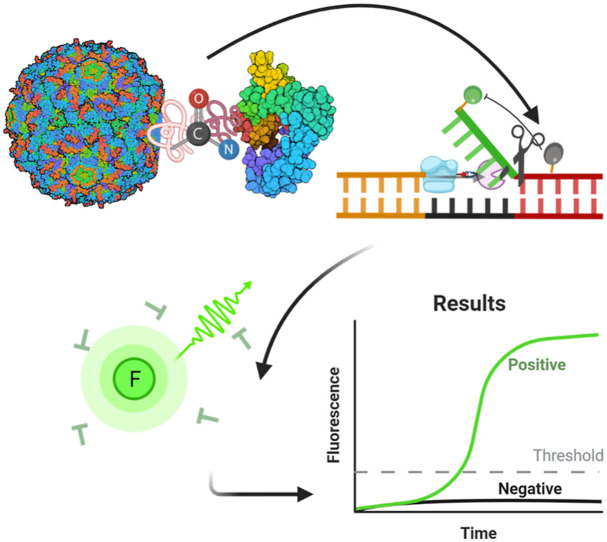
Abstract
The COVID-19 pandemic has unfortunately demonstrated how easily infectious diseases can spread and harm human life and society. As of writing, pandemic has now been on-going for more than one year. There is an urgent need for new nucleic acid-based methods that can be used to diagnose pathogens early, quickly, and accurately to effectively impede the spread of infections and gain control of epidemics. We developed a flap probe-based isothermal nucleic acid amplification method that is triggered by recombinant FEN1-Bst DNA polymerase, which—through enzymatic engineering—has both DNA synthesis, strand displacement and cleavage functions. This novel method offers a simpler and more specific probe–primer pair than those of other isothermal amplifications. We tested the method's ability to detect SARS-CoV-2 (both ORF1ab and N genes), rotavirus, and Chlamydia trachomatis. The limits of detection were 10 copies/μL for rotavirus, C. trachomatis, and SARS-CoV-2 N gene, and 100 copies/μL for SARS-CoV-2 ORF1ab gene. There were no cross-reactions among 11 other common pathogens with characteristics similar to those of the test target, and the method showed 100% sensitivity and 100% specificity in clinical comparisons with RT-PCR testing. In addition to real-time detection, the endpoint could be displayed under a transilluminator, which is a convenient reporting method for point-of-care test settings. Therefore, this novel nucleic acid senor has great potential for use in clinical diagnostics, epidemic prevention, and epidemic control.
Keywords: Nucleic acid diagnosis, Isothermal amplification, COVID-19, Enzymes, Fluorescent probes
Graphical abstract
1. Introduction
Infectious diseases are caused by pathogenic organisms such as bacteria, viruses, fungi, or parasites that spread from person to person, which can lead to a pandemic (Libertucci and Young, 2019). The severity of symptoms caused by different types of pathogens, some of which endanger the patient's life, varies significantly (Daszak et al., 2000; Fauci and Morens, 2012). Coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), has been rapidly spreading since it was first identified in early December 2019 (Wu et al., 2020; Zhu et al., 2020). According to the most recent report from the World Health Organization, there were a total of 180,817,269 confirmed cases of COVID-19 worldwide, including 3,923,238 deaths globally (World Health Organization Coronavirus disease situation dashboard, June 28, 2021). Both the number infections and the number of deaths are still increasing (Organization, 2021).
Pathogen transmission routes include through the respiratory tract by droplets, the fecal–oral route, and by sexual contact. Rotavirus and Chlamydia trachomatis are transmitted through the fecal–oral route and by sexual contact, respectively. Rotavirus, an RNA virus that resembles a wheel when viewed under an electron microscope (Boudreaux et al., 2015), is one of the most common causes of diarrhea in infants and children worldwide, killing approximately 215,000 people a year (Zhu et al., 2017). C. trachomatis, with a DNA genome, mainly affects young women, but can infect both men and women in all age groups and lead to serious health problems without appropriate treatment (Paavonen, 2012; Senyonjo et al., 2018).
When the spread of an infectious disease reaches pandemic level, using rapid mass screening to determine the source of infection is a cornerstone of the strategy to control transmission. At present, nucleic acid–based testing is the most effective method to screen and diagnose pathogens such as SARS-CoV-2, especially in the early stages of an epidemic. Isothermal nucleic acid detection is a promising alternative to the standard polymerase chain reaction (PCR) technique used for pathogen diagnosis (Pumford et al., 2020; Wang et al., 2021a). Currently, widely used isothermal nucleic acid detection methods include loop-mediated isothermal amplification (LAMP) (Notomi et al., 2000), recombinase polymerase amplification (Daher et al., 2016), helicase-dependent amplification (Liu et al., 2020), and rolling circle amplification (Mohsen and Kool, 2016). All of these methods amplify multiple types of nucleic acids under isothermal conditions (Li et al., 2018), dispensing with the need for complex thermal cycling equipment by only requiring simple equipment (for example, a simple vacuum flask) (Hu et al., 2017; Yin et al., 2019) and generating ultrasensitive detection (Gu et al., 2019). Thus, these methods are suitable for large-scale use. However, the use of isothermal amplification methods is limited by high false-positive rates (Ding et al., 2021), and a lack of highly-specific probe-based methods because of the drawbacks of its DNA polymerase (which lackscleavage activity) (Li et al., 2021; Zeng et al., 2020). To increase specificity and sensitivity, an invasive reaction, and the CRISPR-Cas system were added to an existing isothermal amplification system (Jang et al., 2019; Xiong et al., 2021; Zou et al., 2011); however, these methods require multiple steps and additional probes.
