The current pandemic (coronavirus disease 2019 [COVID-19]) has caused more than 135 million infections and more than 3 million deaths worldwide. Angiotensin-converting enzyme 2 (ACE2) plays a substantial role for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to gain entry into the host cells and has been recognized since the beginning of the pandemic with more than 6000 publications (PubMed as of April 9, 2021), whereas the role of transmembrane protease, serine 2 (TMPRSS2) is of equal importance though underrecognized with only one-fifth of the publications (1939). In the pathogenicity of SARS-CoV-2, in addition to ACE2 and TMPRSS2 other host factors such as FURIN and neuropilin 1 are important. However, in this perspective we discuss the critical role of TMPRSS21 , 2 and highlight that both ACE2 and TMPRSS2 are equally important. The coexpression3 of both ACE2 and TMPRSS2 in the host target cells is important for the SARS-CoV-2 entry into the target cells. It is important to consider strategies to target TMPRSS2 to control the SARS-CoV-2 infection and the pandemic4 until a safe and effective vaccine against SARS-CoV-2 and its variants becomes available.
All cellular proteins, to sustain life, are subject to a permanent turnover process (apoptosis) that requires de novo synthesis and subsequent degradation. Protein degradation requires proteases to hydrolyze peptide bonds—the bonds that connect amino acids within proteins to make them stable. In 1836, Theodor Schwann introduced the concept of proteolysis and purified an enzyme (later called proteases) pepsin capable of fragmenting peptide bonds. Proteases not only play a substantial role in the hydrolysis of peptide bonds but also reduce the requirement of energy required for such interactions. To date, most of the proteases (>500) are soluble proteins that are active in an aqueous environment within cells, tissues, or body fluids. One hundred fifty years later, in 1997 it became evident that proteolysis also occurs within the cell membranes (integral membrane proteins)5 that allows passage of larger or charged molecules such as glucose and ions. Proteolysis or cleavage of the “integral membrane proteins” liberates protein fragments on both sides of the membrane and are capable of transmitting signals across membranes.
The details of the structure and functions of the proteases are beyond the scope of this review. On the basis of the structure and catalytic processes, proteases are classified as serine, threonine, cysteine, aspartyl, and metalloproteases. Most of the proteases are in solution (soluble) and secreted. However, serine proteases can be membrane bound through their transmembrane domain and divided into 3 groups: carboxy terminal (type I), amino terminal (type II), and glycosylphosphatidylinositol (type III). The largest group of membrane-bound serine proteases is type II, with 17 known members subdivided into 4 phylogenetically distinct subfamilies: matriptase, hepsin/transmembrane protease serine (TMPRSS), corin, and human airway trypsin–like proteases. Most of the group of membrane-bound serine proteases were characterized recently, and their specific physiological role remains unknown.6 However, they appear to play a role in digestion, coagulation, iron hemostasis, spermatogenesis, epithelial barrier integrity, fluid balance, hormone processing, wound healing, embryonic development, immune response, inflammation, tumor growth, and metastases. Serine proteases activate through autocatalytic cleavage.6 Originally type II transmembrane-bound serine protease (TMPRSS2) was a small intestine–associated protease.7 It gained interest because of its highly localized expression in the prostate epithelium and its potential role in carcinogenesis as overexpressed in prostate cancer. However, the function of TMPRSS2 remains unclear as life could be normal in its absence, as the TMPRSS2 (−/−) knockout mouse studies reported normal development, growth, organ function, and survival.8 Furthermore, in the TMPRSS2 knockout mouse, H3N2 influenza strain was found to be avirulent9 and a weakened or delayed inflammatory response with SARS-CoV-2 infection.10
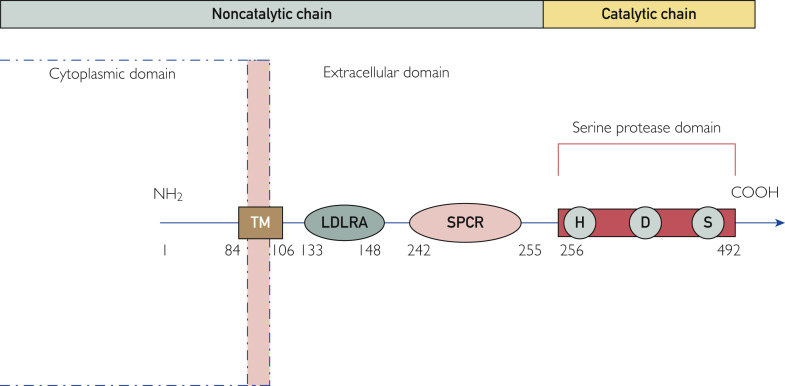
TMPRSS2 gene is widely conserved and has 2 isoforms—TMPRSS11A and TMPRSS11B—both being autocatalytically activated from the inactive zymogen form.11 It is found at human chromosome 21q22.3 and encodes a protein of 492 amino acids (Figure 1 ). It contains 2 chains: a noncatalytic chain formed by amino acids 1 to 255 and a catalytic chain containing remaining amino acids from 256 to 492. It contains type II transmembrane domain, low-density lipoprotein receptor class A domain that forms the binding site for calcium, scavenger receptor cysteine–rich domain involved in binding to other cell surfaces, and serine protease domain, with S1 family that cleaves at arginine or lysine residues. Based on its domain structure, it could work as a receptor for specific ligand(s)-mediating signals from the environment into the cells. In humans, it is expressed in the epithelial cells of the nose, trachea, bronchial epithelial cells, digestive tract, and prostate. The expression of TMPRSS2 is developmentally regulated and increases with age12 , 13; although androgen dependent, it does not appear to differ in the lungs between men and women. However, highest nasal gene expression is seen in Black individuals as compared with Asian, Latino, and White individuals.14 In addition, its expression is up-regulated in smokers.13
Figure 1.
