Abstract
Objectives
We aimed to develop and validate a prediction model, based on clinical history and examination findings on initial diagnosis of coronavirus disease 2019 (COVID-19), to identify patients at risk of critical outcomes.
Methods
We used data from the SEMI-COVID-19 Registry, a cohort of consecutive patients hospitalized for COVID-19 from 132 centres in Spain (23rd March to 21st May 2020). For the development cohort, tertiary referral hospitals were selected, while the validation cohort included smaller hospitals. The primary outcome was a composite of in-hospital death, mechanical ventilation, or admission to intensive care unit. Clinical signs and symptoms, demographics, and medical history ascertained at presentation were screened using least absolute shrinkage and selection operator, and logistic regression was used to construct the predictive model.
Results
There were 10 433 patients, 7850 in the development cohort (primary outcome 25.1%, 1967/7850) and 2583 in the validation cohort (outcome 27.0%, 698/2583). The PRIORITY model included: age, dependency, cardiovascular disease, chronic kidney disease, dyspnoea, tachypnoea, confusion, systolic blood pressure, and SpO2 ≤93% or oxygen requirement. The model showed high discrimination for critical illness in both the development (C-statistic 0.823; 95% confidence interval (CI) 0.813, 0.834) and validation (C-statistic 0.794; 95%CI 0.775, 0.813) cohorts. A freely available web-based calculator was developed based on this model (https://www.evidencio.com/models/show/2344).
Conclusions
The PRIORITY model, based on easily obtained clinical information, had good discrimination and generalizability for identifying COVID-19 patients at risk of critical outcomes.
Keywords: COVID-19, Critical illness, Easily obtained clinical variables, Initial assessment, Prognostic models
Introduction
Coronavirus disease 2019 (COVID-19) has spread globally, with a clinical spectrum ranging from an asymptomatic state to critical illness [[1], [2], [3]]. Notably, Spain was one of the countries with the highest incidence of COVID-19 during the first pandemic peak [4]. To optimize the use of limited healthcare resources, it is essential to identify, as early as possible, those patients who are at high risk of progressing to critical illness.
To date, studies of COVID-19 prognostic factors have focused on laboratory measurements and radiological examinations obtained following admission [[5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15]], which are not available in outpatient or resource-limited settings. Recently published well-developed models tend not to include clinical variables obtained from history and examination carried out on initial assessment [[9], [10], [11], [12], [13]]. Where one machine-learning model has addressed basic clinical features, it has narrowed down the prediction to the mortality outcome only and lacks wider generalizability [16]. Furthermore, a critical appraisal of the COVID-19 models has shown poor reporting and high risk of bias [14].
Prediction models based on easy-to-collect data have previously been developed for other infectious diseases, e.g. meningitis and pneumonia [[17], [18], [19]]. As a global health emergency, management of COVID-19 would benefit from a prediction model that could be readily applied for initial diagnosis. Therefore, we developed and externally validated a prediction model—based on easily obtainable clinical measures at presentation with confirmed COVID-19 diagnosis—to identify patients at risk of developing critical outcomes.
Methods
Study design and data source
This study was based on the SEMI (Sociedad Española de Medicina Interna) COVID-19 Registry [20]. It is an ongoing multicentre nationwide cohort of consecutive patients hospitalized for COVID-19 across Spain. Eligibility criteria were age ≥18 years, confirmed diagnosis of COVID-19 (defined as a positive result on real-time reverse-transcription-PCR (RT-PCR) for the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in nasopharyngeal swab specimens or sputum samples), first hospital admission for COVID-19, and hospital discharge or in-hospital death [20]. The SEMI-COVID-19 Registry was approved by the Provincial Research Ethics Committee of Málaga (Spain) and the Institutional Research Ethics Committees of each participating hospital.