A novel nucleic acid diagnostic system that is highly specific and highly sensitive is urgently needed, especially given the current COVID-19 pandemic (Wang et al., 2021c; Xiong et al., 2021; Zhou et al., 2021). Here, we developed a flap probe-based isothermal nucleic acid amplification technique based on a novel multifunctional enzyme (recombinant FEN1-Bst DNA polymerase). This novel multifunctional enzyme confers the functions of DNA synthesis, strand displacement, and cleavage activity, and along with the introduction of a highly-specific hydrolysis probe method, significantly improved the specificity of the reaction. We have successfully applied this method to the diagnosis of different infectious diseases (SARS-CoV-2, rotavirus, and C. trachomatis).
2. Material and methods
2.1. Materials and equipment
Plasmids containing conserved rotavirus sequences, plasmids containing conserved C. trachomatis sequences, pseudotype viruses carrying the SARS-CoV-2 ORF1ab gene, and pseudotype viruses carrying the SARS-CoV-2 N gene were each synthesized (Shanghai Sangon Biotech). The sequences of all of the flap-probe primers (fluorescently labeled) and reverse primers (Shanghai Sangon Biotech) that were used in this study are listed in the Appendix (Table S1). Additional materials included sodium dodecyl sulfate polyacrylamide gels electrophoresis (SDS-PAGE) gel and agarose gel detection kits (ThermoFisher Scientific), reaction buffers (New England BioLabs), a nucleic acid extraction kit (Shanghai Suxin Biotech), and an isothermal amplification real-time fluorescence detection system (Bioer Technology).
2.2. Handling of the samples
Clinical samples (stool samples, genital secretion swabs, or nasopharyngeal swabs) of rotavirus, C. trachomatis, and SARS-CoV-2 (ORF1ab and N genes), diagnosed by approved reverse transcription (RT)-PCR kits, were collected by hospitals, Shanghai Centers for Disease Control and Prevention, and customs. Each sample contained only a single type of pathogen. Samples were stored in a refrigerator at ˗80 °C until use. The Ethics Committee of Fudan University and the Ethics Committee of Hubei Academy of Preventive Medicine approved the sample collection process (2020-023-01). All experiments were completed in a Biosafety Level 2 laboratory.
2.3. Protein recombination
Protein ligation was performed using Bst DNA polymerase (SpyTag) and FEN1 (SpyCatcher) sequences, which are listed in the Appendix (Table S2). The synthetic genes were each transformed into pET-30a vectors (i.e., from expression plasmids to Escherichia coli expression strains). We selected induction conditions to yield a high amount of soluble expression for subsequent experiments and used SDS-PAGE to detect target protein expression to determine the amount of soluble expression (based on the cell lysis supernatant) (Appendix, Figs. S1 and S2). After shake-flask fermentation, the target protein was purified using nickel column affinity chromatography, ion exchange, and hydrophobic chromatography. Finally, Bst DNA polymerase and FEN1 were recombined using SpyTag and SpyCatcher, respectively, which were solubilized in a buffer (containing 10 mM Tris, 50 mM KCl, 0.1 mM EDTA, 1% Triton X-100, 1 mM DTT, pH7.4). SDS-PAGE and high-performance liquid chromatography (HPLC) were used to assess whether the two enzymes had successfully been combined. The chemistry involved in the protein bioconjugation mediated by the SpyTag/Catcher system was as previously described (Wang and Zhang, 2019).
The recombinant enzyme alone was tested to determine whether it still exhibited the enzyme activity of each monomer by performing classic isothermal amplification reactions and flap structure cleavage reactions. The flap structure was formed with three single-stranded nucleic acids; flap structure sequences are listed in the Appendix (Table S3). The formation of the flap structure was confirmed using electrophoresis (Appendix, Fig. S3).
2.4. Optimization
The feasibility of this novel isothermal amplification method was first verified by detecting rotavirus and C. trachomatis plasmids. The method's parameters were then systematically optimized (flap sequence, flap, amount of the recombinant enzyme, amount of the flap-probe primer, and reaction temperature). Real-time fluorescence curves and agarose gel electrophoresis of the amplified products were used to assess the reactions. The parameter values of the optimized reaction system are listed in Table S4. The optimized reaction system was incubated at 63 °C for 60 min, and the fluorescence signals were collected every minute to record the real-time progress of the reaction.
