Abstract
Control of the rapid spread of the SARS-CoV-2 virus requires efficient testing. We collected paired nasopharyngeal swab (NPs) and saliva samples from 303 subjects (52.8% symptomatic) at a drive-through testing center; 18% of whom tested positive. The NPs, salivas and five saliva pools were tested for SARS-CoV-2 RNA using the Aptima™ assay and a laboratory-developed test (LDT) on the Panther-Fusion™ Hologic® platform. The saliva sensitivity was 80% (LDT) and 87.5% (Aptima™) whereas that of NPs was 96.4% in both assays. The pooled saliva sensitivity of 72.7% (LDT) and 75% (Aptima™) was not significantly different of that of individual saliva testing. Saliva specimens appear to be suitable for sensitive non-invasive assays to detect SARS-CoV-2 nucleic acid; pooling them for a single test will improve laboratory throughput.
Keywords: SARS-CoV-2, Saliva, Nasopharyngeal swabs, Pooling, COVID-19, RT-PCR, TMA
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected over 175 million individuals causing more than 3.8 million of deaths as of June 24, 2021 (Coronavirus 2020). Many governments have used various testing strategies to contain the pandemic and reduce the spread of the virus. The gold standard test assays SARS-CoV-2 RNA in nasopharyngeal swabs (NPs) (COVID-19 Resources and Guidelines for Labs and Laboratory Workers, 2020; WHO 2020). However, the NPs samples are obtained using invasive method that requires trained healthcare personnel and suitable protective equipment. Many patients find it uncomfortable, which dissuades others and reduces compliance.
Saliva sampling is non-invasive, painless and much more acceptable. It does not use swabs, and patients can easily collect their own samples, so reducing crowding at testing sites. However, the results of studies that used saliva samples for SARS-CoV-2 diagnosis have been variable. Some reported that saliva samples were inappropriate (Jamal et al., 2021; Landry et al., 2020; Skolimowska et al., 2020), while others found them acceptable or even provided greater sensitivity (Azzi et al., 2020; Berenger et al., 2021; Migueres et al., 2020; Pasomsub et al., 2021; Rao et al., 2020; Williams et al., 2020; Wyllie et al., 2020). These differences could be due to the sampling protocol used (saline gargle and spit, posterior oral saliva, crude saliva, oral swabs) or the individuals tested (hospitalized patients or health-care workers in most cases). Few studies assessed the performance of saliva samples for SARS-CoV-2 testing according to the time from symptom onset, or in asymptomatic non-hospitalized individuals (Nacher et al., 2021; Rao et al., 2020; Torres et al., 2021; Yokota et al., 2020). In addition, little is known of the suitability of pooled saliva samples for increasing laboratory throughput, which optimizes reagent use and reduces cost.
This prospective study of outpatients examined SARS-CoV-2 detection in individual and pooled saliva specimens, and NP swabs. SARS-CoV-2 RNA was detected using two assays: the AptimaTM SARS-CoV-2 transcription-mediated amplification (TMA) (AptimaTM) and a laboratory-developed test (LDT) that uses RT-PCR on the Panther FusionTM Hologic® platform.
2. Materials and methods
2.1. Study subjects
Patients who came to the Toulouse University Hospital drive-through testing center 27 to 29 October 2020 during the second epidemic wave in France were enrolled prospectively. They were informed of and consented to the test before they provided a saliva specimen plus a NP swab sample. The saliva samples were self-collected under the supervision of a health care worker (HCW) and the NPs was collected by a HCW following CDC specimen collection guidelines (Coronavirus Disease 2019 COVID-19 Internet, 2020).
2.2. Specimen collection and processing
NPs were collected on flocked swabs and placed in 3.5mL of virus transport medium (VTM) (Virus sampling kit, Yocon, Beijing, China). Saliva (1mL) was collected after the subjects had swilled their saliva around their mouths for at least 30 seconds and then spitting into a sterile container (Migueres et al., 2020). The saliva samples were diluted 3 fold dilution in Minimum Essential Media (MEM) and all samples, NPs and saliva, were tested in the clinical laboratory within 24 hours.
Pooled saliva (5 sequential individual samples) and individual patient salivas were used to evaluate pooling.
