Abstract
Background:
The pathophysiology underlying the progression and development of autoimmune conditions, such as Rheumatoid Arthritis (RA), is a result of dysregulations of the immune system. Research has explored the genetic alterations present in RA; however, limited studies have examined the role of Killer cell Immunoglobulin-like Receptors (KIR) and Human Leukocyte Antigen (HLA) molecules in RA. Therefore, the aim of this study was to examine KIR genes, their HLA ligands, and KIR-HLA compounds in patients with RA.
Methods:
In this case-control study, a total of 50 patients with RA and 100 healthy individuals were enrolled. DNA samples were evaluated using PCR with sequence specific Primers (PCR-SSP). Odds ratio (OR) with a 95% confidence interval (CI) were reported.
Results:
Among the KIR genes examined, KIR2DLA (p= 0.0255, OR= 0.389, 95% CI= 0.210-0.722) and KIR2DS4-full (p< 0.0001, OR= 6.163, 95% CI= 3.174-11.968) were observed to have a statistically significant correlation with disease susceptibility to RA. As an inhibitory gene, KIR2DLA was observed to have a protective effect against RA while KIR2DS4-full as an activating gene, was found to increase risk for RA. No significant associations were found between any of the other KIR genotypes, HLA ligands, or KIR-HLA compounds examined in this study to RA susceptibility.
Conclusion:
In this study of RA in the Lur population of Iran, KIR2DS4-full was observed to increase susceptibility to RA, while KIR2DL5A was found to act as a protecting factor based on both the cross Table and regression analyses. Further research should focus on repeating this study in additional populations.
Key Words: HLA, KIR, NK cells, Rheumatoid Arthritis
Introduction
Rheumatoid Arthritis (RA) is a common chronic autoimmune disorder characterized by synovitis, chronic hyperplasia, bone erosion, progressive joint damage, irreversible loss of bone shape and mobility problems (1, 2). It is considered a systemic autoimmune disease with multiorgan involvement (3). An estimated 0.5-1 percent of the world's population are affected by RA (4). The prevalence of RA depends on age and gender, occurring three times more often in women than in men (5).
A growing body of literature has found a range of genetic and environmental factors contributing to the incidence of RA including, cigarette smoke, certain ethnicities, age, epigenetic factors, and immunological factors (6). It has been shown that approximately 60% of the genetic contribution to RA susceptibility is present in populations of different ethnicities (2). Human leukocyte antigen (HLA)-DR appears to hold the most significant role among the potential predisposing genetic factors for RA as the example of a gene cluster associated with ethnicities. Some of the most important genes involved in RA have been found to be PTPN22, PAD14, EGFR, MHTFR, IL-23R, CD226, HLA-DR, STST4, TRAF1/C5, CCR6, HLA-G, BACH2, etc. (7). Among the different genes, HLA-DRB1 has the strongest relationship with RA with a sequence of specific amino acids in areas 70-74 of the third variable region called the Shared Epitope (8). Other mechanisms involved in the development and progression of RA include molecular imitation by certain microbial peptides and the activation of inflammatory pathways by T-cell antigen detection and the innate immune response. Among the proposed factors involved in RA, natural killer (NK) cells have been observed to have an important role in disease development and pathogenesis (9).
As a critical part of the immune system, NK cells originate from bone marrow precursors. They have a critical role in the first line of defense against pathogens and inhibiting tumor growth. Two major NK cell subsets exist: CD16+CD56dim with high killing ability and CD16-CD56bright with high cytokine secretion (10, 11). A sharp increase in the CD56bright subtype has been observed in the synovial fluid of patients with RA (12). The activity of NK cells is regulated by cell surface receptors, which can exhibit either inhibitory or activating functions. Two families of NK cells receptors include the immunoglobulin superfamily and the large family of C-type lectins located on chromosomes 12 and 19, respectively (13, 14).
The immunoglobulin superfamily has several members including the Killer cell Immunoglobulin-like Receptors (KIR), Natural Cytotoxicity Receptors (NCR), and the Fc epsilon receptor (FCεR). The KIR holds a key role in the ability of NK cells to recognize self from non-self-ligands (15-20). These receptors are regulatory molecules found on the surface of NK cells and some T cell subgroups, including TCD8+ and TCD4CD28 null. They are located on the 19q13.4 chromosome in the LRC locus (21, 22), which includes a centromeric region and a telomeric region separated by KIR2DL4 in all haplotypes. This cluster is surrounded by KIR3DL2 at the end of the telomere and KIR3DL3 at the end of the centromere (23). The vast diversity of KIR gene loci is a result of gene duplication (24), non-allelic homologous recombination, diverse alleles due to the exchange of amino acids in the polymorphic region, nucleotide mutations, and haplotype variety. These variations create a range of genotypes within the population. This polymorphism of the KIR gene makes it such that it is extremely rare for two unrelated people have the same KIR genes. Some of these KIR variants contribute to the pathogenesis of autoimmune disorders (25, 26). Thus far, fourteen KIR genes and two pseudo genes have been identified in humans including: seven inhibitory genes, six activating genes, and one gene with both inhibitory and activating functions (KIR2DL4). The genes are named based on the structure of the molecules they encode for (27). After binding to their respective ligands, inhibitory KIRs (iKIRs) signal through their Immunoreceptor Tyrosine-based Inhibition Motifs (ITIMs) in their cytoplasmic sequence. Activating KIRs (aKIR) do not carry any signal motifs. They function through the use of membrane-bound signaling adaptor proteins, DAP10 and DAP12 (28). After binding to the receptor through the immunoreceptor tyrosine-based activation motif (ITAM), these molecules trigger an activation signal (29). The most common KIR haplotype in humans is relatively simple and carries five inhibitory genes (KIR2DL1, KIR2DL3, KIR3DL1, KIR3DL2, KIR3DL3), one activating gene (KIR2DS4), one gene with both activating and inhibitory functions (KIR2DL4), and two pseudo genes, commonly known as the group A KIR haplotype (30). The KIR-HLA interactions are involved in the pathogenesis of many diseases, including both autoimmune and inflammatory diseases (31, 32). Both KIR and HLA clusters have a high diversity among different populations that this variation is attractive in bioinformatics and anthropology (33-35).