2.5. Detection performance
The detection sensitivity and the lower limits of detection were determined with 10-fold serial diluted rotavirus plasmids, C. trachomatis plasmids, and RNA extracted from each of the pseudotype viruses carrying the SARS-CoV-2 ORF1ab and N genes. The linear relationship (using simple linear regression) between nucleic acid concentrations and the time-to-threshold (Tt) values, which is the time to a fluorescence threshold that is automatically judged by software, was calculated to determine whether the new method has quantitative potential. The threshold was defined as the fluorescence value at the start of the exponential amplification period. The reaction for each nucleic acid concentration was repeated three times, and average Tt values were used to calculate linearity.
2.6. Evaluation in clinical applications
The sensitivity, specificity, positive coincidence rate, negative coincidence rate, and accuracy were evaluated using qualitative RT-PCR results provided by the hospitals as the reference standard.
3. Results and discussion
3.1. Recombinant FEN1-Bst DNA polymerase
The flap endonuclease 1 protein (FEN1) removes 5′ overhanging flaps, short segments of single-stranded DNA that hang off the end because their nucleotide bases cannot bind to the complementary base pair, regardless of any downstream base pairs) during DNA repair (Guo et al., 2020; Xu et al., 2018). This enzyme's cleavage activity should be a useful property in isothermal amplification. In this study, we first attempted to recombine the FEN1 and Bst DNA polymerase by enzymatic engineering through the Spy Tag/Catcher system. SpyTag forms an amide bond with its protein partner (SpyCatcher) in minutes, which results in a high yield after simple mixing, and the peptide bond is not reversed by boiling or by competing peptides. The robust reaction conditions and irreversible linkage of SpyTag and SpyCatcher did not affect the functions of individual enzymes (Keeble et al., 2017; Wu et al., 2018; Zakeri et al., 2012).
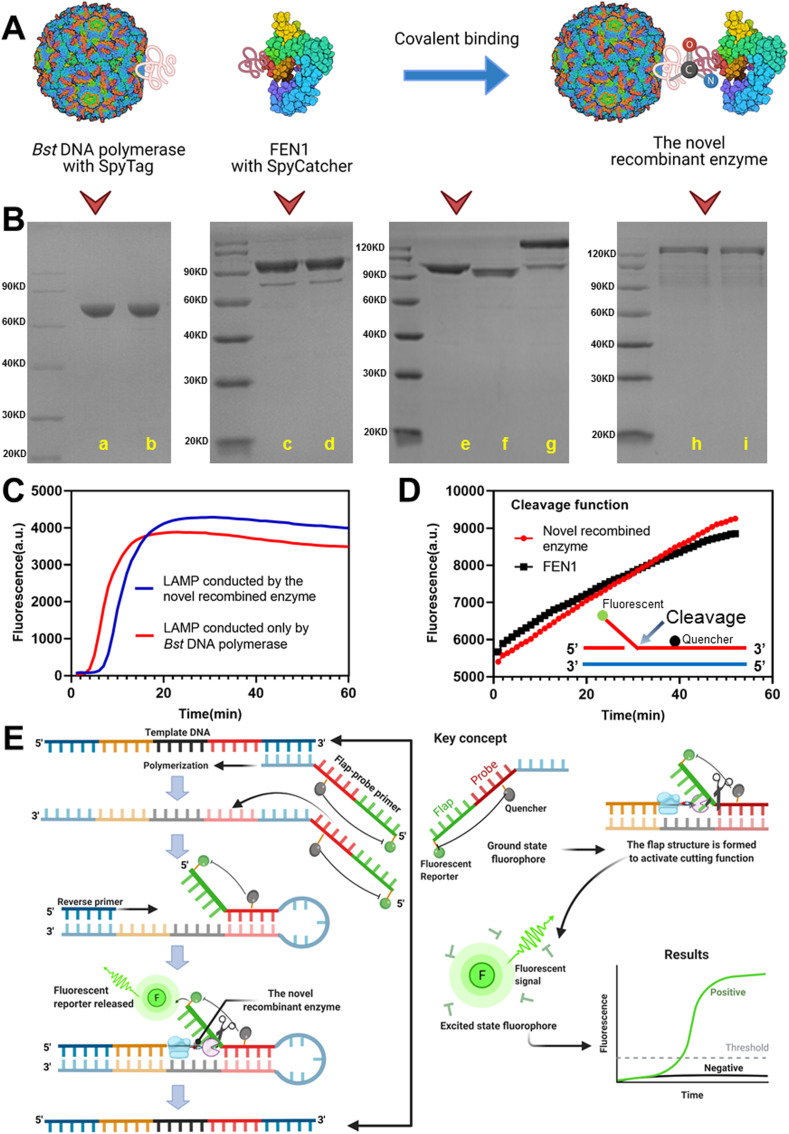
Bst DNA polymerase (large fragment), containing SpyTag, was covalently bound to FEN1, containing SpyCatcher, to form recombinant FEN1-Bst DNA polymerase (Fig. 1 A). Using SDS-PAGE to verify the expression and purification of Bst DNA polymerase with SpyTag, FEN1 with SpyCatcher, and recombinant FEN1-Bst DNA polymerase, we determined that the enzyme proteins had been successfully expressed and purified and that the novel recombinant enzyme had been successfully constructed (Fig. 1B). The reduced state bands were consistent with the non-reduced state bands, which indicated that the enzyme protein states were relatively consistent with one another and that there was no spontaneous homopolymer formation. Using SDS-PAGE and HPLC (Fig. S4), we confirmed successful binding of the two enzyme proteins. In addition, conventional isothermal amplification and flap structure-specific cleavage were successfully triggered by the recombinant enzyme alone (Fig. 1C and D), which indicated that the recombination process (linkage of SpyTag/SpyCatcher) did not affect the enzymes’ individual activities. By creating this multifunctional enzyme, the DNA polymerization, strand displacement, and cleavage functions could exert simultaneous synergistic effects. Using this feature, more strategies can be designed for isothermal nucleic acid amplification, such as introducing a highly-specific hydrolysis probe detection mode.
