PURPOSE
Ultrahigh resolution (UHR) HLA matching is reported to result in better outcomes following unrelated donor hematopoietic cell transplantation, improving survival and reducing post-transplant complications. However, most studies included relatively small numbers of patients. Here we report the findings from a large, multicenter validation study.
METHODS
UHR HLA typing was available on 5,140 conventionally 10 out of 10 HLA-matched patients with malignant disease transplanted between 2008 and 2017.
RESULTS
After UHR HLA typing, 82% of pairs remained 10 out of 10 UHR-matched; 12.3% of patients were 12 out of 12 UHR HLA-matched. Compared with 12 out of 12 UHR-matched patients, probabilities of grade 2-4 acute graft-versus-host disease (aGVHD) were significantly increased with UHR mismatches (overall P = .0019) and in those patients who were HLA-DPB1 T-cell epitope permissively mismatched or nonpermissively mismatched (overall P = .0011). In the T-cell–depleted subset, the degree of UHR HLA mismatch was only associated with increased transplant-related mortality (TRM) (overall P = .0068). In the T-cell–replete subset, UHR HLA matching was associated with a lower probability of aGVHD (overall P = .0020); 12 out of 12 UHR matching was associated with reduced TRM risk when compared with HLA-DPB1 T-cell epitope permissively mismatched patients, whereas nonpermissive mismatching resulted in a greater risk (overall P = .0003).
CONCLUSION
This study did not confirm that UHR 12 out of 12 HLA matching increases the probability of overall survival but does demonstrate that aGVHD risk, and in certain settings TRM, is lowest in UHR HLA-matched pairs and thus warrants consideration when multiple 10 out of 10 HLA-matched donors of equivalent age are available.
INTRODUCTION
Allogeneic hematopoietic cell transplantation (HCT) using HLA-matched unrelated donors (MUDs) is used to treat patients with malignant hematologic disease.1,2 Successful outcome is due in part to donor selection criteria including, but not limited to, compatible HLA types, younger donor age, and cytomegalovirus serostatus matching.3-5 HLA matching is associated with better survival and reduced post-transplant complications including graft-versus-host disease (GVHD).5-7 Minimum HLA matching on the basis of 8 out of 8 high-resolution HLA match (HLA-A, HLA-B, HLA-C, and HLA-DRB1) at the antigen recognition domain (ARD) level is widely accepted,8 whereas a 10 out of 10 HLA match (including HLA-DQB1) high-resolution is preferred.9 Matching or T-cell epitope (TCE) permissive mismatching for HLA-DPB1 is recommended and in common use.10,11
CONTEXT
Key Objective
Are the previously unrecognized mismatches identified by ultrahigh resolution (UHR) HLA typing associated with differences in overall survival (OS) and the risk of complications following unrelated donor hematopoietic cell transplantation?
Knowledge Generated
UHR HLA typing identifies mismatches that were not previously identified. Prior studies reported associations between UHR HLA mismatches and inferior OS. This validation analysis found no associations between UHR mismatching and OS among transplants equivalently matched (10 out of 10) on the basis of conventional high-resolution matching standards. Consideration of UHR differences among equivalently matched donor options was associated with significantly reduced risk of grade 2-4 acute graft-versus-host disease and in some cases transplant-related mortality.
Relevance
This study provides evidence for consideration of UHR matching when multiple 10 out of 10 high-resolution–matched donors are available.
The HLA genes are highly polymorphic, and the widespread adoption of next-generation sequencing (NGS) methods has greatly increased the number of novel HLA alleles identified in recent years. The latest release of the IPD-IMGT/HLA Database includes 28,320 distinct HLA alleles, but potential numbers could be considerably higher.12-14 To date, few studies have attempted to determine the functional and clinical relevance of HLA polymorphism in regions outside of the ARD.15-18
A 2019 study suggested that any variation between the six HLA loci considered for matching correlated with detrimental outcomes post-HCT.19 In a cohort of MUD-HCT pairs from the United Kingdom predominantly receiving in vivo T-cell–depleting serotherapy, recipients receiving a 12 out of 12 ultrahigh resolution (UHR; including HLA-DPB1) HLA-matched graft (defined as matched at a genomic level for HLA class I genes and at an extended coding DNA sequence level for HLA class II) had significantly improved overall survival (OS) than those with any degree of mismatch. In addition, 10 out of 10 UHR-matched, permissively DPB1-mismatched transplant did not have significantly different OS compared with 12 out of 12 UHR-matched, whereas increasing UHR mismatching was associated with increased grade 2-4 acute GVHD (aGVHD). Although possibly controversial,20,21 these findings were partially confirmed in similar, albeit smaller, cohorts.22,23
The aims of this study were to determine whether UHR HLA matching in an independent cohort of MUD-HCT recipients was associated with transplant outcomes, to validate the previous findings of better OS and less aGVHD with UHR matching in a cohort of T-cell–depleted MUD-HCT patients, and to analyze a cohort of T-cell–replete MUD-HCTs.
