Abstract
COVID-19, the disease caused by SARS-CoV-2, has been declared as a global pandemic. Traditional medicinal plants have long history to treat viral infections. Our in silico approach suggested that unique phytocompounds such as emodin, thymol and carvacrol, and artemisinin could physically bind SARS-CoV-2 spike glycoproteins (6VXX and 6VYB), SARS-CoV-2 B.1.351 South Africa variant of Spike glycoprotein (7NXA), and even with ACE2 and prevent the SARS-CoV-2 binding to the host ACE2, TMPRSS2 and neutrapilin-1 receptors. Since Chloroquine has been looked as potential therapy against COVID-19, we also compared the binding of chloroquine and artemisinin for its interaction with spike proteins (6VXX, 6VYB) and its variant 7NXA, respectively. Molecular docking study of phytocompounds and SARS-CoV-2 spike protein was performed by using AutoDock/Vina software. Molecular dynamics (MD) simulation was performed for 50ns. Among all the phytocompounds, molecular docking studies revealed lowest binding energy of artemisinin with 6VXX and 6VYB, with Etotal −10.5 KJ mol−1 and −10.3 KJ mol−1 respectively. Emodin showed the best binding affinity with 6VYB with Etotal −8.8 KJ mol−1and SARS-CoV-2 B.1.351 variant (7NXA) with binding energy of −6.4KJ mol−1. Emodin showed best interactions with TMPRSS 2 and ACE2 with Etotal of −7.1 and −7.3 KJ mol−1 respectively, whereas artemisinin interacts with TMPRSS 2 and ACE2 with Etotal of −6.9 and −7.4 KJ mol−1 respectively. All the phytocompounds were non-toxic and non-carcinogenic. MD simulation showed that artemisinin has more stable interaction with 6VYB as compared to 6VXX, and hence proposed as potential phytochemical to prevent SARS-CoV-2 interaction with ACE-2 receptor.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s40495-021-00259-4.
Keywords: COVID-19, Antimalarial drugs, Chloroquine, Artemisinin, Phytocompounds, Emodin, Thymol, In silico, MD simulation
Introduction
COVID-19 caused by a member of family Coronaries (CoV) has threatened the survival of human beings on the Earth and it has been declared as global health emergency by the World Health Organization (WHO) [1••]. Coronaviruses (CoV) are known for their ability to cause illness, with severe diseases such as Middle East respiratory syndrome (MERS-CoV) and severe acute respiratory syndrome (SARS-CoV). Though there are other coronaviruses known to infect humans (hCoVsOC43 and 229E), but they are mild pathogens responsible for common cold. In Latin, Corona means “halo” or “crown”; thus, the name represents the structure of the virus which consists of crown like projections on its surface [2]. In 1937, coronavirus was isolated from an infectious bronchitis virus in birds which was responsible to ruin the poultry stocks [3]. Coronaviruses are the type of viruses that directly affect the respiratory tract. These are associated with the common cold, pneumonia, gut, and severe acute respiratory syndrome. Coronaviruses are zoonotic, which means they are transmitted between animals and humans [4]. A new strain, novel coronavirus (nCoV) has come into knowledge since 2019 and has emerged as a threat to mankind.
The first case of Middle East respiratory syndrome (MERS-CoV) was seen in the year 2012, a businessman in Saudi Arabia who died from viral pneumonia [5]. In 2016, a report on 1998 was published by the World Health Organization (WHO) regarding the confirmed cases of MERS-CoV infection and the death rate was approximately 36% (Middle East respiratory coronavirus (MERS-CoV) [5]. The biggest outbreak with first ever confirmed case of this disease came into existence in the year 2015 in South Korea. Including the China, the confirmed cases extend to 186 with total 36 deaths [6, 7]. Cases regarding the novel coronavirus came in to existence among the population of Wuhan, China, on December 8, 2019. Pneumonia was the first symptom of infection and most of the cases were linked to a local fish and animal market. During the research, it was seen that 2019 novel coronavirus was recognized as pathogenic agent responsible for evolution of pneumonia [8]. On January 20, 2020, laboratory in Korea confirmed the first case of coronavirus. On 23 January, 2020, the government of China announced total shutdown of country and advised the people for undergoing personal isolation. In the USA, there are five variants of SARS-Cov-2. B.1.1.7: This variant was discovered for the first time in December 2020 in the USA. It was first discovered in the UK. B.1.351: This variant was discovered for the first time in the USA at the end of January 2021. It was first discovered in December 2020 in South Africa. P.1: In January 2021, this variant was discovered for the first time in the USA. B.1.427 and B.1.429: These two variants were discovered in February 2021 in California (https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant.html).
SARS-CoV-2 consists of four structural proteins: spike (S), membrane (M), envelop (E), and nucleocapsid (N) proteins [9]. Among all, S protein plays an important role in viral attachment, fusion, entry, and also act as a target for development of antibodies, entry inhibitors, and vaccines [10, 11]. The S1 domains of coronaviruses contain receptor-binding domains (RBDs) that directly bind to the cellular receptors [12, 13]. In general, SARS-CoV surface exhibits two components: S1, which contains the receptor binding domain (RBD); and S2, which contains the fusion peptide. SARS-CoV gains entry into cells through interaction of the SARS-SRBD with the cell surface receptor angiotensin-converting enzyme 2 (ACE2) [14, 15]. These interactions are followed by endocytosis, and at the low pH in endosomes, SARS-S is cleaved by a cellular protease called cathepsin L, thereby exposing the S2 domain of the spike protein for membrane fusion [16, 17]. The minimal RBD of SARS-CoV S protein is located in the S1 subunit (AA 318–510) and is responsible for viral binding to host cell receptors [18, 19]. Besides the main receptor for the angiotensin-converting enzyme 2, there are several alternative receptors, such as dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin and liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin [20]. SARS-CoVs recognizes angiotensin-converting enzyme 2 (ACE2) as its receptor, whereas MERS-CoV recognizes dipeptidyl peptidase 4 (DPP4) as its receptor [21, 22]. Two residues (AA 479 and AA 487) in RBD determine SARS progression and tropism, and their mutations may enhance animal-to-human or human-to-human transmission [13]. Some residues (AA 109, 118, 119, 158, 227, 589, and 699) in S protein are critical strategies against this deadly viral agent, especially in high-risk groups, including people of every age group [23]. According to the previous data, the ACE2 receptor expressing cell fused with SARS-S-expressing cells adds to the cell surface by pH-independent mechanism [19]. It enhances the cell stress responses and apoptosis [24]. Binding is very critical for pathogenesis and if the binding of SARS-S with ACE2 receptor is blocked, infection can be stopped. Traditional medicinal plants produce large number of compounds which are used as therapeutics to kill the pathogens [25]. In the recent years, many reports published on antimicrobial activity of the medicinal plants [25–27]. It is expected that plant extracts and phytocompounds showing the target site other than antibiotics, a very little information is available on this type of activity of medicinal plants [26, 27]. Extracts of medicinal plants have been used from ancient times and these plants are known for their antiviral properties and less side effects. Traditionally, thyme was acclimated to treat asthma and loosen congestion in the throat and stomach [28]. The pharmacological manuscript of Chailander medical codex (fifteenth and sixteenth centuries) mentions the utilizations of wild thyme for the treatment of headaches caused by cold and laryngitis [29]. During the Renaissance period (sixteenth and seventeenth centuries), wild thyme was utilized internally to treat malaria and epilepsy [30]. Traditionally in many countries, areal part of T. serpyllum is utilized as anthelmintic, a vigorous antiseptic, an antispasmodic, a carminative, deodorant, diaphoretic, disinfectant, expectorant, sedative, and tonic. Thymus serpyllum additionally used to treat respiratory quandaries [29]. In western Balkans, thymus species used to amend blood circulation and as anticholesterolemic, immunostimulant [31]. Carvacrol and thymol are isomers, belonging to the group of monoterpenic phenols with potent antiseptic properties. Chauhan et al. [32] reported thymol (25–200 mg kg−1) as immunomodulatory in cyclosporine A-treated Swiss albino mice by enhancing the expression of cluster of differentiation 4 (CD4), cluster of differentiation 8 (CD8), and Th1 cytokines via upregulation of IFN-4 expression and enhanced secretion of interleukin-12 (IL-12).