Fig. 1.
Recombinant FEN1-Bst DNA polymerase and a schematic diagram of flap probe-based isothermal nucleic acid amplification. (A) Schematic diagram of recombination via covalent binding of the SpyTag/SpyCatcher system. (B) SDS-PAGE verification of Bst DNA polymerase with SpyTag, FEN1 with SpyCatcher, and the novel recombinant enzyme; reduced (lane a) and non-reduced (lane b) Bst DNA polymerase (SpyTag) protein; reduced (lane c) and non-reduced (lane d) FEN1 (SpyCatcher) protein; Bst DNA polymerase (SpyTag) (lane e) and FEN1 (SpyCatcher) (lane f), forming the novel recombinant enzyme protein (lane g); and reduced (lane h) and non-reduced (lane i) novel recombinant enzyme protein. (C) Conventional LAMP reaction using the novel recombinant enzyme. (D) Cleavage reaction of the flap nucleic acid structure using the novel recombinant enzyme. (E) The mechanism and key concepts including polymerization, flap structure formation, cleavage to release the fluorescent reporter, and real-time fluorescence detection. Notes: LAMP, loop-mediated isothermal amplification.
3.2. Flap probe-based isothermal nucleic acid amplification by the recombinant enzyme
This flap probe-based isothermal nucleic acid detection method is based on the activity of recombinant FEN1-Bst DNA polymerase, which combines the isothermal DNA polymerization, strand displacement, and endonuclease cleavage functions of the two separate enzymes (Fig. 1E). Specifically, the probe-primer system comprises a flap-probe primer and a reverse primer. The flap-probe primer comprises a nucleic acid flap region that does not match the template (green sequence) and a probe region that recognizes the template (dark red sequence). Each color in the template DNA indicates different segments of DNA. Each color has two shades, dark and light, to represent complementary sequences. First, the dark blue in the template is matched with the light blue in the flap-probe primer. After polymerization and strand displacement, the products have complementary sequences inside (dark and light red), which may generate the circular structure in one side. After forming the circular structure, the flap (green sequence does not match) and probe sequence form a recognizable flap structure. When the reverse primer extends to the flap structure, the new recombinant enzyme cleaves the probe and releases fluorescence, which results in exponentially-amplified signals and real-time amplified curves. Under the reaction temperature (63 °C), the double strands of DNA are in a loose single chain state, and in the schematic diagram we show the reaction from one side only. However, the reaction can also take place from the other side, whereby the reverse primer must first match with the complementary template DNA and trigger the same amplification. Consequently, one template can generate two DNA products, leading to cyclic amplification.
With this design, the probe is highly specific, while flap cleavage can be used to detect the signals in real time, which avoids the need for multi-step detection. The specificity of the present method was granted by the probe sequence recognition. Only when the complementary sequence of the probe existed, could the flap structure form. Then, the released fluorescent signal was sequence specific. This method therefore has superior specificity compared with other technologies such as classic LAMP. Classic LAMP typically uses SYBR-Green or SYBR as the fluorescent reporter. However, these reporters generate fluorescent signals when double stranded DNA exists, which means they are non-sequence specific.
