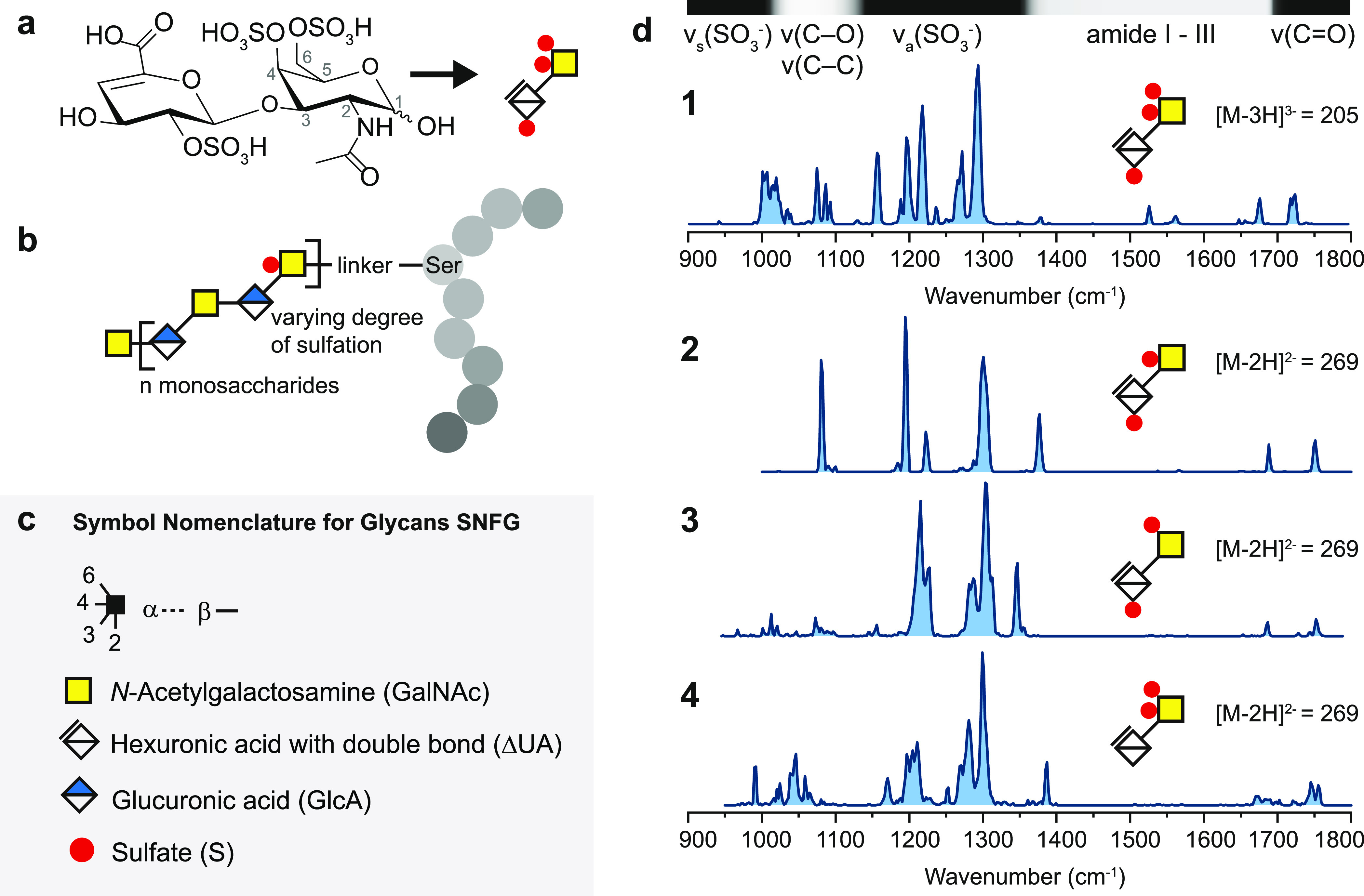
(a) Investigated chondroitin sulfate disaccharide 1 in chemical representation followed by its representation in the symbol nomenclature for glycans (SNFG).37 Both α and β anomers are present in the sample. (b) Representative structure of the proteoglycan bikunin, which carries one site for an O-linked chondroitin sulfate chain.5,38−40 The protein chain is depicted with gray circles and the three-letter code is used to highlight serine. (c) SNFG. (d) Cryogenic IR spectra of triply sulfated disaccharide 1 investigated as a [M – 3H]3– anion with a mass-to-charge ratio (m/z) of 205 and doubly sulfated disaccharides 2–4 investigated as [M – 2H]2– isomeric anions with m/z of 269. On top of the first spectrum, the main vibrational features for these ions are qualitatively assigned in a horizontal bar.