Antiviral property of Thymus serpyllum [33] and thymol is already reported [34]. Pilau et al. [35] reported the antiviral activity of carvacrol from Lippia graveolens against human and animal virus (herpes simplex virus, acyclovir-resistant herpes simplex virus 1, bovine herpesvirus 2, respiratory syncytial virus; human rotavirus, bovine viral diarrhea virus). Antiviral nature of Emodin was also reported in several studies [36, 37]. Study from Efferth et al. [38] showed in vitro antiviral properties of artemisinin against hepatitis B virus, hepatitis C virus, and bovine viral diarrhea. Keeping in view the antiviral potential of Himalayan herbs, the current study was focused on the identification of potent phytocompounds from Himalayan herbs (Rheum emodi, Thymus serpyllum, and Artemisia annua) to cure a dangerous COVID-19.
Material and Methods
Bioinformatics Tools
Open Babel GUI [39], UCSF Chimera 1.8.1 [40], Pubchem (www.pubchem.com), RCSB PDB (http://www.rscb.org/pdb), PDBsum (www.ebi.ac.uk/pdbsum), and Autodock/vina software [41, 42] were used in the present investigation.
Ligand Preparation
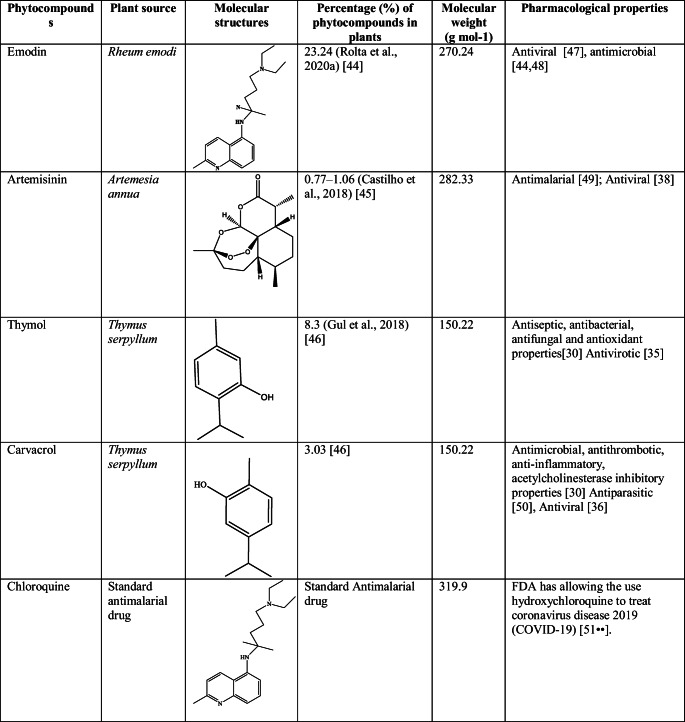
Four major phytocompounds of three medicinal plants—emodin of Rheum emodi, thymol and carvacrol of Thymus serpyllum, and artemisinin of Artemisia annua—were used for the docking studies. The 3-dimensional structures of all the phytocompounds and chloroquine were obtained from PubChem (www.pubchem.com) in .sdf format. The .sdf file of the phytocompounds was converted into PDB and pdbqt format by using the Open Babel tool [43]. Table 1 shows molecular structure, molecular weight, pharmacological properties, plant source, and percentage of phytocompounds in the respective plants and antimalarial drug chloroquine. Targets of phytocompounds and standard drug chloroquine were predicted by using SwissADME online server.
Table 1.
Molecular structure, molecular weight, pharmacological properties, plant source, and percentage of selected phytocompounds and chloroquine
Protein Preparation
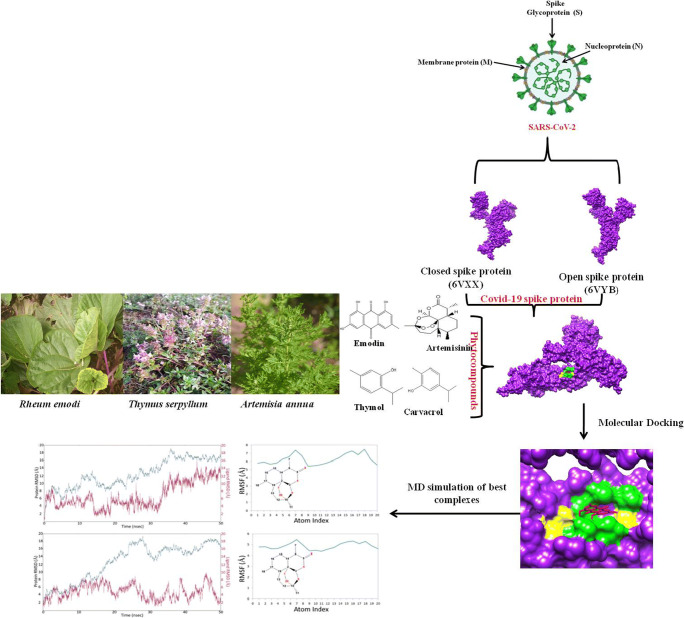
Two spike proteins of SARS-CoV-2 spike glycoprotein (PDB ID: 6VXX, closed conformation), SARS-CoV-2 spike ectodomain structure (PDB ID: 6VYB, open conformation) [52] and one mutated variant of SARS-CoV-2 B.1.351 (South African variant) variant of Spike glycoprotein (PDB ID: 7NXA) [53] and two receptor of SARS-CoV-2 (Human TMPRSS2 (PDB ID: 7MEQ) [54], Angiotensin-converting enzyme-2 (ACE2 PDB ID: 6M1D)) [55••], and neuropilin-1 (PDB ID: 4DEQ) were used to analyze the interactions of major phytocompounds of R. emodi, T. serpyllum, and A. annua. It has been shown that SARS CoV-2 SB open protein conformation is necessary for binding with ACE2 at host surface; and coronavirus with open surface S-glycoprotein trimers found to be highly pathogenic to human [56••].
The 3-dimensional structures of selected target proteins were retrieved from the Protein Data Bank (PDB) (http://www.rscb.org/pdb). Non-essential water molecules, including heteroatoms, were removed from the target receptor molecule and hydrogen atoms were added to the target receptor molecule. Binding site of both the target proteins of COVID-19 (SARS-CoV-2 spike glycoprotein (PDB ID: 6VXX), SARS-CoV-2 spike ectodomain structure (PDB ID: 6VYB)), SARS-CoV-2 B.1.351 variant Spike glycoprotein (PDB ID: 7NXA), Human TMPRSS2 (7MEQ), Angiotensin-converting enzyme-2, ACE2 (PDB ID: 6M1D), and neuropilin-1 (PDB ID: 4DEQ) was determined by grid box generation. Grid box was generated by adjusting the grid parameter x, y, z coordinate values; grid value for 6VYB and 6VXX was center x: −189.229, y: −255.9, z: 229.87 Å; 7NXA was x: −14.806, y: −19.528, z: −51.972 Å; 7MEQ was x: −1.028, y: −0.352, z: 10.912; and 6MID was x: 126.806, y: 133.196, z:121.533. Size of the grid was same for all the target proteins (i.e., x—40, y—40, z—40 Å) using AutoDock software. The grid values were recorded in the config.txt file format [57••].
Prediction of Drug Likeness of Selected Phytocompounds
The aim of the drug scan was to see whether selected phytochemicals met the drug-likeness criteria. Lipinski’s filters using Molinspiration (http://www.molinspiration.com) were applied for examining drug-likeness attributes, including quantity of hydrogen acceptors (should not be more than 10), quantity of hydrogen donors (should not be more than 5), molecular weight (mass should be more than 500 Daltons), and partition coefficient log P (should not be less than 5). The smiles format of each of the phytochemical was uploaded for the analysis [58].
ADME and Toxicity Prediction of Selected Phytocompounds
Absorption, distribution, metabolism, excretion, and toxicity (ADMET) screening was done to determine the absorption, toxicity, and drug-likeness properties of the selected ligands. The 3-dimensional structures of ligands such as emodin, thymol, carvacrol, artemisinin, and chloroquine were saved in .smiles format and chloroquine was uploaded on SWISSADME (Molecular Modeling Group of the SIB (Swiss Institute of Bioinformatics), Lausanne, Switzerland), admetSAR (Laboratory of Molecular Modeling and Design, Shanghai, China), and PROTOX web servers (Charite University of Medicine, Institute for Physiology, Structural Bioinformatics Group, Berlin, Germany) for ADMET screening. SWISSADME is a web tool used for the prediction of ADME and pharmacokinetic properties of a molecule. The predicted results consist of lipophilicity, water solubility, physicochemical properties, pharmacokinetics, drug likeness, medicinal chemistry, and Brain or Intestinal Estimated permeation method (blood-brain barrier and PGP ± prediction). AdmetSAR provides ADMET profiles for query molecules and can predict about fifty ADMET properties. Toxicity classes are as follows: (i) category I contains compounds with LD50 values ≤ 50 mg kg−1, (ii) category II contains compounds with LD50 values > 50 mg kg−1 but 500 mg kg−1 but 5000 mg kg−1 (Cheng, 2020). PROTOX is a rodent oral toxicity server predicting LD50 value and toxicity class of query molecule. The toxicity classes are as follows: class 1: fatal if swallowed (LD50 ≤5), class 2: fatal if swallowed (55000) [59].
