Abstract
Patients with bipolar disorder (BD) often show increased risk-taking propensity, which may contribute to poor clinical outcome. While these two phenotypes are genetically correlated, there is scarce knowledge on the shared genetic determinants. Using GWAS datasets on BD (41,917 BD cases and 371,549 controls) and risk-taking (n = 466,571), we dissected shared genetic determinants using conjunctional false discovery rate (conjFDR) and local genetic covariance analysis. We investigated specificity of identified targets using GWAS datasets on schizophrenia (SCZ) and attention-deficit hyperactivity disorder (ADHD). The putative functional role of identified targets was evaluated using different tools and GTEx v. 8. Target druggability was evaluated using DGIdb and enrichment for drug targets with genome for REPositioning drugs (GREP). Among 102 loci shared between BD and risk-taking, 87% showed the same direction of effect. Sixty-two were specifically shared between risk-taking propensity and BD, while the others were also shared between risk-taking propensity and either SCZ or ADHD. By leveraging pleiotropic enrichment, we reported 15 novel and specific loci associated with BD and 22 with risk-taking. Among cross-disorder genes, CACNA1C (a known target of calcium channel blockers) was significantly associated with risk-taking propensity and both BD and SCZ using conjFDR (p = 0.001 for both) as well as local genetic covariance analysis, and predicted to be differentially expressed in the cerebellar hemisphere in an eQTL-informed gene-based analysis (BD, Z = 7.48, p = 3.8E−14; risk-taking: Z = 4.66, p = 1.6E−06). We reported for the first time shared genetic determinants between BD and risk-taking propensity. Further investigation into calcium channel blockers or development of innovative ligands of calcium channels might form the basis for innovative pharmacotherapy in patients with BD with increased risk-taking propensity.
Subject terms: Genetic markers, Bipolar disorder
Introduction
Bipolar disorder (BD) is a severe psychiatric disorder characterized by the recurrence of depressive and manic or hypomanic episodes, alternating with intervals of well-being [1]. With a prevalence of 1% of the population and being associated with relevant illness-related disability, high prevalence of medical comorbidities characterized by premature mortality and a risk to die by suicide 20–30 times higher compared to the general population [2], BD exerts a substantial socio-economic impact [3]. The diagnosis of BD requires a comprehensive clinical assessment and no reliable biomarker is currently available. Besides mood episodes of mania and depression, which represent primary features of this disorder, a subset of patients with BD might also present deficits across multiple domains of cognitive function [2, 4–6]. These impairments are not exclusively observed in patients with BD but also in other psychiatric disorders such as e.g. schizophrenia (SCZ) [7] and attention-deficit hyperactivity disorder (ADHD) [8, 9], with the latter being specifically characterized by symptoms in the inattentive or hyperactive and impulsive domains, or both [10].
Among deficits in executive function, patients with BD may exhibit abnormalities in impulsivity, sub-optimal decision-making and increased propensity for risk-taking behaviors. Risk-taking behaviors can be described as activities with high potential for negative consequences and may be linked to abnormal processing of reward-predicting stimuli [11–14]. Excessive involvement in activities with high potential for negative consequences is one of the seven symptoms included in the DSV-IV or DSM-V diagnostic criteria for a manic episode (at least three symptoms must be present during a period characterized by persistently elevated, expansive, or irritable mood and increased energy) [15, 16]. However, in patients with BD increased risk-taking propensity can also be present during remission and may contribute to poor clinical outcome, being linked to increased prevalence of substance abuse and suicide [17, 18]. Risk-taking propensity in BD is still understudied, as can be inferred from a recent meta-analysis which included only six studies [19]. In this meta-analysis, a nonsignificant trend for impairment in risk-behavior was observed when considering all studies in a sample with high heterogeneity (p = 0.06; I2 = 81.3%), while significant impairment was observed in a more homogenous subgroup of BD type I and euthymic patients with no heterogeneity (I2 = 0%, standardized mean difference = 0.92; p < 0.0001).
The neurobiological determinants of impairments in decision-making and, specifically of increased risk-taking propensity, are still largely unknown. A recent study suggested risk-taking to be negatively associated with white matter integrity in the right cingulum in both patients with BD and controls, while white matter alterations in the left inferior fronto-occipital fasciculus were specifically implicated in risk-taking behavior in patients with BD [20]. In addition, risk-taking propensity has been associated with alterations in reward salience in the frontostriatal pathway in patients with BD [21], reduced gray matter volume in the amygdala and hippocampus in humans [22] and increased hippocampal glutamate and monoamine levels in preclinical studies and in humans [23–25]. The mood stabilizer lithium, which represents the gold-standard in the maintenance of BD, being able to reduce recurrences and suicide risk [2, 26], has been suggested to be able to reduce risk-taking behaviors in preclinical models of mania [27, 28], although the underlying molecular mechanisms are not known.
Specific subgroups of patients with BD might show increased predisposition to risk-taking propensity. Indeed, a recent study including 54 euthymic BD type 1 patients who underwent cognitive testing and resting state neuroimaging identified three main clusters using hierarchical cluster-analysis on executive function scores [29]. One of these clusters was characterized by increased risk-taking propensity during the Cambridge Gambling Task [29]. Increased risk-taking predisposition in a subset of patients with BD might be at least partly explained by shared genetic determinants, as also supported by the observation of poorer adjustment in risk-taking behavior measured with the Balloon Analogue Risk Task in both patients with BD and their first-degree relatives compared to healthy controls [30]. BD has a strong genetic component which has recently started to be elucidated by genome-wide association studies (GWAS) that identified multiple single nucleotide polymorphisms (SNP) associated with this disorder [31, 32]. Similarly, a recent GWAS identified several genetic variants associated with risk-taking [33] and also showed this trait to be positively genetically correlated with BD (rg = 0.21), SCZ (rg = 0.17), and ADHD (rg = 0.25), using linkage disequilibrium (LD) score regression [33]. However, the specific casual genes and biological mechanisms, as well as the potential shared genetic factors between BD and risk-taking propensity, have not been investigated. It is well known that several genetic variants exhibit allelic pleiotropy, i.e. are associated with more than one phenotype [34]. For instance, BD has been shown to share part of its genetic architecture with other psychiatric disorders [35–37]. The identification of shared genetic variants can improve our understanding of the biological underpinnings of two phenotypes as well as lay the basis to develop improved treatment strategies [34]. Recently, the conditional false discovery rate (condFDR)/conjunctional false discovery rate (conjFDR) method allowed to identify novel loci associated with BD leveraging pleiotropic association with SCZ [38], ADHD [39], and intelligence [40]. However, this approach has never been applied to leverage pleiotropic association between BD and risk-taking propensity.
Aims of our study were to (1) identify genetic variants and genes that influence both susceptibility to BD and risk-taking propensity, (2) investigate whether genetic data on risk-taking propensity may improve discovery of loci associated with BD, and (3) evaluate the potential functional role, functional enrichment, and suitability as drug targets of genomic loci associated with these two traits. In addition, we explored which of the identified genetic loci might be specifically shared between risk-taking and BD and which represent instead cross-disorders markers, being also shared between risk-taking propensity and either SCZ or ADHD. These two disorders were selected based on previous reports of positive genetic correlation with risk-taking propensity [33] as well as studies supporting their association with executive function disturbances [7–9].
Material and methods
GWAS samples
We conducted a cross-trait analysis using the largest publicly available datasets for BD and risk-taking propensity. GWAS summary statistics for BD were obtained from the Psychiatric Genomics Consortium (PGC) BD group (freeze 3) [41]. The BD sample included 41,917 cases from 57 cohorts collected in Europe, North America, and Australia and 371,549 controls of European origin [41]. GWAS summary statistics on risk-taking propensity were obtained from the Social Science Genetic Association Consortium [33]. The sample included 466,571 participants from UK Biobank and replication cohorts. General risk tolerance was coded as a categorical variable based on the answer to the question: “Would you describe yourself as someone who takes risks?”. After exclusion of variants ambiguous (A/T and C/G) or located in regions characterized by strong LD such as the Major Histocompatibility Complex region (6:25119106-33854733), chromosome 8p23.1 (chr8:7200000–12500000) and gene MAPT, (chr17:40000000–47000000), 6,346,208 variants common to the two datasets were retained.
In order to verify whether observed associations between BD and risk-taking propensity were specific for BD or common to other psychiatric disorders, we also conducted cross-trait analyses using large publicly available datasets on SCZ (dataset from the PGC, freeze 2, including data 35,476 patients with SCZ or schizoaffective disorder and 46,839 controls [42]) and ADHD (PGC dataset including data for 19,099 cases and 34,194 controls of European ancestry from 10 cohorts [43]). After exclusion of ambiguous variants and variants located in regions characterized by strong LD, analyses were conducted on 8,176,252 and 5,448,916 variants common between risk-taking and SCZ or ADHD, respectively. For all GWAS datasets, quality control procedures, including adjustment for population stratification, were performed by the original studies [33, 42–44].
Conditional and conjunctional false discovery rate analysis
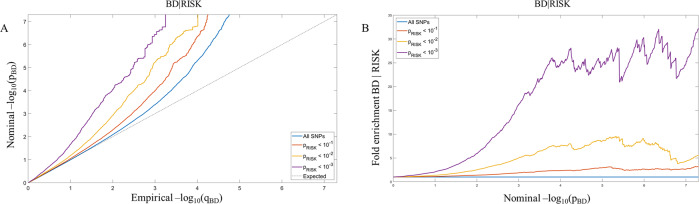
To identify shared loci between risk-taking propensity and psychiatric traits we used the condFDR/conjFDR method implemented in pleioFDR [38, 45], which allows to re-adjust the GWAS statistics in a primary phenotype (e.g. BD) by leveraging pleiotropic enrichment with a GWAS in a secondary phenotype (e.g. risk-taking). For each p value in the primary phenotype, condFDR estimates are obtained by calculating the stratified empirical cumulative distribution function of the p values [34]. The strata are obtained by the enrichment of SNP associations depending on increased p values in a secondary phenotype [34]. False discovery rate (FDR) allows to control for the expected proportion of false discoveries among all significant discoveries [46]. In the procedure first proposed by Benjamini and Hochberg [46], p values are ordered from smallest to largest, assigned a corresponding rank i and compared to a critical value calculated as (i/m)*q where i is the rank, m is the number of tests and q the FDR chosen. The largest p value lower than this critical value and all p values smaller than it are considered to be significant. The condFDR represents a Bayesian extension of FDR that allows to incorporate prior information on each SNP to improve power. In the presence of pleiotropy, stratification of test statistics in a primary phenotype based on the association with a second trait will result in a reduction in the FDR at a given nominal p value relative to the FDR computed from the unstratified distribution of the primary phenotype p values [34]. We first constructed conditional QQ plots, which extend the standard QQ plots to visualize the cross-trait polygenic enrichment. The plot is constructed by creating subsets of SNPs based on the level of association with the secondary phenotype (using three thresholds p ≤ 0.10, p ≤ 0.01, and p ≤ 0.001). Under the null hypothesis, nominal p values follow the straight line, while under cross-trait polygenic enrichment they show leftward deflections as levels of SNP association with the secondary phenotype increase. We also constructed fold-enrichment plots, in which the fold enrichment is calculated as the ratio between the −log10(p) cumulative distribution for a given stratum and the cumulative distribution for all SNPs. The conjFDR method is an extension of condFDR aimed at discovering SNPs associated with two phenotypes simultaneously. After inverting the roles of the primary and secondary phenotypes, the conjFDR is defined as the maximum of the two condFDR values. Thresholds for significant condFDR and conjFDR associations were set at 0.05 and 0.01 as in previous publications [34, 47–49]. Proportion of variance in phenotype explained by SNPs associated with both BD and risk-taking propensity at a conjFDR < 0.05 was estimated as in [50]. As the PGC BD freeze 3 dataset contains a cohort part of the UK Biobank (including 1454 BD cases and 58,113 controls) we checked for correlation of Z scores among intergenic SNPs using the function implemented in pleioFDR. While we detected low correlation coefficients between risk-taking and BD (0.054), SCZ (0.042), or ADHD (0.047), we still controlled results for sample overlap using the function implemented in pleioFDR consisting in decorrelation of vectors of Z scores based on the Mahalanobis Transformation [45].