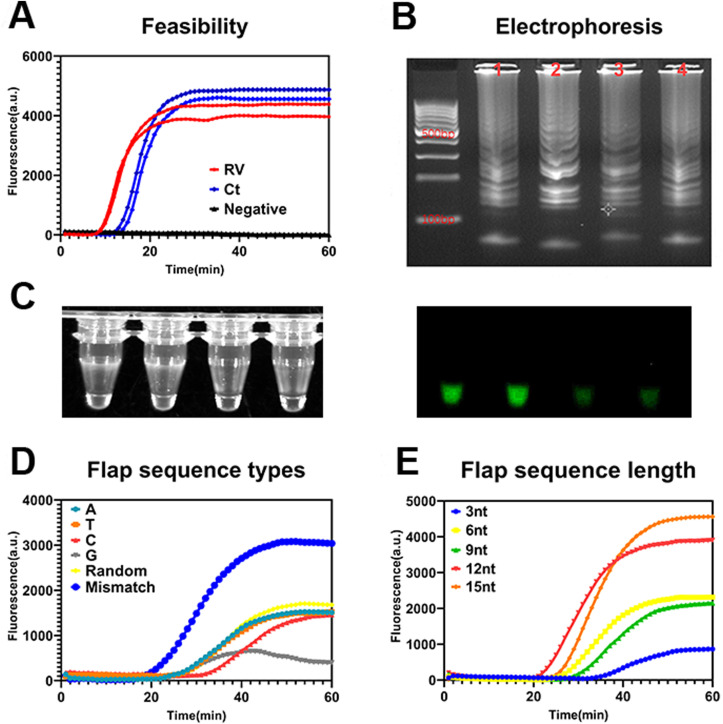
After theoretically designing the method, to verify its feasibility, we designed a suitable flap sequence. First, random nucleotide probe primers were designed for C. trachomatis and rotavirus. Fig. 2 A shows that amplification can be successfully achieved. The products of amplification exhibited classic ladder-like bands upon electrophoresis (Fig. 2B), which was consistent with Bst DNA polymerase-induced amplification (Shi et al., 2016; Tomita et al., 2008) and the product patterns may be determined by the inherent properties of the Bst DNA polymerase (Zyrina et al., 2007). As shown in Fig. 2C, significant green fluorescence is evident under ultraviolet light for positive products, indicating that this method can be used for real-time monitoring and the endpoint can be read by the naked eye, providing a convenient reporting method under the point-of-care test settings. Second, we investigated the optimal flap sequence type including different types of random nucleotides, mismatched nucleotides, and all adenines, thymines, cytosines, or guanines. The flap primer with mismatched nucleotide sequences showed the best amplification effect due to, the formation of a full flap structure between the mismatch nucleotides and the template (Fig. 2D). The recombinant enzyme could mediate full cleavage when it encounters a complete flap structure; however, cleavage may be weaker for the other five types of flap sequences because some nucleotides match the template. Fig. 2E shows that the flap primer with different length mismatched nucleotides, and the 12 mismatched nucleotides, had the best cutting function when compared with other flap sequence lengths. These results suggested that the cleavage function is related to the flap length. The cleavage function was found to be weak when the flap length was relatively short. Consequently, when designing the flap area (green area) on the probe, it should totally mismatch with the template and should be 12 mismatched nucleotides in length. The results of the other parameter optimizations are shown in the Appendix (Figs. S5–S7).
Fig. 2.
Optimization of the flap probe-based isothermal nucleic acid amplification. (A) Feasibility verification of fluorescence; (B) agarose electrophoresis results (corresponding to the curves in A) for rotavirus (line 1 and 3) and Chlamydia trachomatis (line 2 and 4) amplicons; (C) photographs of the amplified products under white light (left) and ultraviolet light (right); (D) fluorescence by flap sequence type; (E) fluorescence by flap sequence length. Notes: A, adenine; C, cytosine; G, guanine; T, thymine; Ct, Chlamydia trachomatis; RV, rotavirus.
3.3. Detection performance of flap probe-based isothermal nucleic acid amplification
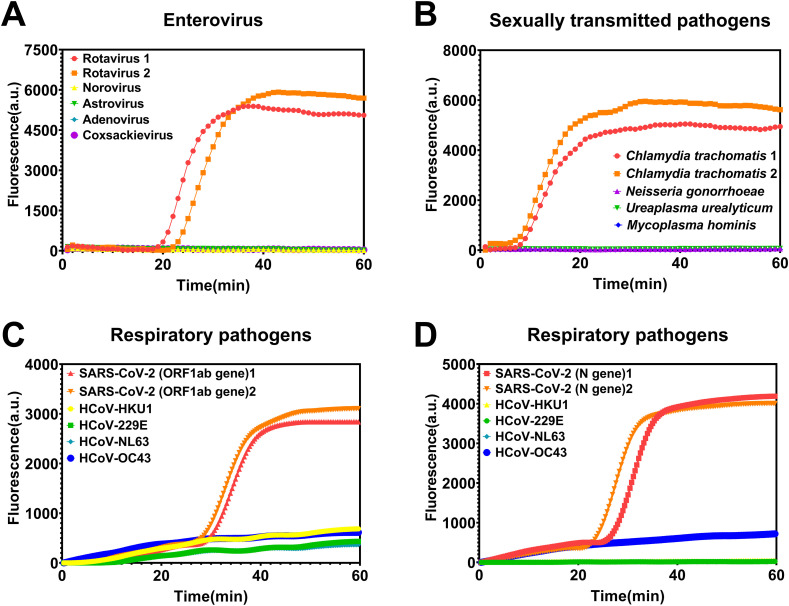
Specificity was determined using three additional types of pathogens with the sites of infection common to each of the three target pathogens: enteroviruses (norovirus, astrovirus, adenovirus, and coxsackievirus) were used to assess the detection specificity of rotavirus; sexually transmitted pathogens (Neisseria gonorrhoeae, Ureaplasma urealyticum, and Mycoplasma hominis) were used to assess the detection specificity of C. trachomatis; and respiratory pathogens (HCoV-HKU1, HCoV-229E, HCoV-NL63, and HCoV-OC43) were used to assess the detection specificity of SARS-CoV-2 (both ORF1ab and N genes). The novel flap probe-based isothermal amplification exhibited perfect specificity (Fig. 3 ); a signal was only generated if the target pathogen was present among the pathogens with similar infection sites and clinical symptoms. These results show that this method can perform accurate differential diagnoses, without false positives, which is a basic requirement in infectious disease diagnosis and control. Given that the specificity was tested among pathogens that included bacteria, viruses (both DNA and RNA types), and mycoplasma, the method has the potential for broad application.