For the study, we retrieved from the Registry clinical baseline data, history of previous medication, and comorbidities collected on admission, as well as complications during hospitalization and status at discharge. We used data from patients admitted to 132 hospitals between 23rd March and 21st May 2020. We chose hospital complexity as the criterion to assess the transportability of the prognostic model in a setting other than the one in which it was derived [21,22]. Patients admitted to tertiary referral hospitals (≥300 beds, according to the Ministry of Health of Spain [23]) were selected for the development cohort, while patients from smaller hospitals (<300 beds) were included in a separate validation cohort.
Outcome description
The primary outcome, critical illness during hospitalization, was defined as the composite of in-hospital death, mechanical ventilation or admission to the intensive care unit (ICU), according to previously published studies [10,24,25].
Potential predictors
To develop a predictive model based only on easily measurable variables registered at admission, we considered clinical signs and symptoms, demographic variables, and medical history. An initial list of 29 candidate variables was selected based on review of the existing evidence [[5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16]], clinical plausibility, and relevance to clinical care. To improve consensus on model applicability, a one-round online questionnaire was conducted among a multidisciplinary panel of 24 physicians involved in the clinical management of COVID-19 at nursing homes, emergency departments, primary care centres and hospitalization wards (six per each setting). The panellists were asked to rate (on a nine-point Likert scale) the availability/reliability of each predictor, its ability to predict the outcome, the best way to merge predictors of rare occurrence, and the maximum number of variables the model should contain. Agreement was considered when seven or fewer panellists rated outside the three-point region containing the median [26].
Statistical analysis
The predictive model, called PRIORITY, was presented as the formula for estimating the probability of COVID-19 critical illness outcome, as well as an associated web-based calculator. Patients' characteristics were summarized as frequencies and percentages or means and standard deviations. Statistical analysis was performed using R v4.0.0, with mice, mfp, glmnet, pROC, rms and rmda packages.
Model development
Missing values in the potential predictors were imputed using single imputation, a reasonable alternative to multiple imputation when dealing with relatively few missing values [27]. A stochastic single imputation dataset was created for both cohorts (development and validation) through multiple imputation by chained equations. Quantitative variables were kept as continuous to avoid loss of prognostic information, and non-linear relationships were modelled by multivariate fractional polynomials with a maximum of two degrees of freedom [28]. The least absolute shrinkage and selection operator (LASSO) was the feature selection method used to reduce the number of predictors down to the maximum agreed by the expert panel [29]. Briefly, the potential predictors were entered into the LASSO regularization process, which penalizes the coefficients by gradually shrinking them to zero. We selected the penalty parameter (λ) that minimized the deviance within the given maximum number of predictors. Those variables with non-zero coefficients were retained for risk estimation using a logistic regression model. Coefficients were presented as odds ratios (ORs) and 95% confidence intervals (95%CIs).
Model performance
Nagelkerke's R2 and Brier score were used as overall performance measures. We assessed the discriminative ability of the model using the C-statistic, calculated as the area under the receiver operating characteristic curve, with 95%CI. Calibration of the model was visually assessed by plotting deciles of predicted versus observed probabilities, and the calibration slope with 95%CI was calculated [22].
Model validation
Internal validation was carried out to assess optimism-corrected performance by repeating the entire model development over 1000 bootstrap samples drawn from the development cohort [27]. We externally validated the model in a separate cohort of patients admitted at smaller hospitals to evaluate model transportability [21]. Within this validation cohort, we reassessed model performance and compared its discrimination ability with models based on oxygen saturation and/or age, the most discriminating univariate predictors for in-hospital mortality previously reported [15]. We also undertook a decision curve analysis, a method to ascertain the adequacy of prediction models based on the relative value of benefits (true positives) and harms (false positives) [30]. We plotted the net benefit of the models for the full range of critical illness probability thresholds.
Sensitivity analysis
To assess the impact of assumptions adopted in the model development, we carried out a complete-case analysis using only those patients with complete data in the potential predictors. We also developed models without restricting the maximum number of predictors (λ at one-standard-error of the minimum) or using linear continuous predictors instead of non-linear terms.