Previous studies exploring the relationship between KIR and RA have conflicting and ambiguous results. Due to the inconsistent findings, the limited number of studies examining KIR-HLA and RA, and the diverse range of ethnicities in Iran, the aim of this study was to explore the relationship between KIR and their respective HLA ligands in RA susceptibility among the Lur population of Iran.
Materials and Methods
Design and patients
In this case-control study, a total of 50 patients with RA (35 women and 15 men) were enrolled. Participant demographics included an average age of 46.5 years, in which the mean age of women was 44 years and men was 52.4 years. Patients with RA were initially diagnosed according to the criteria of the American Rheumatology Association and referred to the hospitals of Shahid Rahimi and Shohadaye Ashayer of Khorramabad, Lorestan Province, Iran. Exclusion criteria included patients who were not of Lur heritage and those with additional comorbidities, such as malignancies. Genomic DNA of the participants was extracted from the white blood cells of peripheral blood samples. A total of 100 healthy controls were selected from a previous study by Shahsavar et al. (36). All controls were of Lur heritage and have lived in Lorestan.
KIR genotyping
The KIR-TYPE kit (BAG Health Care GmbH, Germany) was used to determine the expression of KIR genes using polymerase chain reaction technology and sequence-specific primers (PCR-SSP). Our PCR products included 20 KIR genes, 2DL1, 2DL2, 2DL3, 2DL4, 2DL5 (with two form 2DL5A and 2DL5B), 2DS1, 2DS2, 2DS3, 2DS4-full, 2DS4-del, 2DS4-del(*008), 2DS5, 3DL1, 3DL2, 3DL3, 3DS1, 2DP1, 3DP1-full, and 3DP1-del for patients with RA and healthy controls according to the kit's brochure. In order to prepare the mixture of reaction, 180 µl of molecular grade water, 26 µl of master mix (10 x PCR buffer) and 2.1 µl of Taq DNA polymerase enzyme (500 U) was transferred to a 2 ml microtube. Then, 10 µl of the prepared mixture was transferred to strip number 1 (negative control). Next, 52 µl of DNA (25-40 ng/µl) was added to the solution prepared in the microtube. After a few seconds of microfusion, 10 µl of the prepared mixture was transferred to strip numbers 2-22.
HLA genotyping
The EPITOP-TYPE kit (BAG Health Care GmbH, Germany) based on the PCR-SSP method was used to determine the presence of HLA alleles. In this study, we evaluated 5 HLA-ligand alleles including HLA-C1, HLA-C2, HLA-Bw4T, HLA-Bw4I, and HLA-Bw4A according to the kit brochure. In order to prepare the mixture of reaction, 55 µl of ordinary grade water, 8 µl of master mix (10 x buffer) and 0.64 µl of Taq DNA polymerase enzyme (500 U) was transferred to a 2 ml microtube. Then, 10 µl of the prepared mixture was transferred to strip number 6 (negative control). Next, 16 µl of DNA (25-40 ng/µl) was added to the solution prepared in the microtube (first stage). After a few seconds of microfusion, 10 µl of the prepared mixture was transferred to strip numbers 1-5.
Gel electrophoresis
Following PCR, gel electrophoresis was performed using horizontal electrophoresis and immersion of the gel in 1 X TBE buffer and Erythrogel dye solution. The results were reviewed and recorded using the GelDoc device (Fig. 1).
Fig. 1.
Gel electrophoresis of the PCR products. A) KIR genes including 1: 2DL1, 2: 2DL2, 3: 2DL3, 4: 2DL4, 5: 2DL5A, 6: 2DL5B, 7: 2DS1, 8: 2DS2, 9: 2DS3, 10: 2DS4-full, 11: 2DS4-del, 12: 2DS4-del(008), 13: 2DS5, 14: 3DL1, 15: 3DL2, 16: 3DL3, 17: 3DS1, 18: 2DP1, 19: 3DP1-full, 20: 3DP1-del. B) HLA genes including 1: C1, 2: C2, 3: Bw4T, 4: Bw4I, 5: A Bw4.
Statistical analysis
Chi-square test with Yate's correction and multivariable logistic regression were used to analyze the data. After calculating the odds ratio (OR) with 95% confidence interval (CI), the results were reported at a significance level of 0.05.
Ethical considerations
Approval of the methods for this study was provided by the ethics committee of Lorestan University of Medical Sciences (registration number IR.LUMS.REC.1396.375). Informed consent was obtained from each participant prior to taking part in the study.
