Abstract
Large-scale genetic screening has identified KMT5B (SUV420H1), which encodes a histone H4 K20 di- and tri-methyltransferase highly expressed in prefrontal cortex (PFC), as a top-ranking high-risk gene for autism. However, the biological function of KMT5B in the brain is poorly characterized, and how KMT5B deficiency is linked to autism remains largely unknown. Here we knocked down Kmt5b in PFC and examined behavioral and electrophysiological changes, as well as underlying molecular mechanisms. Mice with Kmt5b deficiency in PFC display social deficits, a core symptom of autism, without the alteration of other behaviors. Kmt5b deficiency also produces deficits in PFC glutamatergic synaptic transmission, which is accompanied by the reduced synaptic expression of glutamate receptor subunits and associated proteins. Kmt5b deficiency-induced reduction of H4K20me2 impairs 53BP1-mediated DNA repair, leading to the elevation of p53 expression and its target gene Ddit4 (Redd1), which is implicated in synaptic impairment. RNA-sequencing data indicate that Kmt5b deficiency results in the upregulation of genes enriched in cellular stress response and ubiquitin-dependent protein degradation. Collectively, this study has revealed the functional role of Kmt5b in the PFC, and suggests that Kmt5b deficiency could cause autistic phenotypes by inducing synaptic dysfunction and transcriptional aberration.
Subject terms: Autism spectrum disorders, Preclinical research
Introduction
Autism is a prevalent and devastating neurodevelopmental disorder that affects social and communicative behaviors. Strong genetic factors contribute to the etiology of autism [1]. One of the top-ranking autism risk factors (FDR < 0.01) identified by large-scale whole-genome sequencing of ASD patients across multiple cohorts is KMT5B (SUV420H1) [2–7]. KMT5B mutations discovered in ASD patients, such as substitution splicing variants [4], missense mutations [5], frameshift deletions [3], and nonsynonymous substitutions [6], all result in loss of function. However, the biological function of KMT5B in the brain, as well as how KMT5B haploinsufficiency contributes to ASD symptoms, is largely unknown.
Chromatin structure, and consequently the transcriptional accessibility of the associated DNA, can be transiently modified by histone modifications, such as histone methylation and acetylation [8]. KMT5B is a histone methyltransferase that mediates di-methylation of histone 4 lysine 20 (H4K20me2) [9, 10]. This histone modification has been implicated in the repair of DNA damage, such as double-strand breaks (DSBs) [11, 12], which can be induced by both endogenous (e.g., gene transcription) [13] and exogenous stimuli (e.g., chemo, radiation). KMT5B also works with KMT5C to catalyze tri-methylation of histone 4 lysine 20 (H4K20me3) [11], which is implicated in gene silencing [14–16].
KMT5B expression is enriched in the prefrontal cortex (PFC) [17, 18], a brain region that plays a major role in regulating higher-level executive functions and social cognition [19, 20]. The PFC is also highly implicated in ASD pathology [21, 22]. Synaptic dysfunction of PFC pyramidal neurons has been observed in human ASD patients [22] and in multiple ASD mouse models [23–26]. The refinement of PFC circuitry, as well as the significant development of sociability and other cognitive and emotional behaviors regulated by PFC, starts around 3–5 weeks postnatal and continues for a few weeks into adolescence (~7 weeks) [27–30]. Thus, we induced PFC-specific knockdown of Kmt5b during this critical developmental period, and examined behavioral and physiological consequences. We also revealed the molecular and genomic mechanisms underlying the social and synaptic deficits caused by Kmt5b deficiency in PFC. These results should improve our understanding on the in vivo biological function of Kmt5b and the mechanisms by which its disruption may drive autistic phenotypes.
Materials and methods
Animals and human samples
All mice (both male and female, C57BL6/J) were group-housed with 1–3 gender-matched conspecifics and provided standard enrichment. All animals were maintained on a 12-h light (6:00 a.m. to 6:00 p.m.)/dark (6:00 p.m. to 6:00 a.m.) cycle. All animal studies were performed with the approval of the Institutional Animal Care and Use Committee of the State University of New York at Buffalo. Postmortem human brain tissue samples (Broadman’s area 9) were received from NIH NeuroBioBank.
Animal surgery
Mice (3–4-week-old) received bilateral stereotaxic injection of GFP-tagged Kmt5b shRNA AAV or a scrambled (scr.) control shRNA AAV. Stereotaxic injection of the virus (1.5-μl total volume, bilateral) to the medial PFC region was performed as described previously [31]. In brief, mice were anesthetized and placed on a stereotaxic apparatus (David Kopf Instruments). The injection was performed with a Hamilton syringe (gauge 31) at a speed of 0.2 μl/min, and the needle was kept in place for an additional 5 min. The virus was delivered bilaterally to the target area using the following coordinates: 2.0-mm anterior to bregma; 0.25-mm lateral; and 2.0-mm dorsal to ventral. The viral expression sites usually include the whole medial PFC (cingulate cortex, prelimbic, and infralimbic). All behavioral, biochemical, and electrophysiological tests were conducted 14–20 days after surgery.
Virus generation
We inserted Kmt5b shRNA (GTGTCAACTGGTCGAGATA) into the GFP-tagged AAV vector (#85741, Addgene, U6 promoter), and verified the construct by sequencing. The Kmt5b shRNA plasmid was transfected into 80–90% confluent N2a cells with lipofectamine 2000. After 48-h transfection, we collected cells, extracted RNA, and used qPCR to measure the knockdown effect. After knockdown verification, the KMT5B shRNA plasmid was sent to the viral core facility of Emory university (http://neurology.emory.edu/ENNCF/viral_vector/) for the generation of AAV.
Immunohistochemistry
Mice were anesthetized and transcardially perfused with PBS, followed by 4% paraformaldehyde (PFA) before brain removal. Brains were postfixed in 4% PFA for 2 days and cut into 30-μm slices. Slices were cut coronally and washed and blocked for 1 h in PBS containing 5% donkey serum and 0.3% Triton for permeabilization. After washing, slices were incubated with the primary antibody against KMT5B (Novus Biologicals, NBP1-97312), H4K20me2 (Abcam, ab9052), or p53 (Santa Cruz, sc-6243) for 48 h at 4 °C. After washing three times (30 min with gentle shaking) in PBS, slices were incubated with secondary antibody (Alexa Fluor 568, Invitrogen, A10037 or ab175470, 1:1000) for 1 h at room temperature, followed by three washes with PBS. Slices were mounted on slides with Vectashield mounting media with DAPI (Vector Laboratories). Images were acquired using a Leica DMi8 fluorescence microscope. All specimens were imaged under identical conditions and analyzed with identical parameters using ImageJ software.
Behavioral testing
All behavioral testing was performed on at least three independent cohorts. The sequence of behavioral tests was randomized between litters. All behavioral tests were performed 24 h apart, with one test performed per day. Sample sizes were determined based on power analyses and were similar to those reported in previous works [32]. All behavioral testing apparatuses were wiped down with 75% ethanol between animals and trials. All testing (unless otherwise indicated) was performed in a dimly lit room. The Anymaze behavior tracking software (Stoelting, Wood Dale, IL) was used to record footage of behavior tests. All scoring was performed by Anymaze or manually by researchers blind to both genotypes and treatments.