Cryogenic IR spectra in the mid-IR range were recorded at a minimum from 1000 to a maximum of 1800 cm–1, as shown in Figures 1d and 2. The assignment of spectral ranges to vibrational features is based on previous theoretical calculations of sulfated mono- and disaccharides.23,25 In brief, the spectral range below 1100 cm–1 is most characteristic for symmetric stretching of the charged sulfate νs(SO3–) and overlaps the spectral range from 1000 to 1150 cm–1 in which ν(C–O) and ν(C–C) are found. From 1150 to 1350 cm–1, strong antisymmetric stretching vibrations of the charged sulfate νa(SO3–) typically dominate the IR signature in sulfated glycosaminoglycans. Multiple minor vibrational features between 1200 and 1500 cm–1 correspond to C–H and O–H bending modes. Furthermore, above 1300 up to 1700 cm–1, the amide vibrations I to III are found in N-acetylated monosaccharides, of which amide I between 1600 and 1700 cm–1 is usually the most intense. Minor contributions of ν(C=C) in the range of amide I are characteristic for GAGs derived from lyase digestion. Above 1700 cm–1, ν(C=O) indicates the absence of a charged site at the carboxyl functional group. Carboxylate anions on the other hand yield an intense vibrational feature around 1650 cm–1, corresponding to the antisymmetric stretch νa(COO–), whereas the potentially weaker νs(COO–) is typically found between 1300 and 1400 cm–1.
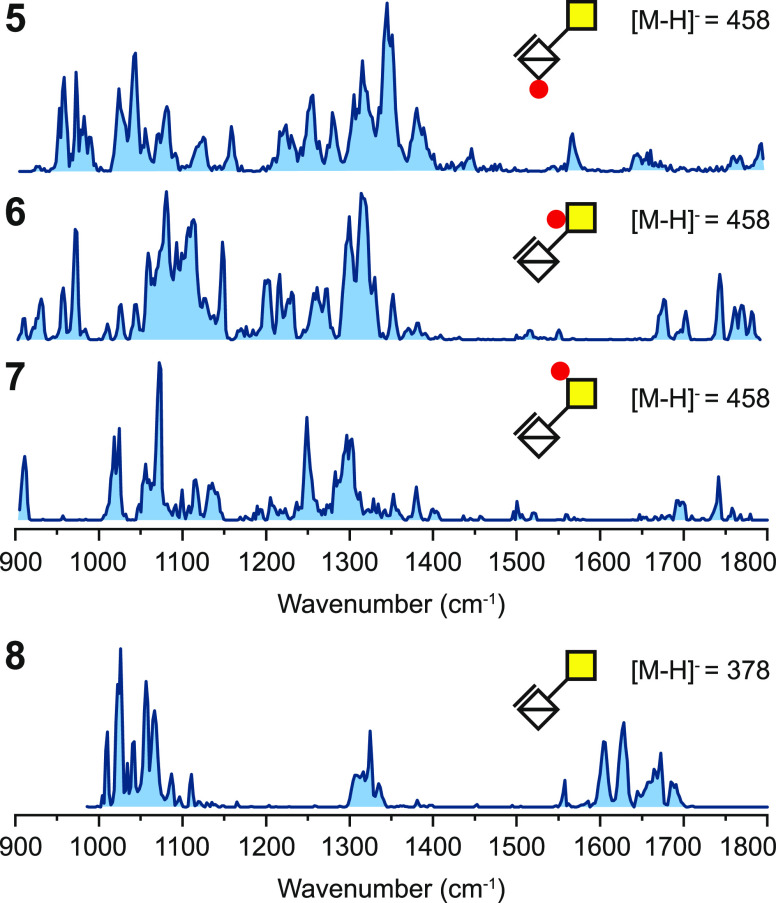
Figure 2.
Cryogenic IR spectra of sulfated disaccharides 5–7 investigated as [M – H]− isomeric anions with m/z of 458 and nonsulfated disaccharide 8 investigated as a [M – H]− anion with m/z of 378.
The IR signature of the triply sulfated disaccharide 1 exhibits a unique absorption pattern with the strongest feature centered at 1292 cm–1 in the spectral range of νa(SO3–). Single bands are, in some cases, close to the bandwidth of the free-electron laser (FWHM ca. 5 cm–1, i.e., 0.3–0.5% of the respective wavenumber). The IR signatures of the isomeric disaccharides 2 to 4 show a variety of well-resolved bands and are unambiguously distinguishable from each other. Especially, the IR signatures of disaccharides 2 and 3 with O-sulfation at C2 exhibit surprisingly narrow spectral features for ions of this size.