Results
In our study population, a total of 22 genotypes were observed in the case and control groups. As expected, most of the participants had the AA genotype (cAcA-tAtA) (28% of the cases and 29% of the controls). No significant differences were observed among the genotypes (Table 1).
Table 1.
Genotypes of KIR among the participants.
| Genotype group | Genotype number | Genotype | A Score | B score | KIR genes | Frequency in Controls N (%) | Frequency in Cases N (%) | p Value | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inhibitory | Activating | Pseudo | |||||||||||||||||||||||
| 2DL1 | 2DL2 | 2DL3 | 2DL4 | 2DL5 | 3DL1 | 3DL2 | 3DL3 | 2DS1 | 2DS2 | 2DS3 | 2DS4 | 2DS5 | 3DS1 | 2DP1 | 3DP1 | Inhibitory | Activating | ||||||||
| AA | 1 | cAcA-tAtA | 4 | 0 | * | * | * | * | * | * | * | * | * | 6 | 1 | 29 | 14(28) | 0.8984 | |||||||
| BX | 2 | cAcA-tAtB | 2 | 1 | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 4 | 12 | 4(8) | 0.6401 | ||||
| 3 | cAcB-tAtA | 3 | 0 | * | * | * | * | * | * | * | * | * | * | * | 7 | 2 | 10 | 5(10) | 1 | ||||||
| 4 | cAcB-tAtA | 3 | 1 | * | * | * | * | * | * | * | * | * | * | * | * | * | 8 | 3 | 7 | 2(4) | 0.7154 | ||||
| 5 | cAcB-tAtB | 2 | 2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 8 | 6 | 7 | 2(4) | 0.7154 | |
| 2 | cAcB-tAtB | 2 | 1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 8 | 5 | 7 | 4(8) | 0.8247 | ||
| 7 | cAcB-tAtB | 2 | 1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 8 | 5 | 6 | 2(4) | 0.4651 | ||
| 8 | cBcB-tAtA | 1 | 1 | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 3 | 4 | 4(8) | 0.5206 | |||||
| 9 | cBcB-tAtB | 1 | 2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 6 | 3 | 3(6) | 0.6585 | ||
| 10 | cAcB-tAtB | 2 | 0 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 8 | 0 | 2 | 0(0) | 0.8013 | |||
| 11 | cBcB-tAtA | 1 | 0 | * | * | * | * | * | * | * | * | * | * | * | 6 | 2 | 4 | 0(0) | 0.8013 | ||||||
| 12 | cAcB-tBtB | 1 | 3 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 5 | 1 | 0(0) | 0.478 | |||
| 13 | cBcB-tBtB | 0 | 3 | * | * | * | * | * | * | * | * | * | * | * | * | * | 6 | 5 | 1 | 3(6) | 0.2097 | ||||
| 14 | cAcA-tBtB | 1 | 2 | * | * | * | * | * | * | * | * | * | * | * | 6 | 3 | 1 | 0(0) | 0.478 | ||||||
| 15 | cAcB-tBtB | 1 | 2 | * | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 4 | 1 | 1(2) | 0.6147 | ||||
| 16 | cAcB-tAtB | 2 | 1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 8 | 4 | 1 | 1(2) | 0.6147 | |||
| 17 | cAcA-tAtB | 2 | 0 | * | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 4 | 1 | 0(0) | 0.478 | ||||
| 18 | cAcA-tAtB | 2 | 1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 5 | 1 | 1(2) | 0.6147 | |||
| 19 | cAcA-tAtB | 2 | 1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 5 | 1 | 0(0) | 0.478 | |||
| 20 | cAcA-tAtB | 2 | 0 | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 3 | 1 | 1(2) | 0.6147 | |||||
| 21 | cBcB-tAtB | 1 | 1 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 5 | 1 | 1(2) | 0.6147 | |||
| 22 | cBcB-tAtA | 1 | 0 | * | * | * | * | * | * | * | * | * | * | * | * | * | 7 | 4 | 1 | 2(4) | 0.5362 | ||||
Our findings show that the inhibitor gene, KIR2DL5A (p= 0.0255, OR= 0.389, 95% CI= 0.210-0.722), and the activating gene, KIR2DS4-full (p< 0.0001, OR= 6.163, 95% CI= 3.174-11.968), had a significant association with RA susceptibility. No significant relationship was observed between other KIR genes and susceptibility to RA. After Bonferroni correction, only the relationship between KIR2DS4-full and increased risk for RA remained statistically significant (Table 2). For the examination of the HLA ligand alleles, our findings showed no significant relationship between any of the examined HLA alleles and RA susceptibility (Table 3). No significant differences were found for any of the examined combinations of HLA-KIR (Table 4). Haplotypes of KIR were studied according to previously reported methods (37). Protection against RA was found to only be associated with the centromeric A haplotype, however, after Yate’s correction, this difference did not remain statistically significant (Table 5). The mean of B score (the count of B haplotype in a genotype from 0 to 4) was observed to be higher in the group with RA (1.40 vs 1.17), although this was not statistically significant (p= 0.201, independent t test) (Table not shown).
Table 2.