Definition of genomic loci using FUMA
Independent significant genomic loci were defined according to the FUMA protocol [51]. Lead SNPs were defined by double clumping (a clumping of SNPs significant and independent at r2 < 0.6, and a secondary clumping of these SNPs at r2 < 0.1). Loci separated by a distance lower than 250 kb were merged. 1000 genome phase 3 was used as a reference panel to compute LD in FUMA. Variants were considered novel if they have not been reported within ±500 Kb of a significant variant within previous studies investigating BD or risk-taking propensity. Loci jointly associated with risk-taking and BD were considered to be specific in case they were at a distance of at least ±500 Kb from loci jointly associated with risk-taking and either SCZ or ADHD. Loci associated with BD conditioning on risk-taking were considered specific in case when at a distance at least ±500 kb from loci associated with SCZ or ADHD conditioning on risk-taking or with risk-taking and SCZ or ADHD jointly. Finally, loci associated with risk-taking conditioning on BD were considered specific when at a distance at least ±500 kb from loci associated with risk-taking conditioning on SCZ or ADHD. The direction of allelic effects for significant variants was evaluated by comparing betas reported in the original GWAS.
Functional annotation
Positional and functional annotation of lead SNPs associated with BD (condFDR < 0.01), risk-taking (condFDR < 0.01), or both phenotypes (conjFDR < 0.05) was performed using different tools. Nearest gene and functional category were annotated using FUMA [44, 51]. The Combined Annotation Dependent Depletion (CADD) score [52], which predicts how deleterious a variant is on protein structure/function by contrasting variants that survived natural selection with simulated mutations, was computed in FUMA. Higher scores indicate more deleterious SNPs, with a suggested threshold of 12.37 for a SNP to be considered deleterious [53]. RegulomeDB rank (from 1 to 7, with 1 being associated with highest evidence of functional effects) was calculated using RegulomeDB [54] based on known and predicted regulatory elements including regions of DNase hypersensitivity, binding sites of transcription factors and promoter regions. We searched whether SNPs acted as expression quantitative trait loci (eQTL) based on genotyping and gene expression data (obtained from a range of 114–209 samples) from Genotype-Tissue Expression (GTEx) v.8 in brain regions and whole blood. In the GTEx project, gene expression was measured with Illumina TrueSeq RNA sequencing or Affymetrix Human Gene 1.1 ST Expression Array, while genotyping data were obtained with whole genome sequencing, whole exome sequencing, Illumina OMNI 5 M, 2.5 M or Exome SNP arrays [55]. We reported cis eQTLs in a ±1 Mb cis window around the transcription start site and significant based on FDR.
Genes in which significantly associated variants were located or nearest genes were searched in the Drug Gene Interaction Database (DGIdb) [56] to assess whether they are known targets of existing drugs (drug-gene interactions) or ‘potentially druggable’ based on their involvement in selected pathways, molecular functions or gene families (druggable genome). According to this definition, the genes included in the druggable genome have some properties that make them suitable for drug targeting, even in absence of a drug currently targeting them [56]. The DGIdb database classifies genes in categories based on information retrieved from different drug target repositories (DrugBank, PharmGKB, Chembl, Drug Target Commons, Therapeutic Target Database and others).
Functional enrichment
Functional enrichment of genes in which variants jointly associated with BD and risk-propensity in the conjFDR analysis were located (excluding intergenic variants) was investigated using different approaches. Namely, functional enrichment analyses for GO terms and KEGG pathways were conducted with WebGestalt [57] with default options, adjusting results according to FDR. Functional enrichment for targets of drugs classified based on clinical indication according to Anatomical Therapeutic Chemical Classification System (ATC) or International Classification of Diseases 10 (ICD10) diagnostic codes was conducted using genome for REPositioning drugs (GREP) [58]. In addition, we searched for upstream regulators of our genes of interest using Ingenuity Pathway Analysis (IPA, Ingenuity System Inc, USA). Upstream regulators are defined as genes, microRNAs, transcription factors or chemical compounds that affect the genes of interest through effects on expression, transcription, activation, molecular modification, transport or binding events according to the Ingenuity Knowledge Base, a large collection of observations in various experimental contexts [59]. A p value of overlap <0.01 was set as the significant threshold as default.
Local genetic covariance analysis using SUPERGNOVA
As a complementary approach to investigate pleiotropy between risk-taking propensity and psychiatric traits, we estimated local genetic covariance using SUPERGNOVA [60]. This tool estimates the genetic similarity of complex traits in specific genomic regions using GWAS summary statistics and is robust to sample overlap [60]. Genetic covariance between two traits is estimated by minimizing the distance between the empirical covariance of Z scores. LD was estimated using the 1000 genomes project reference panel [61]. In order to control for sample overlap, the first Ki eigenvectors are used to transform and decorrelate Z scores in any given region i, where Ki is determined adaptively in SUPERGNOVA. After decorrelation, local genetic covariance is estimated using a weighted least squares regression in each region [60]. The software identifies genomic regions characterized by a significant local genetic covariance between two traits. P values were adjusted based on FDR. As in the case of analyses with pleioFDR, ambiguous variants or variants located in regions with strong LD were excluded.
eQTL-informed gene-based analysis
In order to investigate the functional effects of genetics variants associated with BD and risk-taking propensity, we conducted eQTL-informed gene-based analysis using eMAGMA [62]. This method leverages tissue-specific eQTL information across multiple human tissues to identify putative casual genes for a phenotype. eMAGMA provides tissue-specific annotation files while gene-based statistics were computed using MAGMA (v. 1.09) [63]. SNPs were assigned to genes based on their association with gene expression in the 13 brain tissues included in GTEx (v. 8) as well as in whole blood. Bonferroni-adjusted p value thresholds were set accordingly to the number of genes included in MAGMA (p = 0.05/19427 = 2.6E−06). Heatmaps based on expression levels of genes significantly associated with BD or risk-taking propensity in at least one brain region or in whole blood were produced based on data from GTEx (v. 8) using the ComplexHeatmap R package [64]. Preliminary results of analyses conducted with eMAGMA were presented at the 40th Italian Society of Pharmacology conference [65].
Results
Genetic overlap between risk-taking propensity and psychiatric disorders
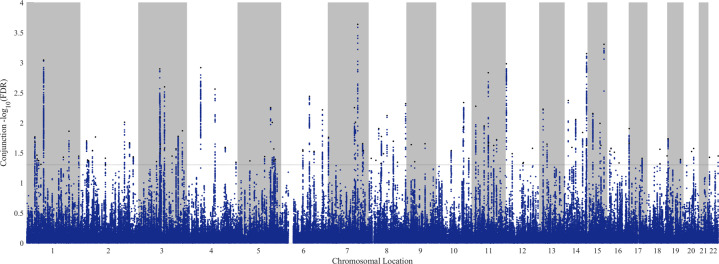
The conditional QQ plot and the fold-enrichment plot showed significant cross-trait enrichment in variants associated with BD when conditioning on risk-taking propensity (Fig. 1). We identified 102 independent genomic loci associated with both BD and risk-taking at a conjFDR < 0.05 (Fig. 2, Table 1 and Supplementary Table 1). Notably, 89 of these loci (87%) showed the same direction of effect on BD and risk-taking based on betas reported in the original GWAS (i.e. a variant associated with increased risk-taking propensity was also associated with increased predisposition to BD) (Table 1). Fifty of these SNPs were located in introns (49.0%), 4 in UTR or downstream regions (3.9%), 2 in exonic regions (2.0%), 34 in intergenic regions (33.3%) and 12 in noncoding RNAs (11.8%) (Table 1). In order to assess specificity of loci shared between BD and risk-taking, conjFDR analyses were also conducted between risk-taking and SCZ or ADHD (Supplementary Table 2). Among the 102 loci, 62 (61%) were specifically shared between risk-taking propensity and BD, while the others were also shared between risk-taking and SCZ (n = 30), ADHD (n = 4), or both (n = 6) (Table 1 and Supplementary Table 2). Proportion of variance of the BD trait explained by single SNPs ranged from 0.06% to 0.01%, with the total variance explained by all 102 SNPs being estimated at 1.6%.
Fig. 1. Pleiotropic enrichment between bipolar disorder and risk-taking propensity.
A Conditional QQ plot. The progressive leftward deflection from the null line as levels of SNP associations with the secondary phenotype increase shows significant cross-trait enrichment between BD (primary phenotype) and risk-taking propensity (secondary phenotype). B Fold-enrichment plot. The fold enrichment, calculated as the ratio between the −log10(p) cumulative distribution for a given stratum and the cumulative distribution for all SNPs, shows a significant enrichment for variants associated with BD conditioning on risk-taking propensity.
Fig. 2. Manhattan plot showing genomics loci associated with BD and risk-taking propensity.
The figure shows 102 independent genomic loci associated with both BD and risk-taking propensity at a conjunctional false discovery rate <0.05.
Table 1.
Independent genomic loci significantly associated with BD and risk-taking propensity at a conjunctional false discovery rate <0.05.