Docking of COVID-19 Receptors and Phytocompounds
The docking of selected ligands to the catalytic triad of protein was performed by using AutoDock/Vina [41]. Docking was performed to obtain populations of conformations and orientation for ligands at binding sites. Docking was performed to study the interactions between SARS-CoV-2 receptors such as 6VXX (closed state), 6VYB (open state), 7NXA (SARS-CoV-2 B.1.351 variant), 7MEQ, 6MID, and neuropilin-1 (4DEQ) with major phytocompounds of R. emodi, T. serpyllum, and A. annua and .pdb file of proteins-ligand complexes was generated. All the bonds of ligands were set to be rotatable. All calculations for protein-ligand docking were performed using the Lamarckian genetic algorithm (LGA) method. The best conformation was chosen with the lowest docked energy, after the completion of docking search. The .pdb complex of protein and ligand was analyzed by using Discovery Studio (https://discover.3ds.com/d) to study the list of interactions between protein and ligand complex. Detailed visualization and comparison of the docked sites of target proteins and ligands were done by using Chimera [60].
Molecular Dynamics (MD) Simulation
In order to further verify the accuracy of docking observations, the two best complexes of S-protein were selected for extensive MD simulation for 50ns. Both the complexes were introduced into Desmond software to study the binding stability of the ligand within the binding site of S-protein [60]. Both complexes were prepared prior to simulation to remove any structural error as described earlier [61, 62]. Both the complexes were solvated in TIP3P water model and 0.15 M NaCl to mimic a physiological ionic concentration. This MD simulation was performed with OPLS3e force field.
Results
Drug Likeness of Selected Phytocompounds
Early preclinical production is supported by drug-likeness filters, which help to prevent expensive late-stage preclinical and clinical failure. The Lipinski rule of 5 was used to evaluate the drug-likeness properties of the emodin, thymol, carvacrol, artemisinin, and chloroquine. The emodin, thymol, carvacrol, and artemisinin followed the Lipinski’s rule of five, whereas chloroquine showed one violation (Table 2).
Table 2.
Prediction of drug-likeness activity of selected phytocompounds
| Phytocompounds | miLogP | TPSA | natoms | MW | nON | nOHNH | nviolations |
|---|---|---|---|---|---|---|---|
| Emodin | 3.01 | 94.83 | 20 | 270.24 | 5 | 3 | 0 |
| Thymol | 3.34 | 20.23 | 11 | 150.22 | 1 | 1 | 0 |
| Carvacrol | 3.81 | 20.23 | 11 | 150.22 | 1 | 1 | 0 |
| Artemisinin | 3.32 | 54.01 | 20 | 282.34 | 5 | 0 | 0 |
| Chloroquine | 5 | 28.16 | 22 | 319.88 | 3 | 1 | 1 |
ADMET Prediction and Toxicity Analysis of Selected Phytocompounds
The comparative ADME properties predicted by SwissADME of emodin, thymol, carvacrol, artemisinin, and chloroquine are summarized in Table 3. Consensus Log Po/w value of ˂5 indicates good aqueous solubility, which means that an adequate amount of drug can reach and be maintained inside the body through oral administration. The emodin, thymol, carvacrol, artemisinin, and chloroquine showed consensus Log Po/w value of < 5 (Table 3). TPSA indicates permeability of compounds into the cells. A TPSA value of < 140 Å2 is required for good permeation of compound into the cell membrane and value < 90 Å2 is required to permeate through blood-brain barrier. All the selected phytocompounds showed TPSA value < 90 Å2, except 94.83 Å2 for emodin, indicating good permeability of selected phytocompounds into the cell as well as through blood-brain barrier. Lipinski’s rule of five helps to determine drug likeness of the compound and orally active drug should not violate the Lipinski’s rule. The emodin, thymol, carvacrol, artemisinin, and chloroquine followed Lipinski’s rule of five (Table 3). The predicted cellular targets of emodin, thymol, carvacrol, artemisinin, and chloroquine are shown in Table 4.
Table 3.
ADME properties of selected phytocompounds and chloroquine predicted by SwissADME
| Phytocompounds | SwissADME | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Consensus Log PO/W | Water solubility | GI absorption | TPSA (Å2) | Lipinski’s rule | Ghose rule | Veber rule | Egan rule | Muegge rule | |
| Emodin | 1.87 | Soluble | High | 94.83 | Yes | Yes | Yes | Yes | Yes |
| Thymol | 2.8 | Soluble | High | 20.23 | Yes | No | Yes | Yes | No |
| Carvacrol | 2.82 | Soluble | High | 20.83 | Yes | No | Yes | Yes | No |
| Artemisinin | 2.50 | Soluble | High | 53.99 | Yes | Yes | Yes | Yes | Yes |
| Chloroquine | 4.15 | Moderate soluble | High | 28.16 | Yes | Yes | Yes | Yes | Yes |
Table 4.
Predicted targets of phytocompounds and standard drug chloroquine
| Phytocompounds | Predicted targets |
|---|---|
| Emodin | Estrogen receptor alpha, Estrogen receptor beta, Serine/threonine-protein kinase PIM1, Casein kinase II alpha, Protein-tyrosine phosphatase 4A3 |
| Artemisinin | Cytochrome P450 1A2 |
| Thymol | Transient receptor potential cation channel subfamily A member 1, Cyclooxygenase-1, GABA-A receptor; alpha-1/beta-2/gamma-2, Serotonin 2b (5-HT2b) receptor, GABA-A receptor; alpha-1/beta-3/gamma-2 |
| Carvacrol | Cyclooxygenase-1, Transient receptor potential cation channel subfamily A member 1, Serotonin 2b (5-HT2b) receptor, Carbonic anhydrase II, GABA-A receptor; alpha-1/beta-3/gamma-2, |
| Chloroquine | Voltage-gated calcium channel alpha2/delta subunit 1, Serotonin 1a (5-HT1a) receptor, Histamine H3 receptor, Histamine N-methyltransferase, Alpha-1d adrenergic receptor |
Toxicity of emodin, thymol, carvacrol, artemisinin, and chloroquine was predicted by PROTOX-II and admetSAR and results are summarized in Table 5. It was observed that emodin, thymol, carvacrol, artemisinin, and chloroquine are non-carcinogenic and non-cytotoxic in nature and are safe to administer. However, LD50 value of emodin (5000 mg kg−1) and artemisinin (4228 mg kg−1) calculated from Protox II was higher than that of all other phytocompounds and chloroquine. This suggests that natural phytocompounds are safer even at higher dosage than that of chemically synthesized chloroquine.
Table 5.
Toxicity prediction of phytocompounds and chloroquine by admetSAR and PROTOX-II softwares
| Compounds | admet SAR | Protox II | ||
|---|---|---|---|---|
| Carcinogenic nature | Rate acute toxicity (LD50) kg mol−1 | LD50 (mg kg−1) | Cytotoxicity | |
| Emodin | Non-carcinogen | 2.5826 (III) | 5000 (class 5) | Inactive |
| Thymol | Non-carcinogen | 2.202 (III) | 640 (class 4) | Inactive |
| Carvacrol | Non-carcinogen | 2.531 (III) | 1190 (class 4) | Inactive |
| Artemisinin | Non-carcinogen | 1.79 (V) | 4228 (class 5) | Inactive |
| Chloroquine | Non-carcinogen | 2.684 (II) | 311 (class 4) | Inactive |
Molecular Docking (MD) Analysis of Phytocompounds and Chloroquine With Spike Protein and Its Variant of SARS-CoV-2
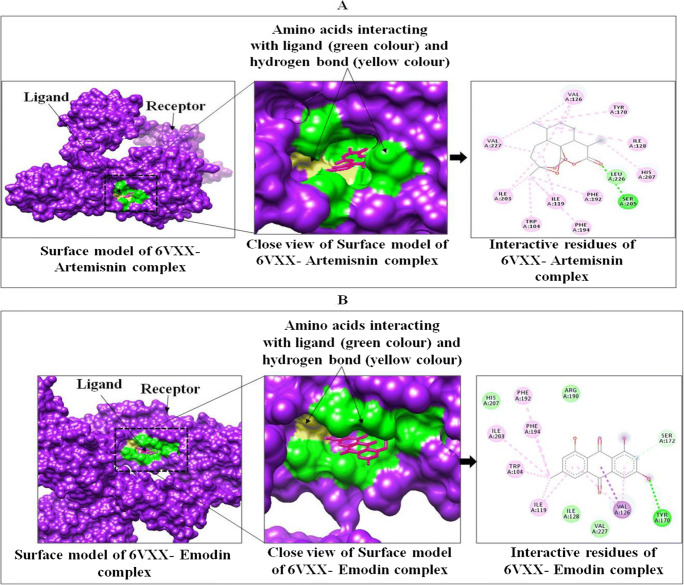
Docking results of phytocompounds of medicinal plants showed good binding affinity and modes of interaction with both the spike protein of SARS-CoV-2 (6VXX closed state and 6VYB open state), SARS-CoV-2 B.1.351 variant, Human TMPRSS2 (7MEQ), Angiotensin-converting enzyme-2, ACE2, and neuropilin-1 as compared to chloroquine. Among all the selected phytocompounds, artemisinin showed the best binding affinity (−10.5 kcal mol−1), followed by thymol (−6.9 kcal mol−1), carvacrol (−6.8 kcal mol−1), emodin (−6.4 kcal mol−1), and chloroquine (−5.6 kcal mol−1) with SARS-CoV-2 spike glycoprotein (6VXX). Artemisinin makes week hydrogen bonds with SER 205, HIS 207, and hydrophobic interactions with ILE 119, VAL 126, ILE 128, PHE 192, PHE 194, ILE 203, LEU 226, and VAL 227 (Table 6, Fig. 1, and Figure S1).