Three-chamber social preference
The three-chamber social preference test was performed as previously described [23, 25, 31, 33]. Briefly, the test mouse was first placed into a Plexiglass arena (L: 101.6 cm, W: 50.8 cm, H: 50.8 cm) containing two empty inverted pencil cups for a 10-min habituation period. On the following day, the mouse was reintroduced to the apparatus for a 10-min trial in which the pencil cups contained two identical objects. The animal was then returned to its home cage for 5 min. The animal was then placed into the apparatus for a 10-min trial (social preference test), in which one cup contained a novel object (nonsocial stimulus) while the other contained an age- and gender-matched WT mouse (social stimulus). The amount of time spent interacting with each stimulus was recorded. The preference index was calculated as (social time − nonsocial time)/(social time + nonsocial time).
Social approach
The test animal was habituated in a rectangular apparatus (L: 67.7 cm, W: 50.8 cm, H: 50.8 cm) containing a capsule (inverted pencil cup, placed in the center) for 10 min, then returned to its home cage. A social stimulus (an age- and sex-matched mouse) was then placed inside the capsule. The test animal was placed into the apparatus to explore for 10 min. The amount of time the test animal spent interacting with the social stimulus was measured.
Self-grooming
Self-grooming is a rodent behavior linked to repetitive behaviors in neuropsychiatric models including autism [34]. The test mouse was placed individually into a clean cage containing a thin layer (~1 cm) of bedding for 30 min. During the first 20 min, the mouse was allowed to habituate to the environment. Grooming behavior was assessed manually in the final 10-min period. For scoring purposes, self-grooming behavior was defined as any repetitive grooming motion (i.e., licking, brushing) directed at the front paws, head/ears, or abdomen.
Rotarod
The test mouse was placed onto an accelerating rotarod (SD Instruments, San Diego, CA), which slowly accelerated from 4 to 40 revolutions per minute over a 5-min test session. The mouse was required to walk forward in order to remain on top of the rotating cylinder rod [31]. After a practice trial in which the mouse was allowed to familiarize with the apparatus, two test trials were conducted. For each trial, the amount of time that the mouse remained on the rotarod before falling off (latency to fall) was recorded.
Novel-object recognition
The test mouse was placed on a circular open white platform (2-foot diameter) for 5 min for habituation, then returned to its home cage. The mouse was then placed on the platform with two identical objects for 5 min, then returned to its home cage again. The mouse was then allowed to explore the platform containing one of the original objects (familiar) and a new object (novel) for 5 min, during which the amount of time spent interacting with each object was scored. Discrimination ratio was calculated as: (novel-object time − familiar-object time)/(novel-object time + familiar-object time).
Locomotion
Locomotor activity was measured in a rectangular apparatus (40 × 40 × 30 cm) equipped with photo beam monitors (AccuScan Intruments). Total distance traveled, along with the time in center and distance traveled in center, was recorded during a 60-min test session and divided into 30-min bins.
Startle response/pre-pulse inhibition (PPI)
PPI was assessed as previously described [26]. The acoustic startle response test was performed in an SR-LAB startle chamber (San Diego Instruments, San Diego, CA). The test consisted of three trial types: null trials with only a white noise background (66 dB), pulse-only trials with 40 ms white noise stimulus (90, 100, 110, and 120 dB), and pre-pulse trials, where the 120-dB startle stimulus was preceded 100 ms earlier by a 20-ms pre-pulse stimulus that was either 70, 76, or 85 dB. Mice were placed into the plexiglass holder and habituated to the apparatus for 5 min before testing. Each test consisted of 74 trials with 30 pulse-only trials, 11 null trials, and 33 pre-pulse trials. Trial order was the same for all animals, with ten pulse-only trials followed by combinations of the pre-pulse and null trials, then terminating with ten pulse-only trials. The maximum startle response was measured for each trial. PPI (percentage) = (1 − pre-pulse trial response/pulse-only trial response) × 100.
Electrophysiological recordings
Whole-cell voltage-clamp recording was used to measure synaptic currents in layer V mPFC (prelimbic and infralimbic) pyramidal neurons as previously described [23, 35, 36]. Mouse brain slices (300 µm) were positioned in a perfusion chamber attached to the fixed stage of an upright microscope (Olympus) and submerged in continuously flowing oxygenated ACSF (in mM: 130 NaCl, 26 NaHCO3, 1 CaCl2, 5 MgCl2, 3 KCl, 1.25 NaH2PO4, 10 glucose, pH 7.4, 300 mOsm). For EPSC recordings, bicuculline (10 µM) was added to ACSF. The pipette contained the following solution (in mM: 130 cesium-methanesulfonate, 10 CsCl, 4 NaCl, 10 HEPES, 1 MgCl2, 5 EGTA, 2 QX-314, 12 phosphocreatine, 5 MgATP, 0.5 Na3GTP, 0.1 leupeptin, pH 7.2–7.3, 265–270 mOsm). Evoked synaptic currents were generated with a pulse from a stimulation isolation unit controlled by an S48 pulse generator (Grass Technologies, West Warwick, RI). A bipolar stimulating electrode (FHC, Bowdoinham, ME) was placed ~100 μm from the neuron under recording. Stimulation pulses (0.4 ms, 70 μA) were delivered at 0.05 Hz. AMPAR-EPSC was first recorded at −70 mV, then the mixture of AMPAR- and NMDAR-EPSC was recorded at +40 mV with the same stimulation. The peak of NMDAR-EPSC was calculated at 40 ms from the onset of the EPSC mixture. Data analyses were performed with the Clampfit 10.0.7 software (Molecular Devices, Sunnyvale, CA, USA).
Western blotting
Western blots were performed using antibodies against KMT5B (1:500, Novus Biologicals, NBP1-97312), H3 (1:1000, Cell Signaling, 4499), GluR1 (1:500, Santa Cruz, SC-13152), GluR2 (1:500, Abcam, Ab20673), NR1 (1:1000, Cell Signal, D65B7), NR2A (1:500, Millipore, 07-632), NR2B (1:1000, Millipore, 06-600), PSD-95 (1:1000, Neuromab, 75-028), Tubulin (1:10000, Sigma, T9026), VAMP2 (1:1000, Proteintech, 10135-1-AP), SNAP25 (1:1000, Proteintech, 60169-1-Ig), Synaptophysin (1:500, BD Biosciences, 611880), SynGAP1 (1:1000, Cell Signaling Technology, D88G1), γH2AX (Biolegend, #613405), and Ddit4 (1:500, Proteintech, 10638-1-AP). Nuclear extraction was performed as previously described [23–26].
Quantitative real-time RT-PCR
Total RNA was isolated from mouse PFC punches (GFP+ regions) using Trizol reagent (Invitrogen) and treated with DNase I (Invitrogen) to remove genomic DNA. Then the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen) was used to reverse-transcribe mRNA into cDNA, followed by the treatment with RNase H (2 U/l) for 20 min at 37 °C. Quantitative real-time RT-PCR was performed using the iCycler iQ™ Real-Time PCR Detection System and iQ™ Supermix (Bio-Rad), according to the manufacturer’s instructions. In brief, GAPDH was used as the housekeeping gene for quantitation of the expression of target genes in samples from GH vs. SI group. Fold changes in the target genes were determined by: fold change = 2−Δ(ΔCT), where ΔCT = CT(target) − CT(GAPDH), and Δ(ΔCT) = ΔCT(SI group) − ΔCT(GH group). CT (threshold cycle) is defined as the fractional cycle number at which the fluorescence reaches 10× the standard deviation of the baseline. A total reaction mixture of 20 µl was amplified in a 96-well thin-wall PCR plate (Bio-Rad) using the following PCR cycling parameters: 95 °C for 5 min followed by 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s. The primer of DNA-damage-inducible transcript 4 (DDIT4) is: forward: CTGGACAGCAGCAACAGTGG, reverse: AGCAGCTGCATCAGGTTGG.
