Abstract
Neurodevelopmental disorders (NDDs) are a group of diseases characterized by high heterogeneity and frequently co-occurring symptoms. The mutational spectrum in patients with NDDs is largely incomplete. Here, we sequenced 547 genes from 1102 patients with NDDs and validated 1271 potential functional variants, including 108 de novo variants (DNVs) in 78 autosomal genes and seven inherited hemizygous variants in six X chromosomal genes. Notably, 36 of these 78 genes are the first to be reported in Chinese patients with NDDs. By integrating our genetic data with public data, we prioritized 212 NDD candidate genes with FDR < 0.1, including 17 novel genes. The novel candidate genes interacted or were co-expressed with known candidate genes, forming a functional network involved in known pathways. We highlighted MSL2, which carried two de novo protein-truncating variants (p.L192Vfs*3 and p.S486Ifs*11) and was frequently connected with known candidate genes. This study provides the mutational spectrum of NDDs in China and prioritizes 212 NDD candidate genes for further functional validation and genetic counseling.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12035-021-02377-y.
Keywords: Neurodevelopmental disorders, Targeted sequencing, De novo variants, Candidate genes
Introduction
Neurodevelopmental disorders (NDDs) are complex and heterogeneous disorders characterized by impaired motor function, learning, and verbal and non-verbal communication resulting from dysfunction during brain development [1–4]. NDDs span a very wide range of neurological and psychiatric disorders [1, 5] and affect more than 3% of children worldwide [6]. Although each diagnosis is distinct in clinical settings, in general, NDDs are characterized by high clinical and genetic heterogeneity. Previous studies have estimated the heritability of NDDs, with the highest being up to ~ 80% [7–10]. Multiple phenotype-genotype correlation studies have revealed that patients carrying deleterious variants in the same risk gene have manifested broad clinical phenotypes, including impaired social interaction, developmental delay, regression, and repetitive behavior [11–13]. The high level of heterogeneity and similar pathogenetic mechanisms present challenges for clinical diagnosis and treatment.
Recent studies have reported that targeted sequencing is a powerful and cost-effective tool for discovering new genetic risk genes in many diseases, particularly diseases with high genetic heterogeneity [4, 14, 15]. Both rare inherited and de novo variants (DNVs) have been demonstrated to contribute to NDDs [16–18]. Stessman et al. sequenced 208 candidate genes in patients with NDDs and identified 91 risk genes with locus-specific significance for disruptive variants in 5.7% of patients [4]. Wang et al. performed targeted sequencing of 189 autism spectrum disorder (ASD) candidate risk genes in Chinese patients and found that genes identified in European ASD cohorts were highly relevant in Chinese cohorts [19]. Guo et al. expanded the above study to a larger cohort of patients and provided support for a multifactorial model of ASD risk [20]. Some studies have shown that there are multiple risk genes that are shared between neurodevelopmental disorders, and integrating datasets at multiple levels is beneficial for the etiology of NDDs [21, 22]. Takata et al. combined published DNV data and found that integrative analyses was conducive to identifying significant genes and extending ASD-related molecular and brain networks [23]. Gonzalez-Mantilla et al. used multilevel data-integration approach and identified novel candidate genes for developmental brain disorders [24]. We have previously demonstrated that DNVs, gene set enrichment analysis, and protein-protein interaction (PPI)/co-expression analysis can provide new insights into the genetic mechanisms underpinning NDDs [25–28]. However, only a few known pathogenetic genes have been described to explain the genetic causes of NDDs, and the etiology behind NDDs remains unclear.
Here, we used targeted sequencing to examine 547 genes from 1102 Chinese patients with NDDs. We identified potential functional variants and explored the patterns of DNVs in Chinese patients with NDDs. We then integrated our dataset with public datasets of neurodevelopmental disorders from the Gene4Denovo [29] database to prioritize NDD candidate genes and discover novel candidate genes. Finally, we investigated whether novel candidate genes were functionally associated with known candidate genes using multilevel bioinformatics analysis.
Materials and Methods
Panel Gene Design and Targeted Sequencing
In this study, potential NDD risk genes are collected based on the following criteria: (1) genes recorded in the Online Mendelian Inheritance in Man (OMIM) (https://omim.org/) are associated with NDDs, mainly including autism, cerebral palsy, mental retardation, and epilepsy; (2) candidate genes in the NPdenovo database [28]; (3) strong risk genes from the SFARI Gene database (https://gene.sfari.org/) [30]; and (4) genes prioritized in our previous NDD-related studies [25, 26, 31, 32]. After removing duplicated genes, 547 target genes were selected (Table S1).
A total of 935 unrelated trios (probands and their unaffected parents) and 167 probands without parents were recruited from China. Genomic DNA (1 μg) extracted from whole blood was sheared and assembled into a DNA library prior to targeted sequencing. The Illumina X10 sequencing system (Illumina, San Diego, CA, USA) was used to generate paired-end raw data. This panel resulted in an average depth of 181.46× in target regions, and 98.82% of target bases were covered with depth ≥ 10× on average. This study was approved by the Institutional Review Board of the State Key Laboratory of Medical Genetics, School of Life Sciences, Central South University, Changsha, Hunan, China. All subjects who participated in this study provided informed consent prior to sample collection.
Variation Detection and Annotation
Quality control of the sequencing data was performed using Cutadapt [33] and FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to remove adapter and unqualified sequences, respectively. BWA-MEM [34] was employed to align the clean reads to the human reference genome (hg19). Samtools [35] utilities were used to mark duplicate reads and generate position-sorted files, while the Genome Analysis Toolkit HaplotypeCaller was used to call variants. Comprehensive annotation of all variants was performed using ANNOVAR [36], including functional implications (gene region, functional effect, mRNA GeneBank accession number, amino acid change, cytoband, etc.), functional predictions for missense variants, and allele frequencies of gnomAD, ExAC, and in-house data (1113 WES samples and 2469 WGS samples). Deleterious missense variants (Dmis) were predicted by ReVe [37] and missense variants with ReVe ≤ 0.7 were excluded. Only protein-truncating variants (PTVs, including stop-gain, stop-loss, frameshift, and splicing) and Dmis with minor-allele frequencies ≤ 0.1% were defined as potential functional variants and selected for further analysis. Sanger sequencing was used to validate all potential functional variants in our study.
Prioritization of Candidate Genes
Using de novo PTVs, together with Dmis and background DNV rates, we employed the TADA classification tool [38] to prioritize candidate genes, with gene exhibiting a false discovery rate (FDR) < 0.1 defined as candidates. First, DNVs from 935 trios, inherited variants from 1102 probands, and genetic variants from 3582 in-house Chinese controls (Table S6) were applied to TADA model, and we prioritized 17 candidate genes with FDR values < 0.1. To increase the statistical power, we integrated DNVs from 16,807 probands with different types of NDDs and included 3391 controls from the Gene4Denovo database (version 1.0) [39] and 208 candidate genes with FDR values < 0.1 for further analysis. All samples integrated in our study have been carefully deduplicated.
To discover novel candidate genes, we collected known candidate genes from previous studies and related databases, and excluded them from the list of candidate genes prioritized by TADA. Known candidate genes were defined as follows: (1) genes defined as risk genes in any ten recent publications involving large-scale exome, whole-genome, or targeted sequencing (De Rubeis et al. [17], Sanders et al. [40], Lelieveld et al. [41], McRae et al. [42], Stessman et al. [43], Yuen et al. [44], Nguyen et al. [22], Takata et al. [23], Coe et al. [21], and Satterstrom et al. [45]); (2) genes collected from OMIM that were associated with neurodevelopmental disorders, including intellectual developmental disorder, autism, mental retardation, epileptic encephalopathy, and schizophrenia; (3) genes belonging to the category of syndromic gene or score category of 1 or 2 in SFARI Gene [30]. Genes that met any of the above conditions were classified as known candidate genes. In contrast, genes that failed to meet any of the above conditions above and had no obvious genetic evidence in association with NDDs in PubMed were classified as novel candidate genes.