| SNP | Position | Nearest gene (Kb) | Functional category | A1/A2 | Beta BD | p BD | Beta risk | p risk | conjFDR | Specificity |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1746662 | 1:33337632 | FNDC5 | Intronic | T/C | 0.05 | 1.5E−04 | 0.01 | 2.9E−05 | 0.017 | BD |
| rs12138864 | 1:33803148 | PHC2 | Intronic | T/C | 0.03 | 2.2E−04 | 0.01 | 1.7E−06 | 0.021 | BD |
| rs12096927 | 1:42841841 | RIMKLA (4.6) | Intergenic | T/C | 0.04 | 5.3E−04 | 0.01 | 5.8E−06 | 0.035 | BD |
| rs2367724 | 1:44107428 | KDM4A (8.4) | Intergenic | T/C | −0.04 | 2.6E−04 | −0.01 | 4.7E−04 | 0.040 | BD, SCZ, ADHD |
| rs1417364 | 1:52263082 | NRD1:RP4-657D16.3 | ncRNA | A/G | 0.06 | 3.7E−04 | 0.01 | 5.2E−04 | 0.043 | BD |
| rs182823 | 1:61680266 | NFIA | Intronic | T/C | −0.03 | 9.8E−04 | 0.01 | 3.6E−04 | 0.049 | BD |
| rs11210099 | 1:73429567 | RP4-660H19.1 (64.7) | Intergenic | T/C | 0.05 | 1.1E−06 | 0.01 | 2.0E−07 | 0.001 | BD, SCZ, ADHD |
| rs34194740 | 1:182658151 | RGS8 (4.4) | Intergenic | T/C | 0.04 | 2.6E−04 | 0.01 | 4.3E−04 | 0.037 | BD |
| rs823130 | 1:205714372 | NUCKS1 | Intronic | T/C | 0.04 | 9.2E−05 | 0.01 | 8.7E−05 | 0.014 | BD, SCZ |
| rs4146671 | 1:243505047 | SDCCAG8 | Intronic | A/G | 0.03 | 5.6E−04 | 0.01 | 1.1E−09 | 0.036 | BD, SCZ |
| rs11124327 | 2:22450730 | AC068490.2 | ncRNA | T/C | 0.03 | 2.0E−04 | 0.01 | 7.6E−06 | 0.020 | BD, SCZ, ADHD |
| rs35605321 | 2:27038638 | CENPA (14.7) | Intergenic | T/G | −0.03 | 7.3E−04 | −0.01 | 2.6E−04 | 0.041 | BD, SCZ |
| rs2194464 | 2:31328651 | GALNT14 | Intronic | T/C | −0.03 | 7.7E−04 | −0.01 | 2.8E−04 | 0.043 | BD |
| rs55951536 | 2:44770733 | CAMKMT | Intronic | T/C | 0.04 | 3.8E−04 | 0.01 | 6.2E−05 | 0.029 | BD |
| rs7591022 | 2:55151266 | EML6 | Intronic | T/C | 0.04 | 1.5E−04 | 0.01 | 8.2E−05 | 0.017 | BD |
| rs1433309 | 2:104059283 | AC092568.1 (51.4) | Intergenic | A/G | −0.03 | 5.7E−04 | −0.01 | 3.7E−04 | 0.039 | BD |
| rs73041394 | 2:185479385 | ZNF804A | Intronic | A/G | −0.04 | 5.6E−05 | −0.01 | 4.3E−05 | 0.010 | BD, SCZ |
| rs55811672 | 2:210269767 | MAP2 (19.0) | Intergenic | A/G | −0.04 | 1.7E−04 | −0.01 | 1.8E−04 | 0.022 | BD |
| rs2047134 | 2:225347713 | CUL3 | Intronic | A/C | 0.04 | 6.8E−05 | 0.01 | 4.1E−04 | 0.037 | BD, SCZ |
| rs1288974 | 3:71128028 | FOXP1 | Intronic | A/G | −0.04 | 6.9E−04 | 0.01 | 4.1E−04 | 0.044 | BD |
| rs9831123 | 3:85052150 | CADM2 | Intronic | T/C | 0.04 | 1.8E−06 | 0.01 | 8.0E−07 | 0.001 | BD, SCZ, ADHD |
| rs9681407 | 3:87210062 | MIR4795 (65.3) | Intergenic | T/C | −0.04 | 3.4E−04 | −0.01 | 8.5E−05 | 0.027 | BD |
| rs836927 | 3:107201428 | RP11-115H18.1 (18.7) | Intergenic | A/C | −0.04 | 9.9E−05 | 0.01 | 2.6E−04 | 0.027 | BD, SCZ |
| rs326359 | 3:107820619 | CD47 (10.8) | Intergenic | A/G | 0.05 | 4.0E−07 | 0.01 | 6.1E−06 | 0.003 | BD |
| rs12054405 | 3:139446347 | RP11-442N1.1 (38.2) | Intergenic | A/G | 0.05 | 5.6E−04 | −0.01 | 2.5E−04 | 0.036 | BD |
| rs359544 | 3:155268588 | PLCH1 | Intronic | T/C | 0.05 | 9.5E−05 | 0.01 | 2.6E−04 | 0.027 | BD |
| rs4350923 | 3:163721718 | RP11-208P4.1 (1.5) | Intergenic | T/C | −0.04 | 1.5E−04 | −0.01 | 9.0E−05 | 0.017 | BD |
| rs4434184 | 3:181422854 | SOX2-OT | ncRNA | A/G | −0.05 | 1.0E−04 | −0.02 | 1.4E−10 | 0.014 | BD |
| rs535066 | 4:46240287 | RP11-320H14.1 (5.3) | Intergenic | T/G | 0.05 | 1.7E−06 | 0.01 | 8.1E−07 | 0.001 | BD, SCZ |
| rs2647256 | 4:106201556 | TET2 (0.6) | Downstream | T/C | 0.05 | 6.1E−06 | 0.02 | 3.3E−10 | 0.003 | BD |
| rs11737121 | 4:147268639 | SLC10A7 | Intronic | A/G | −0.06 | 1.8E−05 | −0.01 | 2.3E−04 | 0.026 | BD |
| rs7696225 | 4:186782340 | SORBS2 | Intronic | A/C | −0.04 | 8.5E−04 | −0.01 | 7.9E−05 | 0.045 | BD |
| rs201587781 | 5:49478612 | EMB (213.4) | Intergenic | A/G | 0.09 | 7.8E−04 | 0.02 | 7.2E−05 | 0.043 | BD, SCZ |
| rs13163662 | 5:113756805 | KCNN2 | Intronic | A/G | 0.04 | 4.5E−05 | −0.01 | 4.1E−04 | 0.036 | BD |
| rs13169274 | 5:137855305 | ETF1 | Intronic | T/C | −0.04 | 2.0E−05 | −0.01 | 9.7E−06 | 0.006 | BD, SCZ |
| rs76157183 | 5:145833478 | TCERG1 | Intronic | T/C | 0.09 | 9.5E−06 | 0.02 | 5.1E−05 | 0.010 | BD |
| rs10053762 | 5:152276650 | AC091969.1 | ncRNA | A/C | −0.03 | 3.4E−04 | −0.01 | 2.2E−04 | 0.027 | BD, SCZ |
| rs2195450 | 5:152871009 | GRIA1 | Intronic | A/G | 0.04 | 7.9E−04 | 0.01 | 5.7E−06 | 0.043 | BD |
| rs10068495 | 5:158232847 | EBF1 | Intronic | A/G | −0.04 | 7.0E−04 | −0.01 | 1.8E−04 | 0.041 | BD |
| rs852944 | 6:72164390 | RP1-288M22.2 | ncRNA | T/C | 0.03 | 3.6E−04 | −0.01 | 2.7E−04 | 0.028 | BD |
| rs1487445 | 6:98565211 | RP11-436D23.1 | ncRNA | T/C | 0.07 | 1.5E−15 | 0.01 | 1.1E−05 | 0.004 | BD |
| rs7739294 | 6:117786542 | GOPC | Intronic | T/C | 0.04 | 3.3E−04 | 0.01 | 3.0E−04 | 0.030 | BD |
| rs6557271 | 6:153437735 | RGS17 | Intronic | T/C | 0.05 | 6.2E−06 | 0.01 | 2.4E−05 | 0.006 | BD |
| rs11768212 | 7:1992582 | MAD1L1 | Intronic | A/C | −0.04 | 1.6E−04 | −0.01 | 7.2E−05 | 0.017 | BD, ADHD |
| rs117450257 | 7:100446237 | SLC12A9:RP11-126L15.4 | ncRNA | A/G | −0.11 | 2.6E−06 | −0.02 | 2.1E−05 | 0.006 | BD, SCZ |
| rs2470943 | 7:104583843 | RP11-325F22.2 | ncRNA | A/G | 0.04 | 5.6E−05 | 0.01 | 4.4E−05 | 0.010 | BD, SCZ |
| rs10251192 | 7:115026459 | RP11-222O23.1 (117.0) | Intergenic | T/C | −0.05 | 1.2E−07 | −0.01 | 1.9E−07 | 0.000 | BD, SCZ |
| rs7785663 | 7:137070298 | DGKI | UTR3 | A/G | −0.04 | 2.4E−04 | −0.01 | 3.0E−05 | 0.022 | BD, SCZ |
| rs80274100 | 7:140122339 | RAB19 | Intronic | A/G | 0.04 | 1.5E−04 | 0.01 | 2.8E−04 | 0.029 | BD |
| rs2924726 | 8:4842010 | CSMD1 | Intronic | A/G | 0.04 | 1.6E−04 | 0.01 | 4.5E−04 | 0.039 | BD |
| rs10106054 | 8:16991254 | RP11-468H14.2 | ncRNA | A/G | −0.04 | 4.1E−04 | −0.01 | 5.1E−04 | 0.042 | BD |
| rs78035175 | 8:26093490 | RP11-98P2.1 (19.0) | Intergenic | A/G | 0.10 | 6.5E−05 | −0.02 | 7.4E−05 | 0.012 | BD |
| rs16883443 | 8:34884281 | AC098612.1 (56.4) | Intergenic | T/G | 0.03 | 2.5E−04 | 0.01 | 6.3E−04 | 0.048 | BD |
| rs11777067 | 8:38298647 | FGFR1 | Intronic | T/C | −0.04 | 1.2E−04 | −0.01 | 1.2E−04 | 0.017 | BD, SCZ |
| rs10957894 | 8:51291872 | SNTG1 | Intronic | A/G | −0.03 | 4.8E−04 | −0.01 | 1.8E−04 | 0.033 | BD |
| rs7813444 | 8:65437506 | RP11-21C4.4 (29.3) | Intergenic | A/G | 0.04 | 3.5E−05 | 0.01 | 8.5E−06 | 0.008 | BD, SCZ |
| rs4623479 | 8:93038708 | RUNX1T1 | Intronic | T/C | 0.04 | 1.6E−04 | 0.01 | 1.6E−04 | 0.020 | BD, ADHD |
| rs7011741 | 8:110812770 | RP11-25D10.2 (18.3) | Intergenic | A/G | 0.03 | 8.5E−04 | 0.01 | 6.7E−05 | 0.045 | BD |
| rs34853464 | 8:143363277 | TSNARE1 | Intronic | T/C | 0.04 | 1.6E−05 | 0.01 | 3.2E−07 | 0.005 | BD, SCZ |
| rs6474852 | 9:14738019 | FREM1 | Intronic | A/G | −0.04 | 2.6E−04 | −0.01 | 1.2E−04 | 0.023 | BD |
| rs10967586 | 9:26895808 | RN7SL100P | ncRNA | A/G | −0.06 | 9.4E−05 | −0.01 | 5.6E−04 | 0.044 | BD, SCZ |
| rs10821122 | 9:96157754 | RNU6-829P (19.8) | Intergenic | T/C | −0.04 | 1.7E−04 | −0.01 | 1.9E−04 | 0.022 | BD, SCZ |
| rs9888039 | 10:56657419 | PCDH15 | Intronic | T/C | −0.04 | 2.0E−04 | −0.01 | 2.8E−04 | 0.029 | BD |
| rs7085104 | 10:104628873 | C10orf32-ASMT | Intronic | A/G | 0.04 | 8.0E−05 | −0.01 | 5.2E−05 | 0.012 | BD, SCZ |
| rs12761679 | 10:106512727 | SORCS3 | Intronic | A/C | 0.05 | 1.5E−05 | 0.01 | 4.1E−06 | 0.005 | BD, SCZ, ADHD |
| rs12359871 | 10:127111493 | RPS27P18 (50.8) | Intergenic | T/C | −0.06 | 4.3E−04 | −0.01 | 4.5E−04 | 0.039 | BD |
| rs10082688 | 11:13266951 | ARNTL (31.3) | Intergenic | T/C | 0.04 | 1.9E−05 | 0.01 | 1.6E−05 | 0.005 | BD |
| rs11038655 | 11:45807100 | CTD-2210P24.4 (13.2) | Intergenic | T/C | −0.07 | 7.2E−05 | 0.02 | 3.0E−05 | 0.011 | BD |
| rs11227478 | 11:66173400 | RP11-867G23.10 (3.2) | Intergenic | A/G | −0.06 | 2.3E−06 | −0.01 | 1.3E−06 | 0.001 | BD |
| rs4988321 | 11:68174189 | LRP5 | Exonic | A/G | 0.08 | 2.2E−04 | −0.02 | 1.7E−04 | 0.021 | BD |
| rs10831015 | 11:88264454 | GRM5 | Intronic | A/C | 0.04 | 2.7E−04 | 0.01 | 7.2E−05 | 0.024 | BD |
| rs7932899 | 11:99152997 | CNTN5 | Intronic | A/G | 0.05 | 4.5E−05 | 0.01 | 1.5E−04 | 0.019 | BD |
| rs61909095 | 12:2301189 | CACNA1C | Intronic | T/C | −0.08 | 9.4E−15 | −0.01 | 1.6E−06 | 0.001 | BD, SCZ |
| rs10842271 | 12:24233843 | SOX5 (129.9) | Intergenic | T/C | −0.03 | 4.1E−04 | −0.01 | 3.4E−04 | 0.033 | BD |
| rs7959452 | 12:69735492 | LYZ (6.6) | Intergenic | A/G | 0.03 | 2.2E−04 | 0.01 | 6.2E−04 | 0.047 | BD |
| rs11178282 | 12:70941195 | PTPRB | Intronic | T/C | 0.05 | 7.7E−04 | 0.01 | 4.0E−04 | 0.046 | BD |
| rs3764002 | 12:108618630 | WSCD2 | Exonic | T/C | −0.04 | 3.3E−04 | −0.01 | 9.7E−05 | 0.027 | BD, ADHD |
| rs3885907 | 13:31314455 | ALOX5AP | Intronic | A/C | −0.04 | 2.3E−05 | −0.01 | 6.7E−06 | 0.006 | BD |
| rs7139704 | 13:47726258 | GNG5P5 (155.0) | Intergenic | A/G | 0.04 | 2.5E−04 | 0.01 | 1.5E−04 | 0.023 | BD |
| rs34012672 | 14:33394294 | NPAS3 (9.8) | Intergenic | T/C | −0.06 | 1.3E−05 | −0.01 | 1.3E−05 | 0.004 | BD, SCZ, ADHD |