2.3. Laboratory testing
All specimens (NPs, individual and pooled salivas) were tested simultaneously. We used the Aptima™ SARS-CoV-2 transcription mediated amplification assay (TMA) (Hologic®, San Diego, California) and a laboratory-developed test (LDT) based on real-time RT-PCR on a Panther Fusion™ module (Hologic®, San Diego, California) that has been validated on saliva specimens (Migueres et al., 2020). The Aptima™ assay targets 2 sequences on the virus ORF1ab gene. The LDT targets 2 sequences on the virus RNA-dependent RNA-polymerase (RdRp) gene (IP2, IP4, Institut Pasteur, Paris, France) (Trémeaux et al., 2020). An internal control was included in both assays.
Sample processing was the same for both methods. 500 µL of VTM for NPs or 500µL of diluted saliva were placed in Aptima lysis tubes (Hologic®, San Diego, California). Saliva pools were prepared by mixing 100µL diluted saliva from each of five subjects directly in an Aptima lysis tube.
Samples with invalid result by either assay were re tested after adding 500µL MEM directly to the Aptima lysis tube.
2.4. Statistical analysis
We calculated the sensitivity of NPs and saliva by using the total number of positive patients diagnosed by either test as the reference standard. The sensitivity differences between individual and pooled saliva were calculated by dividing the number of patients missed with pooling compared to individual treatment by the number of positive patients diagnosed with saliva. The 95% confidence intervals (CI 95%) were calculated by the Clopper and Pearson method using GraphPad Prism. The saliva sensitivity between asymptomatic and symptomatic individuals was compared using Fisher's exact tests. Pooled saliva sensitivity and individual saliva sensitivity were compared using Fisher's exact tests. Statistically significant difference was defined as P < 0.05.
Cycle threshold (Ct) values for saliva and NPs samples were compared using Student's paired t-test. The Ct values of NPs from individual groups (symptomatic and asymptomatic, or paired NPs and saliva from positive individuals and only NPs from positive individuals) were compared using the Mann-Whitney U-test.
3. Results
A total of 606 NPs and saliva samples were collected from 303 subjects (average age: 33 years; range: 03 to 77 years; 52.5% female). The 303 subjects included 160 (52.8%) who were mildly symptomatic and 143 (47.2%) who were asymptomatic at the time of sampling.
Two saliva samples gave invalid results even after additional dilution and repeated testing with both assays. Thus, 301 paired NPs-saliva samples were analysed. We tested 61 saliva pools, 60 pools of five samples and one of three.
-
1.
Detection of SARS-CoV-2 RNA in individual specimens with the LDT
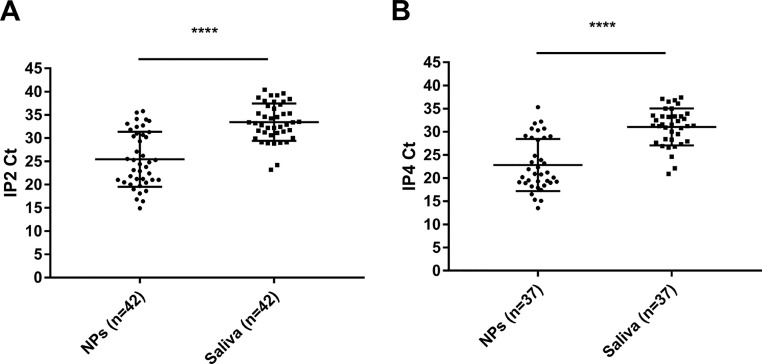
Overall, 55 subjects tested positive using NPs or saliva specimens and 42 tested positive with both samples (Table 1 ). Global sentivity of saliva and NPs were 80% [95% CI: 67.0%-89.6%] and 96.4% [95% CI: 87.5%-99.6%] respectively. The mean difference between paired positive NPs and/or saliva samples using the IP2 target was 8 Ct [95% CI: 6.57-9.43] and that between these samples on the IP4 target was 8.2 Ct [95% CI: 6.47-9.99] (P < 0.001) (Fig. 1 ).
Table 1.
Qualitative results for saliva and NPs using the LDT assay.
| Saliva |
||||||
|---|---|---|---|---|---|---|
| Positive | Negative | ∑ | ||||
| NPs | Positive | 42 | 11 | 53 | ||
| Negative | 2 | 246 | 248 | |||
| ∑ | 44 | 257 | 301a | |||
2 specimens gave invalid result.