METHODS
Study Design
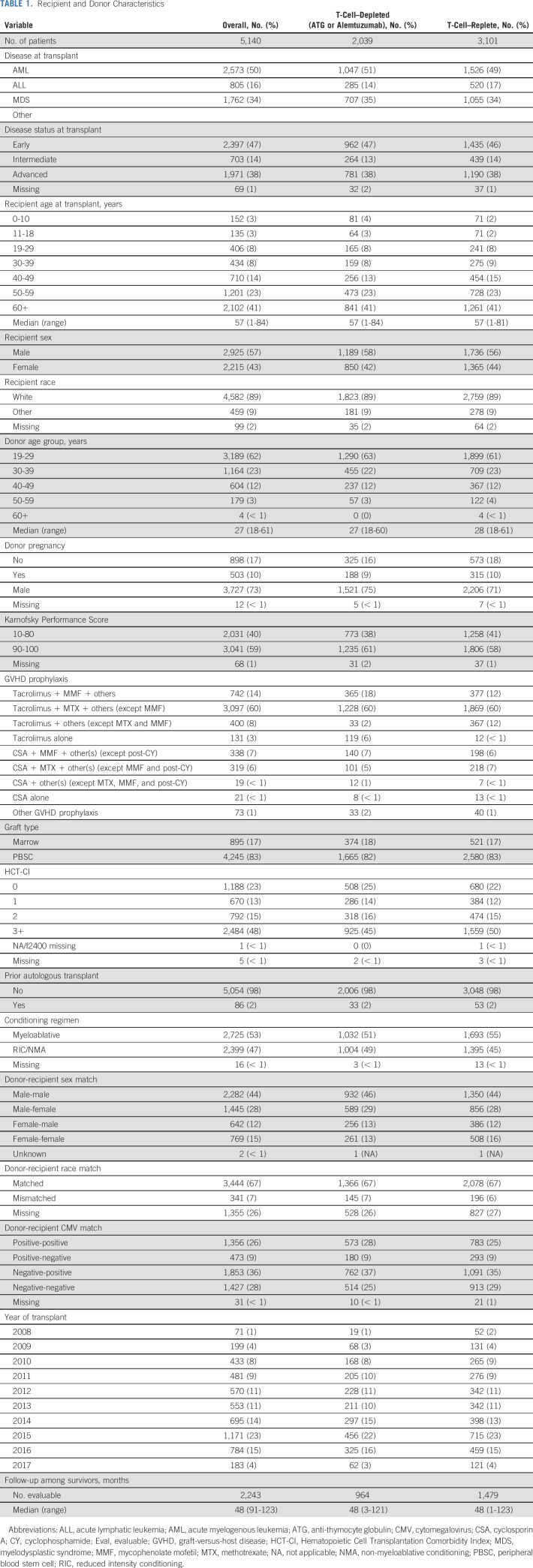
UHR HLA typing and clinical data were available on a cohort of 5,140 first allogeneic MUD-HCT donor and recipient pairs in the Center for International Blood and Marrow Transplant Research (CIBMTR). Patients received a high-resolution, 10 out of 10 HLA-matched graft at ARD level between 2008 and 2017 for acute myelogenous leukemia, acute lymphatic leukemia, or myelodysplastic syndrome (Table 1). Recipients of umbilical cord blood grafts, ex vivo T-cell–depleted grafts, or post-transplant cyclophosphamide for GVHD prophylaxis were excluded. Subgroup analyses included those receiving in vivo T-cell–depleted (antithymocyte globulin or alemtuzumab, n = 2,039) and T-cell–replete transplants (n = 3,101). The National Marrow Donor Program Institutional Review Board approved this study. Informed written consent was obtained from all participants, in accordance with the Declaration of Helsinki.
TABLE 1.
Recipient and Donor Characteristics
HLA Typing
UHR HLA typing was performed by a commercial vendor using Pacific Biosciences' single-molecule real-time DNA sequencing method. Typing was completed for the six classical HLA loci (class I: HLA-A, HLA-B, and HLA-C; class II: HLA-DRB1, HLA-DQB1, and HLA-DPB1). The NGS typing methodology included all exons and the intervening introns for the classical HLA class I genes and exons 2 and 3 only for the class II genes.
Determining HLA Matching
Because of the timing of the typing, different ambiguous HLA typing results were obtained. All HLA typing data were reviewed, curated, and normalized to ensure consistency. Matching was determined according to criteria used in a previous study.19 Analyses that included the HLA-DPB1 TCE (DP-TCE) matching algorithm used the model proposed by Crivello et al.10
Sixteen pairs were removed from the analysis because of an inability to assign DP-TCE groups or where UHR matching identified uncharacterized null alleles resulting in ARD matches < 10 out of 10.
Models of UHR HLA Matching Analysis
Several models of HLA matching were tested to replicate those used previously:19 model 1, compared 12 out of 12 UHR HLA-matched individuals and those mismatched at any level, at any loci (12 out of 12 v ≤ 11 out of 12 matched); model 2, tested the degree of UHR HLA matching (12 out of 12 v 11 out of 12 v 10 out of 12 v ≤ 9 out of 12); and model 3, 10 out of 10 UHR HLA-matched pairs were subdivided into 12 out of 12 UHR HLA-matched and TCE permissively mismatched or nonpermissively mismatched.
Statistical Methods
The primary study outcome was OS, defined as the time from HCT to death from any cause. Surviving patients were censored at the time of last follow-up. Secondary outcomes included aGVHD, chronic GVHD, relapse, transplant-related mortality (TRM), disease-free survival (DFS), and neutrophil engraftment. aGVHD and chronic GVHD, relapse, and neutrophil engraftment were per CIBMTR definitions. TRM was death without recurrent malignancy. DFS was defined as the time to treatment failure because of death or relapse. HCT Comorbidity Index (HCT-CI) values were calculated using the model proposed by Sorror et al.24
Death was considered a competing risk event for all outcomes except OS, DFS, and TRM, whereas relapse was considered a competing risk for TRM. Patients were censored at time of second HCT or if alive at last follow-up.
Multivariate analyses for OS, aGVHD, relapse, DFS, TRM, and neutrophil engraftment were performed using Cox proportional hazards models. All variables were tested for the affirmation of the proportional hazards assumption. Factors violating the proportional hazards assumption were adjusted through stratification. A stepwise variable selection procedure was then used to select adjusted covariates and develop multivariable models for the primary and secondary outcomes. Seven UHR matching variables were tested separately for association with each outcome. For the association test of UHR matching in the T-cell–depleted cohort, a P value of < .05 was considered significant as this was a validation analysis of Mayor et al.19 For the main hypothesis of 12 out of 12 versus ≤ 11 out of 12 in the full cohort, P < .01 was considered significant. To adjust for multiple testing, a significance level of P < .01/6 = .0017 was used for all other UHR matching variables. All P values were raw and two-sided. Statistical analyses were done using SAS version 9.4 (Cary, NC).
RESULTS
Patient Characteristics
Median time of follow-up was 48 months (range 1 to 123; Table 1). Fifty percent of the patients had acute myelogenous leukemia, and 47% were transplanted with early disease stage. Fifty-three percent received myeloablative conditioning. Patients were predominantly Caucasian (89%), had a median age of 57 years, and mainly had young (19 to 29 years, 62%), male (73%) donors. The majority of patients received peripheral blood stem-cell grafts (83%). Nearly half of the patients had an HCT-CI of three or more (48%). The characteristics of the T-cell–depleted and T-cell–replete subgroups were well-balanced.