Permutation Test
Spatial and temporal expression data of the human brain were downloaded from BrainSpan (http://www.brainspan.org/). For ethical reasons, we removed samples during fetal stages and selected postnatal cortical samples for further study. We calculated the Pearson correlation coefficients between any two genes based on their expression levels. Gene pairs with |R| > 0.7 were regarded as being co-expressed in the human brain. The permutation test was performed to compare the novel and the known candidate gene sets, in order to evaluate their functional connections. In brief, we compared the number of co-expressed genes within the novel and the known candidate gene sets and their connections with a random gene set of 1,000,000 random iterations.
Functional Network Analysis
We downloaded PPI data from IntAct Molecular Interaction database (https://www.ebi.ac.uk/intact/) for analysis and considered gene pairs with an intact-miscore ≥ 0.45 to be interacting genes. We then constructed a network based on gene pairs selected from our PPI and co-expression analyses. Novel candidate genes were defined as “seed genes” that were directly connected and used to form an interconnected functional network. Known NDD candidate genes directly connected to at least two novel candidate genes were added to the above network. To further investigate the functional pathways of novel candidate genes, we performed GO enrichment analysis using MetaScape (https://metascape.org). Similar pathways were merged into a single cluster. Network figures were drawn using Cytoscape v.3.7.2.
Results
DNVs in Our Chinese Cohort
In this study, we sequenced 547 target genes in 1102 Chinese patients with NDDs and identified a set of predicted potential functional variants, including PTVs and Dmis (Fig. 1 and Table S1). Using Sanger sequencing, we successfully validated 1271 potential functional variants, including 108 de novo variants (54 de novo PTVs and 54 de novo Dmis), 975 inherited variants (156 inherited Dmis and 819 inherited PTVs), and 188 undetermined variants (36 PTVs and 152 Dmis) (Fig. S1 and Table S2). We found that 108 DNVs in 78 genes appeared in approximately 10.91% (102/935) of patients in our Chinese cohort. Among these 78 genes, 36 genes with DNVs are the first to be reported in Chinese patients. Furthermore, we revealed that 21 genes carried multiple DNVs, including 28 de novo PTVs and 23 de novo Dmis (Table 1). Both SCN2A and MECP2 were the most frequently mutated genes, each carrying one de novo PTV and three de novo Dmis (Table 1). Five genes (MED13L, GRIN2B, KCNQ2, CTNNB1, and TCF20) carried three DNVs in our Chinese cohort, while another 14 genes (ASH1L, SATB2, NRXN1, BCL11A, ADNP, SHANK3, MSL2, SYNE1, SYNGAP1, BRAF, GATAD2B, LLGL1, SLC2A1, and KDM5C) carried two DNVs (Table 1). We manually collected 805 known candidate genes that have either been reported in studies on large NDD cohorts or have strong evidence associating them with NDDs in the OMIM or SFARI Gene databases [17, 21–23, 40–45] (Table S3). We found that of 21 genes with multiple DNVs that we identified, 20 are classified as known candidate genes (Table S4). For example, TCF20, which carries three de novo PTVs (p.S1803Vfs*6, p.C1795Wfs*13, and p.R1907X), was first reported in a Chinese cohort. Interestingly, we identified a potential novel NDD risk gene (MSL2) that carries two de novo PTVs (p.S560Ifs*11 and p.L266Vfs*3). This study is the first to report an association between MSL2 and NDDs.
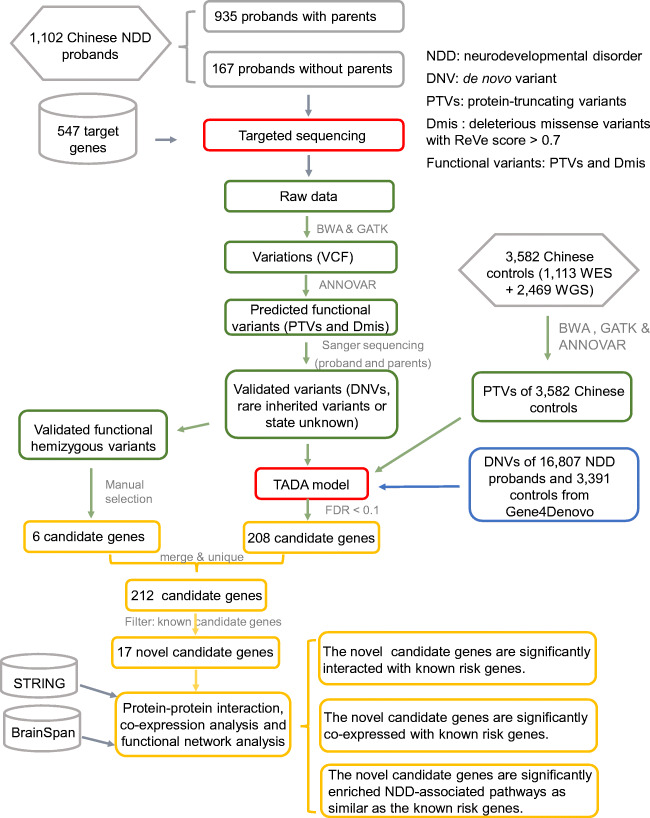
Fig. 1.
Study workflow. This study consisted of four parts: (1) sample collection; (2) identification and validation of variants; (3) prioritization of candidate genes; (4) functional network analysis of novel and known candidate genes. PTVs, protein-truncating variants; Dmis, deleterious missense variants
Table 1.