Table 6.
E-total of ligands (emodin, thymol, carvacrol, artemisinin, and chloroquine) with targets of SARS-CoV-2using Autodock/vina software
| Receptor | Ligands | E total (kcal mol−1) | Interacting amino acids | |
|---|---|---|---|---|
| H-bonding | Hydrophobic interaction | |||
| SARS-CoV-2 spike glycoprotein (6VXX, closed state) | Emodin | −6.4 | TYR 170 (weak) | TRP 104, ILE 119, VAL 126, PHE 192, PHE194, ILE 128, SER 172, VAL 227 |
| Thymol | −6.9 | - | TRP 104, ILE 119, ASN 121, PHE 192, PHE 194, ILE 203, LEU 226 | |
| Carvacrol | −6.8 | HIS 203 (moderate) | TRP 104, ILE 119, ASN 121, ARG 190, PHE 192, PHE 194, ILE 203 | |
| Artemisinin | −10.5 | SER 205, HIS 207 (week) | ILE 119, VAL 126, ILE 128, PHE 192, PHE 194, ILE 203, LEU 226, VAL 227 | |
| Chloroquine | −5.6 | - | TRP 104, ILE 119, ASN 121, VAL 126, SER 172, ARG 190, PHE 192, PHE 194, ILE 203, HIS 207, LEU 226, VAL 227 | |
| SARS-CoV-2 spike ectodomain structure (6VYB, open state) | Emodin | −8.8 | TYR 170, SER 172 (weak), ARG 190 (moderate) | TRP 104, ILE 119, VAL 126, ILE 128, TYR 170, SER 172, ARG 190, PHE 192, ILE 203, VAL 227 |
| Thymol | −6.7 | SER 730, THR 778 (moderate) | LEU 865, PRO 863, PHE 782, ILE 870, ALA 1056, GLY 1059 | |
| Carvacrol | −6.8 | ARG 190 (moderate) | TRP 104, ILE 119, ILE 128, ARG 190, PHE 192, ILE 203, SER 205, HIS 207 LEU 226 | |
| Artemisinin | −10.3 | SER 730, THR 778 (weak) | TRP 104, ILE 119, ILE 126, VAL 128, PHE 194, VAL 227 | |
| Chloroquine | −5.9 | - | TRP 104, ILE 119, ASN 121, VAL 126, ILE 128, TYR 170, ARG 190, PHE 192, PHE 194, ILE 203, SER 205, HIS 207, LEU 226, VAL 227 | |
| SARS-CoV-2 B.1.351 variant Spike glycoprotein (7NXA) | Emodin | −6.4 | GLU 6, GLN 111, LYS 207 (strong) | SER 7, GLY 9, VAL 92, GLY 112, THR 113, PRO 155, PRO 208, |
| Thymol | −4.4 | LEU 114 (strong) | VAL 92, GLN 111, GLY 112, THR 113, PRO 155, THR 157, LYS 207 | |
| Carvacrol | −4.7 | - | SER 7, GLY 8, GLY 9, GLY 10, GLY 112, THR 113, LEU 114, PRO 155, THR 157, PRO 208 | |
| Artemisinin | −5.9 | - | LEU 45, TYR 94, TRP 109, GLY 110, GLN 111 | |
| Chloroquine | −4.9 | - | VAL 92, GLY 112, THR 113, LEU 114, TYR 151, GLU 154, PRO 155VAL 156, THR 157, ALA 174, LEU 184 | |
| Human TMPRSS2 (7MEQ) | Emodin | −7.1 | ASN 277 (moderate) | HIS 274, GLN 276, VAL 278, TRP 306, THR 309, PHE 311, TYR 322, GLN 323, ALA 324, GLY 325, GLN 327 |
| Thymol | −5.5 | - | GLY 312, ILE 314, LEU 315, ARG 316, PHE 319 | |
| Carvacrol | −5.3 | GLN 276 (moderate) | HIS 274, VAL 275, ASN 277, TRP 306, HIS 307, THR 309, PHE 311, TYR 322, GLY 323, ALA 324, GLY 325 | |
| Artemisinin | −6.9 | SER 358 (moderate) | LYS 342, SER 436, CYS 437, GLN 438, GLY 439, ASP 440, THR 459, SER 460, TRP 461, GLY 462, GLY 464, CYS 465 | |
| Chloroquine | −5.7 | - | GLY 235, HIS 274, VAL 275, GLN 276, ASN 277,THIS 307, THR 309, PHE 311, TYR 322, GLY 323, GLN 352 | |
| Angiotensin-converting enzyme-2, ACE2 (6MID) | Emodin | −7.3 | TYR 488, GLU 553 (moderate) | SER 487, SER 491, ILE 492, GLY 591, VAL 587, VAL 586, ALA 590, VAL 552, PHE 549 |
| Thymol | −6.9 | PHE 279 (moderate) | TYR 129, PHE 277, PHE 283, PHE 48, SER 431, CYS 49, SER 280, LEU 281, LEU 494, VAL 125, GLY 490, PHE 279 | |
| Carvacrol | −6.1 | GLU 89 (moderate) | LEU 88, GLU 501, VAL 505, ILE 92, MET 502, LEU 85, ILE 287 | |
| Artemisinin | −7.4 | - | ILE 545, ALA 273, LEU 495, SER 487, LEU 269, SER 491, ILE 492, ASP 270, TYR 488, VAL 552, PHE 549, GLU 553 | |
| Chloroquine | −6.5 | - | PHE 546, ILE 545, LEU 495, ILE 492, GLY 490, ALA 273, LEU 269, PHE 277, ARG 57, TYR 488, SER 491, SER 487, ASP 270, ASP 486, GLN 274 | |
| Neuropilin-1 (4DEQ) | Emodin | −6.6 | ILE 142 (moderate) | TYR 24, TRP 28, THR 43, ASP 47, THR 76, LYS 78, TYR 80, GLY 141 |
| Thymol | −5.6 | LYS 78 (moderate) | TRP 24, TRP 28, THR 43, ASP 47, LYS 78 | |
| Carvacrol | −5.8 | THR 43 (moderate) | TYR 24, TRP 28, ASP 47, SER 73, THR 76, TYR 80, GLY141, ILE 142 | |
| Artemisinin | −5.8 | TYR 24 (moderate) | TRP 28, THR 43, THR 76, LYS 78, TYR 80 | |
| Chloroquine | −4.9 | TYR 80 (moderate) | TYR 24, TRP 28, THR 43, LYS 45, GLU 46, ASP 47 | |
Fig. 1.