Fig. 3.
Specificity of the flap probe-based isothermal nucleic acid amplification. (A) Detection of rotavirus among multiple enteroviruses; (B) Detection of Chlamydia trachomatis among multiple sexually transmitted pathogens; (C) Detection of SARS-CoV-2 ORF1ab gene among multiple respiratory pathogens; (D) Detection of SARS-CoV-2 N gene among multiple respiratory pathogens.
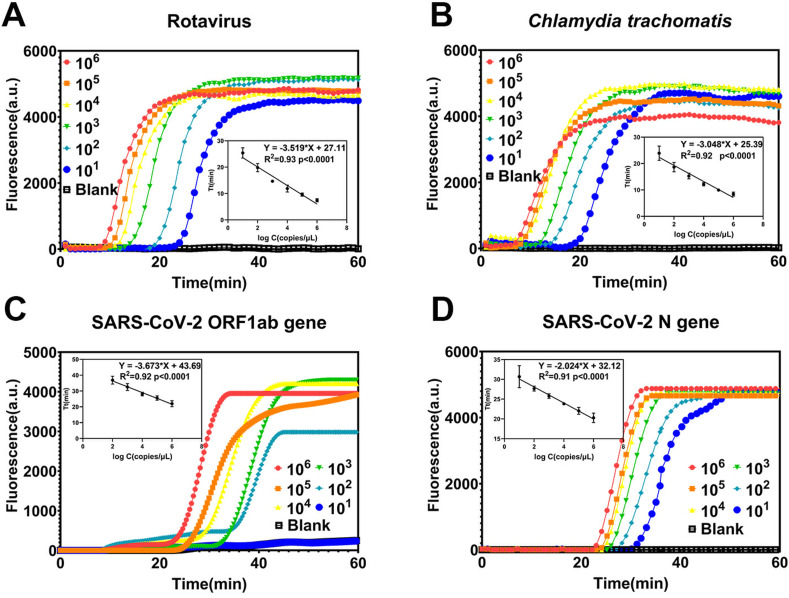
As shown in Fig. 4 , the lower limit of detection for rotavirus, C. trachomatis, and SARS-CoV-2 N gene was 10 copies/μL. The lower limit of detection for SARS-CoV-2 ORF1ab gene was 100 copies/μL. The lower limits of detection are comparable with traditional PCR and CRISPR-Cas12-based detection, which are satisfactory for ensuring that positive detection signals can still be obtained for low infection states (Ma et al., 2020; Wang et al., 2021b). This feature is essential for early-stage infectious disease detection and diagnosis. All four targets demonstrated good linearity (R2 > 0.9, p < 0.0001; Fig. 4) between logarithmically scaled concentrations and Tt values (Table S5), which suggests that the method has potential semi-quantitative capability. The Tt values of the amplified results could be used to infer the pathogen load, which would help doctors judge the infection stage or evaluate clinical treatment effects. The reproducibility of the method is also important. Different batches of the recombinant enzyme were used to detect a rotavirus sample, with eight replications. The amplification curves and the Tt values are provided in Fig. S8 and Table S6 and the results demonstrated good reproducibility. The results only showed a slight difference between two batches of the recombinant enzyme. The coefficient of variation (CV) of the two batches of experiments was 3% and 2.8%, respectively, indicating the good reproducibility of the method.
Fig. 4.
Limits of detection and linearity between logarithmically-scaled concentrations and time-to-threshold (Tt) values of the flap probe-based isothermal nucleic acid amplification: (A) rotavirus, (B) Chlamydia trachomatis, (C) SARS-CoV-2 ORF1ab gene, and (D) SARS-CoV-2 N gene.
3.4. Comparison with RT-PCR
Samples from 120 patients were divided into three categories: stool samples (samples 1–50) from patients with gastrointestinal infections, swab samples (samples 51–90) from patients with urogenital tract infections, and nasopharyngeal swab samples (samples 91–120) from patients with respiratory tract infections. These categorized samples were used to evaluate clinical testing for rotavirus, C. trachomatis, and SARS-CoV-2 (ORF1ab and N genes), respectively. The target-positive samples and target-negative samples were determined by the hospital using RT-PCR (gold standard method), in each category. The results shown in Table 1 demonstrated that the sensitivity and specificity were both 100% for each of the four target pathogens (the amplification curves and the exact Tt values are provided in the Appendix, Fig. S9, and Table S7, respectively). These results provide support that this isothermal amplification method could be used clinically, given that its diagnostic capabilities are comparable to those of the standard method.