RNA sequencing (RNAseq) and bioinformatic analysis
PFC samples were obtained from three mice injected with a scr. shRNA AAV and three mice injected with Kmt5b shRNA AAV. We generated strand-specific RNA libraries from 1 μg purified RNA using TruSeq stranded total RNA plus Ribo-zero kits (Illumina). The sequencing was performed at the Genomics and Bioinformatics Core of State University of New York at Buffalo. Single end reads per sample were obtained using the HiSeq 2500 platform from Illumina. Reads were first trimmed using Cutadapt to remove the 3′ end adapters and trailing sequences, followed by aligning to mouse RefSeq mRNAs using TopHat2. Transcript counts were estimated using HTSeq. Differences in gene expression levels between samples were assessed with edgeR and calculated as log2 fold change. DEGs were selected based on the criteria: p (adj) < 0.01, |FC| (fold change) > 1.5, and average count per million > 300. Functional protein classification analyses were undertaken using Panther. Cytoscape software was used to generate interactome networks.
Statistical analyses
All statistical analyses were performed with GraphPad Prism and Minitab 18. Experiments with more than two dependent variables were subjected to two-way ANOVA with Bonferroni correction for multiple post hoc comparisons. Experiments with two groups were analyzed statistically using two-tailed unpaired t-tests. All data are presented as the mean ± SEM. Data points identified as statistically significant outliers (determined by the Grubb’s test, p < 0.05) were removed from the analyses. Sample sizes were determined based on power analyses and were similar to those reported in previous works [23, 25, 26, 32, 33, 35–37].
Results
Kmt5b deficiency in PFC induces social deficits
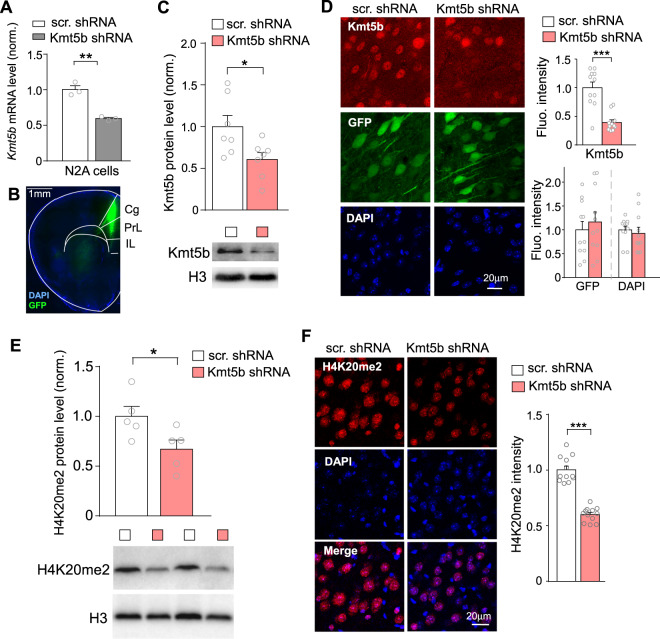
To examine the biological function of KMT5B in the brain, we generated Kmt5b shRNA to knockdown Kmt5b expression in the PFC of mice at juvenile age (~3–4 weeks), a critical period to initiate the maturation of PFC structure and the development of sociability [27–30]. The knockdown effect of Kmt5b shRNA was first confirmed in the mouse neuronal N2A cell line, in which we observed a ~40% reduction of Kmt5b mRNA expression (Fig. 1A, n = 3/group, t(4) = 7.25, p < 0.01, t-test). The AAV carrying GFP-tagged Kmt5b shRNA was then injected into the PFC of wild-type mice (Fig. 1B). After 2–3 weeks of viral expression, Western blotting and immunostaining were performed to verify the in vivo knockdown. A significant reduction of Kmt5b protein level was observed in the nuclear fraction of PFC from mice injected with Kmt5b shRNA AAV, compared to those injected with a scr. control shRNA AAV (Fig. 1C, n = 7 pairs, t(6) = 7, p < 0.05, t-test). The knockdown of Kmt5b expression was also evident in GFP-positive cells in PFC slices from Kmt5b shRNA-injected mice (Fig. 1D, n = 11 slices/3 mice/group, t(14) = 5.58, p < 0.001, t-test), while no differences were observed in the fluorescence intensity of GFP (p = 0.56, t-test) or DAPI (p = 0.62, t-test). Moreover, the level of H4K20me2, the primary histone modification catalyzed by Kmt5b, was significantly reduced in PFC cells injected with Kmt5b shRNA AAV, as shown by Western blotting (Fig. 1E, n = 5 mice/group, t(8) = 2.4, p < 0.05, t-test) and immunostaining (Fig. 1F, n = 12 slices/3 mice/group, t(17) = 10.6, p < 0.001, t-test), suggesting that Kmt5b shRNA effectively exerted the transcriptional repression and functional inhibition of Kmt5b.
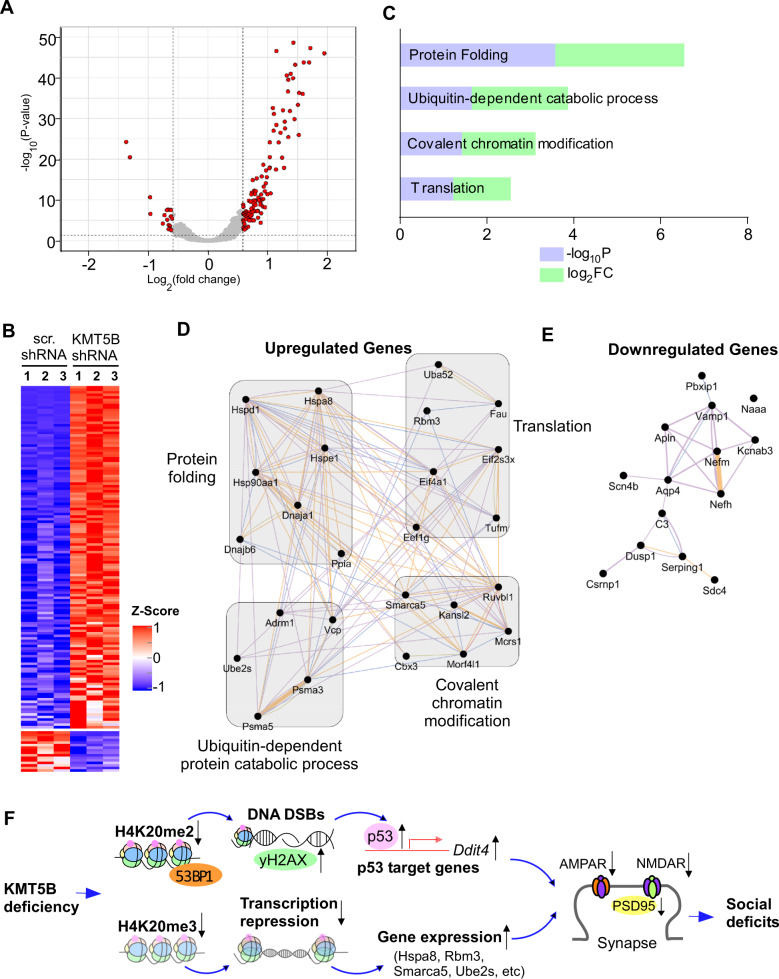
Fig. 1. KMT5B knockdown in PFC.