Fig. 1.
IP2 (A) or IP4 Ct values (B) for paired NPs and saliva samples. Data are means + SD. ****P ≤ 0.0001 (Student's paired t test).
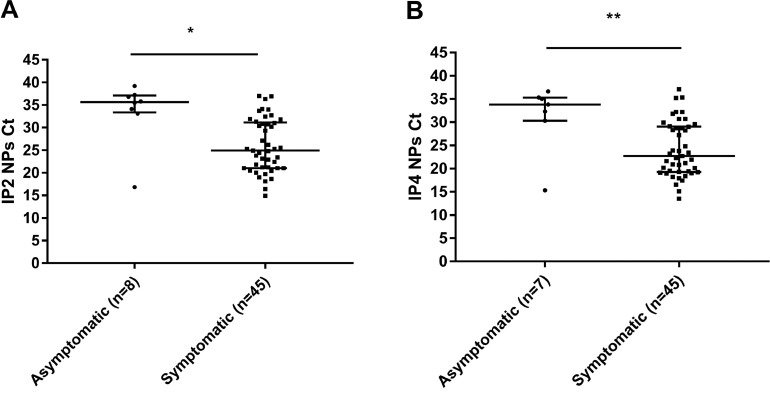
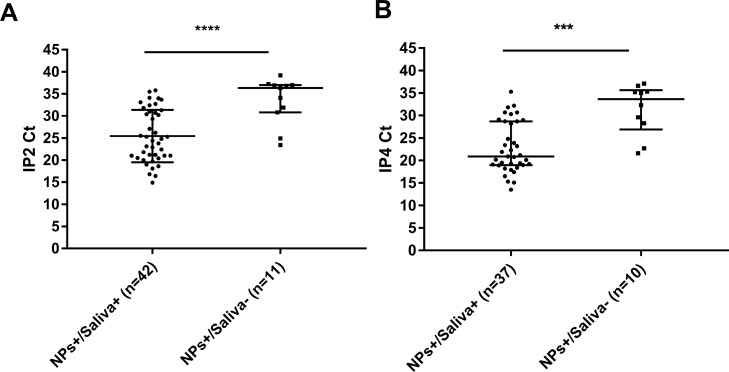
The 55 COVID-19 positive patients included 9 who were asymptomatic and 46 who were mildly symptomatic. Of the symptomatic patients, 41 were tested during the first week of symptoms and 5 were tested later. We found a higher saliva sensitivity for symptomatic people (84.8% [95% CI: 71.1%-93.7%]) than for asymptomatic ones (55.6% [95% CI: 21.2%-86.3%]) (Table 2 ). The median Ct values for NPs from asymptomatic individuals were significantly higher (IP2 Ct: 35.6; IP4 Ct: 33.8) than those for symptomatic subjects (IP2 Ct: 24.9, IP4 Ct: 22.7) (P = 0.019 (IP2) and P = 0.009 (IP4)) (Fig. 2 ). The median Ct values for patients whose NP was positive and their saliva was negative (IP2 Ct: 36.3 and IP4 Ct: 33.6) were higher than those of patients whose NP and saliva were positive (IP2 Ct: 24.6 and IP4 Ct: 20.9) (P < 0.001 and P < 0.001) (Fig. 3 ).
-
2.
SARS-CoV-2 RNA detection in individual samples with the AptimaTM assay
Table 2.
Qualitative results for saliva and NPs with both assays according to patient's category.
| Saliva +ve/ NP +ve patients | Saliva +ve/NP-ve patients | Saliva –ve/ NP+ve patients | Saliva –ve/NP-ve patients | |
|---|---|---|---|---|
| LDT assay (N = 301a) | ||||
| Asymptomatic (N = 141a) | 4 | 1 | 4 | 132 |
| Symptomatic (N = 160) | 38 | 1 | 7 | 114 |
| Aptima™ assay (N = 301a) | ||||
| Asymptomatic (N = 141a) | 5 | 1 | 4 | 131 |
| Symptomatic (N = 160) | 42 | 1 | 3 | 114 |
a 2 specimens gave invalid result
Fig. 2.
IP2 (A) or IP4 Ct values (B) for asymptomatic and symptomatic individuals. Data are medians plus interquartile range (IQR). *P ≤ 0.05 **P ≤ 0.01 (Mann-Whitney U-test).