Table 1.
Comparison between flap probe-based isothermal amplification and RT-PCR diagnoses (rotavirus, Chlamydia trachomatis, and SARS-CoV-2).
| RT-PCR method |
|||||
|---|---|---|---|---|---|
| Positive | Negative | Total | |||
| The novel flap-probe-based isothermal amplification | Target: Rotavirus (sample 1–50) |
Positive | 27 | 0 | 27 |
| Negative | 0 | 23 | 23 | ||
| Total | 27 | 23 | 50 | ||
| Target: Chlamydia trachomatis (sample 51–90) |
Positive | 21 | 0 | 21 | |
| Negative | 0 | 19 | 19 | ||
| Total | 21 | 19 | 40 | ||
| Target: SARS-CoV-2 ORF1ab gene (sample 91–120) |
Positive | 12 | 0 | 12 | |
| Negative | 0 | 18 | 18 | ||
| Total | 12 | 18 | 30 | ||
| Target: SARS-CoV-2 N gene (sample 91–120) |
Positive | 12 | 0 | 12 | |
| Negative | 0 | 18 | 18 | ||
| Total | 12 | 18 | 30 | ||
4. Conclusions
In conclusion, we successfully recombined Bst DNA polymerase and FEN1 enzymes via the SpyTag/SpyCatcher system. This recombinant enzyme exhibits DNA synthesis functions, cleavage functions, and strand displacement activity, on the basis of which we designed a flap probe-based isothermal amplification method, with simple and specific primers. In addition to real-time analysis, this novel nucleic acid sensor method can also display the endpoint under a transilluminator, which is suitable for point-of-care test. Therefore, this method has great potential for application in clinical diagnosis, epidemic prevention, and epidemic control; however, we acknowledge that large-scale clinical trials are still needed to further evaluate the performance of this method before it can be used for definitive clinical diagnosis.
Author contributions
X. Y. and N. W. contributed equally to this work. All authors have given approval to the final version of the manuscript.
CRediT authorship contribution statement
Xin Ye: Methodology, Formal analysis, Writing – original draft. Ning Wang: Formal analysis. Yang Li: Validation, Software. Xueen Fang: Conceptualization, Writing – review & editing. Jilie Kong: Writing – review & editing, Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the National Natural Science Foundation of China (21974028), the Natural Science Foundation of Shanghai (19441903900), Scientific and Technological Innovation Action Plan (18142201000), and the Shanghai Outstanding Academic Leaders program (19XD1433000, 18QB1403700) for financial support. Some parts in Fig. 1 were created with BioRender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113503.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Boudreaux C.E., Kelly D.F., McDonald S.M. Virology. 2015;477:32–41. doi: 10.1016/j.virol.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher R.K., Stewart G., Boissinot M., Bergeron M.G. Clin. Chem. 2016;62:947–958. doi: 10.1373/clinchem.2015.245829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P., Cunningham A.A., Hyatt A.D. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- Ding S., Chen G., Wei Y., Dong J., Du F., Cui X., Huang X., Tang Z. Biosens. Bioelectron. 2021;178:113041. doi: 10.1016/j.bios.2021.113041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci A.S., Morens D.M. N. Engl. J. Med. 2012;366:454–461. doi: 10.1056/NEJMra1108296. [DOI] [PubMed] [Google Scholar]
- Gu C., Kong X., Liu X., Gai P., Li F. Anal. Chem. 2019;91:8697–8704. doi: 10.1021/acs.analchem.9b02510. [DOI] [PubMed] [Google Scholar]
- Guo E., Ishii Y., Mueller J., Srivatsan A., Gahman T., Putnam C.D., Wang J.Y.J., Kolodner R.D. Proc. Natl. Acad. Sci. U.S.A. 2020;117:19415–19424. doi: 10.1073/pnas.2009237117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Kalsi S., Zeimpekis I., Sun K., Ashburn P., Turner C., Sutton J.M., Morgan H. Biosens. Bioelectron. 2017;96:281–287. doi: 10.1016/j.bios.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Jang H., Lee C.Y., Lee S., Park K.S., Park H.G. Nanoscale. 2019;11:3633–3638. doi: 10.1039/c8nr06699j. [DOI] [PubMed] [Google Scholar]
- Keeble A.H., Banerjee A., Ferla M.P., Reddington S.C., Anuar I., Howarth M. Angew Chem. Int. Ed. Engl. 2017;56:16521–16525. doi: 10.1002/anie.201707623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chang J., Gai P., Li F. ACS Appl. Mater. Interfaces. 2018;10:4561–4568. doi: 10.1021/acsami.7b18676. [DOI] [PubMed] [Google Scholar]