A Bar graph of Kmt5b mRNA level in N2A cells transfected with Kmt5b shRNA or a scrambled control shRNA (scr.). **p < 0.01, t-test. B Immunofluorescent image showing the location of Kmt5b shRNA AAV (GFP-tagged) expression in the PFC of a virus-injected mouse. C Quantitation of Kmt5b protein levels (normalized to total H3 levels) in the nuclear fraction of PFC dissected from mice injected with Kmt5b shRNA or scr. shRNA AAV. *p < 0.05, t-test. Insets: representative immunoblots. D Bar graphs of fluorescence intensity of Kmt5b, GFP, and DAPI signal in the PFC of mice injected with Kmt5b shRNA or scr. shRNA AAV. Inset: representative confocal images. ***p < 0.001, t-test. E Quantitation of H4K20me2 levels (normalized to total H3 levels) in the nuclear fraction of PFC dissected from mice injected with Kmt5b shRNA or scr. shRNA AAV. *p < 0.05, t-test. Insets: representative immunoblots. F Bar graph of fluorescence intensity of H4K20me2 signal in the PFC of mice injected with Kmt5b shRNA or scr. shRNA AAV. Inset: representative confocal images. ***p < 0.001, t-test.
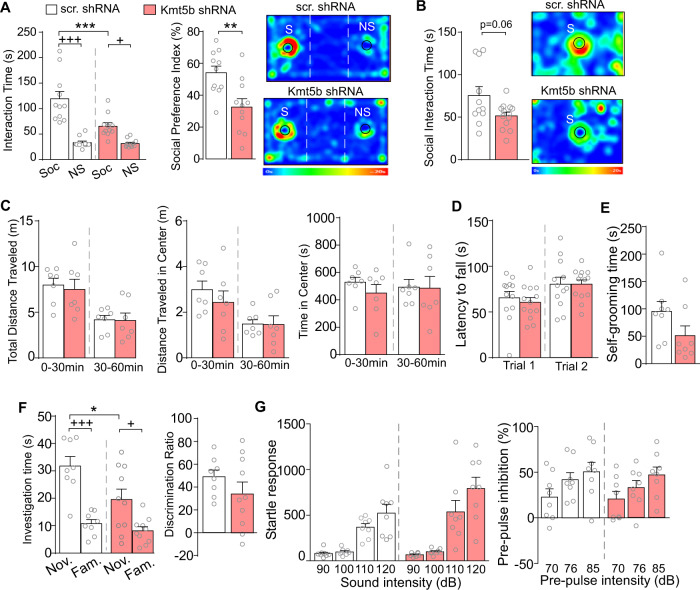
To find out behavioral alterations by PFC knockdown of Kmt5b, we first examined social behavior in Kmt5b shRNA-injected mice via the three-chamber social preference test [33]. As shown in Fig. 2A, Kmt5b shRNA-injected mice spent significantly less time interacting with the social stimulus, compared to the scr. shRNA-injected mice (n = 11 mice/group, F1,40 (interaction) = 10.26, p = 0.003, two-way ANOVA), and displayed a significantly reduced social preference index (t(19) = 3.27, p = 0.004, t-test). In the social approach test, Kmt5b shRNA-injected mice also spent less time interacting with the social stimulus than control animals (Fig. 2B, n = 11 mice/group, t(13) = 2.06, p = 0.06, t-test), albeit being close to but not reaching the defined significance (p < 0.05). In the locomotion test, Kmt5b shRNA-injected mice displayed no change in total distance traveled (Fig. 2C, n = 7 mice/group, F1,12 (treatment) = 0.06, p = 0.81, two-way ANOVA), or activity at the center (distance traveled in the center: F1,12 (treatment) = 0.31, p = 0.59; time spent in the center: F1,12 (interaction) = 0.30, p = 0.59, two-way ANOVA). They also showed no deficits in the rotarod test of motor coordination (Fig. 2D, n = 12–14 mice/group, F1,12 (treatment) = 0.80, p = 0.38, two-way ANOVA).
Fig. 2. Behavioral changes in mice with Kmt5b deficiency in PFC.
A Bar graphs showing the time spent in investigating the social (S) or nonsocial (NS) stimulus and the social preference index in three-chamber social preference tests of mice with PFC injection of scr. or Kmt5b shRNA AAV. Investigation time: ***p < 0.001 (scr. vs. Kmt5b), +++p < 0.001, +p < 0.05 (Soc. vs. NS), two-way ANOVA. Preference index: **p < 0.01, t-test. Inset: representative heat maps. B Bar graphs showing the time spent in investigating the social stimulus (S) in social approach tests of mice with PFC injection of scr. or Kmt5b shRNA AAV. Inset: representative heat maps. C Bar graphs showing the distance traveled (total and in center), as well as the time in center in locomotion tests of mice with PFC injection of scr. or Kmt5b shRNA AAV. Bar graphs showing the latency to fall in rotarod tests (D), the time spent in self-grooming (E), the time spent in investigating novel (Nov.) and familiar (Fam.) objects and the discrimination ratio in novel object recognition (NOR) tests (F) of mice with PFC injection of scr. or Kmt5b shRNA AAV. NOR: investigation time: *p < 0.05 (scr. vs. Kmt5b), +++p < 0.001, +p < 0.05 (Nov. vs. Fam.), two-way ANOVA. G Bar graphs showing startle responses and pre-pulse inhibition of mice with PFC injection of scr. or Kmt5b shRNA AAV.
Self-grooming, a behavior that is thought to model the repetitive behaviors observed in human ASD patients [34], was not altered in Kmt5b shRNA-injected mice (Fig. 2E, n = 11–14 mice/group, t(23) = 0.35, p = 0.73, t-test). In the novel object recognition test, Kmt5b shRNA-injected mice spent less time than controls interacting with the novel object (Fig. 2F, n = 8–10 mice/group, Ftreatment (1,32) = 6.73, p = 0.014, two-way ANOVA), but they maintained a significant preference for the novel object over the familiar object, thus did not differ from controls in the discrimination ratio (t(14) = 1.26, p = 0.23, t-test). In addition, Kmt5b shRNA-injected mice displayed normal startle responses (Fig. 2G, n = 8 mice/group, Finteraction (3,42) = 2.18, p = 0.11, two-way ANOVA) and PPI of startle responses (Finteraction (2,28) = 0.33, p = 0.72, two-way ANOVA), suggesting the lack of sensorimotor gating deficits. Collectively, these data indicate that Kmt5b deficiency in the PFC selectively impairs sociability, a core symptom of autism.
