Abstract
High concentrations of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome have been described in wastewater and sewage sludge. It raises the question of the security of land sludge disposal practices during a pandemic. This study aimed to compare SARS-CoV-2's resistance to the main inactivating factors in sludge treatments, pH and heat, to that of native wastewater somatic coliphages. The latest can be easily used as an indicator of treatment efficiency in the field. The effects of heat treatment and pH on the survival of SARS-CoV-2 and somatic coliphages were investigated in simple media. The T90 value (time required for a 90% reduction in the virus or a 1 × log10 decline) at 50 °C was about 4 min for infectious SARS-CoV-2, and around 133 min for infectious somatic coliphages, with no decrease in SARS-CoV-2 genome. For infectious SARS-CoV-2, a slight decrease (<1 log10 unit) was observed at pH 9 or 10 for 10 min; the decrease was over 5 log10 units at pH 11. However, both SARS-CoV-2 genome and infectious somatic coliphages decreased by less than 1 log10 unit at pH 12. All thermal or pH-based treatments that can remove or significantly reduce infectious somatic coliphages (>4 log10) can be considered efficient treatments for infectious SARS-CoV-2. We concluded that somatic coliphages can be considered highly conservative and easy to use indicators of the inactivation of SARS-CoV-2 during treatments based on heat and alkaline pH.
Keywords: Infectivity, SARS-CoV-2, Somatic coliphages, pH, Heat
Graphical abstract
1. Introduction
Wastewater treatment plants produce large quantities of sewage sludge. The sludge may be incinerated or reused in agricultural applications as a fertiliser. However, raw sludge contains a high quantity of highly diverse pathogenic microorganisms including parasites, bacteria, and viruses (Viau et al., 2011). The main antiviral treatments usually target enteric viruses because of their high degree of resistance to this environment. Enteric viruses are small viruses (20–90 nm in diameter) whose genome is protected only by a proteic capsid. They replicate in the human gut, causing various pathologies (e.g. gastroenteritis, hepatitis, meningitis, heart damage, etc.).
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is responsible for the (Covid-19) coronavirus pandemic that has been raging since December 2019, is an enveloped virus, with a helical nucleocapsid surrounded by lipids. This virus is 60–140 nm in size and its genome is composed of a single RNA-positive strand of around 30 kb (Foladori et al., 2020). SARS-CoV-2 provokes acute pneumonia (cough, fever, rhinorrhoea, dyspnoea) and multiple extra-respiratory signs and, importantly, has also been demonstrated to cause gastrointestinal symptoms (Gu et al., 2020; Pan et al., 2020; Yeo et al., 2020; Zhou et al., 2020). SARS-CoV-2 can productively replicate in human gut enterocytes (Lamers et al., 2020) and the SARS-CoV-2 genome has frequently been detected in the stools of both symptomatic and asymptomatic patients (Wang et al., 2020; Wölfel et al., 2020; Xu et al., 2020; Zhang et al., 2020). It is therefore unsurprising to find high concentrations (until 3 × 103 copies/mL) of the SARS-CoV-2 genome in wastewater (Ahmed et al., 2020; Foladori et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Wu et al., 2020; Wurtzer et al., 2020) and even higher concentrations in sludge (Graham et al., 2021). This suggests the need for a risk assessment of land sludge disposal practices during a pandemic. Yang et al. (2020) separated the risk due to the land disposal of sludge into 3 transmission factors: direct contact, aerosol, and ecological transmission (Yang et al., 2020). Given that aerosols are one of the main modes of transmission (Viau et al., 2011), caution should clearly be taken with SARS-CoV-2, which is a respiratory virus.
Sludge is usually treated using several processes that allow for the complete removal of infectious forms of pathogenic microorganisms (class A biosolids) or their partial removal (class B biosolids) (Viau et al., 2011). The main treatments that inactivate the virus are primarily based on heat and pH (Yang et al., 2020). Temperatures applied during sludge treatments can vary widely depending on treatment type: 35–40 °C in mesophilic digestion, almost 50–55 °C in thermophilic digestion, over 55–65 °C in composting, and up to 70 °C in pasteurization (Alkarimiah and Suja, 2019; Viau et al., 2011). Lime stabilisation leads to a huge increase in pH: it usually raises the pH to higher than 11 and ideally to 12 (Parmar et al., 2001). Viau et al. (2011) have emphasised that a rigorous biosolid pathogen process is the key to reducing pathogen exposure and the risk of infection. Foladori et al. (2020) have reported on the lack of studies on how SARS-CoV-2 behaves in sludge.
The direct detection of SARS-CoV-2 genomes in sludge is not an appropriate way of evaluating treatment efficiency. The presence of the genome does not prove the presence of the infectious virus, since genomes can persist after the virus is no longer infectious. This has been demonstrated to be true of a wide spectrum of viruses and treatments (Gassilloud et al., 2003; Hartard et al., 2018). The presence of viral genome in biosolids may not be a reliable indicator of the presence of the infectious pathogen (Viau et al., 2011). There have been reports of very high concentrations of SARS-CoV-2 genome in stool and wastewater samples, although very few studies have demonstrated the presence of infectious particles (Foladori et al., 2020). However, the absence of the SARS-CoV-2 genome does not prove the absence of contamination because there is no standardised method for quantifying such viruses in sludge and the very small volumes used for detection purposes result in a high detection threshold.