Results
We considered data from 10 433 patients included in the SEMI-COVID-19 Registry. The development cohort included 7850 patients, of whom 1967 (25.1%) presented critical illness: 650 (8.3%) were admitted to the ICU and 1598 (20.4%) died. The mean age was 65.8 ± 16.4 years and 57.2% (4483/7834) were male. Demographics and clinical characteristics for the development cohort are shown on Table 1 .
Table 1.
Demographic and clinical characteristics among patients included in the development and validation cohorts
| Development cohort |
Validation cohort |
||||
|---|---|---|---|---|---|
| No of patients (%) or mean ± SD | Total No (%) | No of patients (%) or mean ± SD | Total No (%) | ||
| Characteristics of the population | |||||
| Critical illness | 1967 (25.1%) | 7850 (100%) | 698 (27.0%) | 2583 (100%) | |
| Age [years] | 65.8 ± 16.4 | 7816 (99.6%) | 69.5 ± 16.0 | 2575 (97.3%) | |
| Male | 4483 (57.2%) | 7834 (99.8%) | 1415 (54.8%) | 2580 (99.9%) | |
| Ethnicity | Caucasian | 6836 (89.1%) | 7677 (98.8%) | 2340 (91.0%) | 2572 (99.6%) |
| Latino | 693 (9.0%) | 193 (7.5%) | |||
| Other | 148 (1.9%) | 39 (1.5%) | |||
| Smoking history | Never | 5270 (70.9%) | 7433 (94.7%) | 1625 (65.7%) | 2475 (95.8%) |
| Former smoker | 1764 (23.7%) | 718 (29.0%) | |||
| Active Smoker | 399 (5.4%) | 139 (5.3%) | |||
| Medical history | |||||
| Obesitya | 1665 (23.7%) | 7012 (89.3%) | 584 (24.3%) | 2401 (93.0%) | |
| Hypertension | 3803 (48.6%) | 7833 (99.8%) | 1444 (56.1%) | 2576 (99.7%) | |
| Diabetes mellitus | 1440 (18.4%) | 7820 (99.6%) | 509 (19.8%) | 2570 (99.5%) | |
| Cardiovascular diseaseb | 1974 (25.3%) | 7800 (99.4%) | 806 (31.7%) | 2545 (98.5%) | |
| Pulmonary diseasesc | 1625 (20.9%) | 7776 (99.1%) | 576 (22.6%) | 2583 (98.9%) | |
| Severe chronic kidney diseased | 488 (6.2%) | 7825 (99.7%) | 163 (6.3%) | 2583 (99.7%) | |
| Malignancye | 793 (10.2%) | 7803 (99.4%) | 259 (10.1%) | 2571 (99.5%) | |
| Immunocompromised statusf | 650 (8.6%) | 7549 (96.2%) | 187 (7.6%) | 2473 (95.7%) | |
| Dependency (moderate/severe)g | 1129 (14.7%) | 7701 (98.1%) | 605 (23.7%) | 2555 (98.9%) | |
| Symptoms at admission | |||||
| Feverh | 5138 (67.0%) | 7663 (97.6%) | 1670 (65.6%) | 2544 (98.5%) | |
| Dyspnoea | 4427 (56.7%) | 7805 (99.4%) | 1523 (59.4%) | 2562 (99.2%) | |
| Clinical signs and physical exploration at admission | |||||
| SBP (mmHg) | 129.0 ± 21.5 | 7430 (94.6%) | 127.6 ± 21.0 | 2451 (94.9%) | |
| HR (beats/minute) | 88.6 ± 17.4 | 7500 (95.5%) | 87.5 ± 17.5 | 2504 (96.9%) | |
| Tachypnoea (>20 breaths/min) | 2271 (29.9%) | 7604 (96.9%) | 879 (35.1%) | 2504 (96.9%) | |
| SpO2 ≤93% or oxygen requirement at presentation | 4152 (52.9%) | 7842 (99.9%) | 1605 (62.1%) | 2583 (100%) | |
| Pulmonary rales | 4630 (60.7%) | 7626 (97.1%) | 1588 (63.6%) | 2495 (96.6%) | |
| Confusion | 849 (11.0%) | 7736 (98.5%) | 384 (15.1%) | 2546 (98.6%) | |
SD, standard deviation; HR, heart rate; SBP, systolic blood pressure; SpO2, peripheral oxygen saturation.