Kmt5b deficiency in PFC impairs glutamatergic transmission
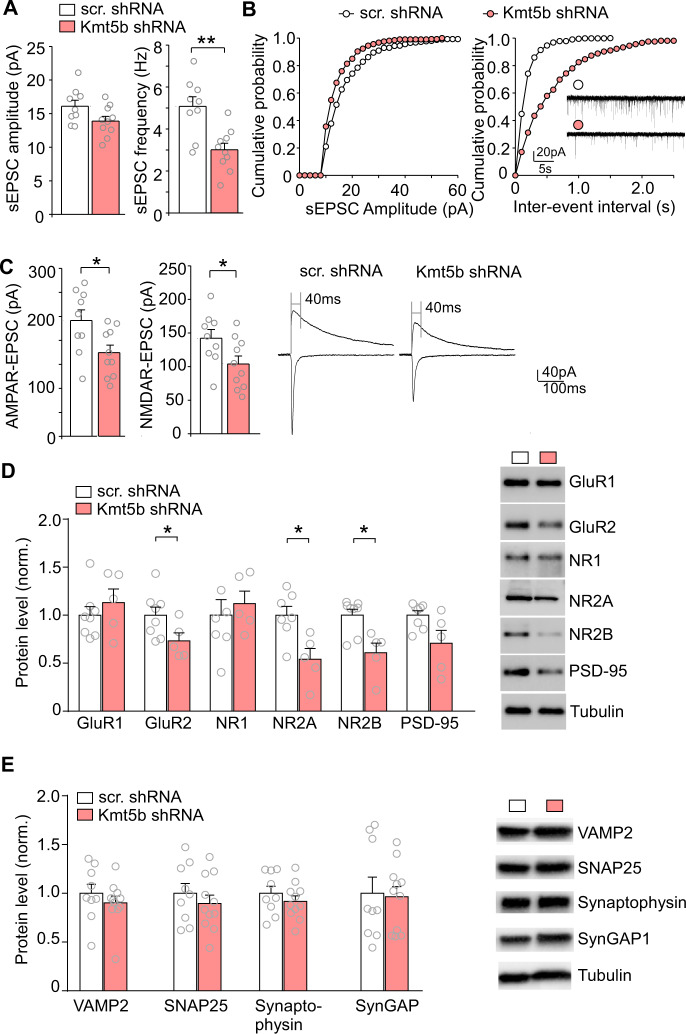
Next, we examined the functional impact of Kmt5b deficiency on synaptic transmission, as glutamatergic neurons in deep layers of PFC are severely impaired in autistic patients [22]. Synaptic dysfunction in PFC has been linked to social deficits in several preclinical autism mouse models [23–26]. To do so, we performed whole-cell patch-clamp recordings in layer V PFC pyramidal neurons of mice injected with Kmt5b shRNA AAV. As shown in Fig. 3A, B, spontaneous excitatory postsynaptic current (sEPSC) frequency was significantly reduced in PFC neurons expressing Kmt5b shRNA, and a trend of reduction was also detected with sEPSC amplitude, compared to those expressing a scr. control shRNA, (scr. shRNA: 16.1 ± 0.9 pA, 5.08 ± 0.46 Hz, n = 9; Kmt5b shRNA: 13.9 ± 0.7 pA, 3.01 ± 0.31 Hz, n = 10; amp, t(15.9) = 2.0, p = 0.07; freq. t(14.4) = 3.8, p = 0.002, t-test). The amplitudes of AMPAR- and NMDAR-mediated EPSC evoked by electrical stimulations were also significantly decreased in Kmt5b-deficient PFC neurons (Fig. 3C, AMPAR-EPSC, scr. shRNA: 241.1 ± 22.3 pA, n = 9, Kmt5b shRNA: 173.5 ± 15.6 pA, n = 10, t(14.6) = 2.5, p = 0.03, t-test; NMDAR-EPSC, scr. shRNA: 142.2 ± 13.0 pA, n = 9, Kmt5b shRNA, 103.5 ± 11.9 pA, n = 10, t(16.6) = 2.3, p = 0.04, t-test).
Fig. 3. Decreased synaptic transmission in Kmt5b-deficient PFC.
Bar graphs and cumulative probability plots of spontaneous EPSC (sEPSC) amplitude (A) and frequency (B) recorded from pyramidal neurons in the PFC of mice injected with Kmt5b shRNA or scr. shRNA AAV. Inset: representative sEPSC traces. C Bar graphs of evoked AMPAR- and NMDAR-EPSC amplitudes recorded from pyramidal neurons in the PFC of mice injected with Kmt5b shRNA or scr. shRNA AAV. Inset: representative eEPSC traces. D, E Bar graphs showing the expression level of synaptic proteins (normalized to tubulin) in the PFC from mice injected with Kmt5b shRNA or scr. shRNA AAV. Inset: representative Western blotting images. In all figures, *p < 0.05, **p < 0.01, t-test.
Considering the impairment of glutamatergic synaptic function in Kmt5b shRNA-infected neurons, we further assessed the expression of several synaptic proteins in the PFC with Kmt5b deficiency. As shown in Fig. 3D, the level of AMPAR subunit GluR2, NMDAR subunits NR2A and NR2B was significantly reduced in Kmt5b shRNA-injected mice (n = 5–8 mice/group, GluR2: t(10) = 2.28, p = 0.046; NR2A: t(9) = 3.17, p = 0.012; NR2B: t(7) = 3.24, p = 0.015; t-test). A trend of reduction was also observed with the postsynaptic density marker PSD-95 (t(5) = 2.05, p = 0.097, t-test). However, the level of presynaptic markers, VAMP2, SNAP25, and Synaptophysin, as well as the PSD protein SynGAP, was unchanged in Kmt5b shRNA-injected mice (Fig. 3E, n = 9–11 mice/group, VAMP2: t(18) = 0.86, p = 0.40; SNAP25: t(18) = 0.79, p = 0.44; Synaptophysin: t(18) = 0.91, p = 0.37; SynGAP: t(18) = 0.19, p = 0.85; t-test). These data indicate that Kmt5b deficiency in the PFC leads to the diminished glutamate receptor expression and glutamatergic synaptic transmission in pyramidal neurons.
Kmt5b deficiency in PFC induces alterations in DNA repair
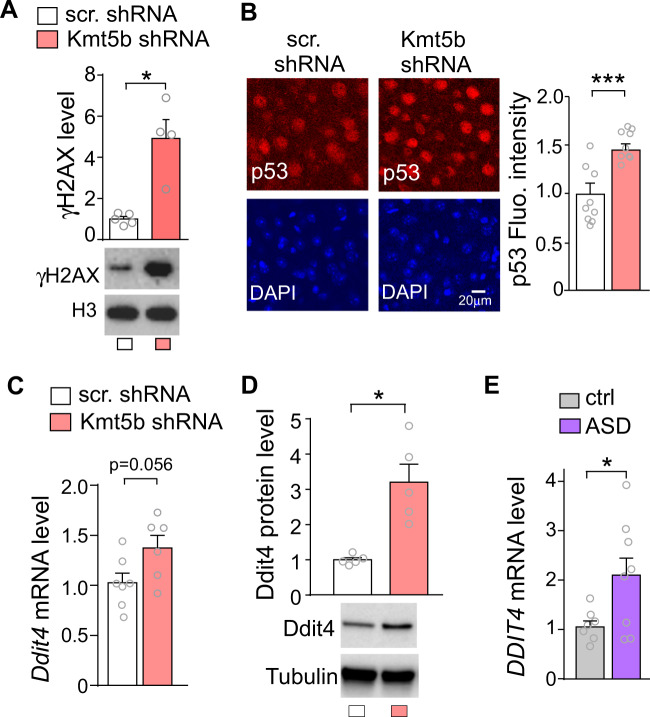
KMT5B is a histone methyltransferase that mediates H4K20me2 [9, 10], which is implicated in DNA damage response by providing a binding platform for the p53-binding protein 1 (53BP1) to facilitate DNA repair [11, 12]. Kmt5b deficiency-induced decrease of H4K20me2 may impair the DNA repair process by reducing 53BP1 foci formation [38, 39]. To test this, we examined the level of γH2AX (the Ser-139 phosphorylated form of histone variant H2AX), which is a highly specific and sensitive molecular marker for monitoring DNA damage and repair [40]. As shown in Fig. 4A, the level of γH2AX in PFC from Kmt5b shRNA-injected mice was significantly higher than that from scr. shRNA-injected mice (n = 4–5/group, t(3.1) = 4.2, p < 0.05, t-test), suggesting that Kmt5b deficiency disrupts the DNA repair and causes the accumulation of DNA DSBs.