Fig. 3.
IP2 (A) or IP4 Ct values (B) for paired positive NPs and saliva individuals and NPs positive only individuals. Data are medians and IQR. ****P ≤ 0.0001 ***P ≤ 0.001 (Mann- Whitney U-test).
The NPs or saliva samples from 56 subjects tested positive, and both saliva and NP samples from 47 subjects tested positive (Table 3 ). Saliva and NPs sensitivity were 87.5% [95% CI: 75.9%-94.8%] and 96.4% [95% CI: 87.7%-99.6%] respectively. Among these 56 COVID-19 positive patients, 10 were asymptomatic and 46 mildly symptomatic. Saliva sensitivity was higher for symptomatic people (93.5% [95% CI: 82.1%-98.6%]) than for asymptomatic ones (60% [95% CI: 26.2%-87.8%]) (P = 0.015) (Table 2).
-
3.
SARS-CoV-2 RNA detection in pooled saliva samples
Table 3.
Qualitative results for saliva and NPs using the Aptima™ assay.
| Saliva |
||||||
|---|---|---|---|---|---|---|
| Positive | Negative | ∑ | ||||
| NPs | Positive | 47 | 7 | 54 | ||
| Negative | 2 | 245 | 247 | |||
| ∑ | 49 | 252 | 301a | |||
2 specimens gave invalid result
The 61 saliva pools included 31 that tested positive and 30 that tested negative in the LDT assay. Among the 31 positive pools, 28 included at least 1 positive specimen looking at individual saliva results: 18 with only 1 positive sample, 8 pools with 2 positive samples and 2 pools with 3 positive samples. Therefore, 40 subjects were diagnosed using the pooling strategy giving an overall sensitivity of 72.7% [95% CI: 59.0%-83.9%] not significantly different of that of individual testing (P = 0.5) (Table 4 ). With 4 patients missed with this strategy compared to individual saliva testing, we observed a loss of sensitivity of 9.1% [95% CI: 2.5%-21.7%]. Three pools tested positive whereas the individual saliva results with the LDT assay were all negative. These three pools all contained a single saliva specimen that tested positive only in the Aptima™ assay and whose paired NPs tested positive in both assay. Potential RT-PCR inhibitors that would have been diluted thanks to the pooling strategy could explain the negative result of these saliva samples in individual treatment with the LDT assay.
Table 4.
Qualitative results for five-saliva pools and individual saliva using the LDT assay.
| Pooled saliva |
||||||
|---|---|---|---|---|---|---|
| Positive | Negative | ∑ | ||||
| Individual saliva | Positive | 40 | 4 | 44 | ||
| Negative | 3 | 254 | 257 | |||
| ∑ | 43 | 258 | 301a | |||
2 specimens gave invalid result
With the AptimaTM assay, 29 of 61 pools tested positive. The 29 positive pools included 18 with only 1 positive sample, 9 pools with 2 positive samples, and 2 pools with 3 positive samples. Thus, with 42 subjects detected the overall sensitivity of pooled saliva of 75.0% [95% CI: 61.6%- 85.6%] was not significantly different of that of individual saliva (P = 0.15). The loss of sensitivity compared to individual saliva testing was 14.3% [95% CI: 5.9%-27.2%] with 7 patients missed (Table 5 ).
Table 5.
Qualitative results for pools of five saliva specimens and individual saliva using the Aptima™ assay.
| Pooled saliva |
||||||
|---|---|---|---|---|---|---|
| Positive | Negative | ∑ | ||||
| Individual saliva | Positive | 42 | 7 | 49 | ||
| Negative | 0 | 252 | 252 | |||
| ∑ | 42 | 259 | 301a | |||
2 specimens gave invalid result
4. Discussion
The emergence of new more infective strains of virus has made it essential that the SARS-CoV-2 screening strategies should be as effective as possible (Kirby, 2021). Sample pooling seems to be promising strategy (Mutesa et al., 2021) but very few data are available on tests using saliva samples and their sensitivity is still debated. The results of this prospective study provide further evidence that non–invasive saliva sampling is an acceptable alternative to NPs. They effectively identified individuals who were most likely to spread the infection. Moreover, pooling saliva samples may increase laboratory capacity while maintaining a correct sensitivity.