Interactions between targeted protein receptor SARS-CoV-2 spike glycoprotein (PDB ID: 6VXX) with artemisinin (A) and emodin (B) using Chimera
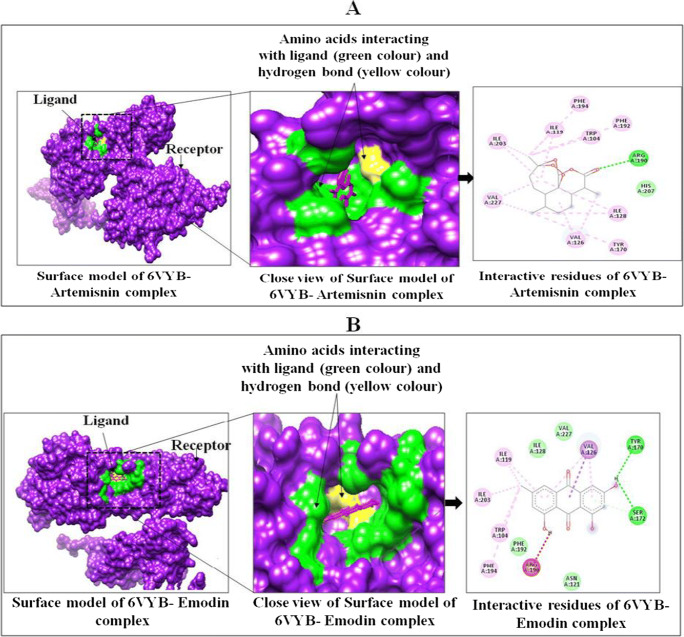
The binding interaction almost follows the similar pattern of interaction with SARS-CoV-2 spike ectodomain structure (6VYB, open state) with a binding affinity of −10.3 kcal mol−1 (artemisinin), −8.8 kcal mol−1 (emodin), −6.7 kcal mol−1 (thymol), −6.8 kcal mol−1 (carvacrol), and −5.9 kcal mol−1 (chloroquine). Artemisinin makes week hydrogen bonds with SER 730 and THR 778 and showed hydrophobic interactions with TRP 104, ILE 119, ILE 126, VAL 128, PHE 194, and VAL 227 (Table 6 and Fig. 2 and S2). Artemisinin in complex with 6VYB makes week hydrogen bonds with SER 730 and THR 778 and showed hydrophobic interactions with TRP 104, ILE 119, ILE 126, VAL 128, PHE 194, and VAL 227 (Table 5 and Fig. 2 and Figure S2).
Fig. 2.
Interactions between targeted protein receptor SARS-CoV-2 spike ectodomain structure (PDB ID: 6VYB) with artemisinin (A) and emodin (B) using Chimera
In case of SARS-CoV-2 B.1.351 variant, the binding affinity was −6.4, −4.4, −4.7, −5.9, and −4.9 kcal mol−1 for emodin, thymol, carvacrol, artemisinin, and chloroquine respectively. Emodin in complex with SARS-CoV-2 B.1.351 variant (7NXA) makes strong hydrogen bonds with GLU 6, GLN 111, and LYS 207 and showed hydrophobic interactions with SER 7, GLY 9, VAL 92, GLY 112, THR 113, PRO 155, and PRO 208 (Table 6 and Fig. S3). Emodin also showed the best interactions (−7.1 kcal mol−1) with Human TMPRSS2 (7MEQ), followed by artemisinin (−6.9 kcal mol−1), chloroquine (−5.7 kcal mol−1), thymol (−5.5 kcal mol−1), and carvacrol (−5.3 kcal mol−1). Emodin makes hydrogen bonds (moderate strength) with ASN 277 and showed hydrophobic interactions with HIS 274, GLN 276, VAL 278, TRP 306, THR 309, PHE 311, TYR 322, GLN 323, ALA 324, GLY 325, and GLN 327 (Table 6 and Fig. S4). Neuropilin-1 interacts with emodin (−6.6 kcal mol−1), carvacrol and artemisinin (−5.8 kcal mol−1), thymol (−5.6 kcal mol−1), and chloroquine (4.9 kcal mol−1). Emodin makes hydrogen bond (moderate strength) with ILE 147 and hydrophobic interactions with TYR 24, TRP 28, THR 43, ASP 47, THR 76, LYS 78, TYR 80, and GLY 141 (Table 6 and Fig. S5). Similarly, Angiotensin-converting enzyme-2, ACE2 showed best interaction with artemisinin (−7.4 kcal mol−1), followed by emodin (−7.3 kcal mol−1), thymol (−6.9 kcal mol−1), chloroquine (−6.5 kcal mol−1), and carvacrol (−6.1 kcal mol−1). Artemisinin do not make any hydrogen bonds with ACE2, but showed hydrophobic interactions with LEU 545, ALA 273, LEU 495, SER 487, LEU 269, SER 491, ILE 492, ASP 270, TYR 488, VAL 552, PHE 549, and GLU 553 (Table 6 and Figure S6).
The nature of hydrogen bonds was determined on the basis of donor-acceptor distances in protein secondary structure elements. Jeffrey and Jeffrey [63••] categorize hydrogen bonds with donor-acceptor distances of 2.2–2.5 Å as “strong,” 2.5–3.2 Å as “moderate” (mostly electrostatic), and 3.2–4.0 Å as “weak, electrostatic.” Hydrogen bond interaction and hydrophobic interactions of these phytocompounds with target proteins were analyzed through discovery studio and results are summarized in Table 6.
Molecular Dynamics Analysis of Complexes of 6VXX and 6VYB With Artemisinin
Among all the selected phytocompounds, artemisinin showed the best binding affinity with both the spike proteins of SARS-CoV-2 (6VXX closed conformation, 6VYB open confirmation) and these complexes were selected for MD simulation [60, 64, 65]. The binding and conformational stability of the artemisinin complex with the spike receptor protein is a major factor to advocate the inhibitory action of the artemisinin against SARS-CoV-2 infection. MD simulations were carried out for 50ns at 300 Kelvin temperature and 1.01325 bar pressure. Both the complexes of artemisinin with spike receptors (PDB ID: 6VXX, and 6VYB) have 6Na+ ions to neutralize complexes. The artemisinin complexes with 6VXX and 6VYB have water molecules 58601 and 72196, respectively. The Na+ and Cl− ions were added in the environment to these complexes, such that artemisinin +6VXX (Cl− 50.573 nM; Na+ 52.435 nM) and artemisinin+6VYB (Cl− 50.620 nM; Na+ 50.620 nM) could mimic physiological ionic concentration.
The Stereochemical Geometry of S-Protein in Complex With Ligand After Molecular Dynamics Simulation
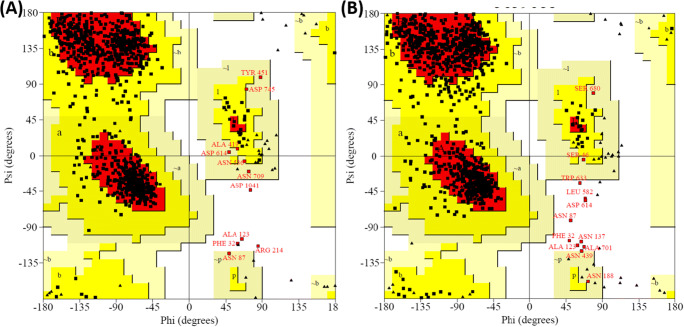
The Ramachandran mapping of S-protein residues after analyzing the stereochemical geometry of artemisinin+6VXX, and artemisinin+6VYB after MD simulation revealed a very acceptable number of residues in favored region (Fig. 3; Table 7). These complexes possess outlier residues within the acceptable range (less than 1.0%). Overall, spike protein in complex with artemisinin showed a sterically acceptable conformation of molecule after MD simulation, indicating the stability of the complexes.
Fig. 3.
Ramachandran plots of S-proteins: (A) 6VXX (closed conformation) complex with artemisinin, and (B) 6VYB (open conformation) complex with artemisinin
Table 7.