Impact of UHR Typing on HLA Matching Status
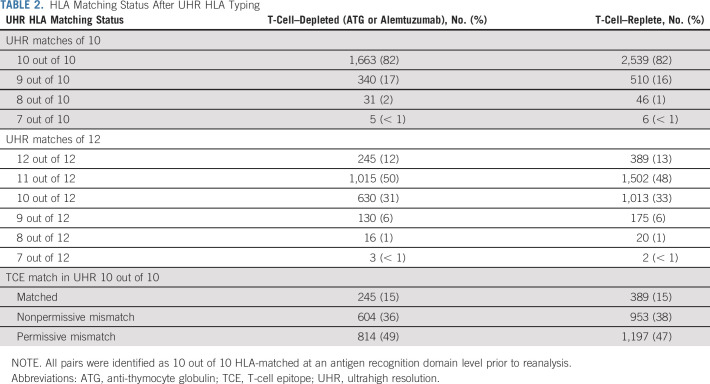
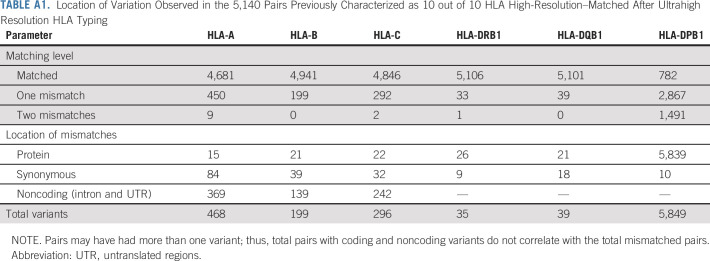
After UHR matching status was determined, only 82% of recipients remained a 10 out of 10 match, with the remainder being 9 out of 10 (17%) or less (1%; Table 2; Appendix Table A1, online only). When matching was extended to include HLA-DPB1, 634 (12%) patients received a 12 out of 12 UHR-matched graft, whereas 2,517 (49%), 1,643 (32%), and 346 (7%) received an 11 out of 12, 10 out of 12, or ≤ 9 out of 12 UHR-matched grafts, respectively. There were no significant differences in UHR matching status between the T-cell–depleted or T-cell–replete subgroups.
TABLE 2.
HLA Matching Status After UHR HLA Typing
In recipients identified as 10 out of 10 UHR-matched (n = 4,202), 634 (15%) were found to be 12 out of 12 HLA-matched, whereas 2,011 were DP-TCE permissively mismatched (48%) and 1,557 were DP-TCE nonpermissively mismatched (37%). As previously, no significant differences between the T-cell–depleted or T-cell–replete subgroups were observed.
Analysis of the Full Cohort
The 4-year OS for the complete cohort was 48% (95% CI, 47 to 49). Adjusting for significant clinical covariates, there were no significant associations between UHR matching and OS in the full cohort or when 12 out of 12 UHR-matched patients were compared with those with any other mismatch (model 1), nor when the degree of mismatch (model 2) was tested. There was no survival advantage for patients who were either 12 out of 12 UHR or HLA-DPB1 nonpermissively mismatched (model 3) when compared with permissively mismatched patients.
The 4-year TRM for the complete cohort was 27% (95% CI, 26 to 28). Adjusting for patient and donor age, disease, and HCT-CI score, UHR HLA matching was not associated with risk of TRM in any of the models tested.
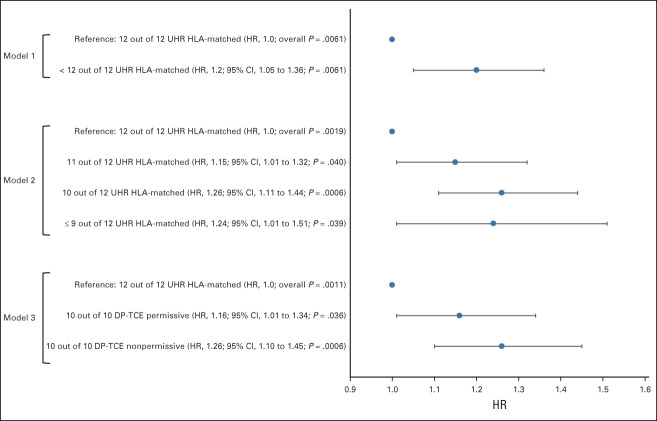
The probability of aGVHD at 100 days for the complete cohort was 43% (95% CI, 41 to 44). Adjusting for significant covariates, UHR HLA matching was associated with the probability of aGVHD (grades 2-4) in the overall cohort. Patients receiving a graft from a ≤ 11 out of 12 UHR-matched donor had a significantly higher risk of aGVHD as compared with those receiving a 12 out of 12 UHR match (model 1: hazard ratio (HR), 1.2; 95% CI, 1.05 to 1.36; P = .0061; Fig 1). This was further refined with UHR matching model 2, where increasing numbers of UHR mismatches were associated with increasing risk of aGVHD when compared with 12 out of 12 UHR-matched patients (11 out of 12 HR: 1.15; 95% CI, 1.01 to 1.32; 10 out of 12 HR: 1.26; 95% CI, 1.11 to 1.44; and ≤ 9 out of 12 HR: 1.24; 95% CI, 1.01 to 1.51; overall P = .0019; Fig 1). Additionally, 10 out of 10 UHR DPB1-TCE permissively mismatched pairs (HR: 1.16; 95% CI, 1.01 to 1.34; P = .036) and DPB1-TCE nonpermissively mismatched patients (HR: 1.26; 95% CI, 1.10 to 1.45; P = .0006; Fig 1) were associated with a higher risk of aGVHD compared with 12 out of 12 UHR-matched pairs (overall P = .0011).
FIG 1.
A forest plot showing the adjusted hazard ratio of aGVHD in the overall cohort (N = 5,140) for the three analysis models. The analysis was stratified by year of transplant and adjusted for disease, GVHD prophylaxis, HCT-CI, use of T-cell depletion, conditioning regimen, graft type, and disease status. aGVHD, acute graft-versus-host disease; HCT-CI, Hematopoietic Cell Transplantation Comorbidity Index; HR, hazard ratio; TCE, T-cell epitope; UHR, ultrahigh resolution.