Although smaller in molecular size compared to the disaccharides with higher sulfation, the singly sulfated disaccharides 5 to 7 show a more complex IR signature, as shown in Figure 2. Especially the range up to 1400 cm–1 is more congested and shows less deviation between weak- and high-intensity signals. In disaccharide 6 carrying O-sulfation at C4, the range between 1650 and 1800 cm–1 shows a multitude of vibrational features for the two strongest potential oscillators in that range, i.e., C=O in the carboxyl and the amide groups. The nonsulfated disaccharide 8 was included to assess the impact of the backbone, and the IR spectrum clearly reveals that this ion carries the charge at the carboxylate.
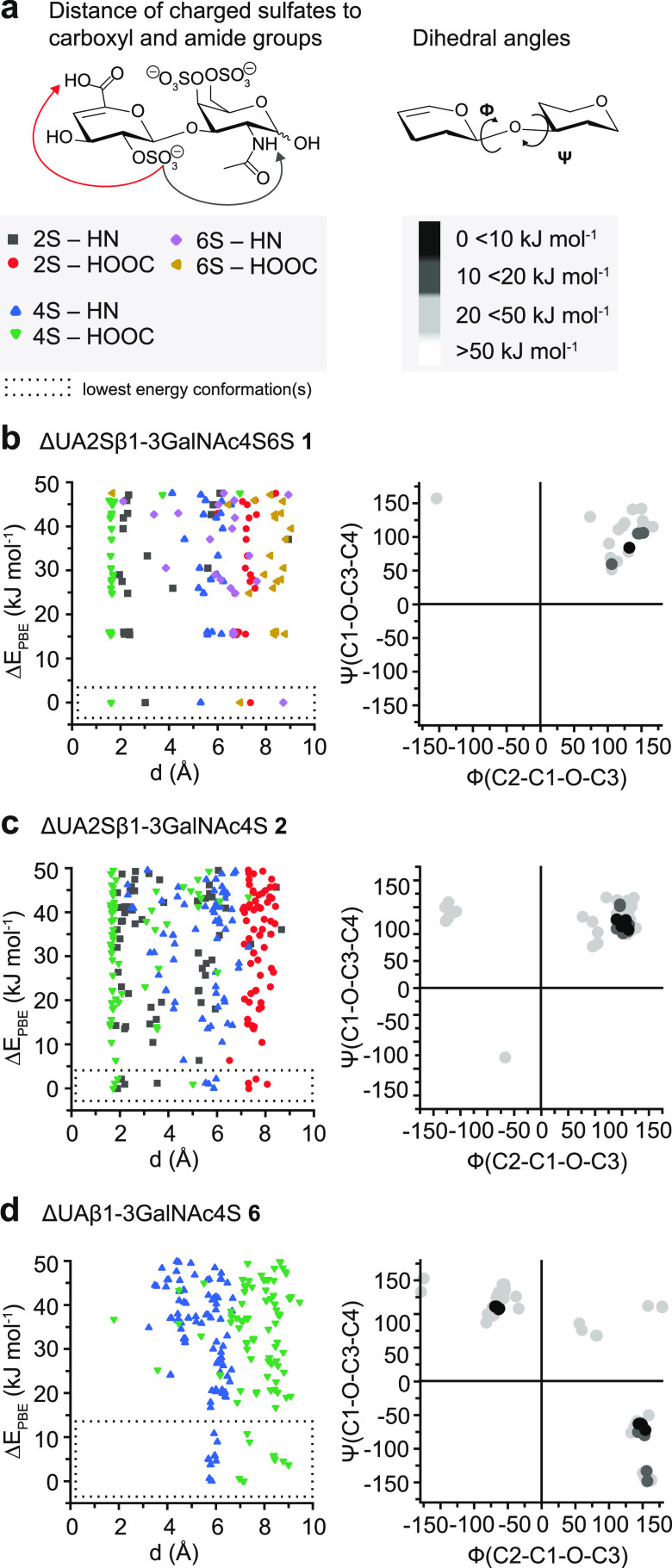
The size of the ensemble of conformations present during the experiment correlates to the congestion of the IR spectrum and the number of vibrational features in a range in which a limited number of functional groups absorb. In our experimental setup, the ensemble of conformations is mainly defined by the temperature of the coolable ion trap (ca. 90 K). When the molecules are picked up by the superfluid helium nanodroplets (0.4 K), the rate of cooling is so fast that ions are kinetically trapped in local conformational minima.41 The IR spectra of the higher sulfated disaccharides are remarkably well resolved, whereas the IR spectra of the singly sulfated disaccharides are highly congested. Our hypothesis is that the higher sulfated and also higher charged disaccharides exhibit only a limited number of low-energy conformers, whilst the singly sulfated and also singly charged disaccharides exhibit a larger ensemble of low-energy conformers. To test this hypothesis, quantum chemical calculations were performed and the conformational space of the sulfated disaccharide β anomers was extensively sampled to yield ca. 250 conformers for each disaccharide. The β anomer was chosen under the premise that the stereocenter at C1 has only a minor influence on the overall conformation of the disaccharide. For each ion, all sampled conformers with relative energies (ΔEPBE) up to 50 kJ mol–1 were selected and characterized using two criteria: (1) the intramolecular distances of charged sulfates toward the hydrogens of the carboxyl and amide groups and (2) the dihedral angles Ψ(C1-O-C3-C4) and Φ(C2-C1-O-C3).