Schematic representation of transmembrane protease, serine 2: structure and domains. The numbers represent the amino acid sequence. COOH, C terminal; H, D, S, catalytic amino acid triad histidine, aspartic acid, and serine; LDLRA, low-density lipoprotein receptor class A; NH2, N terminal; SPCR, scavenger protein cysteine rich; TM, transmembrane domain.
Significance of TMPRSS2 in the COVID-19 Pandemic
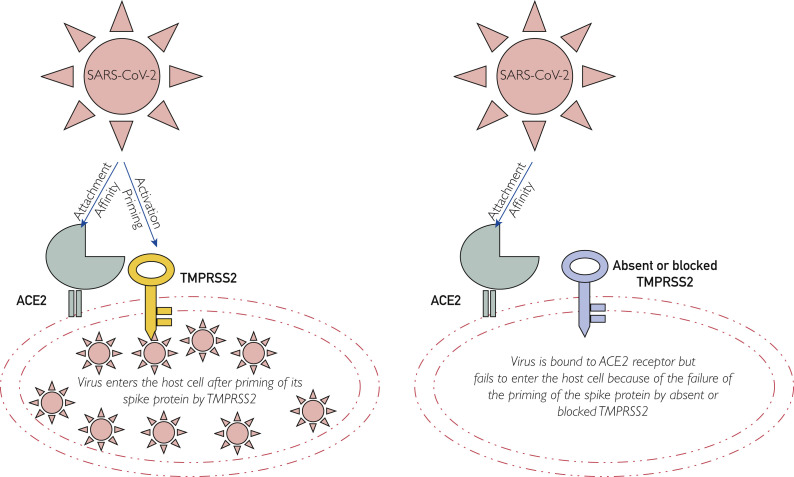
Coronaviruses, including severe respiratory distress syndrome, Middle East respiratory syndrome, and SARS-CoV-2 (COVID-19), belong to the largest RNA viruses of the Coronaviridae family and cause upper and lower respiratory infections. Coronaviruses encode 4 structural proteins: matrix glycoproteins, envelope, nucleocapsid, and spike proteins, and the S protein mediates attachment of the virus to host cell receptors.15 The S protein consists of 2 subunits: S1 (N terminus), which acts as a receptor-binding region, and S2 (C terminus), which serves to promote fusion activity or proteolytic priming.16 As most human coronaviruses, including SARS-CoV-2, exit the producer cells with “unprimed” S proteins,17 their entry into the host target cells depends on both the receptor binding with membrane-bound ACE2 and target cell proteases, mainly TMPRSS2 (Figure 2 ).
Figure 2.
Schematic (simplified) representation of transmembrane protease, serine 2 (TMPRSS2) acting as a key to the entry of the virus into the host cell. ACE2, angiotensin-converting enzyme 2; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Severe acute respiratory syndrome coronavirus 2, like other coronaviruses, after inoculation, enters the target cells in 2 steps. First, it binds to cell receptors, the membrane-bound ACE2, second, followed by S protein priming or cleaving by host cell proteases,18 and this must occur in a specific sequence. The proteolytic cleavage must occur after ACE2 engagement, because if it occurs before ACE2 binding then these proteases may cleave and inactivate the viral spike.19 Although several proteases are available but cannot remain in the vicinity of ACE2 receptors,19 and these may inactivate the viral spike before ACE2 engagement. Therefore, the location and timing of the activating proteases are important for the virus entry into the host target cell and its pathogenicity. Transmembrane protease, serine 2 needs to be anchored in the cell membrane near or coexpressed in the host cells with ACE2 receptors, and the membrane-bound type II transmembrane serine proteases (TMPRSS2) are pivotal in priming of the S protein and entry of the virus into the host target cell as well as in its replication and spread. Severe acute respiratory syndrome coronavirus 2 can use other endosomal cysteine proteases—cathepsin B and L—for priming of the S protein, but priming by TMPRSS2 is essential.10 Cathepsin B and L activity is dispensable. The coexpression of both ACE2 and TMPRSS2 is important for SARS-CoV-2 to enter target cells to cause damage. Furthermore, the expression of TMPRSS2 is developmentally regulated and increases with age,20 which is racially modified being highly expressed in Blacks and up-regulated in smokers, likely contributing to the severity of infection in the elderly Black individuals and smokers.