| rs3007061 | 14:47238606 | MDGA2 (70.2) | Intergenic | T/C | −0.04 | 2.5E−04 | −0.01 | 2.3E−04 | 0.026 | BD, ADHD |
| rs8005321 | 14:62458832 | SYT16 | Intronic | T/G | −0.04 | 2.1E−05 | −0.01 | 4.2E−05 | 0.009 | BD |
| rs72703614 | 14:89860386 | FOXN3 | Intronic | A/G | −0.04 | 1.1E−04 | −0.01 | 3.0E−05 | 0.014 | BD |
| rs12892189 | 14:104319989 | LINC00637 | ncRNA | A/C | 0.05 | 8.2E−07 | 0.01 | 2.4E−07 | 0.001 | BD, SCZ |
| rs4924676 | 15:42710619 | ZNF106 | Intronic | T/C | 0.07 | 2.0E−05 | 0.02 | 2.9E−05 | 0.007 | BD, SCZ |
| rs4327001 | 15:74010301 | CD276 (3.4) | Intergenic | A/G | 0.04 | 4.5E−05 | 0.01 | 5.4E−05 | 0.010 | BD |
| rs12442456 | 15:78751962 | IREB2 | Intronic | T/G | −0.05 | 2.7E−04 | 0.01 | 2.5E−05 | 0.024 | BD, SCZ |
| rs2071382 | 15:91428197 | FES | Intronic | T/C | −0.05 | 4.2E−08 | −0.01 | 5.6E−07 | 0.000 | BD, SCZ |
| rs6500948 | 16:7371458 | RBFOX1 | Intronic | A/G | −0.04 | 4.0E−04 | −0.01 | 1.3E−04 | 0.030 | BD, SCZ |
| rs2352759 | 16:10215483 | GRIN2A | Intronic | T/C | 0.03 | 3.3E−04 | 0.01 | 4.8E−05 | 0.027 | BD |
| rs62029337 | 16:23820102 | PRKCB (27.2) | Intergenic | T/C | 0.06 | 5.4E−05 | 0.01 | 3.1E−04 | 0.031 | BD |
| rs55910718 | 16:58366306 | GINS3 (60.0) | Intergenic | T/C | 0.04 | 8.9E−04 | 0.01 | 1.6E−04 | 0.046 | BD |
| rs7219635 | 17:1270427 | YWHAE | Intronic | T/C | −0.04 | 6.4E−06 | −0.01 | 7.4E−05 | 0.012 | BD, SCZ |
| rs112562460 | 17:61377135 | TANC2:AC037445.1 | ncRNA | T/C | 0.04 | 6.5E−04 | 0.01 | 1.8E−04 | 0.039 | BD |
| rs9636107 | 18:53200117 | TCF4 | Intronic | A/G | −0.03 | 3.5E−04 | −0.01 | 1.1E−07 | 0.028 | BD, SCZ |
| rs12928 | 18:77663863 | PQLC1 | UTR3 | A/G | −0.03 | 5.5E−04 | −0.01 | 3.0E−04 | 0.036 | BD, SCZ |
| rs1736182 | 19:2798686 | THOP1 | Intronic | T/G | −0.04 | 2.8E−05 | 0.01 | 1.4E−04 | 0.018 | BD |
| rs2304204 | 19:50169020 | IRF3:BCL2L12 | UTR5 | T/C | 0.04 | 7.0E−04 | 0.01 | 1.4E−04 | 0.041 | BD, SCZ |
| rs1291112 | 20:35500151 | RN7SL156P (1.4) | Intergenic | T/C | −0.06 | 1.0E−04 | −0.01 | 3.0E−04 | 0.030 | BD |
| rs12624433 | 20:44680853 | SLC12A5 | Intronic | A/G | 0.05 | 3.6E−06 | 0.01 | 2.5E−04 | 0.027 | BD |
| rs404060 | 22:20164360 | XXbac-B444P24.8 (21.9) | Intergenic | T/C | 0.03 | 6.1E−04 | −0.01 | 3.0E−04 | 0.038 | BD |
| rs13055562 | 22:51133518 | SHANK3 | Intronic | A/G | 0.04 | 8.4E−05 | 0.01 | 3.9E−04 | 0.036 | BD |
The table reports 102 linkage disequilibrium independent genomic loci associated with bipolar disorder and risk-taking propensity at a conjFDR < 0.05. Position denotes the chromosome and location of the lead SNP based on the hg19 assembly. Nearest gene and functional category have been annotated using FUMA (for SNPs located in intergenic regions, distance in Kb from the nearest gene is reported). Beta BD and beta risk show the direction of effect of the A1 allele in the original BD and risk-taking propensity GWAS datasets. The last column shows whether the genomic locus has been found to be specific for BD or whether it was also detected in the analyses conducted between risk-taking and either SCZ or ADHD at a conjFDR < 0.05 (see Supplementary Table 2). The Functional characterization of all loci is reported in Supplementary Table 1.
Among the 62 loci specifically shared between BD and risk-taking, 20 SNPs were found to significantly affect gene expression in at least one brain region, whole blood or both (Supplementary Table 1). Sixteen variants were located in (FNDC5, NRD1:RP4-657D16.3, FOXP1, PLCH1, SOX2-OT, KCNN2, EBF1, FREM1, GRM5, PTPRB, GRIN2A, THOP1, and SLC12A5) or near (CD47, PRKCB and CD276) genes part of the druggable genome or clinically actionable, and 17 in or near genes showing drug-gene interactions in DGIdb (Supplementary Table 1).
We identified 128 independent genomic loci associated with BD after conditioning on risk-taking propensity at a condFDR < 0.01 (Supplementary Table 3). Among these, 45 loci (35%) were specific for BD, while 83 loci were also associated with SCZ (n = 76), or SCZ and ADHD (n = 7) after conditioning on risk-taking propensity (Supplementary Tables 2 and 4). Among loci specific for BD, 15 are novel and are reported in Table 2. Eight SNPs were found to act as eQTLs in brain regions, whole blood or both (Table 2). Three variants were located in genes part of the druggable genome (HTR6, CAPN10:GPR35 and ATP2B2), and 4 in or near genes showing drug-gene interactions in DGIdb (Supplementary Table 3).
Table 2.
Novel genomic loci associated bipolar disorder at a condFDR < 0.01 conditioning on association with risk-taking propensity.
| SNP | Position | Nearest gene (Kb) | Functional category | A1/A2 | Beta BD | p BD | condFDR BD|risk | RegDB rank | cadd score | eQTL | Gene | Region |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10917509 | 1:19992066 | HTR6 | UTR5 | T/C | 0.05 | 7.3E−08 | 5.2E−03 | 2b | 7.91 | Yes |
HTR6 NBL1 |
Nucleus accumbens, whole blood Nucleus accumbens |
| rs483252 | 1:108510358 | VAV3-AS1 | ncRNA | A/C | −0.06 | 9.9E−07 | 7.3E−03 | 3a | 5.74 | No | – | – |
| rs67050019 | 1:154267953 | RNU6-239P (0.03) | Upstream | A/G | 0.08 | 3.7E−07 | 4.5E−03 | 5 | 6.76 | No | – | – |
| rs4915346 | 1:199122192 | RP11-382E9.1 | ncRNA | A/G | −0.05 | 1.3E−05 | 6.6E−03 | 5 | 11.13 | No | – | – |
| rs4972439 | 2:175261443 | SCRN3 | Intronic | T/C | 0.06 | 4.3E−07 | 8.0E−03 | 4 | 1.50 | Yes | SCRN3 | Whole blood |
| rs6433891 | 2:181969709 | AC068196.1:AC104820.2 | ncRNA | A/G | 0.05 | 1.6E−07 | 3.1E−03 | 4 | 13.00 | Yes | UBE2E3 | Cerebellum, whole blood |
| rs4676412 | 2:241553492 | CAPN10:GPR35 | Intronic | A/G | 0.06 | 7.6E−08 | 7.8E−03 | 4 | 0.11 | Yes | ANKMY1 | Cortex |
| rs7644022 | 3:10510618 | ATP2B2 | Intronic | A/C | 0.05 | 2.5E−07 | 6.2E−03 | 5 | 3.51 | No | – | – |
| rs78104110 | 3:114133266 | ZBTB20 | Intronic | T/C | −0.13 | 2.1E−07 | 1.4E−03 | 5 | 5.22 | No | – | – |
| rs2651566 | 4:101507968 | EMCN (68.1) | Intergenic | A/G | −0.04 | 4.3E−06 | 9.0E−03 | 7 | 2.23 | No | – | – |
| rs11770210 | 7:29993998 | SCRN1 | Intronic | A/G | 0.07 | 6.4E−08 | 8.6E−03 | 5 | 1.14 | Yes |
PLEKHA8 AC007285.6 WIPF3 SCRN1 |
Cerebellum, whole blood Cerebellum Cortex Whole blood |
| rs10869262 | 9:76102010 | RP11-404E6.1 (11.0) | Intergenic | A/G | 0.05 | 7.8E−08 | 9.0E−03 | 7 | 1.19 | Yes |
ANXA1 ALDH1A1 |
Whole blood Whole blood |
| rs75888683 | 12:79478068 | SYT1 | Intronic | T/G | 0.12 | 1.1E−07 | 6.3E−03 | 6 | 3.32 | Yes | SYT1 | Spinal cord |
| rs8043792 | 16:61660554 | CDH8 (20.6) | Intergenic | T/C | −0.04 | 2.0E−06 | 3.3E−03 | 6 | 4.88 | No | – | – |
| rs4788865 | 17:73115508 | ARMC7 | Intronic | T/G | −0.05 | 2.3E−07 | 6.8E−03 | 4 | 3.35 | Yes |
NUP85 NTC5 ITGB4 MRPS7 SUMO2 GGA3 |
Amygdala, anterior cingulate, caudate, cerebellum, cortex, hippocampus, hypothalamus, whole blood Nucleus accumbens, putamen, spinal cord hypothalamus Putamen Whole blood Whole blood Whole blood |
Novel independent genomic loci associated with bipolar disorder conditioning on risk-taking propensity at a condFDR < 0.01, after exclusion of loci associated with either SCZ (Supplementary Table 5) or ADHD (Supplementary Table 6) after conditioning on risk-taking propensity. The full list of significant loci is reported in Supplementary Table 3. Nearest gene and functional category have been annotated using FUMA (for SNPs located in intergenic regions, distance in Kb from the nearest gene is reported). Beta and p columns show the direction of effect of the A1 allele and p values from the original GWAS dataset. The RegBD rank (from 1 to 7, with 1 being associated with highest evidence of functional effects) was calculated using RegulomeDB based on known and predicted regulatory elements. The CADD score, which predicts how deleterious a variant is on protein structure/function was computed in FUMA. Higher scores indicate more deleterious SNPs, with a suggested threshold of 12.37 for a SNP to be considered deleterious. In case the SNP is reported to be a significant eQTL in GTEx v.8 in brain regions or whole blood, the last two columns report regulated genes and relative regions.
Similarly, we identified significant enrichment in variants associated with risk-taking propensity when conditioning on BD (Supplementary Fig. 1). We reported 79 independent genomic loci associated with risk-taking propensity at a condFDR < 0.01 (Supplementary Table 5). Among these, 26 loci (33%) were specific for risk-taking after conditioning on BD, while 53 were also associated with risk-taking propensity after conditioning on SCZ (n = 18), ADHD (n = 4), or both SCZ and ADHD (n = 31) (Supplementary Table 6). Of the 26 specific loci, 22 are novel and are reported in Table 3. Eight SNPs were found to act as eQTLs in brain regions or both brain tissues and whole blood (Table 3). Three variants are located in genes part of the druggable genome (GRIA1, SLC12A9, and GRM5) and six in genes showing drug-gene interactions in DGIdb (Supplementary Table 5).