To qualitatively assess the conformational diversity, the diagrams relating relative energies to structural parameters and Ramachandran-type plots for glycosidic dihedral angles give insights into the orientation of selected functional groups and the glycan backbone, respectively. Key interactions in these molecules are hydrogen bonds of the charged sulfates toward the hydrogens of the carboxyl and amide groups. For this reason, the x-axis in the energy diagram shows the distances of all charged sulfates toward either the secondary amide of N-acetyl or the neutral carboxyl functional groups and on the y-axis of the relative energies. The different distances in a single conformation are, thus, given from left to right at the same relative energy. The Ramachandran-type plots for glycosidic dihedral angles, analogous to Ramachandran plots of peptides and proteins, show the dihedral angles Ψ(C1-O-C3-C4) and Φ(C2-C1-O-C3) on y- and x-axes, respectively, with dots for single conformers, which are colored according to the relative energy.
The conformers of disaccharide 1, Figure 3b, with three charged sulfate groups show trends in the orientation of the sulfate groups. More importantly, one conformer is with 15 kJ mol–1 significantly lower in energy than the second- and third-lowest-energy conformers. The hydrogen-bonding network is dominated by the interaction between the O-sulfate group at C4 and the neutral carboxyl functional group but also by the O-sulfate group at C2 and the secondary amide. The O-sulfate group at C6 has in all conformers a distance larger than 5 Å toward the amide and carboxyl to minimize Coulombic repulsion with the sulfate groups in their proximity. The Ramachandran-type plot shows a very narrow distribution, indicating similarities in the glycan backbone of the low-energy conformers. Conformers of high similarity are more likely to relax into the same local minimum or the global minimum in the cold ion trap. Disaccharide 2, Figure 3c, with two charged sulfate groups likewise reveals trends in the energy diagram, yet, far more conformers are within 20 kJ mol–1 of the lowest-energy conformer. The distribution in the Ramachandran-type plot is narrow. In contrast, O-sulfate at C4 does not form a hydrogen bond with the neutral carboxyl in the singly sulfated disaccharide 6, Figure 3d. The Ramachandran-type plot shows that two clusters of conformers with large differences in their glycan backbone are present. This can indicate that at least two but probably several local minima are populated that are conformationally constrained to reach the same or global minimum structure. The plots of the disaccharides 3–5 and 7, in Figure S1, further support the observed trend of a growing conformational heterogeneity in chondroitin sulfate ions with a decreasing degree of charged groups, which are sulfate groups in this case.
Figure 3.
(a) Left panel: calculation of the intramolecular distance of selected functional groups in Å in conformers with relative energies ΔEPBE < 50 kJ mol–1. Right panel: calculation of the dihedral angles Ψ(C1-O-C3-C4) and Φ(C2-C1-O-C3) in the degree at the glycosidic bond with respect to the relative energies of the conformers. The results are presented in Ramachandran-type plots for glycosidic linkages. (b–d) Results for the triply sulfated disaccharide 1, doubly sulfated disaccharide 2, and singly sulfated disaccharide 6. Generally, an increase in conformational heterogeneity is observed with a decreasing extent of sulfation and charge.