Genes with multiple DNVs in our probands
| Gene symbol | DNVs (n = 935) | Inherited or state unknown (n = 1102) | DNVs from Gene4Denovo (n = 16,807) | RVIS (percentile) | pLI (percentile) | Gene function |
|---|---|---|---|---|---|---|
| SCN2A | 1 PTV, 3 Dmis | 1 PTV, 3 Dmis | 18 PTVs, 32 Dmis | − 2.51 (1.05%) | 1.00 (0.72%) | Nervous system development |
| MECP2 | 1 PTV, 3 Dmis | 1 Dmis | 9 PTVs, 10 Dmis | − 0.32 (30.25%) | 0.70 (24.62%) | Chromatin binding |
| MED13L | 3 PTVs | 2 Dmis | 18 PTVs, 11 Dmis | − 1.61 (3.34%) | 1.00 (0.57%) | Brain development |
| GRIN2B | 1 PTV, 2 Dmis | 2 Dmis | 4 PTVs, 15 Dmis | − 2.48 (1.10%) | 1.00 (2.19%) | Nervous system development |
| KCNQ2 | 1 PTV, 2 Dmis | 1 PTV, 3 Dmis | 2 PTVs, 25 Dmis | − 1.25 (5.97%) | 1.00 (5.39%) | Regulation of neuronal excitability |
| CTNNB1 | 3 PTVs | 1 Dmis | 19 PTVs | − 1.07 (8.10%) | 1.00 (4.20%) | Wnt signaling |
| TCF20 | 3 PTVs | 3 Dmis | 11 PTVs | − 3.82 (0.34%) | 1.00 (2.84%) | Wnt signaling |
| ASH1L | 2 PTVs | 3 Dmis | 7 PTVs | − 3.89 (0.31%) | 1.00 (0.16%) | Chromatin organization |
| SATB2 | 2 PTVs | - | 11 PTVs, 5 Dmis | − 1.31 (5.37%) | 1.00 (4.97%) | Chromatin binding |
| NRXN1 | 2 Dmis | 3 PTVs, 11 Dmis | 2 PTVs, 5 Dmis | − 1.88 (2.30%) | 1.00 (3.22%) | Nervous system development |
| BCL11A | 2 PTVs | 1 Dmis | 6 PTVs, 3 Dmis | − 1.67 (3.07%) | 0.83 (20.63%) | Brain development |
| ADNP | 2 PTVs | 5 Dmis | 26 PTVs, 1 Dmis | − 1.54 (3.72%) | 1.00 (5.98%) | Chromatin binding |
| SHANK3 | 2 PTVs | 2 PTVs | 9 PTVs | - | 1.00 (4.13%) | Nervous system development |
| MSL2 | 2 PTVs | 1 Dmis | 1 PTV | − 0.72 (15.01%) | 0.89 (18.19%) | Chromatin organization |
| SYNE1 | 2 Dmis | 4 PTVs, 16 Dmis | 3 Dmis | − 1.10 (7.70%) | 3.75E-27 (99.45%) | Nucleotide binding |
| SYNGAP1 | 1 PTV, 1 Dmis | 1 Dmis | 31 PTVs, 1 Dmis | − 2.30 (1.36%) | 1.00 (1.79%) | Postsynaptic signaling |
| BRAF | 2 Dmis | 1 Dmis | 10 Dmis | − 0.97 (9.63%) | 1.00 (2.83%) | Calcium ion binding |
| GATAD2B | 2 PTVs | 2 Dmis | 11 PTVs | − 0.65 (16.92%) | 1.00 (6.34%) | Chromatin remodeling |
| LLGL1 | 2 Dmis | 2 Dmis, 2 PTV | 1 Dmis | − 1.70 (2.94%) | 0.98 (12.10%) | Axon development |
| SLC2A1 | 2 Dmis | 1 Dmis | 4 PTVs, 1 Dmis | − 0.92 (10.49%) | 0.94 (15.56%) | Brain development |
| KDM5C | 2 Dmis | - | 3 PTVs, 2 Dmis | − 2.71 (0.88%) | 1.00 (7.02%) | Chromatin organization |
PTVs, protein-truncating variants, including frameshift, splicing, stop-gain, and stop-loss; Dmis, deleterious missense variants with ReVe score > 0.7
We then employed probability of loss-of-function intolerance (pLI) [46] analysis from ExAC and residual variation intolerance scores (RVIS) [47] to evaluate the functional impact of genes with multiple DNVs. According to the scores of pLI and RVIS, we ranked them and used percentiles as an indicator of gene intolerance. Genes with low-percentile RVIS or pLI were more likely to be intolerant to genetic variants. As expected, it was shown that genes with multiple DNVs both exhibited significantly lower percentile pLI (p = 4.10 × 10−10, two-tailed Wilcoxon rank-sum test) and RVIS (p = 1.55 × 10−10, two-tailed Wilcoxon rank-sum test) than background genes (Fig. S2a). In particular, 20 of the 21 multiple DNV genes ranked in the top 50% of pLI and RVIS (Table 1), suggesting that they are less tolerant of damaging variants.
Inherited X-Linked Hemizygous Variants
We identified seven inherited X-linked hemizygous variants in six genes (Table S5). Among these seven variants, three variants, including two Dmis (p.R270C on PTCHD1 and p.R197H on SLC16A2) and one splicing variant (c.1030-1G>C on SLC9A7), were recorded in the dbSNP or gnomAD database. The remaining four variants, including three Dmis (p.Q153R on ATP6AP2, p.R287C on ARHGEF9, and p.C517Y on PLXNA3) and one frameshift (p.N136Kfs*30 on SLC16A2) are reported here for the first time. Compared with the 805 known candidate genes, we note that five of the above genes are classified as known candidate genes and three (PTCHD1, ATP6AP2, and SLC9A7) are reported for the first time in Chinese patient with NDDs. In addition, we identified a novel candidate gene (PLXNA3) carrying a novel hemizygous missense variant (p.C517Y). A previous study has described an ASD patient from Maghreb carrying a rare inherited missense variant (p.D863E) in PLXNA3 [48], suggesting that it may contribute to autism.
Prioritization of Candidate Genes
Previous studies have demonstrated that both DNVs and rare inherited variants (RIVs) contribute significantly to NDDs [49–54]. Therefore, we integrated data on DNVs and RIVs and employed TADA [38] to prioritize candidate genes in NDDs (Table S6). Based on DNVs and RIVs identified in our study, we prioritized 17 candidate genes (SCN2A, KCNQ2, SATB2, SHANK3, GATAD2B, NRXN1, MED13L, SYNGAP1, BRAF, MECP2, GRIN2B, TCF20, CTNNB1, ASH1L, ADNP, BCL11A, and LLGL1) with FDR values < 0.1 (Table S7). To increase the power of candidate gene detection [17, 22, 43, 55–57], we integrated data from public datasets of NDD cohorts (N = 16,807) with that of controls (N = 3391) from the Gene4Denovo [39] database (Table S8), and prioritized 208 candidate genes with FDR values < 0.1, in which 193 candidate genes reached an FDR < 0.05 (Table S9). Together with the six genes characterized by inherited X-linked hemizygous variants, in total, we prioritized 212 NDD candidate genes, with 17 genes defined as novel candidate genes (including 13 genes with FDR values < 0.05, three genes with 0.05 ≤ FDR < 0.1 (Table 2), and one gene with an inherited X-linked hemizygous variant). We observed that the 212 candidate genes prioritized in our study exhibited significantly lower percentile pLI scores (p < 2.20 × 10−16, two-tailed Wilcoxon rank-sum test) and RVIS (p < 2.20 × 10−16, two-tailed Wilcoxon rank-sum test; Fig. S2b), consistent with the result regarding genes with multiple DNVs. A similar result was observed among the 17 novel candidate genes (p = 0.0394 and p = 1.74 × 10−4; two-tailed Wilcoxon rank-sum test; Fig. S2c), suggesting that these novel NDD candidate genes were likely to be intolerant of functional variants.
Table 2.