The literature on the sensitivity of tests on saliva has not produced unanimous results. Reported sensitivities range from 46 to 100% (Azzi et al., 2020; Byrne et al., 2020; Chau et al., 2020; Chen et al., 2020; Hanson et al., 2020; Iwasaki et al., 2020; Jamal et al., 2021; Landry et al., 2020; McCormick-Baw et al., 2020; Migueres et al., 2020; Nagura Ikeda et al., 2020; Pasomsub et al., 2021; Rao et al., 2020; To et al., 2020; To et al., 2020; Torres et al., 2021; Williams et al., 2020; Wyllie et al., 2020; Yokota et al., 2020). We find overall sensitivities of 80.0% using our LDT and 87.5% with the Aptima assay, indicating that testing saliva is acceptable for SARS-CoV-2 diagnosis in outpatients, which is consistent with the findings of a recent meta-analysis (Bastos et al., 2021). The published discrepancies could be due to differences between the populations studied, the time of sampling and the patients’ virus load, the saliva collection method, or even the amplification assay used. While some studies found higher virus loads in saliva than in NPs (Rao et al., 2020; Wyllie et al., 2020), we and others find higher Ct values in saliva, indicating lower virus loads in saliva (Landry et al., 2020; Migueres et al., 2020; Pasomsub et al., 2021; Williams et al., 2020). This could be due to differences in the saliva collection method. A recent meta-analysis reported that tests using posterior oropharyngeal saliva samples were more sensitive than samples obtained by the swill-and-spit method (Bastos et al., 2021).
We tested saliva samples provided by subjects, both symptomatic and asymptomatic, who came to a drive-through testing center during the second epidemic wave in France, unlike the majority of studies which were done on inpatients or asymptomatic individuals. We find that tests on samples from asymptomatic individuals are less sensitive than are tests on samples from symptomatic ones, as reported by Nacher et al (Nacher et al., 2021). However, others have reported that tests for SARS-CoV-2 using saliva from asymptomatic individuals gave good results (Rao et al., 2020; Wyllie et al., 2020; Yokota et al., 2020), with no relationship between test sensitivity and symptoms (Torres et al., 2021; Bastos et al., 2021). Our finding of lower sensitivity is probably due to the lower virus load in asymptomatic patients (only one asymptomatic patient had an IP2 Ct < 30) and SARS-CoV-2 infections were less common (7%) than in symptomatic ones (28%). The majority of patients who tested positive only for their NPs had a high Ct (IP2 Ct >30), which confirms previous studies showing that saliva sensitivity depends mainly on the patient's virus load and sampling time after infection (Jamal et al., 2021; Migueres et al., 2020), as the virus concentration in the saliva declines more quickly than that in the NP cavity (Iwasaki et al., 2020; Torres et al., 2021). It is now well accepted that patients who have a high Ct (24-35, depending on the study) are not infectious or at least less so (Bullard et al., 2020; La Scola et al., 2020; Singanayagam et al., 2020). Overall our results indicate that SARS-CoV-2 molecular diagnosis on saliva samples is sensitive enough to identify individuals most likely to spread the virus.
We observed a slight loss of sensitivity with saliva samples compared to NPS. Whereas this slight loss can be unimportant if prevalence is 0.1% (few missed cases per 1000 tested) it can be very important if prevalence is 10% (many missed cases per 1000 tested). However we think, given our results, that these missed cases with saliva samples, are less likely to participate to the viral spread of the SARS-CoV-2 due to their low viral loads. Moreover, any risk that this sampling is less sensitive than NPs sampling is offset by the possibility of repeated tests greatly facilitated by the non-invasiveness, the ease of collection, and the low cost of this sampling (reduced material and human resources needed) (Bastos et al., 2021).