For the discussed disaccharides 1, 2, and 6, selected low-energy conformers were reoptimized at a higher level of theory. Theoretical infrared spectra were computed for the lowest-energy conformers and compared to the experimental spectra, Figure S2. The match between computed and experimental spectra is satisfying, yet, anharmonicities challenge the calculations for glycosaminoglycans and extend the computational effort.42
In addition to gas-phase IR spectroscopy, drift tube ion mobility-mass spectrometry (DTIM-MS) experiments were performed to compare the potential of the method for disaccharide differentiation. The rotationally averaged collision cross section (CCS) reflects the shape of an ion and is an instrument-independent physical property. CCS values of the disaccharides, measured in nitrogen and helium, are summarized in Table 1. The most challenging isomeric disaccharides 6 and 7 with O-sulfate either at C4 or at C6 are not distinguishable based on their CCS. The results indicate that differentiating certain isomeric disaccharides in sets 2–4 and 5–7 is possible but may require IMS instrumentation with resolving power on the order of 100 when they are present in a mixture and measurement uncertainties below 0.5% when present alone.
Table 1. Collision Cross Sections (CCSs) of the Investigated Chondroitin Sulfate Disaccharides.
| molecule | z | m/za | DTCCSN2b | DTCCS Heb | |
|---|---|---|---|---|---|
| 1 | ΔUA2Sβ1-3GalNAc4S6S | –3 | 205 | 317 | 160 |
| 2 | ΔUA2Sβ1-3GalNAc4S | –2 | 269 | 241 | 138 |
| 3 | ΔUA2Sβ1-3GalNAc6S | –2 | 269 | 244 | 140 |
| 4 | ΔUAβ1-3GalNAc4S6S | –2 | 269 | 239 | 135 |
| 5 | ΔUA2Sβ1-3GalNAc | –1 | 458 | 190 | 127 |
| 6 | ΔUAβ1-3GalNAc4S | –1 | 458 | 189 | 122 |
| 7 | ΔUAβ1-3GalNAc6S | –1 | 458 | 189 | 122 |
| 8 | ΔUAβ1-3GalNAc | –1 | 378 | 178 | 112 |
Monoisotopic mass-to-charge (m/z) ratio.
In Å2, DT: drift tube.
Conclusions
In summary, we show here that cryogenic IR spectroscopy has the potential to unambiguously differentiate all known sulfation patterns in uronic acids of chondroitin sulfate and dermatan sulfate disaccharides, including the distinction of 4O- and 6O-sulfation, a long-standing challenge in GAG analysis. Identification of disaccharide isomers based purely on their ion mobility-derived CCS values is partially ambiguous and requires outstanding resolving power. Furthermore, we show that for chondroitin sulfate ions with a low charge state and a low degree of sulfation, large ensembles of low-energy conformers are present. Whereas for higher charged and higher sulfated chondroitin sulfate ions, only a few low-energy conformers are present. This conformational constraint in highly charged gas-phase ions has also been described for other biomolecules such as peptides and proteins.43 Yet, additional experiments are necessary to evaluate whether this effect purely results from Coulombic repulsion or also from the nature of the O-sulfate groups, which, in contrast to more localized carboxylates, potentially induce further constraints with their steric repulsion44 and conformational flexibility.
Acknowledgments
The authors gratefully acknowledge the expertise of Sandy Gewinner and Dr. Wieland Schöllkopf of the FHI FEL. K.G. thanks the Fonds National de la Recherche (FNR), Luxembourg, for funding for the project GlycoCat (13549747). This work was financially supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 372486779—SFB 1340, the Danish National Research Foundation (DNFR107), the Carlsberg Foundation (CF20-0412), and the European Union’s Horizon 2020 Research and Innovation Program under grant number 899687 (HS-SEQ).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpca.1c02463.