Analysis of the T-Cell–Depleted Subset
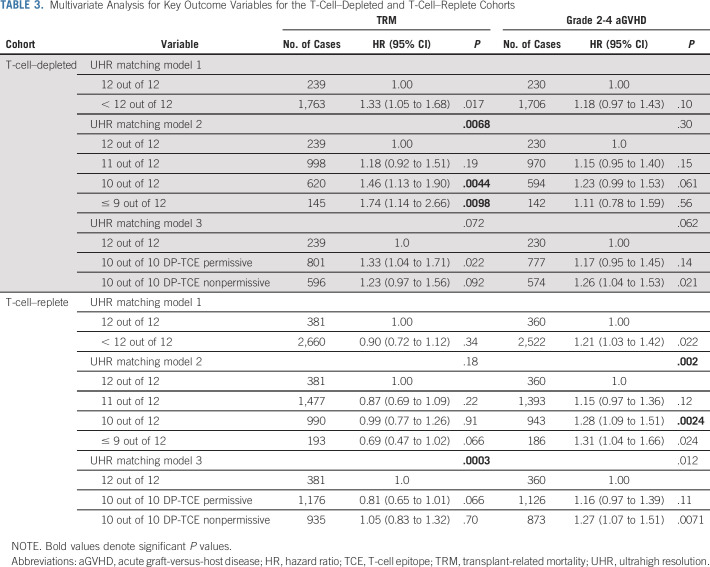
No significant associations with OS were observed with any of the models tested in the T-cell–depleted subgroup (N = 2,039; data not shown). However, UHR mismatches were associated with TRM (Fig 2, Table 3; P = .0068 overall), with patients receiving a 10 out of 12 (HR: 1.46; 95% CI, 1.13 to 1.90; P = .0044) or ≤ 9 out of 12 UHR-matched donor (HR: 1.74; 95% CI, 1.14 to 2.66; P = .0098) at a significantly higher risk. No other UHR matching model had significant correlations with the probability of TRM, nor was UHR HLA matching in any model associated with any outcomes in this T-cell–depleted subset.
FIG 2.
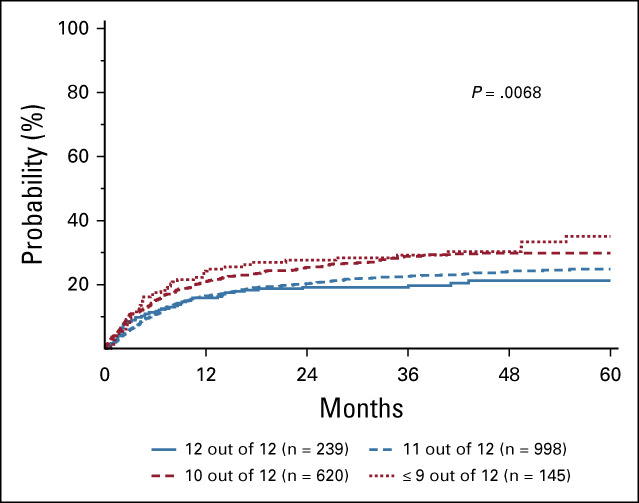
Adjusted cumulative incidence of TRM in a T-cell–depleted cohort. Probability of TRM increases with increasing numbers of ultrahigh resolution HLA mismatches. TRM, transplant-related mortality.
TABLE 3.
Multivariate Analysis for Key Outcome Variables for the T-Cell–Depleted and T-Cell–Replete Cohorts
Analysis of the T-Cell–Replete Subset
No significant associations between any of the UHR HLA matching models and OS were identified in the T-cell–replete subset (data not shown). Compared with 12 out of 12 UHR HLA-matched pairs in model 3, 10 out of 10 DP-TCE nonpermissively mismatched pairs had increased risk for TRM (HR: 1.05; 95% CI, 0.83 to 1.32; P = .70; Table 3), whereas 10 out of 10 DP-TCE permissively mismatched patients had lower risk (HR: 0.81; 95% CI, 0.65 to 1.01; P = .066; overall P = .0003).
The risk of aGVHD (grades 2-4) in the T-cell–replete subset was associated with increasing numbers of UHR mismatches. Compared with a 12 out of 12 UHR HLA match, the probability of aGVHD was higher (overall P = .0020; Table 3) with the use of 10 out of 12 (HR: 1.28; 95% CI, 1.09 to 1.51; P = .0024) and ≤ 9 out of 12 matched donors (HR: 1.31; 95% CI, 1.04 to 1.66; P = .024).
Additional Outcome Analyses
UHR HLA matching was not associated with the risk of grade 3-4 aGVHD, DFS, relapse, and chronic GVHD, nor with neutrophil recovery for any of the models tested in any cohort.
Additional HLA Analyses
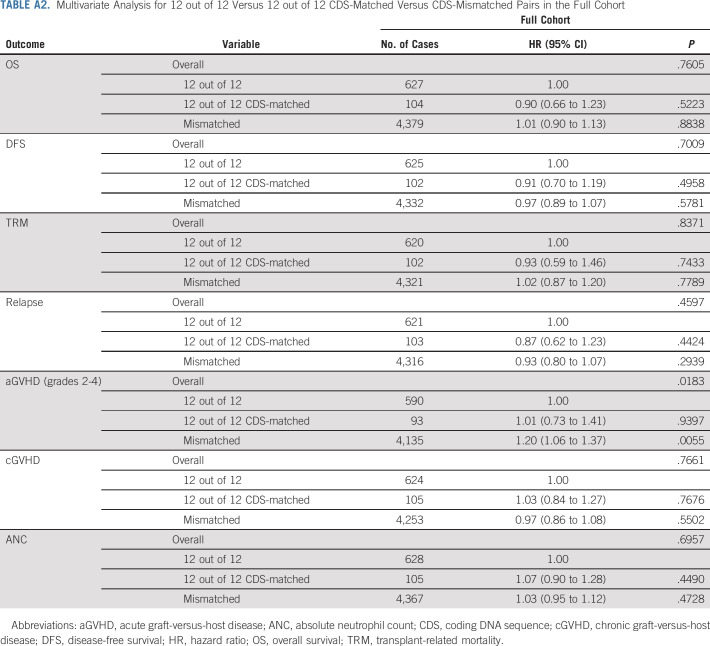
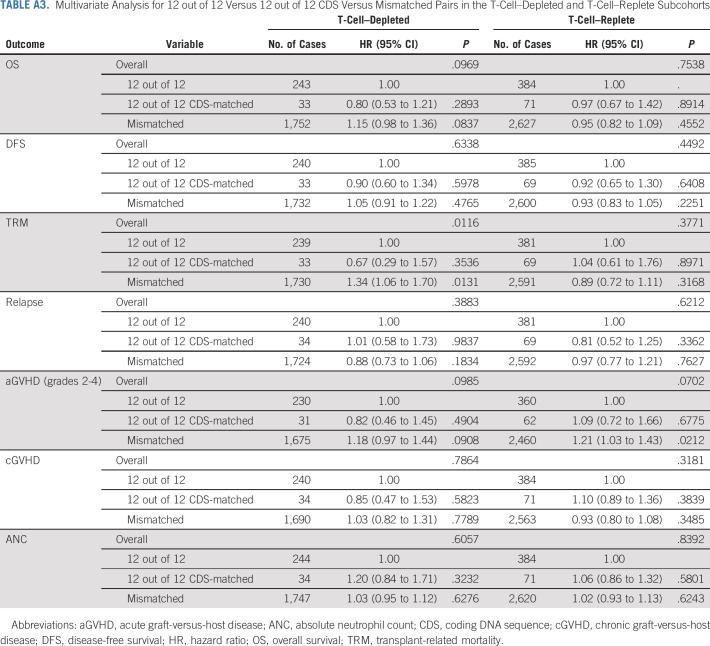
In the study by Mayor et al,19 a matching model that combined UHR HLA matching, the DP-TCE permissive matching algorithm, and cytomegalovirus serostatus matching was proposed. No significant associations were observed in any cohort using the proposed model. Additionally, analyses of noncoding variation found no significant associations with outcomes (Appendix Tables A2 and A3, online only).