Fig. 4. Increased DNA damage, p53, and Ddit4 expression in Kmt5b-deficient PFC.
A Bar graphs and representative Western blots of γH2AX in the PFC of mice injected with Kmt5b shRNA or scr. shRNA AAV. B Immunocytochemical images and quantification of p53 in the PFC of mice injected with Kmt5b shRNA or scr. shRNA AAV. Bar graphs of Ddit4 mRNA (C) and protein (D) in the PFC of mice injected with Kmt5b shRNA or scr. shRNA AAV. E Bar graphs of DDIT4 mRNA in the PFC of postmortem human control vs. ASD patients. In all figures, *p < 0.05, ***p < 0.001, t-test.
DNA DSBs activate p53 [41], one of the major responding molecules to cellular disturbances [42]. p53-dependent biological processes are integrated by the pivotal DSB repair regulator 53BP1 [43]. We found that the level of p53 in PFC was significantly elevated by Kmt5b deficiency (Fig. 4B, n = 8–9 slice/3 mice/group, t(13) = 4.3, p < 0.001, t-test).
Upon activation, p53 acts as a transcription factor to regulate downstream target genes that are involved in various signaling pathways [44, 45]. One downstream target of p53 is DDIT4, which is also called REDD1 (regulated in development and DNA-damage responses 1) [46]. DDIT4 (REDD1) elevation is directly linked to synaptic dysfunction and synaptic loss after chronic stress [47]. We found that Ddit4 mRNA exhibited the trend of increase in Kmt5b-deficient PFC (Fig. 4C, n = 6–7/group, t(10) = 2.17, p = 0.056, t-test), and Ddit4 protein level was significantly elevated by Kmt5b knockdown (Fig. 4D, t(4) = 2.78, p < 0.05, t-test). Interestingly, the mRNA level of DDIT4 (REDD1) was also significantly elevated in the postmortem PFC tissue from ASD human patients (Fig. 4E, n = 7–8/group, t-test, t(8) = 2.34, p < 0.05, t-test). Collectively, these data suggest that KMT5B deficiency-induced loss of H4K20me2 increases DNA damage, leading to the activation of p53 and the increased expression of its target gene Ddit4 (Redd1), which may contribute to synaptic and social deficits.
Kmt5b deficiency in PFC induces transcriptional alterations
KMT5B also catalyzes H4K20me3 [11], a histone modification implicated in transcriptional repression [14–16]. In order to identify the genes regulated by Kmt5b in vivo, we performed RNAseq to assess the downstream transcriptomic changes by Kmt5b deficiency in PFC. Among the differentially expressed genes (DEGs) in PFC with Kmt5b knockdown, 135 were significantly upregulated and 16 were significantly downregulated (Fig. 5A and Supplementary Table 1). The heat maps (Fig. 5B) generated with the expression values for DEGs demonstrated that the Kmt5b shRNA AAV-injected samples clustered together and separated from control samples injected with a scr. shRNA AAV. The majority of DEGs in Kmt5b-deficient PFC displayed the transcriptional upregulation, consistent with Kmt5b’s role as a histone methyltransferase catalyzing the repressive H4K20me3.
Fig. 5. Transcriptomic changes in Kmt5b-deficient PFC.
A Volcano plot showing the transcriptomic distribution in PFC from mice injected with Kmt5b shRNA or scr. shRNA AAV. Significant DEGs are indicated in red circles. B Heat maps representing expression (row z-score) of genes that were significantly up- or downregulated in PFC from mice injected with Kmt5b shRNA or scr. shRNA AAV (n = 3 each group). C Enriched pathways identified via gene ontology analysis of upregulated DEGs in Kmt5b-deficient PFC. Interactome network of upregulated (D) or downregulated (E) genes. Purple lines indicate co-expression; blue lines indicate co-localization. F A schematic model showing the potential mechanism underlying the effects of Kmt5b deficiency in PFC. Kmt5b deficiency-induced loss of H4K20me2 impairs 53BP1-mediated DNA repair, leading to the activation of p53 and its target gene Ddit4 (Redd1). On the other hand, Kmt5b deficiency-induced loss of H4K20me3 reduces transcriptional repression, leading to the upregulation of genes enriched in cellular stress response and ubiquitin-dependent protein degradation. Collectively, it leads to the diminished PFC glutamatergic synaptic transmission and synaptic protein expression. Consequently, mice with Kmt5b deficiency in PFC display social deficits, a core symptom of autism.
To identify functional gene classes regulated by Kmt5b, we performed gene ontology (GO) analysis to classify upregulated DEGs in Kmt5b-deficient PFC. The top-four highly enriched biological process categories included protein folding, translation, covalent chromatin modification, and ubiquitin-dependent catabolic process (Fig. 5C).
To further interrogate the upregulated and downregulated DEGs and their relationships, we created interactome networks based on GO classification with co-expression and co-localization data (Fig. 5D, E). Heat-shock proteins (Hsp), including Hspd1, Hspe1, Hspa8, and Hsp90aa1, which are related to cell stress or damage [48], are the major upregulated hub genes in “protein folding” class. Multiple eukaryotic translation initiation or elongation factors, including Eif4a1, Eif2s3x, and Eef1g, form the upregulated hub genes in “translation” class. Smarca5, a hub gene in “covalent chromatin modification” class, encodes a chromatin remodeling protein that is involved in the regulation of gene transcription. The upregulated hub genes in “ubiquitin-dependent protein catabolic process” class include Ube2s, which encodes an E2 ubiquitin-conjugating enzyme, as well as Psma3 and Psma5, both of which encode essential proteasome subunits that contribute to the complete assembly of 20S proteasome complex.
Discussion
KMT5B is one of the most strongly implicated autism risk factor genes based on large-scale genetic screenings [2–7]. To understand its biological function in the brain related to autism, we have examined the behavioral, physiological, molecular, and genomic consequences of Kmt5b deficiency in mouse PFC, a brain region that exhibits high Kmt5b expression [17, 18] and strongly implicated in autism [22]. We chose to knockdown Kmt5b during the juvenile to adolescent period (3–7 weeks old), because this is a critical time window for the refinement of PFC circuitry, as well as the significant development of sociability and other executive functions mediated by PFC [27–30].