Experimental details on computational methods and coordinates of lowest-energy conformers; the energy diagram and Ramachandran-type plots for disaccharides 3, 4, 5, and 7; and the comparison of experimental and computed infrared spectra for disaccharides 1, 2, and 6 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Varki A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D.; Esko J. D. Demystifying Heparan Sulfate–Protein Interactions. Annu. Rev. Biochem. 2014, 83, 129–157. 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.-Y.; Olson S. K.; Esko J. D.; Robert Horvitz H. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature 2003, 423, 439–443. 10.1038/nature01634. [DOI] [PubMed] [Google Scholar]
- Soares da Costa D.; Reis R. L.; Pashkuleva I. Sulfation of Glycosaminoglycans and Its Implications in Human Health and Disorders. Annu. Rev. Biomed. Eng. 2017, 19, 1–26. 10.1146/annurev-bioeng-071516-044610. [DOI] [PubMed] [Google Scholar]
- Ly M.; Leach F. E.; Laremore T. N.; Toida T.; Amster I. J.; Linhardt R. J. The proteoglycan bikunin has a defined sequence. Nat. Chem. Biol. 2011, 7, 827–833. 10.1038/nchembio.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuppevelt T. H.; Oosterhof A.; Versteeg E. M. M.; Podhumljak E.; van de Westerlo E. M. A.; Daamen W. F. Sequencing of glycosaminoglycans with potential to interrogate sequence-specific interactions. Sci. Rep. 2017, 7, 14785 10.1038/s41598-017-15009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.; Duan J.; Leach F. E.; Toida T.; Higashi K.; Zhang H.; Zhang F.; Amster I. J.; Linhardt R. J. Sequencing the Dermatan Sulfate Chain of Decorin. J. Am. Chem. Soc. 2017, 139, 16986–16995. 10.1021/jacs.7b10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama C. I.; Tully S. E.; Sotogaku N.; Clark P. M.; Rawat M.; Vaidehi N.; Goddard W. A.; Nishi A.; Hsieh-Wilson L. C. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat. Chem. Biol. 2006, 2, 467–473. 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- Leach F. E.; Riley N. M.; Westphall M. S.; Coon J. J.; Amster I. J. Negative Electron Transfer Dissociation Sequencing of Increasingly Sulfated Glycosaminoglycan Oligosaccharides on an Orbitrap Mass Spectrometer. J. Am. Soc. Mass Spectrom. 2017, 28, 1844–1854. 10.1007/s13361-017-1709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.; Wu J.; Tang Y.; Ridgeway M. E.; Park M. A.; Costello C. E.; Zaia J.; Lin C. Characterization and Quantification of Highly Sulfated Glycosaminoglycan Isomers by Gated-Trapped Ion Mobility Spectrometry Negative Electron Transfer Dissociation MS/MS. Anal. Chem. 2019, 91, 2994–3001. 10.1021/acs.analchem.8b05283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach F. E.; Ly M.; Laremore T. N.; Wolff J. J.; Perlow J.; Linhardt R. J.; Amster I. J. Hexuronic Acid Stereochemistry Determination in Chondroitin Sulfate Glycosaminoglycan Oligosaccharides by Electron Detachment Dissociation. J. Am. Soc. Mass Spectrom. 2012, 23, 1488–1497. 10.1007/s13361-012-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmnitzer K.; Riemer T.; Groessl M.; Süβ R.; Knochenmuss R.; Schiller J. Comparison of ion mobility-mass spectrometry and pulsed-field gradient nuclear magnetic resonance spectroscopy for the differentiation of chondroitin sulfate isomers. Anal. Methods 2016, 8, 8483–8491. 10.1039/C6AY02531E. [DOI] [Google Scholar]
- Poyer S.; Lopin-Bon C.; Jacquinet J.-C.; Salpin J.-Y.; Daniel R. Isomer separation and effect of the degree of polymerization on the gas-phase structure of chondroitin sulfate oligosaccharides analyzed by ion mobility and tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2017, 31, 2003–2010. 10.1002/rcm.7987. [DOI] [PubMed] [Google Scholar]
- Solakyildirim K. Recent advances in glycosaminoglycan analysis by various mass spectrometry techniques. Anal. Bioanal. Chem. 2019, 411, 3731–3741. 10.1007/s00216-019-01722-4. [DOI] [PubMed] [Google Scholar]