The gold standard for the evaluation of the efficacy of a virucidal treatment is cell culture. SARS-CoV-2 is easily cultured on different cell lines (Park et al., 2020), but culture conditions in level 3 biosafety labs (BSL-3) and the complexity of the environmental matrix make this an inadequate approach for evaluating sludge treatments. There is an urgent need for SARS-CoV-2 inactivation data, to allow us to evaluate the transmission risk posed by wastewater treatment plants (Yang et al., 2020).
Bacteriophages have been used as viral indicators of the efficiency of water and sludge treatments for years (Martín-Díaz et al., 2020). Quantifying infectious phages is rapid (24 h) and easy. Coliphages (e.g. somatic coliphages or F-specific RNA phages) are most frequently used as their behaviour is representative of pathogenic enteric viruses. Somatic coliphages represent a structurally diverse group that is highly resistant to treatments and to heat and pH in particular. F-specific RNA phages are a homogeneous structural group similar to enteric viruses but slightly less resistant to heat and pH, as well as more resistant to other factors, such as UV light (Simonet and Gantzer, 2006). Coliphages are naturally present in wastewater worldwide (Lucena et al., 2004) at concentrations higher than those observed for SARS-CoV-2 at the height of the pandemic (Bertrand et al., 2021). Coliphages are found in primary sludge at concentrations of 105–107.5 plaque-forming units (PFU)/g of dry matter (Martín-Díaz et al., 2020). Coliphages may therefore be used as a field indicator to evaluate the virological risk associated with the reuse of sludge in agriculture in the context of the SARS-CoV-2 pandemic. However, there is a lack of comparative data on their resistance to sludge treatments.
This study was designed to assess the relative resistance of infectious SARS-CoV-2 and native somatic coliphages in wastewater to the main inactivating factors observed in sludge treatments: pH and heat. Native somatic coliphages were used to reflect their wide diversity. They were compared using the same medium for both viruses.
2. Methods
2.1. Wastewater samples
Wastewater samples were collected at a wastewater treatment plant (WWTP) located in the French “Grand Est” region. The average volume of influent treated in this WWTP was ~88,000 m3/day (~250,000 inhabitants). Average daily samples were taken by an automated sampler following either an initial screening step to separate large solids from liquid (i.e. screened wastewater) or a sedimentation step (settled wastewater). Sampling was based on flow rate and started at 7 am on day 1, finishing at 7 am on day 2. Samples were stored in a refrigerated polyethylene tank at 5 °C. The final volume collected was between 15 and 20 L depending on the flow rate of the entrance of the treatment plant. Samples were then homogenised and allowed to settle into 1 L glass bottles. Upon arrival at the laboratory, the wastewater samples were centrifuged at 6000 ×g for 20 min and the supernatant was filtered through 0.45 μm and 0.22 μm filters (Minisart® Sartorius France S.A.S., Aubagne, France) to remove microorganisms that could interfere with cell culture infectivity assays of coronaviruses.
2.2. Viral SARS-CoV-2 samples
The two different SARS-CoV-2 isolates used in this study were obtained from diagnosed patients, whose samples were investigated at the local university hospital (CHRU Brabois, Nancy, France) in compliance with ethical standards and requirements, i.e. informed consent. The nasopharyngeal samples collected in a universal transport medium (UTM) containing 1 mL of viral transport media (Copan Diagnostics Inc., Murrieta, CA, USA) were tested using reverse transcription - quantitative polymerase chain reaction (RT-qPCR) by the laboratory at the local university hospital and found to be positive for SARS-CoV-2 RNA. The purity of the viral isolates was confirmed by multiplex PCR respiratory panel 2.1 plus (RP2.1 plus) (bioMérieux, Craponne, France).
Vero E6 cells (Merck, France) were kindly supplied by the Institut de Recherche en Infectiologie de Montpellier, France (IRIM, UMR 9004 CNRS Université de Montpellier). The cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, D6046, Sigma-Merck, Saint Quentin, France) supplemented with 1% penicillin, streptomycin, amphotericin (Gibco, Thermo Fischer Scientific, Illkirch Graffenstaden, France) and 10% foetal calf serum (FCS, CVFSVF00-01, Eurobio, Les Ulis, France) (i.e. growth medium) at 37 °C in 5% CO2 and were used to grow stock virus and titrate viral infectivity.
All work with infectious SARS-CoV-2 was conducted in the high-containment laboratory (BSL-3) at the local university hospital. Confluent monolayers of Vero E6 cells were inoculated with the virus, in flasks treated with T-175 cell culture (Corning, Wiesbaden, Germany) at 37 °C in 5% CO2 for 7 days. Seven days after the Vero E6 cells had been inoculated with the virus, distinct cytopathic effects (CPE) consisting of the rounding and detachment of cells were observed throughout the T-175 flasks. Infected cells were centrifuged at 1200 ×g for 5 min to remove cell debris and the culture supernatant was aliquoted and stored at −80 °C until further use.
2.3. Titration of infectious SARS-CoV-2
The infectivity of the viral samples was titrated in triplicate on 96-well microtiter plates containing 100 μL of confluent Vero E6 cells. One hundred μL of serial 10-fold dilutions of the virus from 10−1 to 10−8 in DMEM medium with 2% FCS (i.e. maintenance medium) was added to Vero E6 cells. The infected cells were incubated at 37 °C in 5% CO2 for 7 days. The appearance of CPE was recorded daily. The tissue culture infectious dose (50%) (TCID50), defined as the dilution of the virus required to infect 50% of the cell culture, was determined using the Reed and Muench (1938) method and expressed as TCID50/mL. Titrations were performed in triplicate.