Distribution of KIR genes and their associations with RA.
| KIR gene | Frequency in cases | Frequency in controls | Odds ratio | 95% CI | p value 1 |
|---|---|---|---|---|---|
| 2DL1 | 50 (100%) | 98 (98%) | 5.101 | 0.242-107.624 | 0.8013 |
| 2DL2 | 31 (62%) | 54 (54%) | 1.389 | 0.791-2.442 | 0.4489 |
| 2DL3 | 37 (74%) | 88 (88%) | 0.388 | 0.183-0.822 | 0.0528 |
| 2DL4 | 50 (100%) | 100 (100%) | 1 | 0.019-50.893 | 1 |
| 2DL5 | 30 (60%) | 61 (61%) | 0.959 | 0.544-1.691 | 0.9059 |
| 2DL5A | 11 (22%) | 42 (42%) | 0.389 | 0.210-0.722 | 0.0255* |
| 2DL5B | 23 (46%) | 31 (31%) | 1.896 | 1.064-3.380 | 0.1044 |
| 3DL1 | 47 (94%) | 96 (96%) | 0.653 | 0.178-2.387 | 0.8911 |
| 3DL2 | 50 (100%) | 100 (100%) | 1 | 0.019-50.893 | 1 |
| 3DL3 | 50 (100%) | 100 (100%) | 1 | 0.019-50.893 | 1 |
| 2DS1 | 23 (46%) | 48 (48%) | 0.923 | 0.529-1.608 | 0.9539 |
| 2DS2 | 31 (62%) | 55 (55%) | 1.334 | 0.759-2.346 | 0.5209 |
| 2DS3 | 19 (38%) | 34 (34%) | 1.19 | 0.667-2.121 | 0.7627 |
| 2DS4 | 46 (92%) | 96 (96%) | 0.479 | 0.139-1.646 | 0.5206 |
| 2DS4full | 42 (84%) | 46 (46%) | 6.163 | 3.174-11.968 | 0.0001** |
| 2DS4del | 38 (76%) | 72 (72%) | 1.231 | 0.654-2.320 | 0.7441 |
| 2DS5 | 20 (40%) | 40 (40%) | 1 | 0.568-1.761 | 1 |
| 3DS1 | 23 (46%) | 45 (45%) | 1.041 | 0.597-1.817 | 0.9077 |
| 2DP1 | 50 (100%) | 98 (98%) | 5.101 | 0.242-107.624 | 0.8013 |
| 3DP1 | 50 (100%) | 100 (100%) | 1 | 0.019-50.893 | 1 |
| 3DP1full | 25 (50%) | 39 (39%) | 1.564 | 0.892-2.742 | 0.2674 |
| 3DP1del | 48 (96%) | 90 (90%) | 2.667 | 0.807-8.806 | 0.3382 |
Chi-square with Yate's correction
Significant at 0.05.
Significant at 0.0023 (Bonferroni correction).
Table 3.
Distribution of HLA ligand alleles and their associations with RA.
| HLA ligand | Frequency in cases | Frequency in controls | Odds ratio | 95% CI | p value 1 |
|---|---|---|---|---|---|
| HLA-C1 | 41 (82%) | 75 (75%) | 1.518 | 0.768-3.000 | 0.4482 |
| HLA-C2 | 33 (66%) | 71 (71%) | 0.793 | 0.436-1.1440 | 0.6612 |
| HLA-BW4 | 39 (78%) | 73 (73%) | 1.311 | 0.682-2.504 | 0.6422 |
| HLA-BW4 T | 8 (16%) | 9 (9%) | 1.926 | 0.808-4.592 | 0.3165 |
| HLA-BW4 I | 25 (50%) | 51 (51%) | 0.961 | 0.552-1.673 | 0.9081 |
| HLA-A-BW4 | 18 (36%) | 33 (33%) | 1.142 | 0.637-2.047 | 0.9665 |
Chi-square with Yate's correction.
Table 4.
Distribution of HLA-KIR combinations and their associations with RA.
| HLA-KIR combination | Frequency in cases | Frequency in controls | Odds ratio | 95% CI | p value 1 |
|---|---|---|---|---|---|
| HLA-C1 and KIR2DL2/3 | 41 (82%) | 75 (75%) | 1.518 | 0.768-3.003 | 0.4482 |
| HLA-C2 and KIR2DL1 | 33 (66%) | 69 (69%) | 0.872 | 0.482-1.577 | 0.8527 |
| HLA-Bw4 and KIR3DL1 | 40 (80%) | 69 (69%) | 1.797 | 0.940-3.435 | 0.2184 |
| HLA-C1 and KIR2DS2 | 28 (56%) | 43 (43%) | 1.687 | 0.964-2.951 | 0.1836 |
| HLA-C2 and KIR2DS1 | 14 (28%) | 36 (36%) | 0.691 | 0.380-1.257 | 0.4260 |
| HLA-Bw4 and KIR3DS1 | 18 (36%) | 35 (35%) | 1.044 | 0.585-1.864 | 0.9039 |
Chi-square with Yate's correction.
Table 5.
Distribution of KIR haplotypes and their associations with RA.
| Haplotype | Included genes | Frequency in cases | Frequency in controls | Odds ratio | 95% CI | p-value | p-value (Yate's correction) |
|---|---|---|---|---|---|---|---|
| Centromeric A | 2DL3 | 37 (74%) | 88 (88%) | 0.388 | 0.183-0.822 | 0.0301* | 0.0528 |
| 2DL1 | |||||||
| Centromeric B | 2DS2 | 30 (60%) | 54 (54%) | 1.278 | 0.729-2.239 | 0.4853 | 0.6007 |
| 2DL2 | |||||||
| 2DS3 | |||||||
| 2DL5 | |||||||
| 2DL1 | |||||||
| Telomeric A | 3DL1 | 46 (92%) | 96 (96%) | 0.479 | 0.139-1.646 | 0.3041 | 0.5206 |
| 2DS4 | |||||||
| Telomeric B | 3DS1 | 24 (48%) | 45 (45%) | 1.128 | 0.647-1.967 | 0.7282 | 0.8621 |
| 2DL5 | |||||||
| 3DS3 | |||||||
| 2DS1 | |||||||
| 2DS5 |
Significant at 0.05.