The Ramachandran mapping of S-protein residues for analyzing stereochemical geometry of artemisinin complexes with 6VXX and 6VYB after MD simulation
| Entry | Protein | Favored region | Additional allowed region | Generously allowed region | Outlier region |
|---|---|---|---|---|---|
| 1 | 6VXX (closed) | 717 (85.1%) | 115 (13.6%) | 7 (0.8%) | 4 (0.5%) |
| 2 | 6VYB (open) | 784 (83.5%) | 143 (15.2%) | 4 (0.4%) | 8 (0.9%) |
Conformational Deviation in Cα of S-Protein (6VXX and 6VYB) in Complex With Artemisinin During Molecular Dynamics Simulation for 50ns
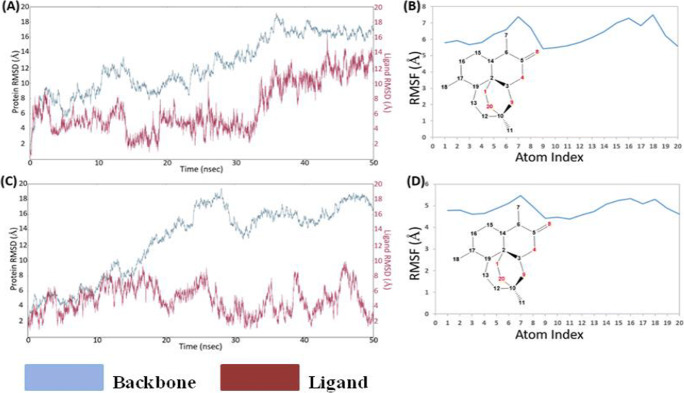
The root mean square deviation (RMSD) of the Cα of the S-protein molecule was analyzed for 50-ns simulation for 6VXX and 6VYB complexed with artemisinin. The Cα RMSD plot for 6VXX (Fig. 4A) gets stabilized at 35ns and remains stabled for the entire simulation period. Ligand RMSD was 4Å at 30ns and then changed to 10Å at 40ns, which is further changed to 14Å at 50-ns time line (Fig. S7). The ligand RMSF plot for artemisinin fit over 6VXX protein showed ligand fluctuation with respect to protein. The high RMSF of atom 7 and 16–18 is mainly due to the exposure to solvent (Fig. 4B). The trajectory analysis reveals conformational shift in artemisinin within binding pocket, which indicates artemisinin is less stable in the binding pocket of 6VXX (closed conformation) complex. It has been proposed that 6VYB (open conformation) is more significant for drug designing. The RMSD plot for 6VYB (Fig. 5C) clearly showed stable interaction between artemisinin and binding pocket residues of 6VYB (open conformation) during the simulation period. Artemisinin showed less fluctuation in open state protein than the closed state protein (Fig. 4C, D).
Fig. 4.
The RMSD and RMSF plots of S-protein (6VXX and 6VYB) in complex with artemisinin. (A) RMSD of artemisinin + 6VXX complex, (B) RMSF of artemisinin + 6VXX complex, (C) RMSD of artemisinin + 6VYB complex, and (D) RMSF of artemisinin + 6VYB complex as indicated for the backbone and ligand
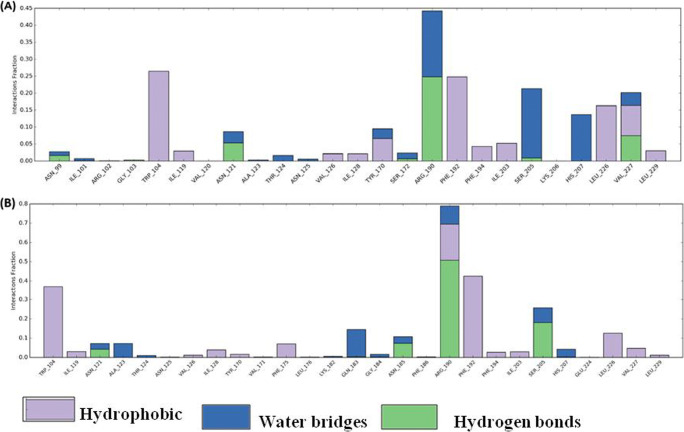
Fig. 5.
Histogram of ligand contacts with amino acid residues of S-protein. (A) Artemisinin + 6VXX (closed conformation) complex, (B) artemisinin + 6VYB (open conformation) complex. Color codes for hydrogen bonds, hydrophobic, and water bridges interactions are as indicated. X-axis showed amino acid residues and y-axis indicated interaction fraction
Furthermore, 6VYBor 6VXXcomplex with artemisinin showed hydrophobic interaction (Trp104, Ile119, Val126, Ile128, Tyr170, Phe192, Phe194, Ile203, Leu226, and Leu229), H-bond interaction (Asn121, Arg192, Ser 205), and water bridges (Asn121, Thr124, Arg192, Ser205, His207) (Fig. 5A, B). There was no salt bridge interaction developed between the 6VYB or 6VXX complex with artemisinin. The observed binding free energy for artimisnin+6VYB complex was −74.54 kcal mol−1, while observed binding free energy for artimisnin+6VXX complex was −55.5 kcal mol−1. The above results clearly advocate the stability of artemisinin in open state protein instead of closed state protein.
Discussion
From the beginning of twenty-first century, three coronaviruses, viz. severe acute respiratory syndrome coronavirus (SARS-CoV) [4], Middle East respiratory syndrome coronavirus (MERS-CoV) [5], and SARS-CoV-2 have crossed the species barrier and resulted in deadly pneumonia in humans [66, 67]. SARS-CoV-2 has caused death of approximately 3.3 million people all around the world. The treatment is symptomatic and oxygen therapy represents the major treatment intervention for patients with severe infection. Mechanical ventilation may be necessary in cases of failure of respiratory, whereas hemodynamic support is essential for managing septic shock. Researchers from Worldwide are continuing to work on developing vaccine againstSARS-CoV-2. Professor Didier Raoult from infectious diseases institute, IHU Méditerranée Infection in Marseille (France), has reported successful results from a new treatment for COVID-19, with early tests suggesting it can stop the virus from being contagious in just 6 days. Chloroquine phosphate and hydroxychloroquine have previously been used to treat coronavirus patients in China, in ongoing COVID-19 clinical trials. Kaletra, a US-based antiviral drug used to treat HIV, is another medicine that is being tested in the fight against the SARS-CoV-2. Emodin has been shown to act as inhibitor of 3a ion channel of SARS-CoV and HCoV-OC43 coronaviruses as well as virus release from HCoV-OC43. It has been reported that emodin is a potent inhibitor of the 3a channel with a K1/2 value of about 20 M. The reduction of extracellular viral RNA copies by emodin reflects inhibition of virus release. At high concentrations of emodin, intracellular levels of viral RNA were reduced suggesting that the high concentrations may also inhibit other stages of the virus life cycle [67]. Ho et al. [68] identified emodin as an effective to block the interaction of the SARS-CoV S protein with the ACE2 and the infection by S protein-pseudo-typed retrovirus. Ahmed et al. [69] reported SARS-CoV-2 spike protein (6VYB) is highly stable protein and it is difficult to un-stabilize the integrity of these proteins by individual drugs. They also reported that inserting of NH2 halogen and vinyl group can increase the binding affinity of coulerpin with 6VYB, while inserting an alkyl group decreases the binding affinity of coulerpin with 6VYB. This work is unique in a way that in silico approach has been utilized to compare the open (6YVB) and closed (6VXX) conformations of spike proteins. It is interesting that open state of the spike protein (6YVB) which is more pathogenic showed more stable interaction with artemisinin as compared to closed state (6VXX) (Data is Fig. 5). Also, artemisinin contacts with amino acid residues of S-protein were different for open and closed conformation (Fig. 5B). Similar to our study, Kumar [60] reported the binding affinity of Nelfinavir (−8.4), Rhein (−8.1), Withanolide D (−7.8), Withaferin A (−7.7), Enoxacin (−7.4), and Aloe-emodin (−7.4) with COVID-19 main protease (6LU7). Rolta et al. [70••] reported the binding affinity of emodin, aloe-emodin, anthrarufin, alizarine, and dantron phytocompound Rheum emodi with three active sites of RNA binding domain of nucleocapsid phospho protein of COVID-19. They reported the binding energies of emodin, aloe-emodin, anthrarufin, alizarine, and dantron were −8.299, −8.508, −8.456, −8.441, and −8.322 Kcal mol−1 respectively with binding site A and −7.714, −6.433, −6.354, −6.598, and −6.99 Kcal mol−1 respectively with binding site B, and −8.299, 8.508, 8.538, 8.841, and 8.322 Kcal mol−1 respectively with binding site C. Similarly, Adem et al. [71] reported khainaoside C, 6-O-Caffeoylarbutin, khainaoside B, khainaoside C, and vitexfolin are potent modulator of open and closed state of SARS-CoV-2 spike S2 proteins. Suravajhala et al. [72] reported the antiviral binding affinity of curcumin with different SARS-CoV-2 Proteins (Spike Glycoprotein-6VYB, nucleocapsid phosphoprotein- 6VYO, membrane glycoprotein-6M17, nsp10-6W4H, and RNA-dependent RNA polymerase- 6M71). Selailia and Chemat [73••] reported hydroxyl chloroquine and artemisinin interact in the same binding pocket of SARS-CoV-2 protein (6LZG); artesunate, artemisinin, and artenimol showed two mode of interaction with LYS 353 and LYS 31; and they also reported the extraction protocol of artemisinin from Artemisia annua. Walls et al. [52] suggested that S-protein is highly pathogenic in human coronaviruses and appears to exist in partially opened states, while S-protein remains largely closed in human coronaviruses that are responsible for common colds. It was also proposed that the S-protein of pathogenic coronaviruses exists in open and close conformation. The current in silico study provides evidence that ligand binding affinity is different for open and closed conformation of S protein and artemisinin interacts more stably with the open conformation of spike protein, than the closed conformation, thus can be used as a potent drug to cure COVID-19. Basu et al. [74••] studied the molecular docking of five phytocompounds (hesperidin, anthraquinone, thein, chrysin, and emodin) with spike protein of SARS-CoV2 and ACE2 receptor. It was shown that hesperidin can bind with ACE2 protein and bound structure of ACE2 protein and spike protein of SARS-CoV2 noncompetitively. The study proposed that the presence of hesperidin, the bound structure of ACE2, and spike protein fragment become unstable. Srivasta et al. [75••] reported the interactions of antimalarial compounds (Mepacrine, Chloroquine, Quinin, Hydroxychloroquine, Artemisinin, Phomarin, and Proguanil) with main protease (PDB ID 6LU7) of SARS-CoV2. They found that mepacrine showed best interactions with 6LU7 (−8.89 kcal mol−1) followed by chloroquine (−8.15 kcal mol−1), quinin (−7.77 kcal mol−1), hydroxychloroquine (−7.62 kcal mol−1), artemisinin (−7.34 kcal mol−1), phomarin (−7.13 kcal mol−1), and proguanil (−6.69 kcal mol−1). Previous studies reported the binding affinity of emodin, artemisinin, and chloroquine against RNA binding domain of nucleocapsid phosphoprotein and main protease of SARS-CoV-2. The current study provides evidence that ligand binding affinity is different for open and closed conformation of S protein. This study also provides the evidence that phytocompounds can inhibit spike protein variant of SARS-CoV-2. Artemisinin interacts more stably with the open conformation of spike protein, than the closed conformation and emodin binds strongly with variant of spike protein SARS-CoV-2; thus, artemisinin and emodin need further attention through in vitro and in vivo studies to be tested to inactivate the SARS-CoV-2.