2.4. SARS-CoV-2 genome quantification
Nucleic acid extraction was performed using NucliSENS® reagents (bioMérieux). Two volumes of NucliSENS® lysis buffer were added to each volume of liquid sample. Following 10 min of incubation at room temperature, nucleic acid extraction was performed using 70 μL of magnetic silica beads and the NucliSENS® easyMAG™ platform (bioMérieux). The extracted nucleic acids were eluted in 100 μL of elution buffer and stored at −80 °C until needed for SARS-CoV-2 genome quantification.
SARS-CoV-2 RNA quantification was performed using E set real-time RT-PCR, developed by Corman et al. (2020) for the envelope protein (E) gene. Quantification was performed using an RNA UltraSense™ One-Step Quantitative RT-PCR system (Applied Biosystem™, Thermo Fischer Scientific) as previously described (Bertrand et al., 2021). The real-time RT-PCR assays were performed using 5 μL of nucleic acid sample in a 25-μL reaction volume to obtain final concentrations of 0.4 μM for each primer and 0.2 μM for the probe. A CFX96 Touch real-time PCR detection system (BioRad, Marnes-la-Coquette, France) was used for the real-time RT-PCR assays. The RT step was performed for 30 min at 50 °C and PCR amplification was performed for 2 min at 95 °C, followed by 45 cycles of 15 s at 95 °C and 30 s at 58 °C. Negative and positive controls were included in each experiment. The nCoV-ALL-Control plasmid (Eurofins genomic) quantified using Qubit 4 fluorometer (Invitrogen) was used for the standard curve which ranged from 1 × 10−1 to 1 × 104 genome copies (gc)/RT-qPCR reaction. The limit of detection was between 1 and 10 gc/RT-qPCR reaction. The Cq values obtained at T0 and time x, and the slope of the standard curve allowed us to determine the loss in genome occurring during time x.
2.5. Titration of somatic coliphages
Somatic coliphages naturally present in the sewage samples were used in these experiments. They were quantified using the E. coli (WG5) bacterial host strain according to the standard ISO 10705-2 method (International Organization for Standardization, 2000). Concentrations were expressed in plaque-forming units (PFU)/mL.
2.6. Effect of heat treatment on the survival of SARS-CoV-2 and somatic coliphages
SARS-CoV-2 samples (105–106 TCID50/mL) were allowed to settle into 5 tubes (300 μL per tube). Separately, with the somatic coliphages, 5 mL of settled wastewater was diluted in 45 mL of DMEM with 2% FCS, to obtain similar conditions to those used with SARS-CoV-2, and decanted into 5 tubes (10 mL per tube). The tubes were kept for 30 min at experimental temperatures of 40, 50, 60 and 70 °C, respectively. The control was kept at room temperature. The concentrations of infectious SARS-CoV-2, SARS-CoV-2 genome and infectious somatic coliphages before (C0) and after (Cx) heating were determined.
2.7. SARS-CoV-2 and somatic coliphages survival in wastewater at 20 °C
One milliliter of virus stock (105–106 TCID50/mL) was spiked into 9 mL aliquots of freshly settled wastewater. A positive control sample for measuring the initial virus concentration in DMEM with 2% FCS, as well as negative control including settled wastewater in DMEM with 2% FCS were included. The samples were held at room temperature (20 °C) for 7 days, without agitation for the duration of the experiment. At each time point (day 1 to 7), samples were taken and assayed for virus infectivity on Vero E6 cell line and expressed in TCID50/mL. In parallel, infectious somatic coliphages were quantified in wastewater every 24 h during storage at room temperature for 7 days.
2.8. Effect of pH on the survival of SARS-CoV-2 and somatic coliphages
Different dilutions of NaOH (1 M) solution in distilled water were prepared in order to achieve final solutions of pH 9, 10, 11 and 12. Fifty μL of stock SARS-CoV-2 virus (105–106 TCID50/mL) were added to 450 μL of different dilutions of NaOH and to distilled water as a control. After 10 min of incubation at room temperature, the dilutions were neutralised to pH 7.5 using HCl (0.1 M). Separately, with the somatic coliphages, settled wastewater was diluted 10-fold in sterile, deionized water. The initial pH was 8.0. A NaOH solution (1 M) was added to produce pH values of 9–12, as with SARS-CoV-2. Following 10 min of incubation at room temperature, the solution was neutralised by adding HCl solution (1 M) until it reached pH 8. Measuring the concentrations of infectious SARS-CoV-2, SARS-CoV-2 genome and infectious somatic coliphages before (C0) and after (Cx) pH treatment of the solutions stored at room temperature allowed us to determine the decay rate of each parameter.
2.9. Data analysis
The samples were analysed using Excel 2007 (Microsoft Corp., San Diego, CA). Regression analysis was used to model the survival of SARS-CoV-2 and somatic coliphages as a function of time. To this end, simple linear regression models were fitted to experimental data and used to estimate T90 reduction times (predicted number of days taken to achieve a 1-log10 reduction). The survival function of each virus under the given experimental conditions (i.e., temperature or pH) was modelled as follows:
where Cx (C0) is viral density at time x (or 0, respectively), expressed in days (t). The slopes (a) calculated for the different viruses under consideration express the corresponding concentration decreases in log10 units per time unit.