Multivariable logistic regression was also used to reach the adjusted effects of KIR genes on disease susceptibility without confounding effects of other KIR genes. Our findings show that KIR2DS4-full exhibited the strongest risk association with RA (p< 0.001, adjusted OR= 18.531, 95% CI= 5.453-62.972) followed by KIR2DS4-del (p= 0.034, adjusted OR= 3.894, 95% CI= 1.107-13.690). KIR3DL1 showed the strongest protecting association with RA (p= 0.037, adjusted OR= 0.073, 95% CI= 0.006-0.850) followed by KIR2DL3 (p= 0.013, adjusted OR= 0.136, 95% CI= 0.028-0.657) and KIR2DL5A (p= 0.021, adjusted OR= 0.195, 95% CI= 0.049-0.783). After Bonferroni correction, only the association of KIR2DS4-full and increased risk for RA remained significant (Table 6 and Fig. 2).
Table 6.
Prediction of RA through complete profile of KIR using multivariable logistic regression.
| KIR genes | Adjusted odds ratio | 95% CI | SE | Z | p value | -log(p) |
|---|---|---|---|---|---|---|
| KIR2DL4 | NA | |||||
| KIR3DL2 | NA | |||||
| KIR3DL3 | NA | |||||
| KIR2DP1 | NA | |||||
| KIR2DL1 | NA | |||||
| KIR2DL2 | 5.284 | 0.292-95.64 | 7.807 | 1.13 | 0.260 | 0.585202 |
| KIR2DL3 | 0.136 | 0.028-0.657 | 0.109 | -2.48 | 0.013* | 1.884818 |
| KIR2DL5A | 0.195 | 0.049-0.783 | 0.138 | -2.31 | 0.021* | 1.674194 |
| KIR2DL5B | 0.83 | 0.311-2.212 | 0.415 | -0.37 | 0.709 | 0.149176 |
| KIR3DL1 | 0.073 | 0.006-0.850 | 0.091 | -2.09 | 0.037* | 1.435487 |
| KIR2DS1 | 0.696 | 0.041-11.707 | 1.002 | -0.25 | 0.801 | 0.096277 |
| KIR2DS2 | 0.301 | 0.016-5.798 | 0.454 | -0.8 | 0.426 | 0.370124 |
| KIR2DS3 | 0.593 | 0.116-3.021 | 0.492 | -0.63 | 0.529 | 0.276502 |
| KIR2DS4full | 18.531 | 5.453-62.972 | 11.565 | 4.68 | <0.001*** | 5.537358 |
| KIR2DS4del | 3.894 | 1.107-13.69 | 2.498 | 2.12 | 0.034* | 1.467455 |
| KIR2DS5 | 0.91 | 0.116-7.131 | 0.956 | -0.09 | 0.929 | 0.032058 |
| KIR3DS1 | 6.873 | 0.625-75.614 | 8.409 | 1.58 | 0.115 | 0.938657 |
| KIR3DP1full | 1.575 | 0.621-3.991 | 0.747 | 0.96 | 0.338 | 0.470567 |
| KIR3DP1del | 1.343 | 0.173-10.412 | 1.404 | 0.28 | 0.778 | 0.109256 |
Nagelkerke R square= 0.429. NA: not applicable because of collinearity.
Significant at 0.05.
Significant at 0.001.
Fig. 2.
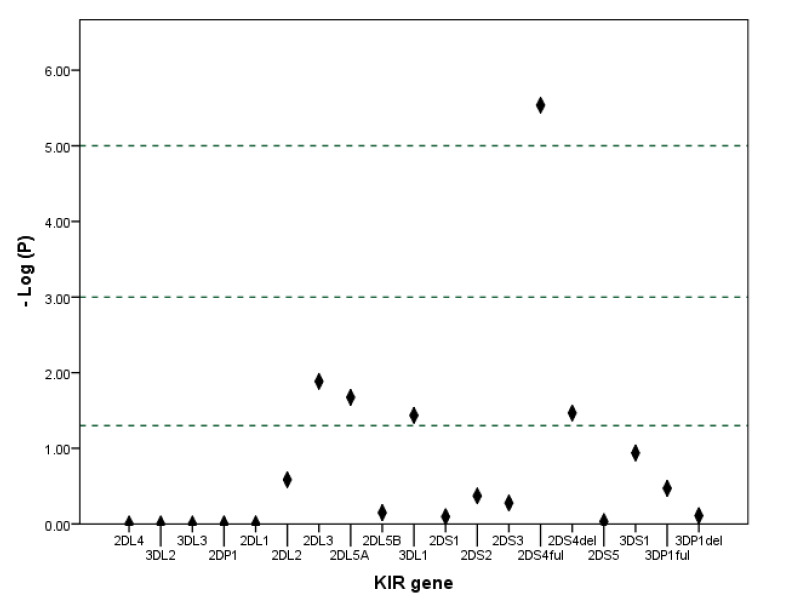
Manhattan plot of the logistic regression model showing –Log10 of P value for each gene. The reference lines indicate significance levels 0.05, 0.001 and 0.00001.