Novel candidate genes prioritized by TADA analysis
| Gene symbol | DNVs (n = 935) | Inherited or state unknown (n = 1102) | DNVs from Gene4Denovo (n = 16,807) | FDR | RVIS (percentile) | pLI (percentile) | Gene summary |
|---|---|---|---|---|---|---|---|
| SMAD6 | - | 1 Dmis | 3 PTVs, 1 Dmis | 4.83E−04 | - | 6.97E−06 (78.89%) | Developmental and cellular process |
| SPG7 | - | 3 PTVs, 3 Dmis | 5 Dmis | 8.89E−04 | − 0.91 (10.60%) | 1.38E−18 (98.28%) | Anterograde axonal transport |
| MSL2 | 2 PTVs | 1 Dmis | 1 PTV | 9.62E−04 | − 0.72 (15.01%) | 0.90 (18.20%) | Chromatin organization |
| CYP27C1 | - | 2 PTVs, 3 Dmis | 1 PTV, 3 Dmis | 1.39E−03 | − 0.89 (11.06%) | 1.85E−04 (70.01%) | Retinal metabolic process; retinol metabolic process |
| ITSN1 | - | 2 Dmis | 3 PTVs, 2 Dmis | 1.89E−03 | − 2.89 (0.76%) | 1.00 (0.87%) | Regulation of modification of postsynaptic actin cytoskeleton |
| PSD3 | - | 1 PTV, 2 Dmis | 3 PTVs, 1 Dmis | 2.11E−03 | 0.09 (56.40%) | 0.85 (19.80%) | Nervous system development |
| POLR3A | 1 Dmis | 5 Dmis | 2 PTVs, 1 Dmis | 2.99E−03 | − 2.26 (1.40%) | 7.39E−14 (96.18%) | Nervous system development |
| UBR3 | - | - | 3 PTVs | 5.41E−03 | - | 1.00 (5.02%) | Chromatin organization; nervous system development |
| GALNT18 | - | 2 Dmis | 2 PTVs, 1 Dmis | 7.34E−03 | − 1.07 (8.11%) | 4.77E−02 (47.21%) | Metabolism of proteins |
| DCX | - | - | 2 PTVs, 1 Dmis | 7.43E−03 | − 0.67 (16.20%) | 0.86 (19.34%) | Axoneme assembly |
| LRRC4 | - | - | 1 PTV, 2 Dmis | 2.56E−02 | − 0.84 (11.90%) | 0.88 (18.68%) | Nervous system development |
| SPAG9 | 1 PTV | 3 Dmis | 1 PTV | 3.15E−02 | − 1.20 (6.59%) | 1.00 (1.67%) | Spinocerebellar ataxia 35 |
| ST3GAL6 | - | - | 2 PTVs | 3.23E−02 | 0.07 (55.10%) | 1.06E−03 (64.45%) | Pre-NOTCH Expression and processing |
| YTHDC1 | - | - | 2 PTVs | 8.48E−02 | − 0.61 (18.16%) | 1.00 (1.0864%) | Chromatin regulation/acetylation |
| RRAGC | - | 1 Dmis | 2 Dmis | 9.00E−02 | − 0.25 (34.05%) | 0.602 (27.41%) | Regulation of TORC1 signaling |
| DNAH17 | 1 Dmis | 3 PTVs, 16 Dmis | 1 PTV, 3 Dmis | 9.58E−02 | - | - | Developmental process |
PTVs, protein-truncating variants, including frameshift, splicing, stop-gain, and stop-loss; Dmis: deleterious missense variants with ReVe score > 0.7
In addition, we found that DNVs and inherited X-linked hemizygous variants in 212 candidate genes were identified in ~ 10.59% (99/935) and ~ 0.94% (7/745) of patients in our study, respectively (Table 3). DNVs and inherited X-linked hemizygous variants in known candidate genes accounted for ~ 10.05% (94/935) and ~ 0.81% (6/745) of patients, whereas in novel candidate genes, these variants accounted for ~ 0.53% (5/935) and ~ 0.13% (1/745) of patients, respectively. Inherited or state unknown variants in all candidate genes were detected in ~ 35.75% (394/1102) of all patients (Table 3).
Table 3.
The contribution of prioritized candidate genes to our Chinese probands
| Class | Known candidate genes (n = 195) | Novel candidate genes (n = 17) | |
|---|---|---|---|
| FDRTADA_combined < 0.05 (n = 193) | 180 | 13 genes: MSL2, SPAG9, POLR3A, CYP27C1, DCX, GALNT18, ITSN1, LRRC4, PSD3, SMAD6, SPG7, ST3GAL6, UBR3 | |
| 0.05 ≤ FDRTADA_combined < 0.1 (n = 15) | 12 | 3 genes: DNAH17, RRAGC, YTHDC1 | |
| Genes with X-linked hemizygous (n = 6) | 5 | 1 gene: PLXNA3 | |
| Chinese probands with DNVs (n = 935) | 94 (10.05%) | 5 (0.53%) | Subtotal: 99 (10.59%) |
| Chinese probands with X-linked hemizygous variants (n = 745) | 6 (0.81%) | 1 (0.13%) | Subtotal: 7 (0.94%) |
| Chinese probands with inherited or state unknown variants (n = 1102) | 345 (31.31%) | 49 (4.45%) | Subtotal: 394 (35.75%) |
PTCHD1 and ARHGEF9 were in two groups, including FDRTADA_combined < 0.05 and genes with X-linked hemizygous
Functional Characteristics of Prioritized Candidate Genes
We next employed MetaScape [58] to perform functional enrichment analysis. As expected, these candidate genes were significantly enriched in NDD-associated pathways, such as synapse organization, covalent chromatin modification, head development, behavior, and regulation of ion transport [17, 59–63] (Fig. S3). Interestingly, the novel candidate genes were also involved in similar biological pathways. For example, MSL2 was implicated in covalent chromatin modification, LRRC4 was involved in chemical synaptic transmission, and PLXNA3 was associated with head development and axonogenesis.
We also performed functional cell-specific enrichment analyses [64, 65] to investigate whether candidate genes were associated with specific tissues or cells. By analyzing the mouse transcriptomic profiling datasets of different developmental stages and brain regions, we found that the 212 NDD candidate genes tended to be enriched in the cortex and striatum during the middle fetal stage (Fig. S4a). In the cell-specific enrichment analyses, we observed a highly significant enrichment in Drd1 + and Drd2 + spiny neurons of neostriatum and rods (Fig. S4b), similar to data reported in a previous study [21]. These results suggest that the 212 NDD candidate genes are functionally associated with the etiology of NDDs.
Functional Network Analysis Between Known and Novel Candidate Genes
To investigate correlations between novel and known candidate genes, we performed a permutation test to estimate the relationship between these genes based on co-expression gene pairs identified from the BrainSpan atlas. We found that 15 of the 17 novel candidate genes (p = 2.35 × 10−3, permutation test, Fig. S5a) were co-expressed with 285 known candidate genes (p = 3.72 × 10−4, permutation test, Fig. S5b), with 523 connections between them (p = 8.80 × 10−5, permutation test, Fig. S5c), suggesting that the novel candidate genes are significantly co-expressed with the known candidate genes and are more likely to be related to the pathology of NDDs.
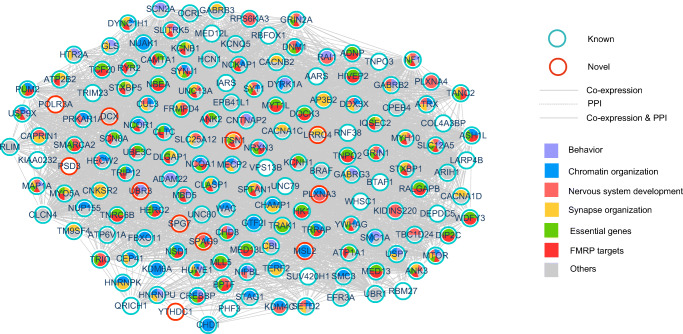
To further investigate the functional relationship between novel and known candidate genes, we constructed a functional network by integrating PPI data from IntAct and brain expression data from BrainSpan. Only known candidate genes directly interacting/co-expressed with at least two novel candidate genes were added to the network. The co-expressed/PPI network encompassed 159 genes, including 11 novel candidate genes and 148 known candidate genes (Fig. 2). These genes were enriched in several biological processes known to be related to NDDs, such as covalent chromatin modification (GO:0016569, p = 2.35 × 10−12, Fisher’s exact test), chemical synaptic transmission (GO:00072686, p = 7.31 × 10−12, Fisher’s exact test), and brain development (GO:0007420, p = 1.23 × 10−7, Fisher’s exact test). Furthermore, these genes showed a significant enrichment of previously reported gene sets: FMRP targets [66] (p < 2.22 × 10−16, Fisher’s exact test), and genes essential in mice [67] (p < 2.22 × 10−16, Fisher’s exact test). It is worth noting that six novel candidate genes (UBR3, PLXNA3, ITSN1, MSL2, LRRC4, and SPAG9) were significantly involved in functional clusters known to be associated with NDDs. For example, MSL2, UBR3, SPAG9, and PLXNA3 are involved in chromatin organization, while ITSN1, SPAG9, and PLXNA3 have been implicated in nervous system development. In addition, we found that nine novel candidate genes showed co-expression/interaction with more than two known candidate genes (Fig. S6). For example, POLR3A, which was the most frequently connected novel gene, was co-expressed/interacted with 99 known candidate genes. Other genes (LRRC4, ITSN1, UBR3, PLXNA3, MSL2, SPAG9, PSD3, and YTHDC1) were connected with more than five candidate genes. These results suggest that novel candidate genes are functionally associated with known risk genes and that these 11 novel candidate genes may have a stronger influence in the etiology of NDDs.