3. Results
3.1. Survival of SARS-CoV-2 and coliphages during heat treatment
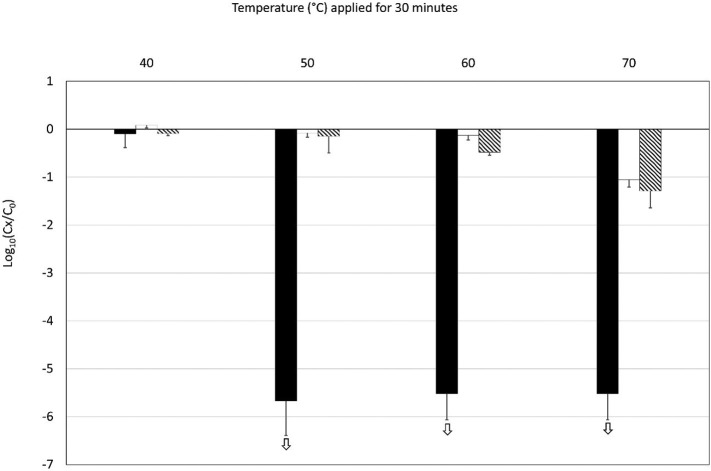
Heat treatment was applied to SARS-CoV-2 and somatic coliphages in triplicate. Temperatures of 40, 50, 60 and 70 °C were applied for 30 min to reflect the range of temperatures usually obtained during the treatment of sludge (Fig. 1 ). The infectivity of SARS-CoV-2, somatic coliphages and SARS-CoV-2 genome before and after heat treatment was estimated. A 30 min exposure to 40 °C produced almost no effect on SARS-CoV-2 or somatic coliphages that could be detected using cell culture or RT-qPCR methods. At 50 °C, inactivation of 5.5 log10 units was observed with infectious SARS-CoV-2, whereas SARS-CoV-2 genome and infectious somatic coliphages remained stable. Due to the initial concentrations of infectious SARS-CoV-2 in our 3 experiments, assessment of inactivation was limited to a maximum of 5.5 log10 units and this level was also reached at 60 and 70 °C. At 70 °C, some inactivation of somatic coliphages was also observed, ranging between 0.9 and 1.6 log10 units (n = 3). A decrease in SARS-CoV-2 genome was only observed at 70 °C: its mean value was 1.5 log10 units.
Fig. 1.
Decrease in infectious SARS-CoV-2 (black bars), SARS-CoV-2 genome (white bars) and infectious coliphages (crosshatched bars) following 30 min heat treatments (at 40, 50, 60 and 70 °C). The experiments were performed in triplicate. The white arrows indicate that the decrease in infectious SARS-CoV-2 was greater than shown, though our estimations were limited by the low initial concentrations.
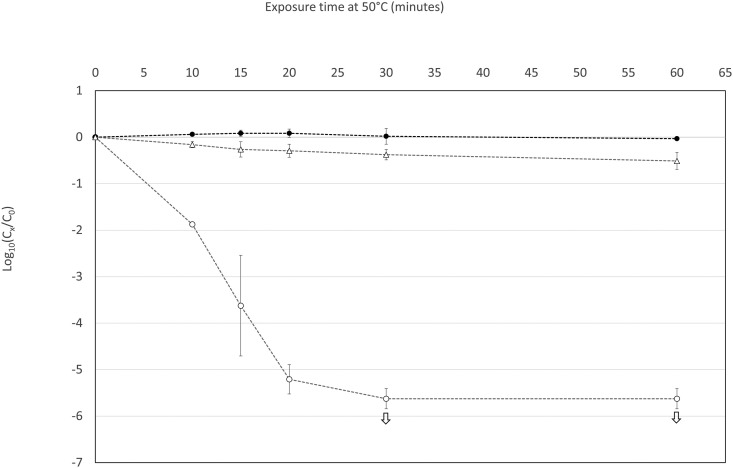
Other experiments were performed by exposing SARS-CoV-2 and somatic coliphages to 50 °C for 10–60 min (Fig. 2 ). The decrease in infectivity observed in SARS-CoV-2 and somatic coliphages allowed us to determine their respective T90 values (time required for a 90% reduction in the virus or a decline of 1 × log10) at this temperature. At 50 °C, infectious SARS-CoV-2 had a T90 value of about 4 min, compared with 133 min for infectious somatic coliphages. No decrease in SARS-CoV-2 genome was observed at this temperature.
Fig. 2.
Decrease in infectious SARS-CoV-2 (blank circles), SARS-CoV-2 genome (filled-in circles) and infectious coliphages (blank triangles) at 50 °C over time. The experiments were performed in triplicate. The white arrows indicate that the decrease in infectious SARS-CoV-2 was greater than shown, though our estimations were limited by the low initial concentrations.
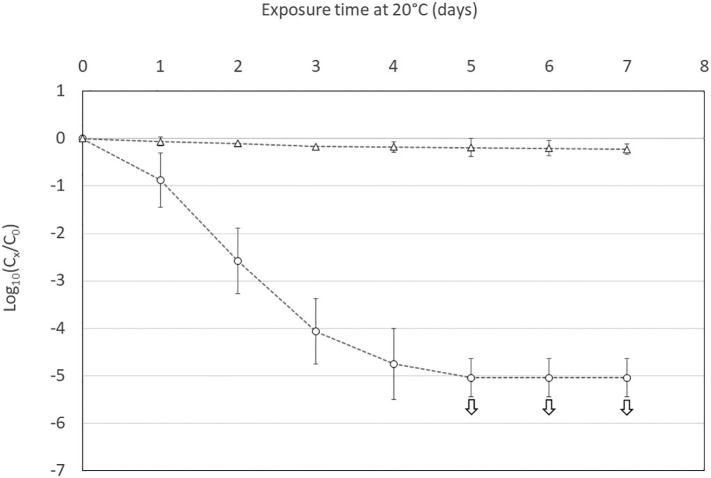
The infectivity of SARS-CoV-2 and somatic coliphages was also monitored in wastewater maintained at 20 °C for 7 days (Fig. 3 ). A constant decrease in infectivity was observed for SARS-CoV-2 and it reached at day 5 the maximum of 5.0 log10 allowed by the initial concentration. From the decrease observed the first four days, it was possible to determine that the T90 value was about 18 h. In accordance with the results previously obtained with higher temperatures, the infectivity of somatic coliphages remained stable during the 7 days of the experiments.