Discussion
This genetic association study was designed to determine the role of KIR genes in susceptibility to RA in the Lur population of Iran. Autoimmune diseases result from the failure of immune tolerance mechanisms to prevent reactions to self-antigens or unregulated activity of the immune system, both leading to tissue damage (38). NK cells detect class I HLA through their KIR cell surface receptors which, upon binding, can have different effects on the immune system in different individuals due to KIR gene polymorphism. RA is one of the most common autoimmune diseases in which overactive NK cells play a major part in the susceptibility and pathogenesis of RA. However, the exact role of NK cells and whether they are beneficial or harmful in disease progression remains controversial (39).
In the present study, KIR2DL3 was observed to be less prevalent in the patient group and, before Yate’s correction, have a statistically significant protective relationship with RA decreasing susceptibility to RA. Although the frequency of KIR2DL5 was the same in both the control and patient groups, one of its subtypes, KIR2DL5A, was significantly lower in the patient group, and statistically, it had a significant protective affect against the development of RA. Among the activating genes, despite the similar presence of KIR2DS4 in both the patient and control groups, one of its subtypes, KIR2DS4-full, had a much higher frequency in the patient group than the control group. This finding indicates a significant relationship between KIR2DS4-ful and increased risk for RA. The results of our study suggest that a reduction in the frequency of KIR2DL3 and KIR2DL5A genes and an increase in the frequency of KIR2DS4-full may predispose an individual to RA. Our logistic regression model showed a similar result.
In a study by Yen et al. (2001) in Finland, they observed an increase in the activating genes, KIR2DS2 and KIR2DS4-full, in patients with RA, corroborating our findings (40). Nevertheless, Kogure et al. (2007) found no correlation between the expression of KIR and disease activity when examining RA in Japanese patients (41). The results of our study were consistent with findings from Yen et al. (2006) in which they observed KIR2DS4 to be a risk factor for RA (42). Our findings were also consistent with a meta-analysis by Xiaona Li et al. (2015) about the protective effect of KIR2DL3 (43). In an Iranian case-control study conducted by Nazari et al. (2015), KIR2DL5A, KIR2DL5B, and KIR2DL2, as well as the activating KIRs, KIR2DS5 and KIR3DS1, were found to have a protective effect against RA (9). A meta-analysis conducted by Aghaei et al. (2019) found that there were significant relationships between KIR2DL3, KIR2DL5, KIR2DS5, and KIR3DL3 and a reduced risk for RA. However, the findings among the different studies were not entirely consistent (2).
With respect to the KIR haplotypes, even though the centromeric A haplotype was more abundant, its association with RA did not remain statistically significant following Yate's correction. Regarding HLA, our study did not show a significant relationship between any of the examined HLA ligands and RA. In previous research studies, Nishimura et al. (2015) in Brazil, observed an association between the presence of HLA-DRB1*1402 and HLA-DRB1*0101 and cutaneous ulcers in patients with arthritis vasculitis (44). Regarding KIR-HLA compounds, although our study found HLA-C1 and KIR2DS2 to be more common in patients with RA, which was consistent with the results of a study by Mc Geough et al. in 2010 in Northern Ireland (45), no statistically significant correlation was observed between these genes and the degree of RA risk in our study. This discrepancy could be due to differences in the type of the study and the type of ethnicity examined. Among different ethnicities, some genes have both haplotypes and genotypes together, resulting in the effects of single genes being hidden under one another. In this study, we overcame this limitation with multiple logistic regression. With respect to the KIR2DS4-full gene after adjustment, the OR almost tripled. In our study, the KIR2DL3 and KIR3DL1 genes were both observed to have protective effects against RA. However, this protection was partially counteracted due to the simultaneous presence of KIR2DS4 in the AA genotype, which may confirm the phenomenon of natural selection during evolution (i.e., the individuals with harmful genes survived if they had some other protecting genes). Our study was consistent with the findings from Yen et al. (2006) related to the presence of KIR2DS4 as a risk factor for RA.
Due to financial limitations, this study had a relatively low sample size and used control samples from previous work. Regardless of the low sample size, our study was able to reveal significant associations among groups, indicating sufficient power. Despite the limitations, this study added this finding that in the Lur population of Iran KIR2DS4-full was the most dominant risk factor for RA among the KIR genes.
In this study, our findings show KIR2DS4-full to increase susceptibility to RA whereas KIR2DL5A was found to be a protective factor based on both cross Table (unadjusted) and regression (adjusted) analyses. Furthermore, KIR2DL3 and KIR3DL1 was observed to have a protective role and KIR2DS4-del was found to increase the risk of RA based on the regression model. However, only the role of KIR2DS4-full remained statistically significant multiple comparison wise. Since the association of these genes with susceptibility to RA is ethnicity dependent, further research should explore these relationships in other populations.
Acknowledgements
We would like to thank Lorestan University of Medical Science for approving this dissertation with grant number 574 as a MSc thesis of Bijan Ansari-Moghaddam under the supervision of Farhad shahsavar.
The authors declare no conflicts of interest in this study.