Fig. 2.
Functional network of novel and known candidate genes. Based on co-expression data from BrainSpan and PPI data from IntAct, 159 candidate genes formed a large interconnected functional network, mainly involving the following major functional clusters: chromatin organization, essential genes, nervous system development, FMRP targets, behavior, and synapse organization. Novel candidate genes are in red circles and known candidate genes are in green circles. Different line types between nodes represent the interactions existing in BrainSpan or IntAct or in both BrainSpan and IntAct
Discussion
Previous studies have provided evidences that targeted sequencing and integrative analyses play a critical role in the discovery of novel candidate genes [21–24]. Based on targeted sequencing of 547 target genes, we identified 108 DNVs, accounting for ~ 10.91% (102/935) of our Chinese cohort. Among the genes with DNVs in our study, most genes have been reported to be associated with NDDs in previous studies, suggesting that genes identified in our Chinese cohort are relevant in other populations. Among the 108 DNVs, 60 DNVs were not recorded in public databases (including dbSNP, gnomAD, Gene4Denovo [39], and PubMed), consistent with the findings of Georgi et al., who reported that recurrently mutated amino acid sites in genes are rarely detected [68]. These results highlight the role of DNVs in the genetic heterogeneity of NDDs. In addition, we identified 21 genes with multiple DNVs present in 51 Chinese patients, and our analyses suggest that these genes display significant intolerance to damaging variants. Consistent with previous ASD study [19], SCN2A is the most frequent gene with multiple DNVs in the Chinese population. Compared with the known candidate genes, 20 of 21 genes with multiple DNVs were classified as known candidate genes. Notably, this study is the first to report TCF20 (with its three de novo PTVs) as a candidate NDD gene in a Chinese cohort.
By integrating information regarding DNVs from public datasets with our results, we prioritized 212 candidate genes, confirming that combing public NDD datasets is beneficial for the discovery of candidate genes [22–24]. Consistent with our analyses of genes with multiple DNVs, the 212 identified candidate genes were more intolerant of damaging variants. In addition, we demonstrated that the 212 candidate genes are closely associated with the etiology of NDDs from the perspective of biological pathway and functional cell-specific enrichment analyses. These results suggest that most of the 212 candidate genes identified in our study truly contribute to NDDs and they are worth validating in genetic functional studies or replicating in cohorts.
In this study, we prioritized 17 novel candidate genes and revealed similar functional characteristics between these novel candidate genes and other known candidate genes. Through functional network analysis, we observed that novel candidate genes frequently interacted/were co-expressed with known candidate genes, and genes in the network were enriched in NDD-associated clusters, as described in previous studies [4, 17, 23, 69, 70]. Interestingly, six novel candidate genes were closely connected with known candidate genes and were involved in NDD-associated clusters, suggesting that these novel candidate genes are more likely to be associated with NDDs. For example, ITSN1 (with an FDR value < 0.01 in the TADA analysis) was involved in nervous system development and synapse organization and connected with 64 known candidate genes. Jakob et al. reported that loss of the signaling scaffold intersectin 1 (ITSN1) in mice led to defective neuronal migration and ablates Reelin stimulation of hippocampal long-term potentiation [71]. Our analyses revealed that SPAG9 carried a de novo PTV in our cohort and it is reportedly overexpressed in human astrocytoma which arises from neural progenitor cells in the central nervous system [72]. We also found that SPAG9 was implicated in chromatin organization and co-expressed/interacted with eight known candidate genes, further supporting a role for this gene in NDDs. Among the novel candidate genes, MSL2 was a particularly interesting gene that carried two de novo PTVs in our Chinese cohort. Except for the multiple DNVs in MSL2, it was the gene that frequently co-expressed/interacted with known candidate genes and was involved in the functional cluster of chromatin organization. Iossifov et al. reported a de novo MSL2 PTV in a patient with autism [69], further suggesting that MSL2 may be strongly linked to NDDs. We expect to have a large cohort study or functional experiments to validate this possibility in future studies.
Despite our best efforts to comprehensively integrate public data with our cohort data to discover novel risk genes, our study was still bound by some limitations. First, while we discovered that novel candidate genes identified in our study are functionally associated with known candidate genes at multiple levels, these novel candidate genes lack strong evidences that support their roles in NDDs. We expect that these novel candidate genes could be validated in larger cohorts or through functional genetic experiments. Second, the sample size of our Chinese cohort is not large enough, and future studies need to recruit more volunteers to improve the statistical power and allow for comparisons with variant patterns of other populations. Third, since NDDs are characterized by high clinical and genetic heterogeneity, and multiple risk factors contribute to the etiology of these disorders, candidate gene analyses can only partially elucidate the processes underlying NDDs. Further studies are needed to integrate the impact of distinct influences such as epigenetic, environmental, and genetic factors.
In summary, our study demonstrates that integrating DNVs from multiple NDD-related studies can help in the identification of risk genes. We highlight that the pattern of DNVs in Chinese cohorts is relevant to other populations. Furthermore, we provide evidence at multiple levels that novel candidate genes are functionally associated with known candidate genes. Finally, our study describes new high-confidence risk genes that should aid the study of the NDD etiology and we expect that these genes will be worth analyzing in large cohorts or being validated in genetic functional experiments.
Supplementary Information
(PDF 147 kb)
(PDF 1817 kb)
(PDF 584 kb)
(PDF 1668 kb)
(PDF 358 kb)
(PDF 96 kb)
(XLSX 49 kb)
(XLSX 269 kb)
(XLSX 83 kb)
(XLSX 23 kb)
(XLSX 12 kb)
(XLSX 174 kb)
(XLSX 156 kb)
(XLSX 322 kb)
(XLSX 81 kb)
(DOCX 21 kb)
Availability of Data and Materials
The datasets supporting the results and conclusions of this manuscript are included within the article and its Supplementary Material. All other datasets used and analyzed during the current study are available from the corresponding authors on reasonable request.
Abbreviations
- ASD
Autism spectrum disorder
- Dmis
Deleterious missense variant
- DNV
De novo variant
- FDR
False discovery rate
- NDD
Neurodevelopmental disorder
- PPI
Protein-protein interaction
- PTV
Protein-truncating variant
- RIV
Rare inherited variant
- WES
Whole-exome sequencing
- WGS
Whole-genome sequencing
Funding
This work was supported by the Key Research and Development Program of Guangdong Province (No. 2018B030335001), the National Natural Science Foundation of China (81730036, 81525007, and 81801133), Young Elite Scientist Sponsorship Program by CAST (YESS) (2018QNRC001), Guangzhou Key Project in “Early diagnosis and treatment of autism spectrum disorders” (202007030002), Science and Technology Major Project of Hunan Provincial Science and Technology Department (2018SK1030), Natural Science Foundation of Hunan Province for outstanding Young Scholars (2020JJ3059), and Innovation-Driven Project of Central South University (20180033040004).
Declarations
Ethics Approval
This study was approved by the Institutional Review Board of the State Key Laboratory of Medical Genetics, School of Life Sciences, Central South University, Changsha, Hunan, China. All subjects who participated in this study provided informed consent prior to sample collection.
Consent to Participate
Not applicable
Consent for Publication
Not applicable
Conflict of Interest
The authors declare no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yi Zhang and Tao Wang contributed equally to this work.
Contributor Information
Kun Xia, Email: xiakun@sklmg.edu.cn.
Jinchen Li, Email: lijinchen@csu.edu.cn.
Zhongsheng Sun, Email: sunzs@biols.ac.cn.