Fig. 3.
Decrease in infectious SARS-CoV-2 (blank circles) and infectious coliphages (blank triangles) in wastewater at 20 °C over time. The experiments were performed in triplicate. The white arrows indicate that the decrease in infectious SARS-CoV-2 was greater than shown, though our estimations were limited by the low initial concentrations.
3.2. The pH-dependent survival of SARS-CoV-2 and coliphages
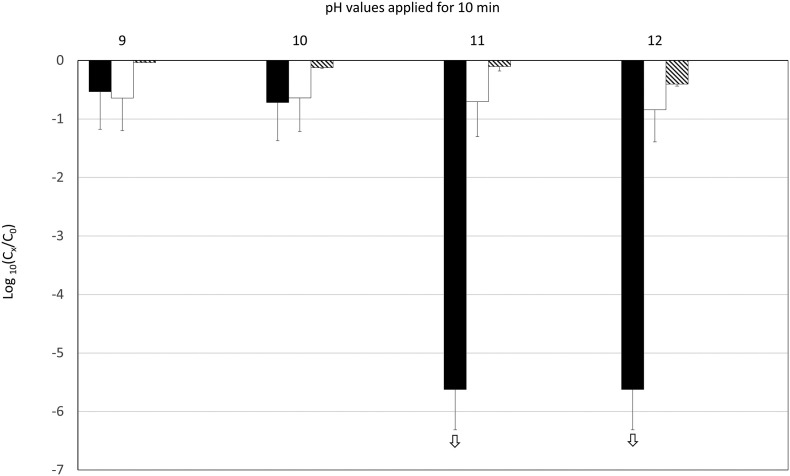
A pH range of 9–12 was applied to SARS-CoV-2 and somatic coliphages for 10 min (Fig. 4 ). A slight decrease (<1 log10 unit) in infectious SARS-CoV-2 was observed at pH 9 or 10. There was a decrease of approximately 5.5 log10 units at pH 11. The decrease was much smaller across all other parameters. There were decreases in SARS-CoV-2 genome and somatic coliphages of less than 1 log10 unit at pH 12.
Fig. 4.
Decrease in SARS-CoV-2 (infectious: black bars; genome: white bars) and coliphages (infectious: crosshatched bars) at different pH values applied for 10 min. The experiments were performed in triplicate. The white arrows indicate that the decrease in infectious SARS-CoV-2 was greater than shown, but our estimations were limited by the low initial concentrations.
Increasing the exposure time at pH 9 or 10 from 10 to 60 min did not affect the inactivation of infectious SARS-CoV-2 (data not shown).
4. Discussion
We compared the inactivation of SARS-CoV-2 and somatic coliphages following thermal and alkaline treatments. The objective was to ascertain whether these bacteriophages are a useful tool for evaluating the effectiveness of SARS-CoV-2 sludge treatments.
The probability that infectious SARS-CoV-2 is present in raw wastewater is still hotly debated. Many studies have demonstrated the presence of SARS-CoV-2 genome in stools and wastewater. A review by Foladori et al. (2020) estimated that SARS-CoV-2 is present in 3.8–80% of stool samples. Viral concentration in the stools of patients, with and without diarrhea, ranged from 5 × 103–107.6 genome copies (gc)/mL (Cheung et al., 2020; Foladori et al., 2020). The maximum concentration in raw wastewater was almost 107 genome copies/L (Wurtzer et al., 2020). Despite these very high genome concentrations, there is very little evidence that infectious SARS-CoV-2 is present in stools or wastewater. Some studies describe the presence of infectious SARS-CoV-2 in stool without providing any details about the procedure used to obtain these results (Wang et al., 2020; Xiao et al., 2020). Zhang et al. (2020) used electron microscopy to obtain an image of the virus after inoculating Vero cells with stool samples, but failed to provide any quantitative data. Environmental studies by Rimoldi et al. (2020) and Wölfel et al. (2020) have not found infectious SARS-CoV-2 in wastewater or stools, despite very high genome concentrations. This evidence, together with our results, clearly shows that, in the case of several different viruses, the presence of viral genome is not proof of infectivity and that an evaluation based only on the detection of SARS-CoV-2 genome can lead to a significant and systematic overestimation of the presence of infectious SARS-CoV-2. The worst case estimates of 107 gc/L SARS-CoV-2 in wastewater at the peak of the first epidemic wave in France therefore suggests a much lower concentration of infectious SARS-CoV-2.
Infectious somatic coliphages are relatively stable in raw wastewater over time. Their concentration in raw wastewater has been shown to vary between 107 and 108 PFU/L worldwide (Lucena et al., 2004). Because of the rich diversity of these phages, the concentration of corresponding genome copies cannot be estimated. Nevertheless, the concentrations are surely much higher than the concentration of infectious SARS-CoV-2, even in the worst-case scenario.