PFC-specific Kmt5b deficiency induces autism-like social deficits, which is linked to glutamatergic synaptic deficits in the PFC, a physiological phenotype commonly found in mouse models of autism [23, 24, 26, 35]. The link is supported by postmortem studies of autistic humans showing the clearest deficits in PFC deep layer pyramidal neuron markers [22]. Moreover, human patients with social dysfunction display reduced activity in the frontal cortex in response to social stimuli [49]. The reduced glutamatergic transmission in Kmt5b-deficient mice is correlated with the diminished expression of AMPAR and NMDAR subunits, including GluR2, NR2A, and NR2B, while some synaptic markers are unchanged, suggesting that Kmt5b-mediated effects may target synaptic and intracellular pools of these glutamate receptors. One possibility is that Kmt5b deficiency causes the change in key regulators that control the synaptic delivery, synthesis, or degradation of glutamate receptors.
One important question is that how Kmt5b deficiency induces the autism-associated physiological and behavioral aberrations. KMT5B catalyzes H4K20me2, which recruits the DNA repair factor 53BP1 to DSBs by forming direct 53BP1/H4K20me2 complex to facilitate DNA repair via nonhomologous end joining [11, 12, 50], one of the DNA repair pathways in nonreplicating neurons. We have found the increased accumulation of DNA DSBs with the molecular marker γH2AX and the increased p53 expression by Kmt5b deficiency, suggesting the critical role of Kmt5b in DNA repair process. The increased P53 is also detected in postmortem brain tissue from idiopathic ASD patients [51–53], implicating the importance of P53 in general pathology of ASD. As a transcription factor, p53 can activate or repress a number of downstream target genes that are involved in various cellular functions [44, 45]. We have found that Kmt5b deficiency induces an increase of the downstream target of p53, Ddit4 (Redd1) [46]. DDIT4 (Ddit4) is elevated in depressed humans and chronically stressed mice, leading to synaptic loss in PFC [47]. It is thus likely that the increased DDIT4 contributes to the loss of synaptic transmission and synaptic proteins, including glutamatergic receptors and the anchoring protein PSD-95, in Kmt5b-deficient PFC.
KMT5B also catalyzes H4K20me3, a histone modification linked to transcriptional repression. Kmt5b deficiency drives the loss of H4K20me3, which could result in the upregulation of genes. Indeed, our RNAseq data indicate that the majority of DEGs are upregulated, with the top GO categories including protein folding, translation, covalent chromatin modification, and ubiquitin-dependent catabolic process. These biological processes are directly involved in the regulation of protein synthesis, gene expression, and protein degradation. Emerging genetic screening and functional studies have linked the compromise of these processes to the manifestation of synaptic and behavioral aberrations associated with ASD [3, 7, 23, 25, 54, 55].
Interactome networks show that hub genes in “protein folding” class are primarily comprised of Hsp, which are broadly implicated in the cellular stress response [48]. Their upregulation suggests that Kmt5b deficiency may compromise cell viability, which is supported by the perinatal lethality of Kmt5b/c homozygous knockout mice [9]. The upregulated hub genes in “translation” class contained multiple eukaryotic translation initiation or elongation factors, as well as Rbm3, a stress-induced RNA-binding protein that has antiapoptotic effects [56]. One upregulated hub gene in “covalent chromatin modification” class was Smarca5, which encodes an ATP-dependent chromatin remodeling protein that is critical for brain development [57] and is closely related to several other SWI/SNF proteins implicated in neurodevelopmental disorders [58]. One upregulated hub gene in “ubiquitin-dependent protein catabolic process” class, Ube2s, encodes an E2 ubiquitin-conjugating enzyme, which is essential for elongating K11-linked polyubiquitin chain on substrates for 26S proteasome-mediated degradation [59]. These altered genes could collectively contribute to the diminished synaptic function and proteins in Kmt5b-deficient PFC.
In summary, the findings reported here suggest that Kmt5b deficiency in the PFC causes the alteration of DNA repair pathway and transcriptional activation of genes involved in cellular stress, resulting in impairment of glutamatergic transmission and social deficits (Fig. 5F). Our results provide a framework for understanding the molecular link between Kmt5b haploinsufficiency and autism spectrum disorders.
Funding and disclosure
We thank Xiaoqing Chen for her excellent technical support. We also thank the support of the Genomics and Bioinformatics Core of State University of New York at Buffalo. This work was supported by grants from the Nancy Lurie Marks Family Foundation and National Institutes of Health (MH112237) to ZY. We also thank EF Trachtman and the Varanasi family for their donations. The authors declare no competing interests.
Supplementary information
Author contributions
Z-JW performed animal surgery, immunocytochemical, biochemical, behavioral experiments and analyzed data. She also participated in bioinformatics analyses, and wrote parts of the draft. BR participated in bioinformatics analyses, some biochemical experiments, and wrote parts of the draft. PZ performed electrophysiological experiments and analyzed data. JW and FY performed bioinformatic analysis of genomic data. QC performed some biochemical experiments. FZ participated in some behavioral experiments. KM and QC generated Kmt5b shRNA AAV plasmid. ZY designed experiments, supervised the project, and wrote the paper.
Data availability
Genomic data will be deposited in a public repository.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41386-021-01029-y.