Table 3.
Novel genomic loci associated with risk-taking propensity at a conditional false discovery rate <0.01 conditioning on association with bipolar disorder.
| SNP | Position | Nearest gene | Functional category | A1/A2 | beta risk | p risk | condFDR risk|BD | RegDB rank | CADD score | eQTL | Gene | Region |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1868402 | 2:68409037 | RP11-474G23.1:PPP3R1 | Intronic | A/G | −0.01 | 8.3E−07 | 3.6E−03 | 1f | 0.15 | Yes |
PNO1 PLEK |
Anterior cingulate, caudate, cerebellum Cerebellum |
| rs545200731 | 2:147644489 | AC062032.1 | ncRNA | T/C | 0.04 | 5.3E−06 | 5.8E−03 | 6 | 1.08 | No | – | – |
| rs34288552 | 2:171661486 | ERICH2 (6.0) | Intergenic | A/G | 0.01 | 2.0E−07 | 5.8E−03 | 5 | 7.33 | Yes |
ERICH2 |
Amygdala, anterior cingulate, cerebellum, cortex, frontal cortex, hypothalamus, susbtantia nigra Anterior cingulate, caudate |
| rs1014959 | 2:185472113 | ZNF804A | Intronic | A/G | −0.01 | 4.0E−05 | 8.3E−03 | 7 | 0.50 | No | – | – |
| rs326353 | 3:107853648 | RP11-861A13.4 | ncRNA | T/C | −0.01 | 6.0E−06 | 2.7E−03 | 3a | 3.28 | Yes |
IFT57 HHLA2 CD47 |
Whole blood Whole blood Whole blood |
| rs7628391 | 3:163680497 | RP11-208P4.1 (38.9) | Intergenic | T/C | 0.01 | 5.8E−06 | 4.0E−03 | 5 | 0.33 | No | – | – |
| rs4696294 | 4:152713089 | RP11-424M21.1 (7.6) | Intergenic | A/C | −0.01 | 2.9E−06 | 9.6E−03 | 6 | 0.02 | Yes |
SH3D19 RP11-164P12.5 FAM160A1 GATB RP11-164P12.3 FAM160A1 |
Caudate, cerebellum Cerebellum, whole blood Cerebellum, whole blood Cerebellum, cortex, frontal cortex, whole blood Cerebellum Cerebellum |
| rs76157183 | 5:145833478 | TCERG1 | Intronic | T/C | 0.02 | 5.1E−05 | 9.6E−03 | 5 | 0.08 | No | – | – |
| rs2195450 | 5:152871009 | GRIA1 | Intronic | A/G | 0.01 | 5.7E−06 | 3.8E−03 | 4 | 16.32 | No | – | – |
| rs852960 | 6:72205635 | RP1-288M22.2 (37.1) | Intergenic | A/G | 0.01 | 1.6E−05 | 5.7E−03 | 5 | 5.28 | No | OGFRL1 | Cerebellum |
| rs7758002 | 6:153440770 | RGS17 | Intronic | T/G | −0.01 | 7.1E−07 | 1.3E−03 | 7 | 1.06 | Yes |
MTRF1L RGS17 |
Anterior cingulate, cerebellum, whole blood Cerebellum |
| rs117450257 | 7:100446237 | SLC12A9:RP11-126L15.4 | ncRNA | A/G | −0.02 | 2.1E−05 | 5.6E−03 | 5 | 0.14 | No | – | – |
| rs80206917 | 7:140159389 | MKRN1 | Intronic | T/C | 0.01 | 2.3E−05 | 6.5E−03 | 2b | 5.92 | No | – | – |
| rs17055053 | 8:26088094 | RP11-98P2.1 (24.3) | Intergenic | T/C | −0.02 | 5.1E−05 | 9.5E−03 | 4 | 14.95 | No | – | – |
| rs7871821 | 9:128992756 | RP11-343J18.1 (39.6) | Intergenic | T/C | 0.01 | 5.5E−06 | 6.9E−03 | 5 | 1.18 | Yes | PBX3 | Cortex |
| rs7111300 | 11:45806624 | CTD-2210P24.4 (12.7) | Intergenic | T/G | 0.02 | 3.0E−05 | 7.0E−03 | 5 | 1.34 | Yes | CTD-2210P24.4 | Caudate, putamen |
| rs11827676 | 11:88263465 | GRM5 | Intronic | A/C | 0.01 | 3.7E−05 | 8.0E−03 | 6 | 3.66 | No | – | – |
| rs3885907 | 13:31314455 | ALOX5AP | Intronic | A/C | −0.01 | 6.7E−06 | 2.9E−03 | 4 | 2.17 | Yes | ALOX5AP | Whole blood |
| rs8005321 | 14:62458832 | SYT16 | Intronic | T/G | −0.01 | 4.2E−05 | 8.6E−03 | 4 | 0.33 | No | – | – |
| rs12927162 | 16:52684916 | CASC16 | ncRNA | A/G | −0.01 | 1.9E−06 | 4.9E−03 | 5 | 21.80 | No | – | – |
| rs72841389 | 17:61437939 | TANC2 | Intronic | A/G | 0.01 | 5.4E−06 | 4.7E−03 | 7 | 4.83 | Yes |
CYB561 TANC2 |
Anterior cingulate, caudate, cortex, frontal cortex, hippocampus, nucleus accumbens, putamen Cerebellum |
| rs6017733 | 20:44712815 | NCOA5 | Intronic | A/G | −0.01 | 2.2E−06 | 5.7E−03 | 6 | 6.25 | Yes | CD40 | Cerebellum |
The Table reports 22 novel independent genomic loci associated with risk-taking propensity conditioning on bipolar disorder at a condFDR < 0.01, after excluding loci associated with risk-taking propensity conditioning on SCZ or ADHD (Supplementary Table 6). The full list of significant loci is reported in Supplementary Table 5. Nearest gene and functional category have been annotated using FUMA (for SNPs located in intergenic regions, distance in Kb from the nearest gene is reported). Beta and p show the direction of effect of the A1 allele and p values from the original GWAS dataset. RegBD rank (from 1 to 7, with 1 being associated with highest evidence of functional effects) was calculated using RegulomeDB based on known and predicted regulatory elements. The CADD score, which predicts how deleterious a variant is on protein structure/function was computed in FUMA. Higher scores indicate more deleterious SNPs, with a suggested threshold of 12.37 for a SNP to be considered deleterious. In case the SNP is reported to be a significant eQTL in GTEx v.8 in brain regions or whole blood, the last two columns report regulated genes and relative regions.
Functional enrichment of genes associated with BD and risk-taking propensity
We next evaluated functional enrichment for KEGG pathways and GO terms for genes in which variants jointly associated with BD and risk-taking propensity were located. Genes in which variants associated with risk-taking propensity and specifically BD were located were enriched for two KEGG pathways (“Glutamatergic synapse” and “Long-term potentiation”), the biological process “Glutamate receptor signaling pathway” GO term and four cellular component GO terms (“Postsynaptic specialization”, “Neuron to neuron synapse”, “Synaptic membrane”, “Neuron spine”) (Supplementary Table 7). In addition, genes in which variants specifically shared between BD and risk-taking were located showed a significant enrichment for drug targets with different clinical indications, including disorders related to the central nervous system (i.e. “mood disorders”, “inflammatory disorders of the central nervous system” and “other degenerative disorders of the nervous system”, Supplementary Table 8).
Using IPA, we identified 161 significant upstream regulators of genes in which variants associated with BD and risk-taking were located (including cross-disorder genes) (Supplementary Table 9). Supplementary Fig. 2 shows a network of these genes including upstream regulators classified as “drugs”. Upstream regulators of genes associated with BD and risk-taking included the antipsychotics haloperidol and flupentixol, as well as psychoactive substances such as cocaine, delta-9-tetrahydrocannabinol and nicotine (Supplementary Fig. 2 and Supplementary Table 9). Several upstream regulators were found to be significantly associated with more than one gene in the network. Supplementary Table 10 shows the list of 134 significant upstream regulators of genes in which variants specifically associated with BD and risk-taking propensity were located.
Local genetic correlation analysis using SUPERGNOVA
Using a complementary approach, we applied SUPERGNOVA to estimate local genetic covariance between BD and risk-taking propensity in specific genomic regions. Seventeen genomic regions characterized by a significant genetic covariance between BD and risk-taking propensity at an FDR < 0.05 are reported in Supplementary Table 11. Fifteen of these regions (88%) were overlapping with genomic loci identified with pleioFDR (Table 1). Ten regions were specifically shared between risk-taking propensity and BD, while seven also showed a significant genetic covariance between risk-taking propensity and SCZ (n = 4), ADHD (n = 2), or both disorders (n = 1) (Supplementary Tables 11 and 12).
eQTL-informed gene-based analysis of variants associated with BD and risk-taking propensity
The eQTL-informed gene-based analysis showed 103 significant eGenes associated with BD in at least one brain region, 74 in whole blood, and 32 in brain and whole blood (Supplementary Table 13). Supplementary Fig. 3 shows levels of expression of these genes in brain regions and whole blood based on GTEx. A total of 28 eGenes were significantly associated with risk-taking propensity in at least one brain region, 14 in whole blood, and 6 in both brain tissues and whole blood (Supplementary Table 14). Levels of expression of these genes in brain regions and whole blood based on GTEx are shown in Supplementary Fig. 4.
CACNA1C was significantly associated with both BD and risk-taking propensity in the cerebellar hemisphere (BD, Z = 6.48, p = 3.8E−14, adj p = 7.4E−10; risk-taking, Z = 4.66, p = 1.6E−06, adj p = 0.033). Notably, a genomic locus located in CACNA1C was also significantly associated with both phenotypes in the conjFDR analysis and in the local genetic covariance analysis, as well as with risk-taking and SCZ (Table 1 and Supplementary Table 9). This gene is part of the druggable genome and a known target of calcium channel blockers (Supplementary Table 1).
Two other genes significantly associated with both BD and risk-taking in the conjFDR analysis were also associated with BD in the gene-based analysis: SLC12A5 in caudate and hippocampus (most significant: hippocampus, Z = 4.87, adj p = 0.11) and FES in whole blood (Z = 4.57, adj p = 0.048) (Supplementary Table 13). In addition, three other genes significantly associated with both BD and risk-taking in the conjFDR analysis were also significantly associated with risk-taking in the eQTL-informed gene-based analysis: CADM2 in several brain regions with the most significant being hypothalamus (Z = 6.26, adj p = 3.8E−06), RGS17 in cerebellum (Z = 4.88, adj p = 0.01) and SDCCAG8 in different brain regions (most significant: cortex, Z = 6.15, adj p = 7.7E−06) and whole blood (Z = 6.27, adj p = 3.6E−06) (Supplementary Table 14).