References
- 1.McInnes IB, Schett G. Cytokines in the pathogenesis of rheumatoid arthritis. . Nat Rev Immunol. . 2007;7(6):429, 42. doi: 10.1038/nri2094. [DOI] [PubMed] [Google Scholar]
- 2.Aghaei H, Mostafaei S, Aslani S, Jamshidi A, Mahmoudi M. Association study between KIR polymorphisms and rheumatoid arthritis disease: an updated meta-analysis. . BMC Med Genet. 2019;20(1):24. doi: 10.1186/s12881-019-0754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deane KD, Holers VM. The natural history of rheumatoid arthritis. . Clin Ther. 2019;41(7):1256, 1269. doi: 10.1016/j.clinthera.2019.04.028. [DOI] [PubMed] [Google Scholar]
- 4.Guo Q, Wang Y, Xu D, Nossent J, Pavlos NJ, Xu J. Rheumatoid arthritis: pathological mechanisms and modern pharmacologic therapies. Bone research. 2018;6(1):15. doi: 10.1038/s41413-018-0016-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlag DM, Raza K, van Baarsen LG, Brouwer E, Buckley CD, Burmester GR, et al. EULAR recommendations for terminology and research in individuals at risk of rheumatoid arthritis: report from the Study Group for Risk Factors for Rheumatoid Arthritis. . Ann Rheum Dis. . 2012;71(5):638–41. doi: 10.1136/annrheumdis-2011-200990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbasi Z, Kazemi Nezhad SR, Pourmahdi-Broojeni M, Rajaei E . Association of PTPN22 rs2476601 Polymorphism with rheumatoid arthritis and celiac disease in Khuzestan province, southwestern Iran. . Iran Biomed J. 2017;21(1):61–6. doi: 10.6091/.21.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sánchez-Hernández P, Muñoz-Valle J, Palafox-Sánchez C, Rosales-Rivera L, García-Iglesias T, Daneri-Navarro A, et al. Associations of killer cell immunoglobulin-like receptor genes with rheumatoid arthritis. . Dis Markers. . 2012;33(4):201–6. doi: 10.3233/DMA-2012-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappelli LC, Dorak MT, Bettinotti MP, Bingham III CO, Shah AA. Association of HLA-DRB1 shared epitope alleles and immune checkpoint inhibitor-induced inflammatory arthritis. . Rheumatology. . 2018;58(3):476–480. doi: 10.1093/rheumatology/key358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nazari M, Mahmoudi M, Rahmani F, Akhlaghi M, Beigy M, Azarian M, et al. Association of killer cell immunoglobulin-like receptor genes in iranian patients with rheumatoid arthritis. . PloS one. 2015;10(12):e0143757. doi: 10.1371/journal.pone.0143757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell JJ, Qin S, Unutmaz D, Soler D, Murphy KE, Hodge MR, et al. Unique subpopulations of CD56+ NK and NK-T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. . J Immunol. 2001;166(11):6477–82. doi: 10.4049/jimmunol.166.11.6477. [DOI] [PubMed] [Google Scholar]
- 11.Fehniger TA, Cooper MA, Nuovo GJ, Cella M, Facchetti F, Colonna M, et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell–derived IL-2: a potential new link between adaptive and innate immunity. . Blood. . 2003;101(8):3052–7. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 12.Dalbeth N, Callan MF. A subset of natural killer cells is greatly expanded within inflamed joints. . Arthritis Rheum. . 2002;46(7):1763–72. doi: 10.1002/art.10410. [DOI] [PubMed] [Google Scholar]
- 13.Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. . 1999;190(10):1505–16. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivori S, Carlomagno S, Pesce S, Moretta A, Vitale M, Marcenaro E. TLR/NCR/KIR: which one to use and when?. . Front Immunol. . 2014;5:105. doi: 10.3389/fimmu.2014.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanier LL. On guard—activating NK cell receptors. Nat Immunol. . 2001;2(1):23–7. doi: 10.1038/83130. [DOI] [PubMed] [Google Scholar]
- 16.McQueen KL, Parham P. Variable receptors controlling activation and inhibition of NK cells. . Curr Opin Immunol. 2002;14(5):615–21. doi: 10.1016/s0952-7915(02)00380-1. [DOI] [PubMed] [Google Scholar]
- 17.Lanier LL. NK cell receptors. Annual review of immunology. 1998;16(1):359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 18.Long EO, Barber DF, Burshtyn DN, Faure M, Peterson M, Rajagopalan S, et al. Inhibition of natural killer cell activation signals by killer cell immunoglobulin-like receptors (CD158). . Immunol Rev. . 2001;181(1):223–33. doi: 10.1034/j.1600-065x.2001.1810119.x. [DOI] [PubMed] [Google Scholar]
- 19.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al., editors. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. . Annu Rev Immunol. . 2001;19(1):197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 20.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. . Annu Rev Immunol. . 2002;20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 21.Lanier LL. NK cell recognition. . Annu Rev Immunol. . 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 22.Kumar V, McNerney ME. A new self: MHC-class-I-independent natural-killer-cell self-tolerance. Nat Rev Immunol. 2005;5(5):363–74. doi: 10.1038/nri1603. [DOI] [PubMed] [Google Scholar]
- 23.Carrington M, Norman P. The KIR gene cluster. 2003 [Google Scholar]