Several studies have shown that pooling of nasopharyngeal samples is efficient and sensitive (Alcoba Florez et al., 2021; Chong et al., 2020; de Salazar et al., 2020; Garg et al., 2021; Lohse et al., 2020 Nov 1) but few have evaluated saliva pooling (Barat et al., 2021; Pasomsub et al., 2020; Watkins et al., 2020). Our pools of 5 saliva samples provided data that agrees well with those obtained from individual saliva samples with both of the assays used. There was a slight loss of sensitivity due to the pooling itself (Barat et al., 2021). The pooling strategy missed 4 or 7 patients (depending on the assay), but they were detected after testing individual salivas. This was undoubtedly due to their low virus loads (IP2 Ct > 30). In contrast, three samples tested positive in the pooled samples but were negative in individual saliva tests. This suggests that some samples should be diluted to reduce the influence of any saliva RT-PCR inhibitors (Ochert et al., 1994). Overall, the slight loss of sensitivity using pooled samples rather than individual tests might be acceptable as it increases laboratory capacity for mass screening and reduces costs.
Most studies assessed the sensitivity of saliva samples using RT-PCR or Reverse Transcription Loop-mediated Isothermal Amplification (RT-LAMP) (Nagura Ikeda et al., 2020; Yokota et al., 2020); only one used a TMA assay on saliva samples (Hanson et al., 2020). Our results demonstrate that the Aptima™ assay is at least as efficient as a classic RT-PCR assay for detecting SARS-CoV-2 in saliva samples. The overall sensitivities were 80% with the LDT and 87.5% with the Aptima assay and the concordance between the two assays was very good with only 10 of 606 discordant results for individual samples.
This study has several limitations. Subjects only self-collected their saliva with the guidance of a health care worker. It would be informative to evaluate unsupervised saliva self-collection by subjects at home; if successful this would increase testing accessibility. This sampling method could also reduce the human resources manning testing centers. These professionals could profitably be employed at laboratory testing platforms or COVID vaccination centers.
We evaluated pools of five saliva samples from subjects at high risk of infection (symptomatic subjects or contact cases) to obtain enough positive subjects to assess pooled saliva sensitivity. This pooling strategy is probably more suitable for mass screening, such as in a city wide survey of mainly asymptomatic individuals and a low prevalence. The pooling of five saliva specimens was effective with an average 30% reduction in testing, despite the relatively high prevalence (18%) and high proportion of positive pools in our study. This strategy would be even more cost effective if it were used after lockdown, when prevalence is low. For example, at a prevalence of 1% or 5%, pooling 5:1 reduces at least (in the worst scenario where each positive are in different pools) the number of samples run per 100 specimens from 100 to 25 or 45 respectively. While pooling is efficient to reduce reagent consuming, its implementation requires good organization with critical steps such as pooling process, tracking positive samples for individual treatment, and results reporting that could be time consuming for technicians and require an automation.
In conclusion, we believe that the good sensitivity of saliva-based tests for SARS-CoV-2 nucleic acid indicates that this medium is a desirable non-invasive way of obtaining samples that is well accepted by patients and allows repeated testing. Pools of 5 saliva samples appear to be suitable for mass screening.
Acknowledgments
Hologic provided the molecular reagents for SARS-CoV-2 detection. The English text was edited by Dr Owen Parkes.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Authors’ statement
M Migueres and C Vellas: Investigation, Methodology, Writing - Original Draft, Statistical analysis. J Izopet: Conceptualization, Methodology, Supervision, Writing - Review & Editing. JM Mansuy, F Abravanel and S Raymond: Supervision and Writing – Review. C Dimeglio: Statistical analysis – Review.I Da Silva and V Ferrer: Methodology and Data acquisition. All authors edited and approved the final manuscript.