Together, these data suggest that the concentration of infectious SARS-CoV-2 in raw wastewater may be considerably lower than the concentration of SARS-CoV-2 genome or of infectious somatic coliphages. However, although the survival rate of infectious virus may be low, we cannot completely rule out the presence of infectious SARS-CoV-2 in raw wastewater given the current state of our knowledge. We may therefore assume that infectious SARS-CoV-2 can be present in primary sludge.
The prevalence of SARS-CoV-2 in sludge has been less well described. Graham et al. (2021) found a higher concentration in sludge than in wastewater, but the concentration in sludge was still less than 104 gc/g. Again, we can assume that genome analysis alone will lead to a huge overestimation of the presence of infectious virus. Infectious somatic coliphages have been estimated to be present in primary sludge at concentrations of 105.2–107.5 PFU/g of dry weight (Martín-Díaz et al., 2020). Once again, these concentration values greatly exceed those of SARS-CoV-2, even when our estimates are based on the presence of viral genome.
Sludge treatments usually rely primarily on heat and alkaline pH to reduce microbiological contamination. The data obtained by our study provide a very clear picture of the impact of such treatments on SARS-CoV-2 and somatic coliphages. There is a 30-fold difference in the survival times of native infectious somatic coliphages and infectious SARS-CoV-2 under the same thermal conditions. The T90 values at 50 °C were almost 4 min and 133 min for SARS-CoV-2 and somatic coliphages, respectively. T90 values ranging from 14 to 17 min have been recently obtained for SARS-CoV-2 inoculated in wastewater stored at 50 °C (Bivins et al., 2020). These data support the hypothesis that coronaviruses are less resistant to such treatments than non-enveloped viruses. Berne virus, TGEV (transmissible gastroenteritis virus), MHV (murine hepatitis virus), SARS-CoV and MERS-CoV all demonstrated less thermal resistance compared to the model provided by Bertrand et al. (2012) for non-enveloped viruses (Quist-Rybachuk et al., 2015; Leclercq et al., 2014; Darnell et al., 2004; Rabenau et al., 2005; Casanova et al., 2010; Casanova et al., 2009; Laude, 1981; Weiss and Horzinek, 1986). This is also true of other enveloped viruses, such as Phi 6 phage and filovirus (Casanova and Weaver, 2015; Bibby et al., 2015; Piercy et al., 2010). Mesophilic and thermophilic anaerobic sludge treatments produce inactivations of around 2 and 4 log10 respectively after 21 days, but lead to an over 6 log10 inactivation of Phi 6 phage in both cases (Pepper et al., 2010; Sassi et al., 2018).
The same significant difference between SARS-CoV-2 and somatic coliphages was observed in pH treatments. At a pH of below 11, little inactivation was observed within 10 min of exposure at room temperature, as previously demonstrated with other coronaviruses, such as PEDV (porcine epidemic diarrhea virus) (Hofmann and Wyler, 1989; Quist-Rybachuk et al., 2015). Nevertheless, at a pH of 11 and above, decreases of 1-log10 and over 5-log10 were observed in somatic coliphages and SARS-CoV-2, respectively.
Synergistic effects between pH and temperature can be observed (Hofmann and Wyler, 1989). For example, at pH 9, PEDV had a T90 value of about 27 min at 40 °C, 18 min at 44 °C and 4 min at 48 °C (Quist-Rybachuk et al., 2015). At pH 10.2, the T90 values were around 14, 8 and 1 min, respectively (Quist-Rybachuk et al., 2015). Some irreversible conformational changes to the viral envelope proteins that prevent cell receptor recognition may occur at an alkaline pH, as described with MHV (Weismiller et al., 1990).
Together, our results allow us to draw important conclusions regarding sludge storage and treatments in the context of land disposal. Taking into account the rapid loss of infectivity of SARS-CoV-2 in wastewater within a few days at 20 °C, and the necessarily long periods of storage of sludge before land spreading, ranging from a few weeks to several months, the presence of infectious SARS-CoV-2 in stored sludge seems very unlikely. Additionally, our results demonstrate that the methods usually applied for sludge hygienization, consisting of heat or pH treatment, will be very efficient in eliminating infectious SARS-CoV-2 particles, if any is still present in sludge. Somatic coliphages are clearly highly conservative indicators of SARS-CoV-2 inactivation. All thermal and pH-based treatments that can remove or significantly reduce somatic coliphages (e.g. ≥6 log10 for a class A reclaimed water (Alcalde-Sanz and Gawlik, 2017)) should be viewed as effective SARS-CoV-2 treatments. Consequently, somatic coliphages constitute a simple, easy-to-implement and cost-effective surrogate to validate the effectiveness of SARS-CoV-2 treatments.
Funding
This study was financially supported by SUEZ (France). It has partially benefited from the financial support of the French Government's Ministry of Higher Education and Research and Innovation (Ministère de l'enseignement supérieur, de la recherche et de l'innovation [MESRI]) for the development of the SARS-CoV-2 quantification method. All the authors' salaries were funded by either Lorraine University (France), the University Hospital of Nancy (France) or SUEZ (France).