References
- 1.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–44. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49:515–26. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.RK CY, Merico D, Bookman M, J LH, Thiruvahindrapuram B, Patel RV, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20:602–11. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satterstrom FK, Kosmicki J, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–84.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–61. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyu G, Guan Y, Zhang C, Zong L, Sun L, Huang X, et al. TGF-beta signaling alters H4K20me3 status via miR-29 and contributes to cellular senescence and cardiac aging. Nat Commun. 2018;9:2560. doi: 10.1038/s41467-018-04994-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorgensen S, Schotta G, Sorensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013;41:2797–806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paquin KL, Howlett NG. Understanding the histone DNA repair code: H4K20me2 makes its mark. Mol Cancer Res. 2018;16:1335–45. doi: 10.1158/1541-7786.MCR-17-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-beta. Nat Neurosci. 2013;16:613–21. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–62. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, Coller HA. H4K20 methylation regulates quiescence and chromatin compaction. Mol Biol Cell. 2013;24:3025–37. doi: 10.1091/mbc.e12-07-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pusalkar M, Suri D, Kelkar A, Bhattacharya A, Galande S, Vaidya VA. Early stress evokes dysregulation of histone modifiers in the medial prefrontal cortex across the life span. Dev Psychobiol. 2016;58:198–210. doi: 10.1002/dev.21365. [DOI] [PubMed] [Google Scholar]
- 18.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 19.Goldman-Rakic PS. Cellular basis of working memory. Neuron. 1995;14:477–85. doi: 10.1016/0896-6273(95)90304-6. [DOI] [PubMed] [Google Scholar]
- 20.Amodio DM, Frith CD. Meeting of minds: the medial frontal cortex and social cognition. Nat Rev Neurosci. 2006;7:268–77. doi: 10.1038/nrn1884. [DOI] [PubMed] [Google Scholar]
- 21.Courchesne E, Mouton PR, Calhoun ME, Semendeferi K, Ahrens-Barbeau C, Hallet MJ, et al. Neuron number and size in prefrontal cortex of children with autism. JAMA. 2011;306:2001–10. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 22.Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, et al. Patches of disorganization in the neocortex of children with autism. N Engl J Med. 2014;370:1209–19. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin L, Ma K, Wang ZJ, Hu Z, Matas E, Wei J, et al. Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat Neurosci. 2018;21:564–75. doi: 10.1038/s41593-018-0110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krueger DD, Osterweil EK, Chen SP, Tye LD, Bear MF. Cognitive dysfunction and prefrontal synaptic abnormalities in a mouse model of fragile X syndrome. Proc Natl Acad Sci USA. 2011;108:2587–92. doi: 10.1073/pnas.1013855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ZJ, Zhong P, Ma K, Seo JS, Yang F, Hu Z, et al. Amelioration of autism-like social deficits by targeting histone methyltransferases EHMT1/2 in Shank3-deficient mice. Mol Psychiatry. 2020;25:2517–33. doi: 10.1038/s41380-019-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapanelli M, Tan T, Wang W, Wang X, Wang ZJ, Zhong P, et al. Behavioral, circuitry, and molecular aberrations by region-specific deficiency of the high-risk autism gene Cul3. Mol Psychiatry 2019; Epub ahead of print. [DOI] [PubMed]
- 27.Semple BD, Blomgren K, Gimlin K, Ferriero DM, Noble-Haeusslein LJ. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol. 2013;106–107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baloch S, Verma R, Huang H, Khurd P, Clark S, Yarowsky P, et al. Quantification of brain maturation and growth patterns in C57BL/6J mice via computational neuroanatomy of diffusion tensor images. Cereb Cortex. 2009;19:675–87. doi: 10.1093/cercor/bhn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, et al. Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci. 1999;2:861–3. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- 30.Sturman DA, Moghaddam B. The neurobiology of adolescence: changes in brain architecture, functional dynamics, and behavioral tendencies. Neurosci Biobehav Rev. 2011;35:1704–12. doi: 10.1016/j.neubiorev.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, et al. Autism-like deficits in shank3-deficient mice are rescued by targeting actin regulators. Cell Rep. 2015;11:1400–13. doi: 10.1016/j.celrep.2015.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuen EY, Wei J, Liu W, Zhong P, Li X, Yan Z. Repeated stress causes cognitive impairment by suppressing glutamate receptor expression and function in prefrontal cortex. Neuron. 2012;73:962–77. doi: 10.1016/j.neuron.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rein B, Ma K, Yan Z. A standardized social preference protocol for measuring social deficits in mouse models of autism. Nat Protoc. 2020;15:3464–77. doi: 10.1038/s41596-020-0382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalueff AV, Stewart AM, Song C, Berridge KC, Graybiel AM, Fentress JC. Neurobiology of rodent self-grooming and its value for translational neuroscience. Nat Rev Neurosci. 2016;17:45–59. doi: 10.1038/nrn.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, Rein B, Zhang F, Tan T, Zhong P, Qin L, et al. Chemogenetic activation of prefrontal cortex rescues synaptic and behavioral deficits in a mouse model of 16p11.2 deletion syndrome. J Neurosci. 2018;38:5939–48. doi: 10.1523/JNEUROSCI.0149-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan T, Wang W, Williams J, Ma K, Cao Q, Yan Z. Stress exposure in dopamine D4 receptor knockout mice induces schizophrenia-like behaviors via disruption of GABAergic transmission. Schizophr Bull. 2019;45:1012–23. doi: 10.1093/schbul/sby163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rein B, Tan T, Yang F, Wang W, Williams J, Zhang F, et al. Reversal of synaptic and behavioral deficits in a 16p11.2 duplication mouse model via restoration of the GABA synapse regulator Npas4. Mol Psychiatry 2020; Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 38.Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, Kelleher NL, et al. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J Biol Chem. 2008;283:12085–92. doi: 10.1074/jbc.M707974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsiao KY, Mizzen CA. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J Mol Cell Biol. 2013;5:157–65. doi: 10.1093/jmcb/mjs066. [DOI] [PubMed] [Google Scholar]
- 40.Sharma A, Singh K, Almasan A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol. 2012;920:613–26. doi: 10.1007/978-1-61779-998-3_40. [DOI] [PubMed] [Google Scholar]
- 41.Nelson WG, Kastan MB. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–23. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 43.Cuella-Martin R, Oliveira C, Lockstone HE, Snellenberg S, Grolmusova N, Chapman JR. 53BP1 integrates DNA repair and p53-dependent cell fate decisions via distinct mechanisms. Mol Cell. 2016;64:51–64. doi: 10.1016/j.molcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis—the p53 network. J Cell Sci. 2003;116:4077–85. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- 45.Tedeschi A, Di Giovanni S. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 2009;10:576–83. doi: 10.1038/embor.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/S1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 47.Ota KT, Liu RJ, Voleti B, Maldonado-Aviles JG, Duric V, Iwata M, et al. REDD1 is essential for stress-induced synaptic loss and depressive behavior. Nat Med. 2014;20:531–5. doi: 10.1038/nm.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol (1985) 2002;92:2177–86. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- 49.Richey JA, Rittenberg A, Hughes L, Damiano CR, Sabatino A, Miller S, et al. Common and distinct neural features of social and non-social reward processing in autism and social anxiety disorder. Soc Cogn Affect Neurosci. 2014;9:367–77. doi: 10.1093/scan/nss146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong S, Napoli E, Krakowiak P, Tassone F, Hertz-Picciotto I, Giulivi C Role of p53, Mitochondrial DNA Deletions, and Paternal Age in Autism: A Case-Control Study. Pediatrics 2016;137. [DOI] [PMC free article] [PubMed]
- 52.Araghi-Niknam M, Fatemi SH. Levels of Bcl-2 and P53 are altered in superior frontal and cerebellar cortices of autistic subjects. Cell Mol Neurobiol. 2003;23:945–52. doi: 10.1023/B:CEMN.0000005322.27203.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheikh AM, Malik M, Wen G, Chauhan A, Chauhan V, Gong CX, et al. BDNF-Akt-Bcl2 antiapoptotic signaling pathway is compromised in the brain of autistic subjects. J Neurosci Res. 2010;88:2641–7. doi: 10.1002/jnr.22416. [DOI] [PubMed] [Google Scholar]
- 54.Sahin M, Sur M. Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders. Science. 2015;350:aab38971–8. doi: 10.1126/science.aab3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bagni C, Zukin RS. A synaptic perspective of fragile X syndrome and autism spectrum disorders. Neuron. 2019;101:1070–88. doi: 10.1016/j.neuron.2019.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wellmann S, Truss M, Bruder E, Tornillo L, Zelmer A, Seeger K, et al. The RNA-binding protein RBM3 is required for cell proliferation and protects against serum deprivation-induced cell death. Pediatr Res. 2010;67:35–41. doi: 10.1203/PDR.0b013e3181c13326. [DOI] [PubMed] [Google Scholar]
- 57.Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci USA. 2003;100:14097–102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodwin LR, Picketts DJ. The role of ISWI chromatin remodeling complexes in brain development and neurodevelopmental disorders. Mol Cell Neurosci. 2018;87:55–64. doi: 10.1016/j.mcn.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 59.Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci USA. 2010;107:1355–60. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data will be deposited in a public repository.