References
- Alcoba Florez J, Gil Campesino H, García Martínez de Artola D, Díez Gil O, Valenzuela Fernández A, González Montelongo R, et al. Increasing SARS-CoV-2 RT-qPCR testing capacity by sample pooling. Int J Infect Dis. 2021;103:19–22. doi: 10.1016/j.ijid.2020.11.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi L, Carcano G, Gianfagna F, Grossi P, Gasperina DD, Genoni A, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat B, Das S, Giorgi VD, Henderson DK, Kopka S, Lau AF, et al. Pooled Saliva Specimens for SARS-CoV-2 Testing. J Clin Microbiol [Internet] 2021;59(3):e02486–20. doi: 10.1128/JCM.02486-20. https://jcm.asm.org/content/early/2020/11/30/JCM.02486-20 [cited 2020 Dec 12]. Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos ML, Perlman-Arrow S, Menzies D, Campbell JR. The sensitivity and costs of testing for SARS-CoV-2 infection with saliva versus nasopharyngeal swabs : a systematic review and meta-analysis. Ann Intern Med. 2021;174(4):501–510. doi: 10.7326/M20-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenger BM, Conly JM, Fonseca K, Hu J, Louie T, Schneider AR, et al. Saliva collected in universal transport media is an effective, simple and high-volume amenable method to detect SARS-CoV-2. Clin Microbiol Infect [Internet] 2021;27(4):656–657. doi: 10.1016/j.cmi.2020.10.035. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7641592 [cited 2020 Nov 27]. Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020;71(10):2663–2666. doi: 10.1093/cid/ciaa638. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7314198 [cited 2020 Dec 12]. Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RL, Kay GA, Kontogianni K, Aljayyoussi G, Brown L, Collins AM, et al. Saliva alternative to upper respiratory swabs for SARS-CoV-2 diagnosis. Emerg Infect Dis. 2020:2770–2771. doi: 10.3201/eid2611.203283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . Centers for Disease Control and Prevention; 2020. Coronavirus Disease 2019 (COVID-19) [Internet]https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html [cited 2021 Jan 10]. Available. [Google Scholar]
- CDC . Centers for Disease Control and Prevention; 2020. COVID-19 Resources and Guidelines for Labs and Laboratory Workers.https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html [Internet][cited 2021 Jan 28]. Available. [Google Scholar]
- Chau NVV, Thanh Lam V, Thanh Dung N, Yen LM, Minh NNQ, Hung LM, et al. The natural history and transmission potential of asymptomatic SARS-CoV-2 infection. Clin Infect Dis[Internet] 2020;71(10):2679-2687. [cited 2020 Jul 17]. Available https://academic.oup.com/cid/article/doi/10.1093/cid/ciaa711/5851471. [DOI] [PMC free article] [PubMed]
- Chen JH K, Yip CC Y, Poon RW S, Chan K H, Cheng VC C, Hung IF N, et al. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):1356–1359. doi: 10.1080/22221751.2020.1775133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong BSW, Tran T, Druce J, Ballard SA, Simpson JA, Catton M. Sample pooling is a viable strategy for SARS-CoV-2 detection in low-prevalence settings. Pathology. 2020;52(7):796–800. doi: 10.1016/j.pathol.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus (COVID-19) Deaths - Statistics and Research [Internet]. Our World in Data. [cited 2020 Nov 28]. Available https://ourworldindata.org/covid-deaths
- de Salazar A, Aguilera A, Trastoy R, Fuentes A, Alados JC, Causse M, et al. Sample pooling for SARS-CoV-2 RT-PCR screening. Clin Microbiol Infect. 2020;26(12):1687.e1–1687.e5. doi: 10.1016/j.cmi.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg J, Singh V, Pandey P, Verma A, Sen M, Das A, et al. Evaluation of sample pooling for diagnosis of COVID-19 by real time-PCR: a resource-saving combat strategy. J Med Virol [Internet] 2021;93(3):1526-1531. [cited 2020 Dec 12];n/a(n/a). Available https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.26475. [DOI] [PubMed]
- Hanson KE, Barker AP, Hillyard DR, Gilmore N, Barrett JW, Orlandi RR, et al. Self-collected anterior nasal and saliva specimens versus health care worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol. 2020 21;58(11):e01824–20. doi: 10.1128/JCM.01824-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Fujisawa S, Nakakubo S, Kamada K, Yamashita Y, Fukumoto T, et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect [Internet] 2020;81(2):e145–e147. doi: 10.1016/j.jinf.2020.05.071. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7270800/ [cited 2020 Jul 17]. Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal AJ, Mozafarihashjin M, Coomes E, Powis J, Li AX, Paterson A, et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis [Internet] 2021;72(6):1064–1066. doi: 10.1093/cid/ciaa848. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7337630/ [cited 2020 Jul 17]. Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir Med [Internet] 2021;9(2):e20–e21. doi: 10.1016/S2213-2600(21)00005-9. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7784534/ [cited 2021 Jan 27]. Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry ML, Criscuolo J, Peaper DR. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol. 2020;130 doi: 10.1016/j.jcv.2020.104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse S, Pfuhl T, Berkó-Göttel B, Rissland J, Geißler T, Gärtner B, et al. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis. 2020;20(11):1231–1232. doi: 10.1016/S1473-3099(20)30362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick-Baw C, Morgan K, Gaffney D, Cazares Y, Jaworski K, Byrd A, et al. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol. 2020;58(8):e01109–20. doi: 10.1128/JCM.01109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueres M, Mengelle C, Dimeglio C, Didier A, Alvarez M, Delobel P, et al. Saliva sampling for diagnosing SARS-CoV-2 infections in symptomatic patients and asymptomatic carriers. J Clin Virol. 2020;130 doi: 10.1016/j.jcv.2020.104580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutesa L, Ndishimye P, Butera Y, Souopgui J, Uwineza A, Rutayisire R, et al. A pooled testing strategy for identifying SARS-CoV-2 at low prevalence. Nature. 2021;589(7841):276–280. doi: 10.1038/s41586-020-2885-5. [DOI] [PubMed] [Google Scholar]
- Nacher M, Mergeay-Fabre M, Blanchet D, Benoit E, Pozl T, Mesphoule P, et al. Prospective comparison of saliva and nasopharyngeal swab sampling for mass screening for COVID-19. Front Med (Lausanne) 2021;8:621160. doi: 10.3389/fmed.2021.621160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagura Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), Direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol. 2020;58(9):e01438–20. doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochert AS, Boulter AW, Birnbaum W, Johnson NW, Teo CG. Inhibitory effect of salivary fluids on PCR: potency and removal. PCR Methods Appl. 1994;3(6):365–368. doi: 10.1101/gr.3.6.365. [DOI] [PubMed] [Google Scholar]
- Pasomsub E, Watcharananan SP, Boonyawat K, Janchompoo P, Wongtabtim G, Suksuwan W, et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2021;27(2):285.e1–285.e4. doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasomsub E, Watcharananan SP, Watthanachockchai T, Rakmanee K, Tassaneetrithep B, Kiertiburanakul S, et al. Saliva sample pooling for the detection of SARS-CoV-2. J Med Virol [Internet] 2020;93(3):1506–1511. doi: 10.1002/jmv.26460. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7461487/ [cited 2020 Dec 12]. Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Rashid FA, Sabri FSAH, Jamil NN, Zain R, Hashim R, et al. Comparing nasopharyngeal swab and early morning saliva for the identification of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis [Internet] 2020 doi: 10.1093/cid/ciaa1156. [cited 2020 Dec 12];(ciaa1156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V, Ellis J, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill [Internet] 2020;25(32):2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7427302/ [cited 2020 Dec 12];25(32). Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolimowska K, Rayment M, Jones R, Madona P, Moore LSP, Randell P. Non-invasive saliva specimens for the diagnosis of COVID-19: caution in mild outpatient cohorts with low prevalence. Clin Microbiol Infect. 2020;26(12):1711–1713. doi: 10.1016/j.cmi.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KK W, Tsang OT Y, Chik Yan Yip C, Chan K H, Wu T C, Chan JMC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KK W, Tsang OT Y, Leung W S, Tam AR, Wu T C, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M, Collins K, Corbit M, Ramirez M, Winters CR, Katz L, et al. Comparison of saliva and nasopharyngeal swab SARS-CoV-2 RT-qPCR testing in a community setting. J Infect [Internet] 2021;82(4):84–123. doi: 10.1016/j.jinf.2020.11.015. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7670897/ [cited 2020 Dec 12]. Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trémeaux P, Lhomme S, Abravanel F, Raymond S, Mengelle C, Mansuy J M, et al. Evaluation of the AptimaTM transcription-mediated amplification assay (Hologic®) for detecting SARS-CoV-2 in clinical specimens. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins AE, Fenichel EP, Weinberger DM, Vogels CBF, Brackney DE, Casanovas-Massana A, et al. Pooling saliva to increase SARS-CoV-2 testing capacity. medRxiv. 2020;2020.09.02.20183830.
- WHO. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. 2020 [cited 2021 Jan 28]. Available https://apps.who.int/iris/handle/10665/331329
- Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8):e00776–20. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383(13):1283–1286. doi: 10.1056/NEJMc2016359. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota I, Shane PY, Okada K, Unoki Y, Yang Y, Inao T, et al. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin Infect Dis. 2020;73(3):e559–e565. doi: 10.1093/cid/ciaa1388. [DOI] [PMC free article] [PubMed] [Google Scholar]