References
- 1.Thapar A, Cooper M, Rutter M. Neurodevelopmental disorders. Lancet Psychiatry. 2017;4(4):339–346. doi: 10.1016/S2215-0366(16)30376-5. [DOI] [PubMed] [Google Scholar]
- 2.Mullin AP, Gokhale A, Moreno-De-Luca A, et al. Neurodevelopmental disorders: mechanisms and boundary definitions from genomes, interactomes and proteomes. Transl Psychiatry. 2013;3:e329. doi: 10.1038/tp.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarlungeanu DC, Novarino G. Genomics in neurodevelopmental disorders: an avenue to personalized medicine. Exp Mol Med. 2018;50(8):100–107. doi: 10.1038/s12276-018-0129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stessman HAF, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, Kvarnung M, Gerdts J, Trinh S, Cosemans N, Vives L, Lin J, Turner TN, Santen G, Ruivenkamp C, Kriek M, van Haeringen A, Aten E, Friend K, Liebelt J, Barnett C, Haan E, Shaw M, Gecz J, Anderlid BM, Nordgren A, Lindstrand A, Schwartz C, Kooy RF, Vandeweyer G, Helsmoortel C, Romano C, Alberti A, Vinci M, Avola E, Giusto S, Courchesne E, Pramparo T, Pierce K, Nalabolu S, Amaral DG, Scheffer IE, Delatycki MB, Lockhart PJ, Hormozdiari F, Harich B, Castells-Nobau A, Xia K, Peeters H, Nordenskjöld M, Schenck A, Bernier RA, Eichler EE. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49(4):515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilfert AB, Sulovari A, Turner TN, Coe BP, Eichler EE. Recurrent de novo mutations in neurodevelopmental disorders: properties and clinical implications. Genome Med. 2017;9(1):101. doi: 10.1186/s13073-017-0498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BWM, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, Leach R, Klein R, Tearle R, Bo T, Pfundt R, Yntema HG, de Vries BBA, Kleefstra T, Brunner HG, Vissers LELM, Veltman JA. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511(7509):344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 7.Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A. The heritability of autism spectrum disorder. JAMA. 2017;318(12):1182–1184. doi: 10.1001/jama.2017.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tick B, Bolton P, Happe F, et al. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J Child Psychol Psychiatry. 2016;57(5):585–595. doi: 10.1111/jcpp.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 10.Chen T, Giri M, Xia Z, Subedi YN, Li Y. Genetic and epigenetic mechanisms of epilepsy: a review. Neuropsychiatr Dis Treat. 2017;13:1841–1859. doi: 10.2147/NDT.S142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Jia X, Wu H, Xun G, Ou J, Zhang Q, Li H, Bai T, Hu Z, Zou X, Xia K, Guo H. Genotype and phenotype correlations for SHANK3 de novo mutations in neurodevelopmental disorders. Am J Med Genet A. 2018;176(12):2668–2676. doi: 10.1002/ajmg.a.40666. [DOI] [PubMed] [Google Scholar]
- 12.Stessman HAF, Willemsen MH, Fenckova M, Penn O, Hoischen A, Xiong B, Wang T, Hoekzema K, Vives L, Vogel I, Brunner HG, van der Burgt I, Ockeloen CW, Schuurs-Hoeijmakers JH, Klein Wassink-Ruiter JS, Stumpel C, Stevens SJC, Vles HS, Marcelis CM, van Bokhoven H, Cantagrel V, Colleaux L, Nicouleau M, Lyonnet S, Bernier RA, Gerdts J, Coe BP, Romano C, Alberti A, Grillo L, Scuderi C, Nordenskjöld M, Kvarnung M, Guo H, Xia K, Piton A, Gerard B, Genevieve D, Delobel B, Lehalle D, Perrin L, Prieur F, Thevenon J, Gecz J, Shaw M, Pfundt R, Keren B, Jacquette A, Schenck A, Eichler EE, Kleefstra T. Disruption of POGZ is associated with intellectual disability and autism spectrum disorders. Am J Hum Genet. 2016;98(3):541–552. doi: 10.1016/j.ajhg.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, Schuurs-Hoeijmakers JH, Fichera M, Bosco P, Buono S, Alberti A, Failla P, Peeters H, Steyaert J, Vissers LELM, Francescatto L, Mefford HC, Rosenfeld JA, Bakken T, O’Roak BJ, Pawlus M, Moon R, Shendure J, Amaral DG, Lein E, Rankin J, Romano C, de Vries BBA, Katsanis N, Eichler EE. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158(2):263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou WZ, Zhang J, Li Z, Lin X, Li J, Wang S, Yang C, Wu Q, Ye AY, Wang M, Wang D, Pu TZ, Wu YY, Wei L. Targeted resequencing of 358 candidate genes for autism spectrum disorder in a Chinese cohort reveals diagnostic potential and genotype-phenotype correlations. Hum Mutat. 2019;40(6):801–815. doi: 10.1002/humu.23724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees E, Carrera N, Morgan J, Hambridge K, Escott-Price V, Pocklington AJ, Richards AL, Pardiñas AF, McDonald C, Donohoe G, Morris DW, Kenny E, Kelleher E, Gill M, Corvin A, Kirov G, Walters JTR, Holmans P, Owen MJ, O’Donovan MC, Alizadeh BZ, van Amelsvoort T, Bartels-Velthuis AA, van Beveren NJ, Bruggeman R, Cahn W, de Haan L, Delespaul P, Meijer CJ, Myin-Germeys I, Kahn RS, Schirmbeck F, Simons CJP, van Haren NE, van Os J, van Winkel R, Luykx JJ. Targeted sequencing of 10,198 samples confirms abnormalities in neuronal activity and implicates voltage-gated sodium channels in schizophrenia pathogenesis. Biol Psychiatry. 2019;85(7):554–562. doi: 10.1016/j.biopsych.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Carrera N, Humphreys I, Johnson JS, Roussos P, Barker DD, Banks E, Milanova V, Grant SG, Hannon E, Rose SA, Chambert K, Mahajan M, Scolnick EM, Moran JL, Kirov G, Palotie A, McCarroll SA, Holmans P, Sklar P, Owen MJ, Purcell SM, O’Donovan MC. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirzaa GM, Chong JX, Piton A, et al. De novo and inherited variants in ZNF292 underlie a neurodevelopmental disorder with features of autism spectrum disorder. Genet Med. 2020;22(3):538–546. doi: 10.1038/s41436-019-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang T, Guo H, Xiong B, Stessman HAF, Wu H, Coe BP, Turner TN, Liu Y, Zhao W, Hoekzema K, Vives L, Xia L, Tang M, Ou J, Chen B, Shen Y, Xun G, Long M, Lin J, Kronenberg ZN, Peng Y, Bai T, Li H, Ke X, Hu Z, Zhao J, Zou X, Xia K, Eichler EE. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun. 2016;7:13316–13316. doi: 10.1038/ncomms13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H, Wang T, Wu H, Long M, Coe BP, Li H, Xun G, Ou J, Chen B, Duan G, Bai T, Zhao N, Shen Y, Li Y, Wang Y, Zhang Y, Baker C, Liu Y, Pang N, Huang L, Han L, Jia X, Liu C, Ni H, Yang X, Xia L, Chen J, Shen L, Li Y, Zhao R, Zhao W, Peng J, Pan Q, Long Z, Su W, Tan J, du X, Ke X, Yao M, Hu Z, Zou X, Zhao J, Bernier RA, Eichler EE, Xia K. Inherited and multiple de novo mutations in autism/developmental delay risk genes suggest a multifactorial model. Molecular autism. 2018;9:64–64. doi: 10.1186/s13229-018-0247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coe BP, Stessman HAF, Sulovari A, Geisheker MR, Bakken TE, Lake AM, Dougherty JD, Lein ES, Hormozdiari F, Bernier RA, Eichler EE. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet. 2019;51(1):106–116. doi: 10.1038/s41588-018-0288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen HT, Bryois J, Kim A, Dobbyn A, Huckins LM, Munoz-Manchado AB, Ruderfer DM, Genovese G, Fromer M, Xu X, Pinto D, Linnarsson S, Verhage M, Smit AB, Hjerling-Leffler J, Buxbaum JD, Hultman C, Sklar P, Purcell SM, Lage K, He X, Sullivan PF, Stahl EA. Integrated Bayesian analysis of rare exonic variants to identify risk genes for schizophrenia and neurodevelopmental disorders. Genome Med. 2017;9(1):114. doi: 10.1186/s13073-017-0497-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takata A, Miyake N, Tsurusaki Y, Fukai R, Miyatake S, Koshimizu E, Kushima I, Okada T, Morikawa M, Uno Y, Ishizuka K, Nakamura K, Tsujii M, Yoshikawa T, Toyota T, Okamoto N, Hiraki Y, Hashimoto R, Yasuda Y, Saitoh S, Ohashi K, Sakai Y, Ohga S, Hara T, Kato M, Nakamura K, Ito A, Seiwa C, Shirahata E, Osaka H, Matsumoto A, Takeshita S, Tohyama J, Saikusa T, Matsuishi T, Nakamura T, Tsuboi T, Kato T, Suzuki T, Saitsu H, Nakashima M, Mizuguchi T, Tanaka F, Mori N, Ozaki N, Matsumoto N. Integrative analyses of de novo mutations provide deeper biological insights into autism spectrum disorder. Cell Rep. 2018;22(3):734–747. doi: 10.1016/j.celrep.2017.12.074. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Mantilla AJ, Moreno-De-Luca A, Ledbetter DH, et al. A cross-disorder method to identify novel candidate genes for developmental brain disorders. JAMA Psychiatry. 2016;73(3):275–283. doi: 10.1001/jamapsychiatry.2015.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Wang L, Guo H, Shi L, Zhang K, Tang M, Hu S, Dong S, Liu Y, Wang T, Yu P, He X, Hu Z, Zhao J, Liu C, Sun ZS, Xia K. Targeted sequencing and functional analysis reveal brain-size-related genes and their networks in autism spectrum disorders. Mol Psychiatry. 2017;22(9):1282–1290. doi: 10.1038/mp.2017.140. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Wang L, Yu P et al (2017) Vitamin D-related genes are subjected to significant de novo mutation burdens in autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet:174 [DOI] [PubMed]
- 27.Zhang Y, Li N, Li C, Zhang Z, Teng H, Wang Y, Zhao T, Shi L, Zhang K, Xia K, Li J, Sun Z. Genetic evidence of gender difference in autism spectrum disorder supports the female-protective effect. Transl Psychiatry. 2020;10(1):4. doi: 10.1038/s41398-020-0699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Cai T, Jiang Y, Chen H, He X, Chen C, Li X, Shao Q, Ran X, Li Z, Xia K, Liu C, Sun ZS, Wu J. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry. 2016;21(2):290–297. doi: 10.1038/mp.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao G, Li K, Li B, Wang Z, Fang Z, Wang X, Zhang Y, Luo T, Zhou Q, Wang L, Xie Y, Wang Y, Chen Q, Xia L, Tang Y, Tang B, Xia K, Li J. Gene4Denovo: an integrated database and analytic platform for de novo mutations in humans. Nucleic Acids Res. 2019;48(D1):D913–D926. doi: 10.1093/nar/gkz923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abrahams BS, Arking DE, Campbell DB, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4(1):36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Hu S, Zhang K, Shi L, Zhang Y, Zhao T, Wang L, He X, Xia K, Liu C, Sun Z. A comparative study of the genetic components of three subcategories of autism spectrum disorder. Mol Psychiatry. 2018;24:1720–1731. doi: 10.1038/s41380-018-0081-x. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Yu P, Jin X, Xu X, Li J, Li Z, Wang M, Wang T, Wu X, Jiang Y, Cai W, Mei J, Min Q, Xu Q, Zhou B, Guo H, Wang P, Zhou W, Hu Z, Li Y, Cai T, Wang Y, Xia K, Jiang YH, Sun ZS. Genomic landscapes of Chinese sporadic autism spectrum disorders revealed by whole-genome sequencing. J Genet Genomics. 2018;45(10):527–538. doi: 10.1016/j.jgg.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal, vol 17, No 1. Next Generation Sequencing Data Analysis. 10.14806/ej.17.1.200
- 34.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Zhao T, Zhang Y, Zhang K, Shi L, Chen Y, Wang X, Sun Z. Performance evaluation of pathogenicity-computation methods for missense variants. Nucleic Acids Res. 2018;46(15):7793–7804. doi: 10.1093/nar/gky678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X, Sanders SJ, Liu L, de Rubeis S, Lim ET, Sutcliffe JS, Schellenberg GD, Gibbs RA, Daly MJ, Buxbaum JD, State MW, Devlin B, Roeder K. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9(8):e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao G, Li K, Li B, Wang Z, Fang Z, Wang X, Zhang Y, Luo T et al (2019) Gene4Denovo: an integrated database and analytic platform for de novo mutations in humans. Nucleic Acids Res. 10.1093/nar/gkz923 [DOI] [PMC free article] [PubMed]
- 40.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, Goldberg AP, Jinlu C, Keaney JF 3rd, Klei L, Mandell JD, Moreno-de-Luca D, Poultney CS, Robinson EB, Smith L, Solli-Nowlan T, Su MY, Teran NA, Walker MF, Werling DM, Beaudet AL, Cantor RM, Fombonne E, Geschwind DH, Grice DE, Lord C, Lowe JK, Mane SM, Martin DM, Morrow EM, Talkowski ME, Sutcliffe JS, Walsh CA, Yu TW, Autism Sequencing Consortium. Ledbetter DH, Martin CL, Cook EH, Buxbaum JD, Daly MJ, Devlin B, Roeder K, State MW. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87(6):1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lelieveld SH, Reijnders MR, Pfundt R, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19(9):1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 42.McRae JF, Clayton S, Fitzgerald TW, et al. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542(7642):433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stessman HA, Xiong B, Coe BP, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49(4):515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.RK CY, Merico D, Bookman M, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20(4):602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, de Rubeis S, An JY, Peng M, Collins R, Grove J, Klei L, Stevens C, Reichert J, Mulhern MS, Artomov M, Gerges S, Sheppard B, Xu X, Bhaduri A, Norman U, Brand H, Schwartz G, Nguyen R, Guerrero EE, Dias C, Betancur C, Cook EH, Gallagher L, Gill M, Sutcliffe JS, Thurm A, Zwick ME, Børglum AD, State MW, Cicek AE, Talkowski ME, Cutler DJ, Devlin B, Sanders SJ, Roeder K, Daly MJ, Buxbaum JD, Aleksic B, Anney R, Barbosa M, Bishop S, Brusco A, Bybjerg-Grauholm J, Carracedo A, Chan MCY, Chiocchetti AG, Chung BHY, Coon H, Cuccaro ML, Curró A, Dalla Bernardina B, Doan R, Domenici E, Dong S, Fallerini C, Fernández-Prieto M, Ferrero GB, Freitag CM, Fromer M, Gargus JJ, Geschwind D, Giorgio E, González-Peñas J, Guter S, Halpern D, Hansen-Kiss E, He X, Herman GE, Hertz-Picciotto I, Hougaard DM, Hultman CM, Ionita-Laza I, Jacob S, Jamison J, Jugessur A, Kaartinen M, Knudsen GP, Kolevzon A, Kushima I, Lee SL, Lehtimäki T, Lim ET, Lintas C, Lipkin WI, Lopergolo D, Lopes F, Ludena Y, Maciel P, Magnus P, Mahjani B, Maltman N, Manoach DS, Meiri G, Menashe I, Miller J, Minshew N, Montenegro EMS, Moreira D, Morrow EM, Mors O, Mortensen PB, Mosconi M, Muglia P, Neale BM, Nordentoft M, Ozaki N, Palotie A, Parellada M, Passos-Bueno MR, Pericak-Vance M, Persico AM, Pessah I, Puura K, Reichenberg A, Renieri A, Riberi E, Robinson EB, Samocha KE, Sandin S, Santangelo SL, Schellenberg G, Scherer SW, Schlitt S, Schmidt R, Schmitt L, Silva IMW, Singh T, Siper PM, Smith M, Soares G, Stoltenberg C, Suren P, Susser E, Sweeney J, Szatmari P, Tang L, Tassone F, Teufel K, Trabetti E, Trelles MP, Walsh CA, Weiss LA, Werge T, Werling DM, Wigdor EM, Wilkinson E, Willsey AJ, Yu TW, Yu MHC, Yuen R, Zachi E, Agerbo E, Als TD, Appadurai V, Bækvad-Hansen M, Belliveau R, Buil A, Carey CE, Cerrato F, Chambert K, Churchhouse C, Dalsgaard S, Demontis D, Dumont A, Goldstein J, Hansen CS, Hauberg ME, Hollegaard MV, Howrigan DP, Huang H, Maller J, Martin AR, Martin J, Mattheisen M, Moran J, Pallesen J, Palmer DS, Pedersen CB, Pedersen MG, Poterba T, Poulsen JB, Ripke S, Schork AJ, Thompson WK, Turley P, Walters RK. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180(3):568–584. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9(8):e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nava C, Lamari F, Héron D, Mignot C, Rastetter A, Keren B, Cohen D, Faudet A, Bouteiller D, Gilleron M, Jacquette A, Whalen S, Afenjar A, Périsse D, Laurent C, Dupuits C, Gautier C, Gérard M, Huguet G, Caillet S, Leheup B, Leboyer M, Gillberg C, Delorme R, Bourgeron T, Brice A, Depienne C. Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl Psychiatry. 2012;2(10):e179–e179. doi: 10.1038/tp.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim ET, Raychaudhuri S, Sanders SJ, et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77(2):235–242. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toma C, Torrico B, Hervas A, et al. Exome sequencing in multiplex autism families suggests a major role for heterozygous truncating mutations. Mol Psychiatry. 2014;19(7):784–790. doi: 10.1038/mp.2013.106. [DOI] [PubMed] [Google Scholar]
- 51.Al-Mubarak B, Abouelhoda M, Omar A, et al. Whole exome sequencing reveals inherited and de novo variants in autism spectrum disorder: a trio study from Saudi families. Sci Rep. 2017;7(1):5679. doi: 10.1038/s41598-017-06033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, Hung WC, Haider S, Zhang J, Knight J, Bjornson RD, Castaldi C, Tikhonoa IR, Bilguvar K, Mane SM, Sanders SJ, Mital S, Russell MW, Gaynor JW, Deanfield J, Giardini A, Porter GA, Jr, Srivastava D, Lo CW, Shen Y, Watkins WS, Yandell M, Yost HJ, Tristani-Firouzi M, Newburger JW, Roberts AE, Kim R, Zhao H, Kaltman JR, Goldmuntz E, Chung WK, Seidman JG, Gelb BD, Seidman CE, Lifton RP, Brueckner M. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sullivan PF, Geschwind DH. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell. 2019;177(1):162–183. doi: 10.1016/j.cell.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krumm N, Turner TN, Baker C et al Excess of rare, inherited truncating mutations in autism. Nat Genet 47(6):582–588 [DOI] [PMC free article] [PubMed]
- 55.Ben-David E, Shifman S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol Psychiatry. 2013;18(10):1054–1056. doi: 10.1038/mp.2012.148. [DOI] [PubMed] [Google Scholar]
- 56.Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci. 2014;17(6):764–772. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L, Sabo A, Neale BM, Nagaswamy U, Stevens C, Lim E, Bodea CA, Muzny D, Reid JG, Banks E, Coon H, DePristo M, Dinh H, Fennel T, Flannick J, Gabriel S, Garimella K, Gross S, Hawes A, Lewis L, Makarov V, Maguire J, Newsham I, Poplin R, Ripke S, Shakir K, Samocha KE, Wu Y, Boerwinkle E, Buxbaum JD, Cook EH, Devlin B, Schellenberg GD, Sutcliffe JS, Daly MJ, Gibbs RA, Roeder K. Analysis of rare, exonic variation amongst subjects with autism spectrum disorders and population controls. PLoS Genet. 2013;9(4):e1003443. doi: 10.1371/journal.pgen.1003443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LCF, Yángüez E, Andenmatten D, Pache L, Manicassamy B, Albrecht RA, Gonzalez MG, Nguyen Q, Brass A, Elledge S, White M, Shapira S, Hacohen N, Karlas A, Meyer TF, Shales M, Gatorano A, Johnson JR, Jang G, Johnson T, Verschueren E, Sanders D, Krogan N, Shaw M, König R, Stertz S, García-Sastre A, Chanda SK. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16(9):551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 60.Mullins C, Fishell G, Tsien RW. Unifying views of autism spectrum disorders: a consideration of autoregulatory feedback loops. Neuron. 2016;89(6):1131–1156. doi: 10.1016/j.neuron.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat Med. 2016;22(4):345–361. doi: 10.1038/nm.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493(7432):327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nanou E, Catterall WA. Calcium channels, synaptic plasticity, and neuropsychiatric disease. Neuron. 2018;98(3):466–481. doi: 10.1016/j.neuron.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 64.Xu X, Wells AB, O'Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34(4):1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38(13):4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Darnell JC, Van Driesche SJ, Zhang C, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Georgi B, Voight BF, Bucan M. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9(5):e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geisheker MR, Heymann G, Wang T, Coe BP, Turner TN, Stessman HAF, Hoekzema K, Kvarnung M, Shaw M, Friend K, Liebelt J, Barnett C, Thompson EM, Haan E, Guo H, Anderlid BM, Nordgren A, Lindstrand A, Vandeweyer G, Alberti A, Avola E, Vinci M, Giusto S, Pramparo T, Pierce K, Nalabolu S, Michaelson JJ, Sedlacek Z, Santen GWE, Peeters H, Hakonarson H, Courchesne E, Romano C, Kooy RF, Bernier RA, Nordenskjöld M, Gecz J, Xia K, Zweifel LS, Eichler EE. Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat Neurosci. 2017;20(8):1043–1051. doi: 10.1038/nn.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zoghbi HY, Bear MF (2012) Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol 4(3). 10.1101/cshperspect.a009886 [DOI] [PMC free article] [PubMed]
- 71.Jakob B, Kochlamazashvili G, Japel M, et al. Intersectin 1 is a component of the Reelin pathway to regulate neuronal migration and synaptic plasticity in the hippocampus. Proc Natl Acad Sci U S A. 2017;114(21):5533–5538. doi: 10.1073/pnas.1704447114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yi F, Ni W, Liu W, Pan X, Han X, Yang L, Kong X, Ma R, Chang R. SPAG9 is overexpressed in human astrocytoma and promotes cell proliferation and invasion. Tumour Biol. 2013;34(5):2849–2855. doi: 10.1007/s13277-013-0845-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 147 kb)
(PDF 1817 kb)
(PDF 584 kb)
(PDF 1668 kb)
(PDF 358 kb)
(PDF 96 kb)
(XLSX 49 kb)
(XLSX 269 kb)
(XLSX 83 kb)
(XLSX 23 kb)
(XLSX 12 kb)
(XLSX 174 kb)
(XLSX 156 kb)
(XLSX 322 kb)
(XLSX 81 kb)
(DOCX 21 kb)