Discussion
This study identified novel genetic variants associated with BD and risk-taking propensity as well as genetic loci shared between these two phenotypes using the condFDR/conjFDR method on large GWAS summary statistics. Notably, the large majority of the identified shared loci (89 out of 102, 87%) showed the same direction of effect, supporting previous evidence suggesting positive genetic correlation between these two phenotypes [33]. Among these loci, 62 (61%) were specifically shared between risk-taking propensity and BD, while the others were also shared between risk-taking and SCZ or ADHD, two other traits previously shown to be genetically correlated with risk-taking [33]. Using complementary approaches, we analyzed local genetic covariance between BD and risk-taking as well as identified tissue-specific genes associated with the two traits through an eQTL-informed gene-based analysis in brain regions and whole blood. The CACNA1C gene at chromosome 12 was the only locus significantly associated with both BD and risk-taking propensity using all analytic approaches. This gene is part of the druggable genome and is a target of calcium channel blockers, a group of medications widely used for different cardiovascular indications such as hypertension and angina pectoris [66]. The locus at CACNA1C was also found to be shared between risk-taking propensity and SCZ, but not ADHD (Table 1). Indeed, besides being previously associated with BD [32], as well as with alcohol dependence in patients with BD [67], CACNA1C has been consistently implicated in different psychiatric disorders such as SCZ [68] and obsessive-compulsive disorder [69], supporting its putative role of a cross-disorder susceptibility gene [70, 71] via modulation of stress-coping behavior [72] and gene-environment interactions in response to adverse life events [73, 74]. The identification of genomic loci shared between BD and risk-taking may have important clinical implications. Namely, patients with BD and increased risk-taking propensity may represent a specific sub-phenotype and benefit of more tailored treatment approaches. CACNA1C, the most robust locus identified in our study, encodes CaV1.2α, the alpha-1 subunit of a voltage-dependent L-type calcium channel [75], which forms the pore through which ions pass into the cell. Therefore, it might be speculated that patients with BD and increased risk-taking propensity might show better response to drugs acting on calcium signaling, such as calcium channel blockers. Increased intracellular calcium ion concentration, with or without stimulation by agonists such as thrombin or serotonin, has been reported in peripheral cells from patients with BD [76] and shown to be normalized by in vitro treatment with lithium [77] or carbamazepine [78]. Recent studies also showed dysregulation in calcium signaling in hippocampal dentate gyrus-like neurons derived from induced pluripotent stem-cells of patients with BD compared with controls [79]. The hyperexcitability phenotype of these neurons was selectively reversed by lithium treatment only in neurons derived from patients who also responded to lithium treatment [79]. In addition, CACNA1C was part of a glutamatergic network suggested to mediate lithium response in a recent epigenome pathway analysis [80] and knockdown of this gene in fibroblasts from patients with BD was found to alter circadian rhythm amplitude and eliminate lithium’s ability to amplify rhythms [81].
Calcium channel blockers have been previously used in BD without clear results. Verapamil has been the first calcium channel blocker suggested to be useful in the treatment of mania either alone or in combination with lithium [82–84]. While a recent meta-analysis did not support clinical efficacy of verapamil in mania, the small number of studies and the lack of high-quality data from randomized trials do not allow to draw definitive conclusions [85]. Similarly, only few studies including scarce numbers of participants investigated the potential clinical utility of nimodipine and isradipine [86, 87]. In the last few years, the interest in therapies targeting calcium channels has grown considerably [85] and new studies are currently being designed to clarify the potential for drug repurposing [88]. Interesting evidence also comes from a recent population study including 142,691 participants from Sweden with a diagnosis of BD, SCZ or nonaffective psychosis [89]. The study found periods of exposure to L-type calcium channel blockers to be associated with reduced rates of psychiatric hospitalization and self-harm in all phenotypes [89]. A recent double-blind pharmacoMRI study recruited healthy men genotyped for the CACNA1C rs1006737 variant who were randomized to a single 60 mg dose of the brain-permeable calcium channel blocker nimodipine or placebo [90]. Participants treated with nimodipine showed decreased frontal cortical and parietal cortical activity and improved working memory performance during the N-back task, with the decrease in frontal cortical activity being more pronounced in carriers of the rs1006737 risk allele [90]. Calcium channel blockers have also started to be studied as potential adjunctive treatment options for cognitive impairment in patients with SCZ. In a recent double-blind, randomized, placebo-controlled trial, patients with SCZ randomized to 5 mg of the nonselective brain-permeable calcium channel blocker isradipine for 6 weeks showed improvement in verbal memory and attention dysfunction measured with the Stroop test compared with patients randomized to placebo [91]. As genetic variants may affect CaV1.2α expression and activity [92], an ongoing trial will evaluate whether CACNA1C polymorphisms and gene expression may affect response to calcium channel blockers in young adults with mood instability [88]. This trial might provide elements to support or confute the hypothesis that participants might show differential response to calcium channel blockers based on their level of risk-taking propensity, as participants will undergo cognitive testing aimed at assessing impulsivity, behavioral inhibition and reward learning [88].
While we found a relevant overlap between genomic loci shared between risk-taking propensity and the three psychiatric disorders we investigated, around half of the loci were specifically shared between risk-taking propensity and BD (Table 1). Similar to cross-disorder genes, these loci were enriched for KEGG pathways related to postsynaptic specialization and long-term potentiation, with GO terms related to postsynaptic specialization and synapses (Supplementary Table 7) as well as for targets of drugs used for mood disorders, substance use disorders and other disorders of the central nervous system (Supplementary Table 8). Among loci specifically shared between risk-taking propensity and BD, the one located in the FNDC5 gene is particularly interesting as the Irisin myokine derived from the transmembrane protein FNDC5, stimulated by physical exercise, has been suggested to mediate antidepressant effects of physical activity and ameliorate cognition impairments in neurodegenerative diseases [93, 94].
Besides the identification of loci jointly associated with BD and risk-taking propensity, another aim of our study was to identify novel loci associated with these phenotypes. By leveraging pleiotropic enrichment between these two traits, we identified 128 loci associated with BD after conditioning on risk-taking propensity, including 15 loci which were novel and specific for BD, and 79 associated with risk-taking propensity conditioning on BD, including 22 novel loci identified when conditioning specifically on BD and not SCZ or ADHD. One novel locus specifically associated with BD was rs10917509 in the UTR5 region of HTR6, which encodes a G protein-coupled serotonin receptor and is a druggable gene as well as a known target of different antipsychotics and antidepressants (Supplementary Table 3). Another locus with rs75888683 as the lead SNP is located in the SYT1 gene which encodes Synaptotagmin-1, a synaptic vesicle membrane protein that serves as a calcium sensor and is implicated in vesicular trafficking and exocytosis [95]. In mouse cortical neurons, a breakpoint mutant version of the DISC1 protein (which is encoded by a known susceptibility gene for BD and SCZ) was shown to disrupt vesicle transport via defective assembly between the kinesin-1 adapter FEZ1 and the cargo protein Synaptotagmin-1 [96], with this effect found to be rescued by in vitro treatment with the mood stabilizer lithium 2 mM for 48 h [96]. Three novel loci specifically associated with BD, four with risk-taking propensity and six with both phenotypes (Tables 1–3) were located in noncoding RNAs. LncRNAs have been increasingly implicated in psychiatric disorders [97] and related phenotypes, such as lithium response [98], highlighting the need to improve our understanding of the role of these molecules in brain processes.
Among loci associated with risk-taking propensity, only 33% were identified when conditioning specifically on BD, while a relevant number of loci (39%) were associated with risk-taking when conditioning on all investigated psychiatric disorders and therefore show extensive pleiotropic profiles. Some of these loci might be worth of investigation as potential therapeutic targets as they were found to be located in genes part of the druggable genome (e.g CGREF1, which has been recently associated with non-response to antidepressants [99], or TET2 which has been implicated in the potential antidepressants effect of metformin in a recent preclinical study [100]). Another interesting locus associated with risk-taking propensity when conditioning on all psychiatric phenotypes was found in the NPAS3 gene, which encodes a transcription factor regulating genes involved in key neuronal processes such as postnatal hippocampal neurogenesis [101]. NPAS3 was first identified as a candidate risk gene for psychiatric disorders through the study of a balanced chromosomal translocation, t(9,14)(q34.2;q13), associated with SCZ and learning disability [102, 103]. Interactions between risk and protective haplotypes at this gene have been suggested to contribute to susceptibility to both SCZ and BD [104] and variants located in this gene have been consistently associated with BD at a nominal p < 0.05 in different GWAS [105]. In addition, a group of SNPs in the first intron (top SNP = rs4982029, p = 3.96E−06) showed pleiotropic effects on BD, SCZ and major depressive disorder [106]. Besides reduced adult hippocampal neurogenesis [107], mice deficient for NPAS3 also show abnormalities in glutamate, dopamine and serotonin neurotransmitter signaling [108]. All these effects might underlie the association we observed between NPAS3 and risk-taking propensity, as this trait has been found to be positively associated with hippocampal glutamate [23, 25] and monoamine levels [24] and negatively associated with gray matter volume in the amygdala and hippocampus [22].
Results from this study have to be interpreted in light of a number of limitations. As in other studies conducted using GWAS, it cannot be excluded that the identified lead SNPs may be in LD with other causal SNPs. No assessment of risk-taking propensity in participants included in the BD GWAS was conducted. Identified SNPs only explained a small proportion of variance of the BD phenotype. Additionally, a cohort from UK Biobank is included in the PGC BD freeze 3 cohort. While we used functions implemented in pleioFDR and SUPERGNOVA to adjust for sample overlap, we cannot exclude that overlapping participants may inflate the cross-trait enrichment statistics. Finally, as our investigation of the potential functional role of identified variants was largely based on in-silico methods, functional characterization based on independent experimental data, as well replication of the loci in independent cohorts, is needed to confirm and further assess the putative role of these variants in the shared polygenic architecture between BD and risk-taking propensity.
In conclusion, we observed pleiotropic enrichment between BD and risk-taking propensity and identified 102 loci shared between these two phenotypes, 87% of which showed the same direction of effect and 61% of which were specifically shared between the two traits. We leveraged pleiotropic enrichment to identify 15 novel loci specifically associated with BD and 22 with risk-taking propensity. Our findings dissect for the first-time genetic factors shared between risk-taking propensity and BD and lay the basis for future investigation of treatment approaches targeting molecular mechanisms involved in both traits.
Funding and disclosure
This study was supported with a grant funded by Fondazione di Sardegna, Call 2019, Project ID: F72F20000260007 and by a grant funded by “Regione Autonoma della Sardegna”, Fondo per lo Sviluppo e la Coesione 2014-2020, 2017 call, ID: F76C18000850002. CP gratefully acknowledges Sardinian Regional Government for the financial support of her PhD scholarship (P.O.R. Sardegna F.S.E.—Operational Programme of the Autonomous Region of Sardinia, European Social Fund 2014-2020—Axis III Education and training, Thematic goal 10, Investment Priority 10ii), Specific goal 10.5. The funders had no further role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication. BTB has consulting roles with the National Health and Medical Research Council, Australia. He has received honoraria from AstraZeneca, Bristol-Myers Squibb, Lundbeck, Pfizer, Servier, Wyeth, Otsuka; Research or grants from private industries or non-profit funds from AstraZeneca, Sanofi-Synthélabo; and research grants from the National Health and Medical Research Council, Australia. He has served on advisory boards for Janssen-Cilag, Lundbeck and received funds from the Fay Fuller Foundation, and James & Diana Ramsay Foundation, Adelaide. The other authors have nothing to disclose.
Supplementary information
Acknowledgements
We thank the Psychiatric Genomics Consortium and the Social Science Genetic Association Consortium for access to the GWAS data.
Author contributions
CP designed the study, undertook the statistical analysis and wrote the first draft of the manuscript. DC, GS, RA, CC and MDZ contributed drafting the manuscript. BTB and AS contributed designing the study, revising and finalizing the manuscript. All authors contributed to and have approved the final manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41386-021-01045-y.