Conclusions
In this study, we are proposing the artemisinin as a lead phytocompound to inactivate the SARS-CoV-2 virus through inhibiting S-protein, especially in open state conformation. The MD simulation for 50ns showed both the S-protein complexes were stable as there are less than 1.0% outlier amino acid residues in Ramachandran plot developed after MD. The RMSD plot for both complexes and ligand RMSF has shown the stability of artemisinin within the binding pocket of S-protein. The present study proposed a safe and less toxic artemisinin for the treatment for SARS-CoV-2 infection, which can be further validated through in vitro and in vivo studies.
Supplementary Information
(DOCX 791 kb)
Acknowledgements
The authors acknowledge Shoolini University, Solan, for providing infrastructure support to conduct the research work. Authors also acknowledge the support provided by Yeast Biology Laboratory, School of Biotechnology, Shoolini University, Solan, India.
Availability of Data and Material
Main data and supporting data are provided in manuscript and supplementary data.
Author Contribution
All the experimental work was done jointly by Rajan Rolta, Deeksha Salaria, and Prem Prakash Sharma. Dr. Brijesh Rathi helped in MD simulations. Er. Bhanu Sharma, Dr. Mansi Verma, Dr. Vikas Kumar, and Dr. David J. Baumler provided the technical inputs in designing and data analysis. Prof. Anuradha Sourirajan and Prof. Kamal Dev conceived the idea and provided guidance to execute the research project. All the authors have read the manuscript.
Declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare that they have no competing interests.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Natural Products: From Chemistry to Pharmacology
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rajan Rolta, PremPrakash Sharma, Bhanu Sharma , and Deeksha Salaria contributed equally to this work.
References
Papers of particular interest, published recently, have been highlighted as: •• Of major importance
- 1.••.Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C. Agha R World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Islam MT. Studies on coronaviruses causing enteric infections in domestic animals in Japan. Graduate School of Veterinary Medicine and Life Science Nippon Veterinary and Life Science University. 2017; 1–91.
- 3.Bracewell CD. Serological studies of avian infectious bronchitis virus. D.V.M., University of Tolima, Colombia. 1977; 1–166.
- 4.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Zaki AM, Van BS, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Bialek SR, Allen D, Alvarado-Ramy F, Arthur R, Balajee A, Bell D, Best S, Blackmore C, Breakwell L, Cannons A, Brown C. First confirmed cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities—May 2014. MMWR Morb Mortal Wkly Rep. 2014;63:431–436. [PMC free article] [PubMed] [Google Scholar]
- 7.Cowling BJ, Park M, Fang VJ, Wu P, Leung GM, Wu JT. Preliminary epidemiologic assessment of MERS-CoV outbreak in South Korea, May–June 2015. Euro surveillance: bulletin Europeensur les maladies transmissibles. Euro Surveill. 2015;20:21175. doi: 10.2807/1560-7917.ES2015.20.25.21163. [DOI] [PubMed] [Google Scholar]
- 8.Zhu YY, Yu G, Wang YY, Xu JH, Xu FZ, Fu H, Zhao YH, Wu J. Antiviral activity and molecular docking of active constituents from the root of Aconitum carmichaelii. Chem Nat Compd. 2019;55:189–193. doi: 10.1007/s10600-019-02651-5. [DOI] [Google Scholar]
- 9.Zhou Y, Jiang S, Du L. Prospects for a MERS-CoV spike vaccine. Expert Rev Vaccines. 2018;17:677–686. doi: 10.1080/14760584.2018.1506702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 2019;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Wong G, Lu G, Yan J, Gao GF. MERS-CoV spike protein: targets for vaccines and therapeutics. Antivir Res. 2016;133:165–177. doi: 10.1016/j.antiviral.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babcock GJ, Esshaki DJ, Thomas WD, Ambrosino DM. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong SK, Li W, Moore MJ, Choe H, Farzan MA. 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. Int J Biol Chem Sci. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 15.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch BJ, Vander ZR, de HAAn CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang ZY, Huang Y, Ganesh L, Leung K, Kong WP, Schwartz O, Subbarao K, Nabel GJ. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitrov DS. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115:652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao X, Chakraborti S, Dimitrov AS, Gramatikoff K, Dimitrov DS. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem Bioph Res Co. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Diego ML, Nieto-Torres JL, Jiménez-Guardeño JM, Regla-Nava JA, Alvarez E, Oliveros JC, Zhao J, Fett C, Perlman S, Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011;7:e1002315. doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD, Thackray LB, Young MD, Mason RJ, Ambrosino DM. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyengar MA. Study of crude drugs. 2. Manipal: College of Pharmaceutical Sciences; 1985. pp. 13–78. [Google Scholar]
- 26.Hasegawa H, Matsumya S, Yamasak K. Reversal of efflux mediated tetracycline resistance in Staphylococcus aureus clinical isolates by Ginseng prosaponenins. Phytother Res. 1995;9:260–263. doi: 10.1002/ptr.2650090406. [DOI] [Google Scholar]
- 27.Lee CK, Kin H, Moon KH, Shun KH. Screening and isolation of antibiotic resistance inhibitors from herb materials - resistance inhibition of volatile components of Korean aromatic herbs. Arch Pharm Res. 1998;21:62–66. doi: 10.1007/BF03216754. [DOI] [PubMed] [Google Scholar]
- 28.Heilmeyer M. Ancient herbs: Getty Publications; 2007.