- Kailemia M. J.; Ruhaak L. R.; Lebrilla C. B.; Amster I. J. Oligosaccharide Analysis by Mass Spectrometry: A Review of Recent Developments. Anal. Chem. 2014, 86, 196–212. 10.1021/ac403969n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaia J.; McClellan J. E.; Costello C. E. Tandem Mass Spectrometric Determination of the 4S/6S Sulfation Sequence in Chondroitin Sulfate Oligosaccharides. Anal. Chem. 2001, 73, 6030–6039. 10.1021/ac015577t. [DOI] [PubMed] [Google Scholar]
- Zaia J.; Li X.-Q.; Chan S.-Y.; Costello C. E. Tandem mass spectrometric strategies for determination of sulfation positions and uronic acid epimerization in chondroitin sulfate oligosaccharides. J. Am. Soc. Mass Spectrom. 2003, 14, 1270–1281. 10.1016/S1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- Bielik A. M.; Zaia J. Multistage tandem mass spectrometry of chondroitin sulfate and dermatan sulfate. Int. J. Mass Spectrom. 2011, 305, 131–137. 10.1016/j.ijms.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamfir A. D.; Flangea C.; Sisu E.; Serb A. F.; Dinca N.; Bruckner P.; Seidler D. G. Analysis of novel over- and under-sulfated glycosaminoglycan sequences by enzyme cleavage and multiple stage MS. Proteomics 2009, 9, 3435–3444. 10.1002/pmic.200800440. [DOI] [PubMed] [Google Scholar]
- Persson A.; Vorontsov E.; Larson G.; Nilsson J. Glycosaminoglycan Domain Mapping of Cellular Chondroitin/Dermatan Sulfates. Sci. Rep. 2020, 10, 3506 10.1038/s41598-020-60526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagnon I.; Schindler B.; Renois-Predelus G.; Daniel R. Lasers and ion mobility: new additions to the glycosaminoglycanomics toolkit. Curr. Opin. Struct. Biol. 2018, 50, 171–180. 10.1016/j.sbi.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Renois-Predelus G.; Schindler B.; Compagnon I. Analysis of Sulfate Patterns in Glycosaminoglycan Oligosaccharides by MSn Coupled to Infrared Ion Spectroscopy: the Case of GalNAc4S and GalNAc6S. J. Am. Soc. Mass Spectrom. 2018, 29, 1242–1249. 10.1007/s13361-018-1955-5. [DOI] [PubMed] [Google Scholar]
- Schindler B.; Barnes L.; Gray C. J.; Chambert S.; Flitsch S. L.; Oomens J.; Daniel R.; Allouche A. R.; Compagnon I. IRMPD Spectroscopy Sheds New (Infrared) Light on the Sulfate Pattern of Carbohydrates. J. Phys. Chem. A 2017, 121, 2114–2120. 10.1021/acs.jpca.6b11642. [DOI] [PubMed] [Google Scholar]
- Khanal N.; Masellis C.; Kamrath M. Z.; Clemmer D. E.; Rizzo T. R. Glycosaminoglycan Analysis by Cryogenic Messenger-Tagging IR Spectroscopy Combined with IMS-MS. Anal. Chem. 2017, 89, 7601–7606. 10.1021/acs.analchem.7b01467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettow M.; Grabarics M.; Greis K.; Mucha E.; Thomas D. A.; Chopra P.; Boons G.-J.; Karlsson R.; Turnbull J. E.; Meijer G.; et al. Cryogenic Infrared Spectroscopy Reveals Structural Modularity in the Vibrational Fingerprints of Heparan Sulfate Diastereomers. Anal. Chem. 2020, 92, 10228–10232. 10.1021/acs.analchem.0c02048. [DOI] [PubMed] [Google Scholar]
- Lettow M.; Grabarics M.; Mucha E.; Thomas D. A.; Polewski Ł.; Freyse J.; Rademann J.; Meijer G.; von Helden G.; Pagel K. IR action spectroscopy of glycosaminoglycan oligosaccharides. Anal. Bioanal. Chem. 2020, 412, 533–537. 10.1007/s00216-019-02327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S. J.; Giles K.; Gilbert T.; Bush M. F. Ion mobility mass spectrometry of peptide, protein, and protein complex ions using a radio-frequency confining drift cell. Analyst 2016, 141, 884–891. 10.1039/C5AN02107C. [DOI] [PubMed] [Google Scholar]
- Gabelica V.; Shvartsburg A. A.; Afonso C.; Barran P.; Benesch J. L. P.; Bleiholder C.; Bowers M. T.; Bilbao A.; Bush M. F.; Campbell J. L.; et al. Recommendations for reporting ion mobility Mass Spectrometry measurements. Mass Spectrom. Rev. 2019, 38, 291–320. 10.1002/mas.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greis K.; Kirschbaum C.; Leichnitz S.; Gewinner S.; Schöllkopf W.; von Helden G.; Meijer G.; Seeberger P. H.; Pagel K. Direct Experimental Characterization of the Ferrier Glycosyl Cation in the Gas Phase. Org. Lett. 2020, 22, 8916–8919. 10.1021/acs.orglett.0c03301. [DOI] [PubMed] [Google Scholar]
- Mucha E.; Gonzalez Florez A. I.; Marianski M.; Thomas D. A.; Hoffmann W.; Struwe W. B.; Hahm H. S.; Gewinner S.; Schollkopf W.; Seeberger P. H.; et al. Glycan Fingerprinting via Cold-Ion Infrared Spectroscopy. Angew. Chem., Int. Ed. 2017, 56, 11248–11251. 10.1002/anie.201702896. [DOI] [PubMed] [Google Scholar]