DISCUSSION
The last 5 years has seen dramatic changes in the methodology routinely used to type HLA genes, particularly within the field of HCT, with increasing numbers of clinical laboratories, MUD registries, and commercial typing providers moving to NGS platforms. There are numerous reasons for this change including in some instances, cost, time, and throughput benefits. A key advantage of NGS protocols is rapid characterization of HLA genes in a timeframe amenable for clinical needs, reducing and often eliminating ambiguous typing results. With this readily available added information about HLA comes the opportunity to identify whether these additional data affect patient outcomes and determine the standard of matching using NGS.
The aim of this retrospective study was to investigate the impact of UHR HLA matching in a large independent data set of unrelated donor (UD) HCT patients from the CIBMTR. Both the full cohort as well as T-cell–depleted and T-cell–replete subgroups were tested. Unfortunately, this study did not confirm the primary hypothesis that UHR HLA matching was associated with improved OS in any of the cohorts tested. However, in the full cohort, UHR mismatching was associated with increased aGVHD risk in all matching models tested. In the T-cell–depleted subgroup, we partly validated the findings of the UK study, with a significant association between UHR HLA mismatching and the probability of TRM. In the T-cell–replete subgroup, increasing numbers of UHR mismatches were associated with increased risk of aGVHD. The probability of TRM was also higher in patients receiving 10 out of 10 UHR-matched, DP-TCE nonpermissively mismatched donors compared with DP-TCE permissively matched donors, but not 12 out of 12 UHR-matched pairs.
This study confirmed that UHR HLA matching is associated with a decreased risk of aGVHD in the T-cell–replete subgroup. aGVHD remains a common, life-threatening condition that can affect patient quality of life after HCT.25 Thus, any means of reducing this risk, especially when achievable within the normal processes surrounding patient and donor testing prior to HCT, should be considered. Abrogation of the graft-versus-leukemia effect could be a concern, but we did not see any association between UHR HLA matching status and relapse or DFS.
A common assumption around NGS typing strategies that deliver UHR HLA typing is that the cost is prohibitive. In fact, today the cost and turnaround time of such typing is comparable with previously used methods, and continued demand for such tests will only continue to decrease costs. Although cost analyses were not within the scope of this study, it should be acknowledged that treatment of patients with aGVHD is also costly26 and not risk-free.
Identifying a 12 out of 12 UHR HLA-matched, or a 10 out of 10 UHR-matched, DP-TCE permissively mismatched donor for patients can be challenging and may not be a possible option. However, studies have shown that in similar populations from the United States, the probability of identifying a 10 out of 10, DP-TCE permissively mismatched donor was 69%, increasing to 80% if additional donors underwent typing, depending on the patient's ethnic origin.27 The resolution of HLA typing used was lower than UHR, which will likely affect the probability of finding such a donor when UHR typing is used, but nonetheless, it suggests that identifying 12 out of 12 or 10 out of 10 DP-TCE permissively matched donors is a realistic possibility. The uptake of UHR typing methods by large UD registries will help facilitate UHR matching in clinical practice.28-30 The potential benefit of using UHR HLA typing for alternative donor sources is yet to be tested.