- Li H., Tang Y., Song D., Lu B., Guo L., Li B. Anal. Chem. 2021;93:3315–3323. doi: 10.1021/acs.analchem.1c00007. [DOI] [PubMed] [Google Scholar]
- Libertucci J., Young V.B. Nat. Microbiol. 2019;4:35–45. doi: 10.1038/s41564-018-0278-4. [DOI] [PubMed] [Google Scholar]
- Liu M., Li C.C., Luo X., Ma F., Zhang C.Y. Anal. Chem. 2020;92:16307–16313. doi: 10.1021/acs.analchem.0c04382. [DOI] [PubMed] [Google Scholar]
- Ma P., Meng Q., Sun B., Zhao B., Dang L., Zhong M., Liu S., Xu H., Mei H., Liu J., Chi T., Yang G., Liu M., Huang X., Wang X. Adv. Sci. 2020;7:2001300. doi: 10.1002/advs.202001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsen M.G., Kool E.T. Acc. Chem. Res. 2016;49:2540–2550. doi: 10.1021/acs.accounts.6b00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization W.H. 2021. WHO Coronavirus (COVID-19) Dashboard. [Google Scholar]
- Paavonen J. Ann. Med. 2012;44:18–28. doi: 10.3109/07853890.2010.546365. [DOI] [PubMed] [Google Scholar]
- Pumford E.A., Lu J., Spaczai I., Prasetyo M.E., Zheng E.M., Zhang H., Kamei D.T. Biosens. Bioelectron. 2020;170:112674. doi: 10.1016/j.bios.2020.112674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyonjo L.G., Debrah O., Martin D.L., Asante-Poku A., Migchelsen S.J., Gwyn S., deSouza D.K., Solomon A.W., Agyemang D., Biritwum-Kwadwo N., Marfo B., Bakajika D., Mensah E.O., Aboe A., Koroma J., Addy J., Bailey R. PLoS Neglected Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0007027. e0007027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Shang F., Zhou M., Zhang P., Wang Y., Ma C. Chem. Commun. 2016;52:11551–11554. doi: 10.1039/c6cc05906f. [DOI] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Wang C., Liu M., Wang Z., Li S., Deng Y., He N. Nano Today. 2021;37:101092. doi: 10.1016/j.nantod.2021.101092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., Zhao M., Qian F., Yu H., Liu Z., Ni T., Zheng Y., Wang Y. Biosens. Bioelectron. 2021;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Zhang W.B. Methods Mol. Biol. 2019;2033:287–300. doi: 10.1007/978-1-4939-9654-4_19. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu D., Lin H., Chen D., Sun J., Xie Y., Wang X., Ma P., Nie Y., Mei H., Zhao B., Huang X., Jiang G., Jiang X., Qu J., Zhao J., Liu J. ACS Chem. Biol. 2021;16:491–500. doi: 10.1021/acschembio.0c00840. [DOI] [PubMed] [Google Scholar]
- Wu X.L., Liu Y., Liu D., Sun F., Zhang W.B. J. Am. Chem. Soc. 2018;140:17474–17483. doi: 10.1021/jacs.8b08250. [DOI] [PubMed] [Google Scholar]
- Wu Y.C., Chen C.S., Chan Y.J. J. Chin. Med. Assoc. 2020;83:217–220. doi: 10.1097/JCMA.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong E., Jiang L., Tian T., Hu M., Yue H., Huang M., Lin W., Jiang Y., Zhu D., Zhou X. Angew Chem. Int. Ed. Engl. 2021;60:5307–5315. doi: 10.1002/anie.202014506. [DOI] [PubMed] [Google Scholar]
- Xu H., Shi R., Han W., Cheng J., Xu X., Cheng K., Wang L., Tian B., Zheng L., Shen B., Hua Y., Zhao Y. Nucleic Acids Res. 2018;46:11315–11325. doi: 10.1093/nar/gky911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K., Pandian V., Kadimisetty K., Ruiz C., Cooper K., You J., Liu C. Theranostics. 2019;9:2637–2645. doi: 10.7150/thno.32224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri B., Fierer J.O., Celik E., Chittock E.C., Schwarz-Linek U., Moy V.T., Howarth M. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Liu M., Xia Y., Jiang X. Analyst. 2020;145:7048–7055. doi: 10.1039/d0an01001d. [DOI] [PubMed] [Google Scholar]
- Zhou H., Liu D., Ma L., Ma T., Xu T., Ren L., Li L., Xu S. Anal. Chem. 2021;93:715–721. doi: 10.1021/acs.analchem.0c03996. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Ding S., Wang P., Wei Z., Pan W., Palm N.W., Yang Y., Yu H., Li H.B., Wang G., Lei X., de Zoete M.R., Zhao J., Zheng Y., Chen H., Zhao Y., Jurado K.A., Feng N., Shan L., Kluger Y., Lu J., Abraham C., Fikrig E., Greenberg H.B., Flavell R.A. Nature. 2017;546:667–670. doi: 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou B., Ma Y., Wu H., Zhou G. Angew Chem. Int. Ed. Engl. 2011;50:7395–7398. doi: 10.1002/anie.201001529. [DOI] [PubMed] [Google Scholar]
- Zyrina N.V., Zheleznaya L.A., Dvoretsky E.V., Vasiliev V.D., Chernov A., Matvienko N.I. Biol. Chem. 2007;388:367–372. doi: 10.1515/BC.2007.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.