- 29.Jaric S, Mitrović M, Pavlović P. Review of ethnobotanical, phytochemical, and pharmacological study of Thymus serpyllum L. Evid. Based Complementary Altern. Med. 2015; 1–10. 10.1155/2015/101978 [DOI] [PMC free article] [PubMed]
- 30.Adams M, Schneider SV, Kluge M, Kessler M, Hamburger M. Epilepsy in the renaissance: a survey of remedies from 16th and 17th century German herbals. J Ethnopharmacol. 2012;143:1–13. doi: 10.1016/j.jep.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Mustafa B, Hajdari A, Pieroni A, Pulaj B, Koro X, Quave CL. A cross-cultural comparison of folk plant uses among Albanians, Bosniaks, Gorani and Turks living in south Kosovo. J Ethnobiol Ethnomed. 2015;11:39. doi: 10.1186/s13002-015-0023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chauhan PS, Satti NK, Suri KA, Amina M, Bani S. Stimulatory effects of Cuminum cyminum and flavonoid glycoside on Cyclosporine-A and restraint stress induced immune-suppression in Swiss albino mice. Chem Biol Interact. 2010;185:66–72. doi: 10.1016/j.cbi.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann EC, Jr, Kucera LS. Antiviral substances in plants of the mint family (Labiatae). III. Peppermint (Menthapiperita) and other mint plants. Exp Biol Med. 1967;124:874–878. doi: 10.3181/00379727-124-31874. [DOI] [PubMed] [Google Scholar]
- 34.Rustaiyan A, Masoudi S, Monfared A, Kamalinejad M, Lajevardi T, Sedaghat S, Yari M. Volatile constituents of three Thymus species grown wild in Iran. Planta Med. 2000;66:197–198. doi: 10.1055/s-0029-1243136. [DOI] [PubMed] [Google Scholar]
- 35.Pilau MR, Alves SH, Weiblen R, Arenhart S, Cueto AP, Lovato LT. Antiviral activity of the Lippia graveolens (Mexican oregano) essential oil and its main compound carvacrol against human and animal viruses. Braz J Microbiol. 2011;42:1616–1624. doi: 10.1590/S1517-83822011000400049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsiang CY, Ho TY. Emodin is a novel alkaline nuclease inhibitor that suppresses herpes simplex virus type 1 yields in cell cultures. Br J Pharmacol. 2008;155:227–235. doi: 10.1038/bjp.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai JP, Wang QW, Su Y, Gu LM, Zhao Y, Chen XX, Chen C, Li WZ, Wang GF, Li KS. Emodin inhibition of influenza A virus replication and influenza viral pneumonia via the Nrf2, TLR4, p38/JNK and NF-kappaB pathways. Molecules. 2017;22:1754. doi: 10.3390/molecules22101754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Efferth T, Romero MR, Wolf DG, Stamminger T, Marin JJ, Marschall M. The antiviral activities of artemisinin and artesunate. Arch Clin Infect Dis. 2008;47:804–811. doi: 10.1086/591195. [DOI] [PubMed] [Google Scholar]
- 39.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Trott O, Olson AJ. AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salaria D, Rolta R, Sharma N, Dev K, Sourirajan A, Kumar V. In silico and In vitro evaluation of the anti-inflammatory and antioxidant potential of Cymbopogon citratus from North-western Himalayas. bioRxiv. 2020; 10.1101/2020.05.31.124982.
- 43.Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, Gu M. MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci. 2009;122:2055–2063. doi: 10.1242/jcs.049080. [DOI] [PubMed] [Google Scholar]
- 44.Rolta R, Kumar V, Sourirajan A, Upadhyay NK, Dev K. Bioassay guided fractionation of rhizome extract of Rheum emodi wall as bio-availability enhancer of antibiotics against bacterial and fungal pathogens. J Ethnopharmacol. 2020a;257:112867. [DOI] [PubMed]
- 45.Castilho PC, Gouveia SC, Rodrigues AI. Quantification of artemisinin in Artemisia annua extracts by 1H-NMR. Phytochem Anal. 2018;19:329–34. 10.1002/pca.1053. [DOI] [PubMed]
- 46.Gul R, Jan SU, Taimor M, Rabbani T, Jahan N. Validation of a high-performance liquid chromatography technique for the investigation of thymol and carvacrol in Thymus serpyllum l. indigenous to balochistan. Indo Am J Pharm. 2018;5:7429–35.
- 47.Schwarz S, Wang K, Yu W, Sun B, Schwarz W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antivir Res. 2011;90:64–9. [DOI] [PMC free article] [PubMed]
- 48.Cao F, Peng W, Li X, Liu M, Li B, Qin R, et al. Emodin is identified as the active component of ether extracts from Rhizoma Polygoni Cuspidati, for anti-MRSA activity. Can J Physiol Pharmacol. 2015;93:485–93. 10.1139/cjpp-2014-0465. [DOI] [PubMed]
- 49.Tu Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat Med. 2011;17:1217–20. [DOI] [PubMed]
- 50.Can Baser KH. Biological and pharmacological activities of carvacrol and carvacrol bearing essential oils. Curr Pharm Des. 2008;14:3106–19. 10.2174/138161208786404227. [DOI] [PubMed]
- 51.••.Jaffe S. Regulators split on antimalarials for COVID-19. Lancet. 2020;395:1179. This reference is relevant to SRAS-COV-2 and published recently. [DOI] [PMC free article] [PubMed]
- 52.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dejnirattisai W, Zhou D, Supasa P, Liu C, Mentzer AJ, Ginn HM, et al. Antibody evasion by the P. 1 strain of SARS-CoV-2. Cell. 2021. [DOI] [PMC free article] [PubMed]
- 54.Fraser B, Beldar S, Hutchinson A, Li Y, Seitova A, Edwards AM, et al. Crystal structure of human TMPRSS2 in complex with Nafamostat: Proten data bank; 2021.
- 55.••.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.••.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94. This reference is relevant to SRAS-COV-2 and published recently. [DOI] [PMC free article] [PubMed]
- 57.••.Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J Med Virol. 2020;92:2105–13. This reference is relevant to SRAS-COV-2 and published recently. [DOI] [PMC free article] [PubMed]
- 58.Pollastri MP. Overview on the Rule of Five. Curr Protoc Pharmacol. 2010;49:9–12. doi: 10.1002/0471141755.ph0912s49. [DOI] [PubMed] [Google Scholar]
- 59.Banerjee P, Eckert AO, Schrey AK. PreissnerR.ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res Spec Publ. 2018;46:W257–W263. doi: 10.1093/nar/gky318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar S, Sharma PP, Shankar U, Kumar D, Joshi SK, Pena L, et al., Discovery of new hydroxyethylamine analogs against 3CLpro protein target of SARS-CoV-2: Molecular docking, molecular dynamics simulation and structure-activity relationship studies. J Chem Inform and Mod. 2020; 60:5754–5770. [DOI] [PubMed]
- 61.Schrödinger Release 2020-2: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2020. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY; 2020.
- 62.Jeffrey GA, Jeffrey GA. An introduction to hydrogen bonding, vol. 12. New York: Oxford university press; 1997. p. 228.
- 63.••.Kumar D, Chandel V, Raj S, Rathi B. In silico identification of potent FDA approved drugs against Coronavirus COVID-19 main protease: a drug repurposing approach. Chem Biol Lett. 2020;7:166–175. [Google Scholar]
- 64.Schrödinger LLC, Schrödinger, LLC; New York, NY. Schrödinger Suite, 2; 2017-1 2017.
- 65.Schrödinger Release 2020-2: Prime, Schrödinger, LLC, New York, NY; 2020.
- 66.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [DOI] [PMC free article] [PubMed]
- 67.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ho TY, Wu SL, Chen JC, Li CC, Hsiang CY. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir Res. 2007;74:92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed SA, Abdelrheem DA, Abd El-Mageed HR, Mohamed HS, Rahman AA, Elsayed KN, et al. Destabilizing the structural integrity of COVID-19 by caulerpin and its derivatives along with some antiviral drugs: an in-silico approaches for a combination therapy. Struct Chem. 2020:1–22. 10.1007/s11224-020-01586-w [DOI] [PMC free article] [PubMed]
- 70.••.Rolta R, Yadav R, Salaria D, Trivedi S, Imran M, Sourirajan A, et al. In silico screening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: an approach to prevent virus assembly. J Biomol Struct Dyn. 2020b:1–18. This reference is relevant to SRAS-COV-2 and published recently. [DOI] [PMC free article] [PubMed]
- 71.Adem S, Eyupoglu V, Sarfraz I, Rasul A, Zahoor AF, Ali M, et al. Caffeic acid derivatives (CAFDs) as inhibitors of SARS-CoV-2: CAFDs-based functional foods as a potential alternative approach to combat COVID-19. Phytomed. 2020:153310. 10.1016/j.phymed.2020.153310. [DOI] [PMC free article] [PubMed]
- 72.Suravajhala R, Parashar A, Malik B, Nagaraj VA, Padmanaban G, KaviKishor P, Polavarapu R, Suravajhala P. Comparative docking studies on curcumin with COVID-19 proteins. Preprints 2020, 2020050439 10.20944/preprints202005.0439.v3.
- 73.••.Sehailia M, Chemat S. Antimalarial-agent artemisinin and derivatives portray more potent binding to Lys353 and Lys31-binding hotspots of SARS-CoV-2 spike protein than hydroxychloroquine: potential repurposing of artenimol for COVID-19. J Biomol Struct Dyn. 2020:1–11. This reference is relevant to SRAS-COV-2 and published recently. [DOI] [PMC free article] [PubMed]
- 74.••.Basu A, Sarkar A, Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci Rep. 2020;10(1):1–15. doi: 10.1038/s41598-020-74715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.••.Srivastava, A.K., Kumar, A., Tiwari, G., Kumar, R. and Misra, N., 2020. In silico investigations on the potential inhibitors for COVID-19 protease. arXiv preprint arXiv:2003.10642. This reference is relevant to SRAS-COV-2 and published recently.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 791 kb)
Data Availability Statement
Main data and supporting data are provided in manuscript and supplementary data.