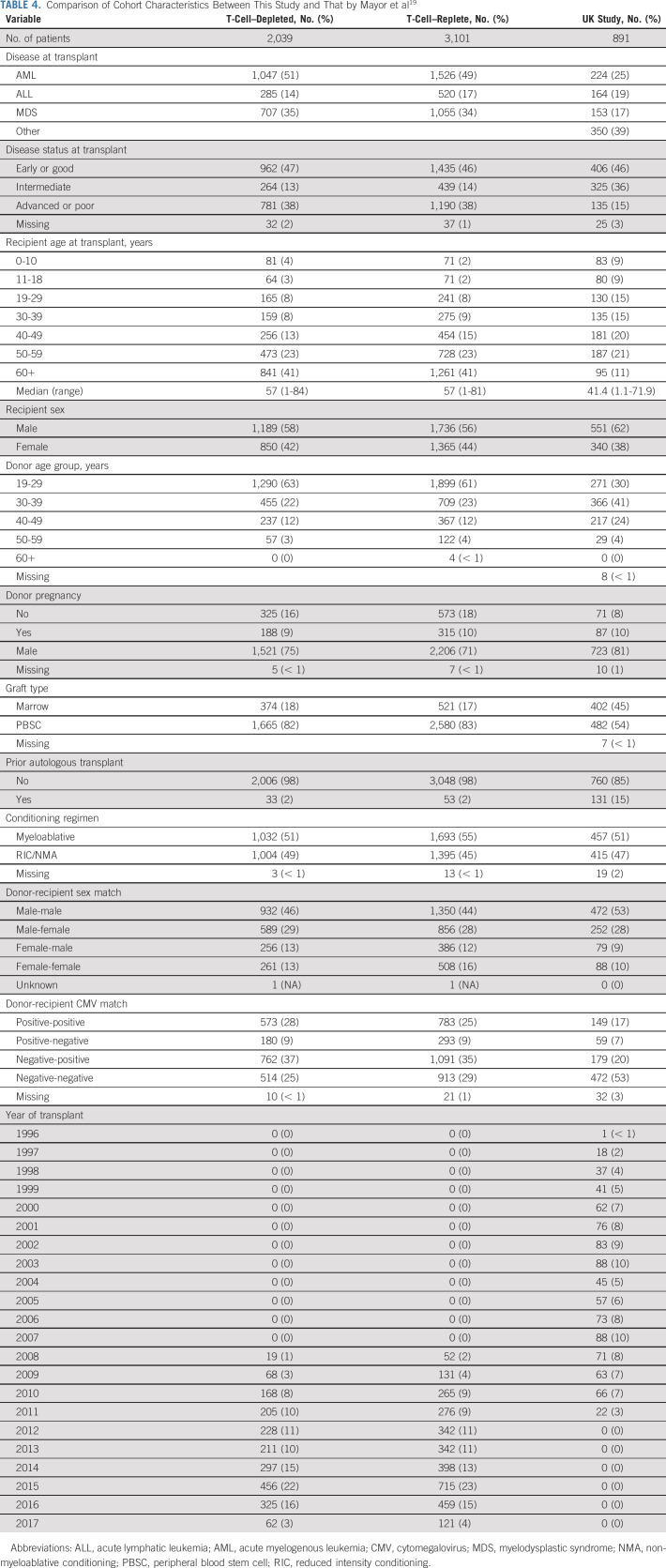
There were important differences between this study and that by Mayor et al,19 which may have contributed to the differing results. Although both studies used Pacific Biosciences' single-molecule real-time sequencing technologies, the extent of the gene coverage differed, with the previous study including all the defined 5′ and 3′ untranslated regions of the class I genes plus additional exons of HLA-DQB1 and HLA-DPB1 (exons 2-5 and 2-4, respectively). We determined that 45 out of 891 patients (5%) of the UK study cohort would have been classified as a different match level than the present analysis on the basis of the additional information obtained from the more extensive typing strategy. If extrapolated to this study cohort, a potential 250 of the 5,140 patients studied may have been classified as less well-matched if the more extensive typing methodology was used. Although it is unlikely that differences in the untranslated regions have functional consequences, they could be suggestive of lesser compatibility over the extended MHC region, resulting in more genetic disparity between patients and donors, and the potential for increased complications post-transplant.17 Further studies at a comparable level of typing will be needed to determine whether this additional information is clinically important.
In addition to the typing differences, the characteristics of the current cohort were notably different (Table 4). Of note, the present study was limited to 10 out of 10 ARD HLA-matched pairs, whereas the previous study included ARD mismatches. Additionally, the years of transplant differed, overlapping by only 4 years (2008-2011). Transplant practice changes over time, which is reflected in additional differences between the two studies (change in disease, use of peripheral blood, older patients, and younger UDs recently). Finally, this study included more advanced disease stage (38% v 15% previously). Although we attempted to mitigate the effects of these differences in the statistical analysis, this variation may have contributed to a different risk profile than that of the UK study. Indeed, HLA mismatching UD-HCT patients at high resolution6 and transplanting patients in older eras31,32 are associated with detrimental outcomes, whereas the adverse effect of HLA mismatching diminishes in patients with advanced disease.7,33 Patients transplanted in more recent years may benefit from changes in practice to the extent that other risk factors such as HLA matching become less marked.
TABLE 4.
Comparison of Cohort Characteristics Between This Study and That by Mayor et al19
Further evidence to support this observation is the surprising lack of association between the DP-TCE matching model and outcomes, which is in contrast to several previous publications. An earlier study from the CIBMTR in a similar cohort of patients reported that either permissive or nonpermissive DPB1 mismatching was associated with increased probability of aGVHD, while nonpermissive DP-TCE mismatching was associated with reduced OS.7 Of note, the cohort studied by Pidala et al resembled the UK UHR cohort: transplants between 1999 and 2011, more bone marrow grafts, more myeloablative conditioning, similar proportions of advanced disease stage, and inclusion of patients with chronic myeloid leukemia. Further studies in more recent cohorts are needed to confirm whether factors previously thought to affect outcome remain as relevant in the current era.
In conclusion, this study did not confirm previous findings that UHR HLA typing was associated with OS in any of the models tested. However, when multiple 10 out of 10 HLA-matched donors of equivalent age are available, the consideration of UHR HLA typing data may identify donors associated with a reduced risk of aGVHD, and in some cases TRM, even if OS does not differ.
ACKNOWLEDGMENT
The authors would like to thank the patients and donors who consented to be part of this study.
Appendix
TABLE A1.
Location of Variation Observed in the 5,140 Pairs Previously Characterized as 10 out of 10 HLA High-Resolution–Matched After Ultrahigh Resolution HLA Typing
TABLE A2.
Multivariate Analysis for 12 out of 12 Versus 12 out of 12 CDS-Matched Versus CDS-Mismatched Pairs in the Full Cohort
TABLE A3.
Multivariate Analysis for 12 out of 12 Versus 12 out of 12 CDS Versus Mismatched Pairs in the T-Cell–Depleted and T-Cell–Replete Subcohorts
Stephanie J. Lee
Honoraria: Wolters Kluwer
Consulting or Advisory Role: Incyte, Pfizer, EMD Serono, Kadmon, MSD Oncology, Sanofi, Genzyme, Regeneron, 4SC
Research Funding: Kadmon, Takeda, Amgen, Bristol-Myers Squibb, EMD Serono, MSD, Novartis, Incyte, Syndax, Pfizer, AstraZeneca
Patents, Royalties, Other Intellectual Property: Patent pending for high-affinity T-cell receptors that target the Merkel polyomavirus
Jeffrey Auletta
Consulting or Advisory Role: MORE Health, AlloVir, AscellaHealth
Vijaya R. Bhatt
Consulting or Advisory Role: Abbvie, Incyte, Agios, Genentech, Rigel, Partnership for Health Analytic Research, LLC, Omeros, Takeda
Research Funding: Incyte, Tolero Pharmaceuticals, National Marrow Donor Program, Abbvie, Pfizer, Jazz Pharmaceuticals
Other Relationship: Oncoceutics, Novartis, Pfizer
Loren Gragert
Other Relationship: National Marrow Donor Program/Be The Match, United Network For Organ Sharing
Yoshihiro Inamoto
Honoraria: Kyowa Kirin Co, Ltd, Astellas Pharma, Sumitomo Dainippon, Novartis, Chugai Pharma, Bristol-Myers Squibb, MSD K.K.