- Thomas D. A.; Mucha E.; Lettow M.; Meijer G.; Rossi M.; von Helden G. Characterization of a trans-trans Carbonic Acid-Fluoride Complex by Infrared Action Spectroscopy in Helium Nanodroplets. J. Am. Chem. Soc. 2019, 141, 5815–5823. 10.1021/jacs.8b13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supady A.; Blum V.; Baldauf C. First-Principles Molecular Structure Search with a Genetic Algorithm. J. Chem. Inf. Model. 2015, 55, 2338–48. 10.1021/acs.jcim.5b00243. [DOI] [PubMed] [Google Scholar]
- Blum V.; Gehrke R.; Hanke F.; Havu P.; Havu V.; Ren X.; Reuter K.; Scheffler M. Ab initio molecular simulations with numeric atom-centered orbitals. Comput. Phys. Commun. 2009, 180, 2175–2196. 10.1016/j.cpc.2009.06.022. [DOI] [Google Scholar]
- Tkatchenko A.; Scheffler M. Accurate molecular van der Waals interactions from ground-state electron density and free-atom reference data. Phys. Rev. Lett. 2009, 102, 073005 10.1103/PhysRevLett.102.073005. [DOI] [PubMed] [Google Scholar]
- Marianski M.; Supady A.; Ingram T.; Schneider M.; Baldauf C. Assessing the Accuracy of Across-the-Scale Methods for Predicting Carbohydrate Conformational Energies for the Examples of Glucose and alpha-Maltose. J. Chem. Theory Comput. 2016, 12, 6157–6168. 10.1021/acs.jctc.6b00876. [DOI] [PubMed] [Google Scholar]
- Marianski M.; Mucha E.; Greis K.; Moon S.; Pardo A.; Kirschbaum C.; Thomas D. A.; Meijer G.; von Helden G.; Gilmore K.; et al. Remote Participation during Glycosylation Reactions of Galactose Building Blocks: Direct Evidence from Cryogenic Vibrational Spectroscopy. Angew. Chem., Int. Ed. 2020, 59, 6166–6171. 10.1002/anie.201916245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A.; Cummings R. D.; Aebi M.; Packer N. H.; Seeberger P. H.; Esko J. D.; Stanley P.; Hart G.; Darvill A.; Kinoshita T.; et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25, 1323–1324. 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepedda A. J.; De Muro P.; Capobianco G.; Formato M. Role of the small proteoglycan bikunin in human reproduction. Hormones 2020, 19, 123–133. 10.1007/s42000-019-00149-x. [DOI] [PubMed] [Google Scholar]
- Lord M. S.; Day A. J.; Youssef P.; Zhuo L.; Watanabe H.; Caterson B.; Whitelock J. M. Sulfation of the bikunin chondroitin sulfate chain determines heavy chain·hyaluronan complex formation. J. Biol. Chem. 2013, 288, 22930–22941. 10.1074/jbc.M112.404186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi L.; Wolff J. J.; Laremore T. N.; Restaino O. F.; Xie J.; Schiraldi C.; Toida T.; Amster I. J.; Linhardt R. J. Structural Analysis of Bikunin Glycosaminoglycan. J. Am. Chem. Soc. 2008, 130, 2617–2625. 10.1021/ja0778500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. A.; Chang R.; Mucha E.; Lettow M.; Greis K.; Gewinner S.; Schöllkopf W.; Meijer G.; von Helden G. Probing the conformational landscape and thermochemistry of DNA dinucleotide anions via helium nanodroplet infrared action spectroscopy. Phys. Chem. Chem. Phys. 2020, 22, 18400–18413. 10.1039/D0CP02482A. [DOI] [PubMed] [Google Scholar]
- Barnes L.; Schindler B.; Allouche A.-R.; Simon D.; Chambert S.; Oomens J.; Compagnon I. Anharmonic simulations of the vibrational spectrum of sulfated compounds: application to the glycosaminoglycan fragment glucosamine 6-sulfate. Phys. Chem. Chem. Phys. 2015, 17, 25705–25713. 10.1039/C5CP02079D. [DOI] [PubMed] [Google Scholar]
- Warnke S.; Hoffmann W.; Seo J.; De Genst E.; von Helden G.; Pagel K. From Compact to String—The Role of Secondary and Tertiary Structure in Charge-Induced Unzipping of Gas-Phase Proteins. J. Am. Soc. Mass Spectrom. 2017, 28, 638–646. 10.1007/s13361-016-1551-5. [DOI] [PubMed] [Google Scholar]
- Queiroz I. N.; Wang X.; Glushka J. N.; Santos G. R.; Valente A. P.; Prestegard J. H.; Woods R. J.; Mourão P. A.; Pomin V. H. Impact of sulfation pattern on the conformation and dynamics of sulfated fucan oligosaccharides as revealed by NMR and MD. Glycobiology 2015, 25, 535–547. 10.1093/glycob/cwu184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.