CRediT authorship contribution statement
M. Varbanov: Methodology, Formal analysis, Investigation, Writing - Original Draft, Supervision, I. Bertrand: Methodology, Formal analysis, Investigation, Writing - Original Draft, Supervision, S. Philippot: Investigation, Writing - Review & Editing, C. Retourney: Investigation, Writing - Review & Editing, M. Gardette: Investigation, Writing - Review & Editing, C. Hartard: Formal analysis, Investigation, Writing - Review & Editing, H. Jeulin: Methodology, Resources, Writing - Review & Editing, Supervision, R.E. Duval: Writing - Review & Editing, J.-F. Loret: Conceptualization, Writing - Review & Editing, Funding acquisition, E. Schvoerer: Methodology, Resources, Writing - Review & Editing, Supervision, C. Gantzer: Conceptualization, Methodology, Writing - Original Draft, Funding acquisition, Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the French scientific interest group “Obépine” for the surveillance of SARS-CoV-2 in wastewater of France. The steering committee of “Obépine” is composed of I. Bertrand and C. Gantzer (Laboratoire de Chimie Physique et Microbiologie pour les Matériaux et l’Environnement, LCPME, UMR 7564 CNRS-Université de Lorraine, Nancy, France), S. Le Guyader (Ifremer Nantes, France), Y. Maday (Institut Carnot Smiles, Sorbonne Université, Laboratoire Jacques-Louis Lions, UMR 7598 and Institut Universitaire de France), V. Maréchal (Sorbonne Université, INSERM, Centre de Recherche Saint-Antoine, 75012, Paris, France), J.-M. Mouchel (Sorbonne Université, CNRS, EPHE, UMR 7619 Metis, Paris, France, e-LTER Zone Atelier Seine), L. Moulin and S. Würtzer (Laboratoire R&D Eau de Paris, France), M. Boni (Val de Grâce, Unité de Virologie de l'IRBA. Institut de Recherche Biomédicale des Armées, Brétigny, France).
Editor: Paola Verlicchi
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Jochen F., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcalde-Sanz L., Gawlik B.M. Publications Office of the European Union; Luxembourg: 2017. Minimum quality requirements for water reuse in agricultural irrigation and aquifer recharge - towards a water reuse regulatory instrument at EU level. JRC Science for Policy Report. EUR 28962 EN. (ISBN 978-92-79-77175-0) [DOI] [Google Scholar]
- Alkarimiah R., Suja F. Effects of technical factors towards achieving the thermophilic temperature stage in composting process and the benefits of closed rector system compared to conventional method – a mini review. Appl. Ecol. Environ. Res. 2019;17(4):9979–9996. doi: 10.15666/aeer/1704_99799996. [DOI] [Google Scholar]
- Bertrand I., Schijven J.F., Sánchez G., Wyn-Jones P., Ottoson J., Morin T., Muscillo M., Verani M., Nasser A., de Roda Husman A.M., Myrmel M., Sellwood J., Cook N., Gantzer C. The impact of temperature on the inactivation of enteric viruses in food and water: a review. J. Appl. Microbiol. 2012;112(6):1059–1074. doi: 10.1111/j.1365-2672.2012.05267.x. [DOI] [PubMed] [Google Scholar]
- Bertrand I., Challant J., Jeulin H., Hartard C., Mathieu L., Lopez S., Schvoerer E., Courtois S., Gantzer C., Scientific Interest Group Obépine Epidemiological surveillance of SARS-CoV-2 by genome quantification in wastewater applied to a city in the northeast of France: comparison of ultrafiltration- and protein precipitation-based methods. Int. J. Hyg. Environ. Health. 2021;233 doi: 10.1016/j.ijheh.2021.113692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Fischer R.J., Casson L.W., Stachler E., Haas C.N., Munster V.J. Persistence of ebola virus in sterilized wastewater. Environ. Sci. Technol. Lett. 2015;2(9):245–249. doi: 10.1021/acs.estlett.5b00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Weaver S.R. Evaluation of eluents for the recovery of an enveloped virus from hands by whole-hand sampling. J. Appl. Microbiol. 2015;118(5):1210–1216. doi: 10.1111/jam.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Jeon S., Rutala W.A., Weber D.J., Sobsey M.D. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl. Environ. Microbiol. 2010;76(9):2712–2717. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., Yip C.C.Y., Leung K.H., Fung A.Y., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J.X., To K.K.W., Chan K.H., Yuen K.Y., Leung W.K. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M.E.R., Subbarao K., Feinstone S.M., Taylor D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Methods. 2004;121(1):85–91. doi: 10.1016/j.jviromet.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassilloud B., Schwartzbrod L., Gantzer C. Presence of viral genomes in mineral water: a sufficient condition to assume infectious risk? Appl. Environ. Microbiol. 2003;69(7):3965–3969. doi: 10.1128/aem.69.7.3965-3969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ Sci Technol. 2021;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020;158(6):1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartard C., Leclerc M., Rivet R., Maul A., Loutreul J., Banas S., Boudaud N., Gantzer C. F-specific RNA bacteriophages, especially members of subgroup II, should be reconsidered as good indicators of viral pollution of oysters. Appl. Environ. Microbiol. 2018;84(1) doi: 10.1128/AEM.01866-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Quantitation, biological and physicochemical properties of cell culture-adapted porcine epidemic diarrhea coronavirus (PEDV) Vet. Microbiol. 1989;20(2):131–142. doi: 10.1016/0378-1135(89)90036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Organization for Standardization . 2000. ISO 10705-2. Water Quality: Detection and Enumeration of Bacteriophages. Part 2: Enumeration of Somatic Coliphages. [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H. Thermal inactivation studies of a coronavirus, transmissible gastroenteritis virus. J. Gen. Virol. 1981;56(Pt 2):235–240. doi: 10.1099/0022-1317-56-2-235. [DOI] [PubMed] [Google Scholar]