It is important to highlight that TMPRSS2 plays a critical role in priming of the S protein of most coronaviruses (including SARS-CoV-2), and its inhibition1 or modification would be an important therapeutic target for intervention in or prevention of COVID-19 by blocking the virus entry into the host target cells. One of the benefits of this approach might be that these proteases are not susceptible to progressive sequence variations that viruses are. The TMPRSS2 inhibitors camostat, nafamostat, and aerosolized aprotinin have been found to attenuate TMPRSS2 activity. There are more than 25 ongoing clinical trials of camostat and 7 of nafamostat around the world for COVID-19 (ClinicalTrials.gov). Other agents such as bromhexine (a mucolytic agent), rimantadine, and plasminogen activator inhibitor type 1 merit evaluation.
Conclusion
Severe acute respiratory syndrome coronavirus 2 infection to date has caused more than 135 million infections and more than 3 million deaths worldwide. In addition to the highly virulent virus, various host factors play an important role in its pathogenicity and it is important to recognize and highlight the equal role played by both ACE2 and TMPRSS2 in the pathogenicity of SARS-CoV-2 infection. The coexpression of both ACE2 and TMPRSS2 in the host target cells is important, so alternate strategies that target TMPRSS2 should be considered and evaluated in a timely fashion to control the pandemic until a safe and effective vaccine against SARS-CoV-2 and its variants becomes available to the population.
Footnotes
Potential Competing Interests: The author reports no competing interests.
Supplemental Online Material
References
- 1.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou Y., Zhao J., Martin W., et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18(1):216. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sungnak W., Huang N., Becavin C., et al. HCA Lung Biological Network. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baughn L.B., Sharma N., Elhaik E., Sekulic A., Bryce A.H., Fonseca R. Targeting TMPRSS2 in SARS-CoV-2 infection. Mayo Clin Proc. 2020;95(9):1989–1999. doi: 10.1016/j.mayocp.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paschkowsky S., Hsiao J.M., Young J.C., Munter L.M. The discovery of proteases and intramembrane proteolysis. Biochem Cell Biol. 2019;97(3):265–269. doi: 10.1139/bcb-2018-0186. [DOI] [PubMed] [Google Scholar]
- 6.Afar D.E., Vivanco I., Hubert R.S., et al. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61(4):1686–1692. [PubMed] [Google Scholar]
- 7.Paoloni-Giacobino A., Chen H., Peitsch M.C., Rossier C., Antonarakis S.E. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3 [published correction appears in Genomics. 2001;77(1-2):114] Genomics. 1997;44(3):309–320. doi: 10.1006/geno.1997.4845. [DOI] [PubMed] [Google Scholar]
- 8.Kim T.S., Heinlein C., Hackman R.C., Nelson P.S. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol Cell Biol. 2006;26(3):965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakai K., Sekizuka T., Ami Y., et al. A mutant H3N2 influenza virus uses an alternative activation mechanism in TMPRSS2 knockout mice by loss of an oligosaccharide in the hemagglutinin stalk region. J Virol. 2015;89(9):5154–5158. doi: 10.1128/JVI.00124-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93(6):e01815–e01818. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thunders M., Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2) J Clin Pathol. 2020;73(12):773–776. doi: 10.1136/jclinpath-2020-206987. [DOI] [PubMed] [Google Scholar]
- 12.Schuler B.A., Christian Habermann A., Plosa E.J., et al. HCA Lung Biological Network Age-related expression of SARS-CoV-2 priming protease TMPRSS2 in the developing lung. https://doi.org/10.1101/2020.05.22.111187 [published online ahead of print May 23, 2020]. bioRxiv.
- 13.Saheb Sharif-Askari N., Saheb Sharif-Askari F., Alabed M., et al. Airways expression of SARS-CoV-2 receptor, ACE2, and TMPRSS2 is lower in children than adults and increases with smoking and COPD. Mol Ther Methods Clin Dev. 2020;18:1–6. doi: 10.1016/j.omtm.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunyavanich S., Grant C., Vicencio A. Racial/ethnic variation in nasal gene expression of transmembrane serine protease 2 (TMPRSS2) JAMA. 2020;324(15):1567–1568. doi: 10.1001/jama.2020.17386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siu Y.L., Teoh K.T., Lo J., et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol. 2008;82(22):11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sui J., Li W., Murakami A., et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A. 2004;101(8):2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai Y. Protease-dependent virus tropism and pathogenicity [published correction appears in Trends Microbiol. 1993;1(4):135] Trends Microbiol. 1993;1(3):81–87. doi: 10.1016/0966-842X(93)90112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci U S A. 2005;102(35):12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuler B.A., Habermann A.C., Plosa E.J., et al. Age-determined expression of priming protease TMPRSS2 and localization of SARS-CoV-2 in lung epithelium. J Clin Invest. 2021;131(1):e140766. doi: 10.1172/JCI140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.