Consulting or Advisory Role: Novartis, Janssen, Meiji Seika Kaisha
Gerald P. Morris
Research Funding: Ferring Inc, Amgen
Travel, Accommodations, Expenses: Thermo Fisher Scientific
Sophie Paczesny
Patents, Royalties, Other Intellectual Property: Dr Paczesny has a patent on “Methods of detection of graft-versus-host disease” licensed to Viracor-IBT Laboratories
Ran Reshef
Consulting or Advisory Role: Atara Biotherapeutics, Novartis, Bristol-Myers Squibb, Gilead Sciences, TScan Therapeutics
Research Funding: Atara Biotherapeutics, Incyte, Pharmacyclics, Shire, Immatics, Takeda, Gilead Sciences, Precision Biosciences, Astellas Pharma, Bristol-Myers Squibb
Bronwen E. Shaw
Honoraria: Therakos
Consulting or Advisory Role: Orcabio
Peter Shaw
Consulting or Advisory Role: Novartis
Other Relationship: Link Pharma
No other potential conflicts of interest were reported.
Listen to the podcast by Dr Majhail at jcopodcast.libsynpro.com
DISCLAIMER
The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, the Health Resources and Services Administration (HRSA), or any other agency of the US Government.
SUPPORT
N.P.M., D.J.M., and S.G.E.M. are funded by the charity Anthony Nolan. The CIBMTR is supported primarily by Public Health Service U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID); U24HL138660 from NHLBI and NCI; U01HL128568 from the NHLBI; HHSH250201700006C and HHSH250201700007C from the Health Resources and Services Administration (HRSA); and N00014-20-1-2705 and N00014-20-1-2832 from the Office of Naval Research; additional federal support is provided by P01CA111412, R01CA152108, R01CA215134, R01CA218285, R01CA231141, R01AI128775, R01HL126589, R01HL129472, R01HL130388, R01HL131731, U01AI069197, U01AI126612, UG1HL06924, and BARDA. Support is also provided by Be the Match Foundation, Boston Children's Hospital, Dana Farber, St Baldrick's Foundation, Stanford University, the Medical College of Wisconsin the National Marrow Donor Program, and the following commercial entities: Actinium Pharmaceuticals Inc; Adienne SA; Allovir Inc; Amgen Inc; Angiocrine Bioscience; Astellas Pharma US; bluebird bio Inc; Bristol Myers Squibb Co; Celgene Corp; CSL Behring; CytoSen Therapeutics Inc; Daiichi Sankyo Co, Ltd; ExcellThera; Fate Therapeutics; Gamida-Cell, Ltd; Genentech Inc; Incyte Corporation; Janssen/Johnson & Johnson; Jazz Pharmaceuticals Inc; Kiadis Pharma; Kite, a Gilead Company; Kyowa Kirin; Legend Biotech; Magenta Therapeutics; Merck Sharp & Dohme Corp; Millennium, the Takeda Oncology Co; Miltenyi Biotec Inc; Novartis Pharmaceuticals Corporation; Omeros Corporation; Oncoimmune Inc; Orca Biosystems Inc; Pfizer Inc; Pharmacyclics, LLC; Sanofi Genzyme; Stemcyte; Takeda Pharma; Vor Biopharma; and Xenikos BV.
AUTHOR CONTRIBUTIONS
Conception and design: Neema P. Mayor, Tao Wang, Stephanie J. Lee, Stephen R. Spellman, Steven G. E. Marsh
Provision of study materials or patients: Bronwen E. Shaw, Stephen R. Spellman
Collection and assembly of data: Neema P. Mayor, Tao Wang, Stephanie J. Lee, Michelle Kuxhausen, Cynthia Vierra-Green, Dominic J. Barker, Stephen R. Spellman, Steven G. E. Marsh
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Impact of Previously Unrecognized HLA Mismatches Using Ultrahigh Resolution Typing in Unrelated Donor Hematopoietic Cell Transplantation
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Stephanie J. Lee
Honoraria: Wolters Kluwer
Consulting or Advisory Role: Incyte, Pfizer, EMD Serono, Kadmon, MSD Oncology, Sanofi, Genzyme, Regeneron, 4SC
Research Funding: Kadmon, Takeda, Amgen, Bristol-Myers Squibb, EMD Serono, MSD, Novartis, Incyte, Syndax, Pfizer, AstraZeneca
Patents, Royalties, Other Intellectual Property: Patent pending for high-affinity T-cell receptors that target the Merkel polyomavirus
Jeffrey Auletta
Consulting or Advisory Role: MORE Health, AlloVir, AscellaHealth
Vijaya R. Bhatt
Consulting or Advisory Role: Abbvie, Incyte, Agios, Genentech, Rigel, Partnership for Health Analytic Research, LLC, Omeros, Takeda
Research Funding: Incyte, Tolero Pharmaceuticals, National Marrow Donor Program, Abbvie, Pfizer, Jazz Pharmaceuticals
Other Relationship: Oncoceutics, Novartis, Pfizer
Loren Gragert
Other Relationship: National Marrow Donor Program/Be The Match, United Network For Organ Sharing
Yoshihiro Inamoto
Honoraria: Kyowa Kirin Co, Ltd, Astellas Pharma, Sumitomo Dainippon, Novartis, Chugai Pharma, Bristol-Myers Squibb, MSD K.K.
Consulting or Advisory Role: Novartis, Janssen, Meiji Seika Kaisha
Gerald P. Morris
Research Funding: Ferring Inc, Amgen
Travel, Accommodations, Expenses: Thermo Fisher Scientific
Sophie Paczesny
Patents, Royalties, Other Intellectual Property: Dr Paczesny has a patent on “Methods of detection of graft-versus-host disease” licensed to Viracor-IBT Laboratories
Ran Reshef
Consulting or Advisory Role: Atara Biotherapeutics, Novartis, Bristol-Myers Squibb, Gilead Sciences, TScan Therapeutics
Research Funding: Atara Biotherapeutics, Incyte, Pharmacyclics, Shire, Immatics, Takeda, Gilead Sciences, Precision Biosciences, Astellas Pharma, Bristol-Myers Squibb
Bronwen E. Shaw
Honoraria: Therakos
Consulting or Advisory Role: Orcabio
Peter Shaw
Consulting or Advisory Role: Novartis
Other Relationship: Link Pharma
No other potential conflicts of interest were reported.