- Leclercq I., Batéjat C., Burguière A.M., Manuguerra J.C. Heat inactivation of the Middle East respiratory syndrome coronavirus. Influenza Other Respir. Viruses. 2014;8(5):585–586. doi: 10.1111/irv.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena F., Duran A.E., Morón A., Calderón E., Campos C., Gantzer C., Skraber S., Jofre J. Reduction of bacterial indicators and bacteriophages infecting faecal bacteria in primary and secondary wastewater treatments. J. Appl. Microbiol. 2004;97(5):1069–1076. doi: 10.1111/j.1365-2672.2004.02397.x. [DOI] [PubMed] [Google Scholar]
- Martín-Díaz J., Lucena F., Blanch A.R., Jofre J. Review: indicator bacteriophages in sludge, biosolids, sediments and soils. Environ. Res. 2020;182 doi: 10.1016/j.envres.2020.109133. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Pan C., Chen L., Lu C., Zhang W., Xia J.A., Sklar M.C., Du B., Brochard L., Qiu H. Lung recruitability in COVID-19-associated acute respiratory distress syndrome: a single-center observational study. Am. J. Respir. Crit. Care Med. 2020;201(10):1294–1297. doi: 10.1164/rccm.202003-0527LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W.B., Kwon N.J., Choi S.J., Kang C.K., Choe P.G., Kim J.Y., Yun J., Lee G.W., Seong M.W., Kim N.J., Seo J.S., Oh M.D. Virus isolation from the first patient with SARS-CoV-2 in Korea. J. Korean Med. Sci. 2020;35(7) doi: 10.3346/jkms.2020.35.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar N., Singh A., Ward O.P. Characterization of the combined effects of enzyme, pH and temperature treatments for removal of pathogens from sewage sludge. World J. Microbiol. Biotechnol. 2001;17:169–172. doi: 10.1023/A:1016606020993. [DOI] [Google Scholar]
- Pepper I.L., Brooks J.P., Sinclair R.G., Gurian P.L., Gerba C.P. Pathogens and indicators in United States class B biosolids: national and historic distributions. J. Environ. Qual. 2010;39(6):2185–2190. doi: 10.2134/jeq2010.0037. [DOI] [PubMed] [Google Scholar]
- Piercy T.J., Smither S.J., Steward J.A., Eastaugh L., Lever M.S. The survival of filoviruses in liquids, on solid substrates and in a dynamic aerosol. J. Appl. Microbiol. 2010;109(5):1531–1539. doi: 10.1111/j.1365-2672.2010.04778.x. [DOI] [PubMed] [Google Scholar]
- Quist-Rybachuk G.V., Nauwynck H.J., Kalmar I.D. Sensitivity of porcine epidemic diarrhea virus (PEDV) to pH and heat treatment in the presence or absence of porcine plasma. Vet. Microbiol. 2015;181(3–4):283–288. doi: 10.1016/j.vetmic.2015.10.010. [DOI] [PubMed] [Google Scholar]
- Rabenau H.F., Cinatl J., Morgenstern B., Bauer G., Preiser W., Doerr H.W. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. 2005;194(1–2):1–6. doi: 10.1007/s00430-004-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27(3):493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi H.P., Ikner L.A., Abd-Elmaksoud S., Gerba C.P., Pepper I.L. Comparative survival of viruses during thermophilic and mesophilic anaerobic digestion. Sci. Total Environ. 2018;615:15–19. doi: 10.1016/j.scitotenv.2017.09.205. [DOI] [PubMed] [Google Scholar]
- Simonet J., Gantzer C. Inactivation of poliovirus 1 and F-specific RNA phages and degradation of their genomes by UV irradiation at 254 nanometers. Appl. Environ. Microbiol. 2006;72(12):7671–7677. doi: 10.1128/AEM.01106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viau E., Bibby K., Paez-Rubio T., Peccia J. Toward a consensus view on the infectious risks associated with land application of sewage sludge. Environ. Sci. Technol. 2011;45:5459–5469. doi: 10.1021/es200566f. [DOI] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismiller D.G., Sturman L.S., Buchmeier M.J., Fleming J.O., Holmes K.V. Monoclonal antibodies to the peplomer glycoprotein of coronavirus mouse hepatitis virus identify two subunits and detect a conformational change in the subunit released under mild alkaline conditions. J. Virol. 1990;64(6):3051–3055. doi: 10.1128/JVI.64.6.3051-3055.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M., Horzinek M.C. Resistance of Berne virus to physical and chemical treatment. Vet. Microbiol. 1986;11(1–2):41–49. doi: 10.1016/0378-1135(86)90005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Mueller M.A., Niemeyer D., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Bruenink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4) doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in wastewater, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill. 2020;25(50) doi: 10.2807/1560-7917.ES.2020.25.50.2000776. (pii=2000776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26(8):1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Kang L., Shen Z., Li X., Wu W., Ma W., Fang C., Yang F., Jiang X., Gong S., Zhang L., Li M. Dynamics of severe acute respiratory syndrome coronavirus 2 genome variants in the feces during convalescence. J. Genet. Genomics. 2020 doi: 10.1016/j.jgg.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Cai C., Dai X. The potential exposure and transmission risk of SARS-CoV-2 through sludge treatment and disposal. Resour. Conserv. Recycl. 2020;162 doi: 10.1016/j.resconrec.2020.105043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5(4):335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020;92(6):680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Zhao N., Shu Y., Han S., Chen B., Shu X. Effect of gastrointestinal symptoms in patients with COVID-19. Gastroenterology. 2020;158(8):2294–2297. doi: 10.1053/j.gastro.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]