- 24.Martin AM, Kulski JK, Gaudieri S, Witt CS, Freitas EM, Trowsdale J, et al. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. . Gene. 2004;335:121–31. doi: 10.1016/j.gene.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. . J Immunol. . 2002;168(5):2307–15. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 26.Berinstein J, Pollock R, Pellett F, Thavaneswaran A, Chandran V, Gladman DD. Association of variably expressed KIR3dl1 alleles with psoriatic disease. . Clin Rheumatol. . 2017;36(10):2261–2266. doi: 10.1007/s10067-017-3784-5. [DOI] [PubMed] [Google Scholar]
- 27.Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. . Hum Immunol. . 2003;64(6):648–54. doi: 10.1016/s0198-8859(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 28.Lanier LL, Bakker AB. The ITAM-bearing transmembrane adaptor DAP12 in lymphoid and myeloid cell function. . Immunol Today. . 2000;21(12):611, 4. doi: 10.1016/s0167-5699(00)01745-x. [DOI] [PubMed] [Google Scholar]
- 29.Rajalingam R. Killer cell immunoglobulin-like receptors influence the innate and adaptive immune responses. Iran J Immunol. 2007;4(2):61, 78. doi: 10.22034/iji.2007.17182. [DOI] [PubMed] [Google Scholar]
- 30.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7(6):753, 63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 31.Shahsavar F, Mapar S, Ahmadi SAY. Multiple sclerosis is accompanied by lack of KIR2DS1 gene: A meta-analysis. . Genom Data. . 2016;10:75, 78. doi: 10.1016/j.gdata.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shahsavar F, Mousavi T, Entezami K, Azargoon A R. Association of KIR-HLA interactions with diseases. . Yafteh. . 2011;13(3):82, 96. [Google Scholar]
- 33.Shapouri-Moghaddam A, Mohammadi M, Rahimi HR, Esmaeili H, Mahmoudi M, Modaghegh M-HS, et al. The Association of HLA-A, B and DRB1 with Buerger's Disease. Rep Biochem Mol Biol. 2019;8(2):153, 160. [PMC free article] [PubMed] [Google Scholar]
- 34.Shahsavar F, Varzi A-M, Ahmadi SAY. A genomic study on distribution of human leukocyte antigen (HLA)-A and HLA-B alleles in Lak population of Iran. . Genom Data. . 2017;11:3, 6. doi: 10.1016/j.gdata.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez-Galarza FF, McCabe A, Santos EJMd, Jones J, Takeshita L, Ortega-Rivera ND, et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. . Nucleic Acids Res. 2020;48(D1):D783, D788. doi: 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shayanrad B, Ghanadi K, Varzi A-M, Birjandi M, Ahmadi SAY, Shahsavar F. Association of KIR genes and their HLA ligands diversity with colorectal cancer in Lur population of Iran. . Meta Gene. . 2019;22:100603. [Google Scholar]
- 37.Leung W. Infusions of allogeneic natural killer cells as cancer therapy. . Clin Cancer Res. 2014;20(13):3390, 400. doi: 10.1158/1078-0432.CCR-13-1766. [DOI] [PubMed] [Google Scholar]
- 38.Toubi E, Vadasz Z. Think autoimmunity, breath autoimmunity, and learn autoimmunity. Clin Rheumatol. 2019;38(5):1227, 1230. doi: 10.1007/s10067-019-04540-2. [DOI] [PubMed] [Google Scholar]
- 39.Shegarfi H, Naddafi F, Mirshafiey A. Natural killer cells and their role in rheumatoid arthritis: friend or foe?. . ScientificWorldJournal. . 2012;2012:491974. doi: 10.1100/2012/491974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen J-H, Moore BE, Nakajima T, Scholl D, Schaid DJ, Weyand CM, et al. Major histocompatibility complex class I–recognizing receptors are disease risk genes in rheumatoid arthritis. J Exp Med. 2001;193(10):1159, 67. doi: 10.1084/jem.193.10.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kogure T, Tatsumi T, Niizawa A, Fujinaga H, Ito T, Shimada Y, et al. No correlation exists between disease activity and the expression of killer-cell immunoglobulin-like receptors in patients with rheumatoid arthritis. . Mediators Inflamm. . 2007;2007:65179. doi: 10.1155/2007/65179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yen JH, Lin CH, Tsai WC, Wu CC, Ou TT, Hu CJ, et al. Killer cell immunoglobulin-like receptor gene's repertoire in rheumatoid arthritis. . Scand J Rheumatol. 2006;35(2):124, 7. doi: 10.1080/03009740500381252. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Xia Q, Fan D, Cai G, Yang X, Wang L, et al. Association between KIR gene polymorphisms and rheumatoid arthritis susceptibility: a meta-analysis. Human Immunology. 2015;76(8):565, 570. doi: 10.1016/j.humimm.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 44.Nishimura WE, Sachetto Z, Costallat LTL, Yazbek MA, Londe ACS, Guariento EG, et al. The role of KIR2DL3/HLA-C* 0802 in Brazilian patients with rheumatoid vasculitis. Clinics. 2015;70(6):408, 12. doi: 10.6061/clinics/2015(06)04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGeough CM, Berrar D, Wright G, Mathews C, Gilmore P, Cunningham RT, et al. Killer immunoglobulin-like receptor and human leukocyte antigen-C genotypes in rheumatoid arthritis primary responders and non-responders to anti-TNF-α therapy. Rheumatology international. . 2012;32(6):1647, 53. doi: 10.1007/s00296-011-1838-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