REFERENCES
- 1.D'Souza A Fretham C Lee SJ, et al. : Current use of and trends in hematopoietic cell transplantation in the United States. Biol Blood Marrow Transplant 26:e177-e182, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Passweg JR Baldomero H Bader P, et al. : Is the use of unrelated donor transplantation leveling off in Europe? The 2016 European Society for Blood and Marrow Transplant activity survey report. Bone Marrow Transplant 53:1139-1148, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kollman C Spellman SR Zhang M-J, et al. : The effect of donor characteristics on survival after unrelated donor transplantation for hematologic malignancy. Blood 127:260-267, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw BE Logan BR Spellman SR, et al. : Development of an unrelated donor selection score predictive of survival after HCT: Donor age matters most. Biol Blood Marrow Transplant 24:1049-1056, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw BE Mayor NP Szydlo RM, et al. : Recipient/donor HLA and CMV matching in recipients of T-cell-depleted unrelated donor haematopoietic cell transplants. Bone Marrow Transplant 52:717-725, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Lee S Klein J Haagenson M, et al. : High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood 110:4576-4583, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Pidala J Lee SJ Ahn KW, et al. : Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood 124:2596-2606, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dehn J Spellman S Hurley CK, et al. : Selection of unrelated donors and cord blood units for hematopoietic cell transplantation: Guidelines from the NMDP/CIBMTR. Blood 134:924-934, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Little AM Green A Harvey J, et al. : BSHI guideline: HLA matching and donor selection for haematopoietic progenitor cell transplantation. Int J Immunogenet 43:263-286, 2016 [DOI] [PubMed] [Google Scholar]
- 10.Crivello P Zito L Sizzano F, et al. : The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 21:233-241, 2015 [DOI] [PubMed] [Google Scholar]
- 11.Fleischhauer K Shaw BE Gooley T, et al. : Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: A retrospective study. Lancet Oncol 13:366-374, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.IPD-IMGT/HLA: Release V3.43.0. https://www.ebi.ac.uk/ipd/imgt/hla/
- 13.Robinson J Barker DJ Georgiou X, et al. : IPD-IMGT/HLA database. Nucleic Acids Res 48:D948-D955, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson J Guethlein LA Cereb N, et al. : Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles. PLoS Genet 13:e1006862, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Hateren A Bailey A Werner JM, et al. : Plasticity of empty major histocompatibility complex class I molecules determines peptide-selector function. Mol Immunol 68:98-101, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersdorf EW, O'hUigin C: The MHC in the era of next-generation sequencing: Implications for bridging structure with function. Hum Immunol 80:67-78, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersdorf EW Malkki M Gooley TA, et al. : MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med 4:e8, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersdorf EW Malkki M O'HUigin C, et al. : High HLA-DP expression and graft-versus-host disease. N Engl J Med 373:599-609, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayor NP Hayhurst JD Turner TR, et al. : Recipients receiving better HLA-matched hematopoietic cell transplantation grafts, uncovered by a novel HLA typing method, have superior survival: A retrospective study. Biol Blood Marrow Transplant 25:443-450, 2019 [DOI] [PubMed] [Google Scholar]
- 20.Hurley CK Spellman S Dehn J, et al. : Regarding “recipients receiving better HLA-matched hematopoietic cell transplantation grafts, uncovered by a novel HLA typing method, have superior survival: A retrospective study”. Biol Blood Marrow Transplant 25:e268-e269, 2019 [DOI] [PubMed] [Google Scholar]
- 21.Mayor NP Hayhurst JD Turner TR, et al. : A reply to Hurley et al. regarding recipients receiving better HLA-matched hematopoietic cell transplantation grafts, uncovered by a novel HLA typing method, have superior survival: A retrospective study. Biol Blood Marrow Transplant 25:e270-e271, 2019 [DOI] [PubMed] [Google Scholar]
- 22.De Santis D Truong L Witt C, et al. : Super high resolution hla matching by next generation sequencing improves the clinical outcomes of haematopoietic stem cell transplantation. HLA 93:276-277, 2019 [Google Scholar]
- 23.Vazirabad I Chhabra S Nytes J, et al. : Direct HLA genetic comparisons identify highly matched unrelated donor-recipient pairs with improved transplantation outcome. Biol Blood Marrow Transplant 25:921-931, 2019 [DOI] [PubMed] [Google Scholar]
- 24.Sorror ML Maris MB Storb R, et al. : Hematopoietic cell transplantation (HCT)-specific comorbidity index: A new tool for risk assessment before allogeneic HCT. Blood 106:2912-2919, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeiser R, Blazar BR: Acute graft-versus-host disease—Biologic process, prevention, and therapy. N Engl J Med 377:2167-2179, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SJ Klar N Weeks JC, et al. : Predicting costs of stem-cell transplantation. J Clin Oncol 18:64-71, 2000 [DOI] [PubMed] [Google Scholar]
- 27.Tram K Stritesky G Wadsworth K, et al. : Identification of DPB1 permissive unrelated donors is highly likely. Biol Blood Marrow Transplant 23:81-86, 2017 [DOI] [PubMed] [Google Scholar]
- 28.Mayor NP Robinson J McWhinnie AJ, et al. : HLA typing for the next generation. PLoS One 10:e0127153, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schöfl G Lang K Quenzel P, et al. : 2.7 million samples genotyped for HLA by next generation sequencing: Lessons learned. BMC Genomics 18:161, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baier DM Hofmann JA Fischer H, et al. : Very low error rates of NGS-based HLA typing at stem cell donor recruitment question the need for a standard confirmatory typing step before donor work-up. Bone Marrow Transplant 54:928-930, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majhail NS Chitphakdithai P Logan B, et al. : Significant improvement in survival after unrelated donor hematopoietic cell transplantation in the recent era. Biol Blood Marrow Transplant 21:142-150, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gooley TA Chien JW Pergam SA, et al. : Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med 363:2091-2101, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw BE Mayor NP Russell NH, et al. : Diverging effects of HLA-DPB1 matching status on outcome following unrelated donor transplantation depending on disease stage and the degree of matching for other HLA alleles. Leukemia 24:58-65, 2010 [DOI] [PubMed] [Google Scholar]