References
- 1.Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet. 2016;387:1561–72. doi: 10.1016/S0140-6736(15)00241-X. [DOI] [PubMed] [Google Scholar]
- 2.McIntyre RS, Berk M, Brietzke E, Goldstein BI, Lopez-Jaramillo C, Kessing LV, et al. Bipolar disorders. Lancet. 2020;396:1841–56. doi: 10.1016/S0140-6736(20)31544-0. [DOI] [PubMed] [Google Scholar]
- 3.Cloutier M, Greene M, Guerin A, Touya M, Wu E. The economic burden of bipolar I disorder in the United States in 2015. J Affect Disord. 2018;226:45–51. doi: 10.1016/j.jad.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarty T, Torres IJ, Su WW, Sawatzky R, Keramatian K, Yatham LN. Cognitive subgroups in first episode bipolar I disorder: Relation to clinical and brain volumetric variables. Acta Psychiatr Scand. 2021;143:151–61.. doi: 10.1111/acps.13245. [DOI] [PubMed] [Google Scholar]
- 5.Depp CA, Mausbach BT, Harmell AL, Savla GN, Bowie CR, Harvey PD, et al. Meta-analysis of the association between cognitive abilities and everyday functioning in bipolar disorder. Bipolar Disord. 2012;14:217–26. doi: 10.1111/j.1399-5618.2012.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kjaerstad HL, Mistarz N, Coello K, Stanislaus S, Melbye SA, Harmer CJ, et al. Aberrant cognition in newly diagnosed patients with bipolar disorder and their unaffected relatives. Psychol Med. 2020;50:1808–19. doi: 10.1017/S0033291719001867. [DOI] [PubMed] [Google Scholar]
- 7.Kuswanto C, Chin R, Sum MY, Sengupta S, Fagiolini A, McIntyre RS, et al. Shared and divergent neurocognitive impairments in adult patients with schizophrenia and bipolar disorder: Whither the evidence? Neurosci Biobehav Rev. 2016;61:66–89. doi: 10.1016/j.neubiorev.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Roshani F, Piri R, Malek A, Michel TM, Vafaee MS. Comparison of cognitive flexibility, appropriate risk-taking and reaction time in individuals with and without adult ADHD. Psychiatry Res. 2020;284:112494. doi: 10.1016/j.psychres.2019.112494. [DOI] [PubMed] [Google Scholar]
- 9.Dekkers TJ, Huizenga HM, Popma A, Bexkens A, Zadelaar JN, Jansen BRJ. Decision-making deficits in adolescent boys with and without attention-deficit/hyperactivity disorder (ADHD): an experimental assessment of associated mechanisms. J Abnorm Child Psychol. 2020;48:495–510. doi: 10.1007/s10802-019-00613-7. [DOI] [PubMed] [Google Scholar]
- 10.Posner J, Polanczyk GV, Sonuga-Barke E. Attention-deficit hyperactivity disorder. Lancet. 2020;395:450–62.. doi: 10.1016/S0140-6736(19)33004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adida M, Jollant F, Clark L, Besnier N, Guillaume S, Kaladjian A, et al. Trait-related decision-making impairment in the three phases of bipolar disorder. Biol Psychiatry. 2011;70:357–65. doi: 10.1016/j.biopsych.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Shakeel MK, Hodgins DC, Goghari VM. A Comparison of Self-Reported Impulsivity in Gambling Disorder and Bipolar Disorder. J Gambl Stud. 2019;35:339–50. doi: 10.1007/s10899-018-9808-5. [DOI] [PubMed] [Google Scholar]
- 13.Sicilia AC, Lukacs JN, Jones S, Perez Algorta G. Decision-making and risk in bipolar disorder: A quantitative study using fuzzy trace theory. Psychol Psychother. 2020;93:105–21. doi: 10.1111/papt.12215. [DOI] [PubMed] [Google Scholar]
- 14.Strakowski SM, Fleck DE, DelBello MP, Adler CM, Shear PK, McElroy SL, et al. Characterizing impulsivity in mania. Bipolar Disord. 2009;11:41–51. doi: 10.1111/j.1399-5618.2008.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American Psychiatric Association. Diagnostic and statistical manual of mental disorders 4. text revision (DSM-IV-TR) ed. Washington, DC: American Psychiatric Press, Inc; 2000.
- 16.Substance Abuse and Mental Health Services Administration (US). Impact of the DSM-IV to DSM-5 Changes on the National Survey on Drug Use and Health. 2016. https://www.ncbi.nlm.nih.gov/books/NBK519704/table/ch3.t9/. Accessed 2 Dec 2020. [PubMed]
- 17.Papolos D, Hennen J, Cockerham MS. Factors associated with parent-reported suicide threats by children and adolescents with community-diagnosed bipolar disorder. J Affect Disord. 2005;86:267–75. doi: 10.1016/j.jad.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Reinharth J, Braga R, Serper M. Characterization of risk-taking in adults with bipolar spectrum disorders. J Nerv Ment Dis. 2017;205:580–84. doi: 10.1097/NMD.0000000000000680. [DOI] [PubMed] [Google Scholar]
- 19.Ramirez-Martin A, Ramos-Martin J, Mayoral-Cleries F, Moreno-Kustner B, Guzman-Parra J. Impulsivity, decision-making and risk-taking behaviour in bipolar disorder: a systematic review and meta-analysis. Psychol Med. 2020;50:2141–53. doi: 10.1017/S0033291720003086. [DOI] [PubMed] [Google Scholar]
- 20.Scholz V, Houenou J, Kollmann B, Duclap D, Poupon C, Wessa M. Dysfunctional decision-making related to white matter alterations in bipolar I disorder. J Affect Disord. 2016;194:72–9. doi: 10.1016/j.jad.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Mason L, Trujillo-Barreto NJ, Bentall RP, El-Deredy W. Attentional bias predicts increased reward salience and risk taking in bipolar disorder. Biol Psychiatry. 2016;79:311–9. doi: 10.1016/j.biopsych.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Xia Y, Zhuang K, Sun J, Chen Q, Wei D, Yang W, et al. Emotion-related brain structures associated with trait creativity in middle children. Neurosci Lett. 2017;658:182–88. doi: 10.1016/j.neulet.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Cortese BM, Mitchell TR, Galloway MP, Prevost KE, Fang J, Moore GJ, et al. Region-specific alteration in brain glutamate: possible relationship to risk-taking behavior. Physiol Behav. 2010;99:445–50. doi: 10.1016/j.physbeh.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermens DF, Chitty KM, Lee RS, Tickell A, Haber PS, Naismith SL, et al. Hippocampal glutamate is increased and associated with risky drinking in young adults with major depression. J Affect Disord. 2015;186:95–8. doi: 10.1016/j.jad.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Pittaras E, Callebert J, Chennaoui M, Rabat A, Granon S. Individual behavioral and neurochemical markers of unadapted decision-making processes in healthy inbred mice. Brain Struct Funct. 2016;221:4615–29. doi: 10.1007/s00429-016-1192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tondo L, Vazquez GH, Baldessarini RJ. Prevention of suicidal behavior in bipolar disorder. Bipolar Disord. 2021;23:14–23. doi: 10.1111/bdi.13017. [DOI] [PubMed] [Google Scholar]
- 27.Shaltiel G, Maeng S, Malkesman O, Pearson B, Schloesser RJ, Tragon T, et al. Evidence for the involvement of the kainate receptor subunit GluR6 (GRIK2) in mediating behavioral displays related to behavioral symptoms of mania. Mol Psychiatry. 2008;13:858–72. doi: 10.1038/mp.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, Wang Y, Tan H, Bharti V, Che Y, Wang JF. Chronic treatment with mood stabilizer lithium inhibits amphetamine-induced risk-taking manic-like behaviors. Neurosci Lett. 2015;603:84–8. doi: 10.1016/j.neulet.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 29.Kollmann B, Yuen K, Scholz V, Wessa M. Cognitive variability in bipolar I disorder: A cluster-analytic approach informed by resting-state data. Neuropharmacology. 2019;156:107585. doi: 10.1016/j.neuropharm.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 30.Hidiroglu C, Demirci Esen O, Tunca Z, Neslihan Gurz Yalcin S, Lombardo L, Glahn DC, et al. Can risk-taking be an endophenotype for bipolar disorder? A study on patients with bipolar disorder type I and their first-degree relatives. J Int Neuropsychol Soc. 2013;19:474–82. doi: 10.1017/S1355617713000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bipolar D, Schizophrenia Working Group of the Psychiatric Genomics Consortium. Electronic address drve, Bipolar D, Schizophrenia Working Group of the Psychiatric Genomics C. Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell. 2018;173:1705–15. doi: 10.1016/j.cell.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson Linner R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–57. doi: 10.1038/s41588-018-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smeland OB, Frei O, Shadrin A, O’Connell K, Fan CC, Bahrami S, et al. Discovery of shared genomic loci using the conditional false discovery rate approach. Hum Genet. 2020;139:85–94. doi: 10.1007/s00439-019-02060-2. [DOI] [PubMed] [Google Scholar]
- 35.Brainstorm C, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360. [DOI] [PMC free article] [PubMed]
- 36.Wu Y, Cao H, Baranova A, Huang H, Li S, Cai L, et al. Multi-trait analysis for genome-wide association study of five psychiatric disorders. Transl Psychiatry. 2020;10:209. doi: 10.1038/s41398-020-00902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee PH, Feng YA, Smoller JW. Pleiotropy and cross-disorder genetics among psychiatric disorders. Biol Psychiatry. 2021;89:20–31. doi: 10.1016/j.biopsych.2020.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andreassen OA, Thompson WK, Schork AJ, Ripke S, Mattingsdal M, Kelsoe JR, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connell KS, Shadrin A, Bahrami S, Smeland OB, Bettella F, Frei O, et al. Identification of genetic overlap and novel risk loci for attention-deficit/hyperactivity disorder and bipolar disorder. Mol Psychiatry. 2019. 10.1038/s41380-019-0613-z. [DOI] [PubMed]
- 40.Smeland OB, Bahrami S, Frei O, Shadrin A, O’Connell K, Savage J, et al. Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry. 2020;25:844–53. doi: 10.1038/s41380-018-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of over 40,000 bipolar disorder cases provides novel biological insights. 2020. 10.1101/2020.09.17.20187054.
- 42.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uffelmann E, Posthuma D. Emerging Methods and Resources for Biological Interrogation of Neuropsychiatric Polygenic Signal. Biol Psychiatry. 2021;89:41–53. doi: 10.1016/j.biopsych.2020.05.022. [DOI] [PubMed] [Google Scholar]
- 45.PleioFDR. https://github.com/precimed/pleiofdr. Accessed 18 March 2021.
- 46.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 47.Smeland OB, Frei O, Kauppi K, Hill WD, Li W, Wang Y, et al. Identification of Genetic Loci Jointly Influencing Schizophrenia Risk and the Cognitive Traits of Verbal-Numerical Reasoning, Reaction Time, and General Cognitive Function. JAMA Psychiatry. 2017;74:1065–75. doi: 10.1001/jamapsychiatry.2017.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smeland OB, Shadrin A, Bahrami S, Broce I, Tesli M, Frei O, et al. Genome-wide Association Analysis of Parkinson’s Disease and Schizophrenia Reveals Shared Genetic Architecture and Identifies Novel Risk Loci. Biol Psychiatry. 2021;89:227–35. doi: 10.1016/j.biopsych.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smeland OB, Wang Y, Frei O, Li W, Hibar DP, Franke B, et al. Genetic Overlap Between Schizophrenia and Volumes of Hippocampus, Putamen, and Intracranial Volume Indicates Shared Molecular Genetic Mechanisms. Schizophr Bull. 2018;44:854–64. doi: 10.1093/schbul/sbx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shim H, Chasman DI, Smith JD, Mora S, Ridker PM, Nickerson DA, et al. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS One. 2015;10:e0120758. doi: 10.1371/journal.pone.0120758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D94. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong S, Boyle AP. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum Mutat. 2019;40:1292–98. doi: 10.1002/humu.23791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Consortium GT. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–D73. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakaue S, Okada Y. GREP: genome for REPositioning drugs. Bioinformatics. 2019;35:3821–23. doi: 10.1093/bioinformatics/btz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–30. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Lu Q, Ye Y, Huang K, Liu W, Wu Y, et al. Local genetic correlation analysis reveals heterogeneous etiologic sharing of complex traits. 2020. 10.1101/2020.05.08.084475. [DOI] [PMC free article] [PubMed]
- 61.Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gerring ZF, Gamazon ER, Derks EM, Major Depressive Disorder Working Group of the Psychiatric Genomics C. A gene co-expression network-based analysis of multiple brain tissues reveals novel genes and molecular pathways underlying major depression. PLoS Genet. 2019;15:e1008245. doi: 10.1371/journal.pgen.1008245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 65.VV AA. The 40th Congress of the Italian Pharmacological Society (SIF). 2021;3.
- 66.Brunton LL, Hilal-Dandan R, Knollmann BC. Goodman & Gilman’s: The Pharmacological Basis of Therapeutics, Thirteenth Edition McGraw-Hill Education; New York. 2018;1–347. 10.36118/pharmadvances.03.2021.01.
- 67.Mosheva M, Serretti A, Stukalin Y, Fabbri C, Hagin M, Horev S, et al. Association between CANCA1C gene rs1034936 polymorphism and alcohol dependence in bipolar disorder. J Affect Disord. 2020;261:181–6. doi: 10.1016/j.jad.2019.10.015. [DOI] [PubMed] [Google Scholar]
- 68.Jaffe AE, Hoeppner DJ, Saito T, Blanpain L, Ukaigwe J, Burke EE, et al. Profiling gene expression in the human dentate gyrus granule cell layer reveals insights into schizophrenia and its genetic risk. Nat Neurosci. 2020;23:510–19. doi: 10.1038/s41593-020-0604-z. [DOI] [PubMed] [Google Scholar]
- 69.Hamidian S, Pourshahbaz A, Bozorgmehr A, Ananloo ES, Dolatshahi B, Ohadi M. How obsessive-compulsive and bipolar disorders meet each other? An integrative gene-based enrichment approach. Ann Gen Psychiatry. 2020;19:31. doi: 10.1186/s12991-020-00280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calabro M, Porcelli S, Crisafulli C, Albani D, Kasper S, Zohar J, et al. Genetic variants associated with psychotic symptoms across psychiatric disorders. Neurosci Lett. 2020;720:134754. doi: 10.1016/j.neulet.2020.134754. [DOI] [PubMed] [Google Scholar]
- 71.Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–79. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ehlinger DG, Commons KG. Cav1.2L-type calcium channels regulate stress coping behavior via serotonin neurons. Neuropharmacology. 2019;144:282–90. doi: 10.1016/j.neuropharm.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bastos CR, Tovo-Rodrigues L, Ardais AP, Xavier J, Salerno PSV, Camerini L, et al. The role of CACNA1C gene and childhood trauma interaction on bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2020;101:109915. doi: 10.1016/j.pnpbp.2020.109915. [DOI] [PubMed] [Google Scholar]
- 74.Dedic N, Pohlmann ML, Richter JS, Mehta D, Czamara D, Metzger MW, et al. Cross-disorder risk gene CACNA1C differentially modulates susceptibility to psychiatric disorders during development and adulthood. Mol Psychiatry. 2018;23:533–43. doi: 10.1038/mp.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clark MB, Wrzesinski T, Garcia AB, Hall NAL, Kleinman JE, Hyde T, et al. Long-read sequencing reveals the complex splicing profile of the psychiatric risk gene CACNA1C in human brain. Mol Psychiatry. 2020;25:37–47. doi: 10.1038/s41380-019-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harrison PJ, Hall N, Mould A, Al-Juffali N, Tunbridge EM. Cellular calcium in bipolar disorder: systematic review and meta-analysis. Mol Psychiatry. 2019. 10.1038/s41380-019-0622-y. [DOI] [PMC free article] [PubMed]
- 77.Andreopoulos S, Wasserman M, Woo K, Li PP, Warsh JJ. Chronic lithium treatment of B lymphoblasts from bipolar disorder patients reduces transient receptor potential channel 3 levels. Pharmacogenomics J. 2004;4:365–73. doi: 10.1038/sj.tpj.6500266. [DOI] [PubMed] [Google Scholar]
- 78.Dubovsky SL, Thomas M, Hijazi A, Murphy J. Intracellular calcium signalling in peripheral cells of patients with bipolar affective disorder. Eur Arch Psychiatry Clin Neurosci. 1994;243:229–34. doi: 10.1007/BF02191579. [DOI] [PubMed] [Google Scholar]
- 79.Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–9. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Higgins GA, Allyn-Feuer A, Barbour E, Athey BD. A glutamatergic network mediates lithium response in bipolar disorder as defined by epigenome pathway analysis. Pharmacogenomics. 2015;16:1547–63. doi: 10.2217/pgs.15.106. [DOI] [PubMed] [Google Scholar]
- 81.McCarthy MJ, Le Roux MJ, Wei H, Beesley S, Kelsoe JR, Welsh DK. Calcium channel genes associated with bipolar disorder modulate lithium’s amplification of circadian rhythms. Neuropharmacology. 2016;101:439–48. doi: 10.1016/j.neuropharm.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garza-Trevino ES, Overall JE, Hollister LE. Verapamil versus lithium in acute mania. Am J Psychiatry. 1992;149:121–2. doi: 10.1176/ajp.149.11.1614-a. [DOI] [PubMed] [Google Scholar]
- 83.Pal Singh G. A double-blind comparative study of clinical efficacy of verapamil versus lithium in acute mania. Int J Psychiatry Clin Pr. 2008;12:303–8. doi: 10.1080/13651500802209670. [DOI] [PubMed] [Google Scholar]
- 84.Wisner KL, Peindl KS, Perel JM, Hanusa BH, Piontek CM, Baab S. Verapamil treatment for women with bipolar disorder. Biol Psychiatry. 2002;51:745–52. doi: 10.1016/S0006-3223(01)01338-5. [DOI] [PubMed] [Google Scholar]
- 85.Cipriani A, Saunders K, Attenburrow MJ, Stefaniak J, Panchal P, Stockton S, et al. A systematic review of calcium channel antagonists in bipolar disorder and some considerations for their future development. Mol Psychiatry. 2016;21:1324–32. doi: 10.1038/mp.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pazzaglia PJ, Post RM, Ketter TA, Callahan AM, Marangell LB, Frye MA, et al. Nimodipine monotherapy and carbamazepine augmentation in patients with refractory recurrent affective illness. J Clin Psychopharmacol. 1998;18:404–13. doi: 10.1097/00004714-199810000-00009. [DOI] [PubMed] [Google Scholar]
- 87.Ostacher MJ, Iosifescu DV, Hay A, Blumenthal SR, Sklar P, Perlis RH. Pilot investigation of isradipine in the treatment of bipolar depression motivated by genome-wide association. Bipolar Disord. 2014;16:199–203. doi: 10.1111/bdi.12143. [DOI] [PubMed] [Google Scholar]
- 88.Atkinson LZ, Colbourne L, Smith A, Harmer CH, Nobre AC, Rendell J, et al. The Oxford study of Calcium channel Antagonism, Cognition, Mood instability and Sleep (OxCaMS): study protocol for a randomised controlled, experimental medicine study. Trials. 2019;20:120. doi: 10.1186/s13063-019-3175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hayes JF, Lundin A, Wicks S, Lewis G, Wong ICK, Osborn DPJ, et al. Association of Hydroxylmethyl Glutaryl Coenzyme A Reductase Inhibitors, L-Type Calcium Channel Antagonists, and Biguanides With Rates of Psychiatric Hospitalization and Self-Harm in Individuals With Serious Mental Illness. JAMA Psychiatry. 2019;76:382–90. doi: 10.1001/jamapsychiatry.2018.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zink CF, Giegerich M, Prettyman GE, Carta KE, van Ginkel M, O’Rourke MP, et al. Nimodipine improves cortical efficiency during working memory in healthy subjects. Transl Psychiatry. 2020;10:372. doi: 10.1038/s41398-020-01066-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vahdani B, Armani Kian A, Esmaeilzadeh A, Zenoozian S, Yousefi V, Mazloomzadeh S. Adjunctive Raloxifene and Isradipine Improve Cognitive Functioning in Patients With Schizophrenia: A Pilot Study. J Clin Psychopharmacol. 2020;40:457–63. doi: 10.1097/JCP.0000000000001274. [DOI] [PubMed] [Google Scholar]
- 92.Yoshimizu T, Pan JQ, Mungenast AE, Madison JM, Su S, Ketterman J, et al. Functional implications of a psychiatric risk variant within CACNA1C in induced human neurons. Mol Psychiatry. 2015;20:162–9. doi: 10.1038/mp.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pignataro P, Dicarlo M, Zerlotin R, Zecca C, Dell’Abate MT, Buccoliero C, et al. FNDC5/Irisin System in Neuroinflammation and Neurodegenerative Diseases: Update and Novel Perspective. Int J Mol Sci. 2021;22:1605. 10.3390/ijms22041605. [DOI] [PMC free article] [PubMed]
- 94.Gruhn K, Siteneski A, Camargo A, Freitas AE, Olescowicz G, Brocardo PS, et al. Physical exercise stimulates hippocampal mTORC1 and FNDC5/irisin signaling pathway in mice: Possible implication for its antidepressant effect. Behav Brain Res. 2021;400:113040. doi: 10.1016/j.bbr.2020.113040. [DOI] [PubMed] [Google Scholar]
- 95.Koh TW, Bellen HJ. Synaptotagmin I, a Ca2+ sensor for neurotransmitter release. Trends Neurosci. 2003;26:413–22. doi: 10.1016/S0166-2236(03)00195-4. [DOI] [PubMed] [Google Scholar]
- 96.Flores R, 3rd, Hirota Y, Armstrong B, Sawa A, Tomoda T. DISC1 regulates synaptic vesicle transport via a lithium-sensitive pathway. Neurosci Res. 2011;71:71–7. doi: 10.1016/j.neures.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yoshino Y, Dwivedi Y. Non-Coding RNAs in Psychiatric Disorders and Suicidal Behavior. Front Psychiatry. 2020;11:543893. doi: 10.3389/fpsyt.2020.543893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hou L, Heilbronner U, Degenhardt F, Adli M, Akiyama K, Akula N, et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. Lancet. 2016;387:1085–93. doi: 10.1016/S0140-6736(16)00143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kang HJ, Kim KT, Yoo KH, Park Y, Kim JW, Kim SW, et al. Genetic Markers for Later Remission in Response to Early Improvement of Antidepressants. Int J Mol Sci. 2020;21:4884. 10.3390/ijms21144884. [DOI] [PMC free article] [PubMed]
- 100.Wang Y, Liu B, Yang Y, Wang Y, Zhao Z, Miao Z, et al. Metformin exerts antidepressant effects by regulated DNA hydroxymethylation. Epigenomics. 2019;11:655–67. doi: 10.2217/epi-2018-0187. [DOI] [PubMed] [Google Scholar]
- 101.Michaelson JJ, Shin MK, Koh JY, Brueggeman L, Zhang A, Katzman A, et al. Neuronal PAS Domain Proteins 1 and 3 Are Master Regulators of Neuropsychiatric Risk Genes. Biol Psychiatry. 2017;82:213–23.. doi: 10.1016/j.biopsych.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kamnasaran D, Muir WJ, Ferguson-Smith MA, Cox DW. Disruption of the neuronal PAS3 gene in a family affected with schizophrenia. J Med Genet. 2003;40:325–32. doi: 10.1136/jmg.40.5.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pickard BS, Malloy MP, Porteous DJ, Blackwood DH, Muir WJ. Disruption of a brain transcription factor, NPAS3, is associated with schizophrenia and learning disability. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:26–32. doi: 10.1002/ajmg.b.30204. [DOI] [PubMed] [Google Scholar]
- 104.Pickard BS, Christoforou A, Thomson PA, Fawkes A, Evans KL, Morris SW, et al. Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:874–84. doi: 10.1038/mp.2008.24. [DOI] [PubMed] [Google Scholar]
- 105.Nurnberger JI, Jr., Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I, et al. Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry. 2014;71:657–64. doi: 10.1001/jamapsychiatry.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang J, Perlis RH, Lee PH, Rush AJ, Fava M, Sachs GS, et al. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167:1254–63. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sha L, MacIntyre L, Machell JA, Kelly MP, Porteous DJ, Brandon NJ, et al. Transcriptional regulation of neurodevelopmental and metabolic pathways by NPAS3. Mol Psychiatry. 2012;17:267–79. doi: 10.1038/mp.2011.73. [DOI] [PubMed] [Google Scholar]
- 108.Brunskill EW, Ehrman LA, Williams MT, Klanke J, Hammer D, Schaefer TL, et al. Abnormal neurodevelopment, neurosignaling and behaviour in Npas3-deficient mice. Eur J Neurosci. 2005;22:1265–76. doi: 10.1111/j.1460-9568.2005.04291.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.