Obesity is defined as medical history or body mass index ≥30 kg/m2.
History of cerebrovascular disease, peripheral arterial disease, myocardial infarction, angina pectoris, heart failure or atrial fibrillation.
Chronic obstructive pulmonary disease, obstructive sleep apnoea/hypopnoea syndrome and asthma.
History of serum creatinine level >3 mg/dL or history of dialysis.
History of solid tumour, leukaemia or lymphoma.
History of autoimmune diseases, solid-organ transplant recipients, HIV infection or previous immunosuppressive treatment including systemic steroids.
Moderate or severe dependency for activities of daily living (Barthel index score <60).
Temperature ≥38°C or history of fever.
Model development and performance
From an initial list of 29 candidate variables, the expert panel forged an agreement on 21 potential predictors for further evaluation in the predictive model. Chronic liver disease, previous medication with angiotensin converting enzyme inhibitors or angiotensin receptor blockers, cough, arthralgia/myalgia, ageusia/anosmia, asthenia/anorexia, headache, gastrointestinal symptoms were excluded. Consensus was achieved for including a range between five and nine variables in the final model. For transparency, univariate analysis is shown in Supplementary Material Table S1, even though it was not part of the model development process. The 21 potential predictors were included in the LASSO selection process, retaining a subset of nine variables as the best predictors of critical illness (Supplementary Material Fig. S1). A multivariable logistic regression model was then fitted with these nine variables. All of them, except for moderate or severe dependency, were statistically significant (Table 2 ).
Table 2.
Multivariate logistic regression of critical illness prediction in coronavirus 2019 (COVID-19)
| Predictors | Odds ratio | 95%CI |
|---|---|---|
| (Age/100)2 Age in yearsa | 14.339 | 10.054, 20.532 |
| Cardiovascular disease | 1.372 | 1.195, 1.573 |
| Severe chronic kidney disease | 1.797 | 1.433, 2.252 |
| Dyspnoea | 1.655 | 1.451, 1.891 |
| 1/(SBP/100)2 SBP in mmHga | 2.326 | 1.837, 2.951 |
| Tachypnoea (>20 breaths/min) | 2.487 | 2.192, 2.824 |
| SpO2 ≤93% or oxygen requirement | 3.320 | 2.889, 3.819 |
| Confusion | 1.976 | 1.642, 2.380 |
| Dependency (moderate or severe) | 1.178 | 0.989, 1.404 |
Predictors in the PRIORITY model retained after LASSO feature selection. Model coefficients were derived from a multivariate logistic regression, and presented as odds ratios (ORs) and 95% confidence intervals (95%CIs). Variables entered into the LASSO feature selection process were: age as a squared term, sex, ethnicity, smoking history, obesity, hypertension, diabetes mellitus, cardiovascular disease, pulmonary diseases, severe chronic kidney disease, malignancy, immunocompromised status, dependency, fever, dyspnoea, systolic blood pressure (SBP) as the inverse of a quadratic term, heart rate (HR) as a cubic term, tachypnoea, peripheral oxygen saturation (SpO2) ≤93% on room air or oxygen requirement at presentation, pulmonary rales, and confusion. All predictors were coded as binary variables (1 when present and 0 when absent) except for age (years), SBP (mmHg) and HR (bpm).
Continuous predictors modelled as fractional polynomial terms, including rescaling when the range of values of the predictor was reasonably large. As interpretability of the effect of non-linear continuous predictors can be difficult, linear local approximations of ORs for 10-unit variations are provided at selected values. ORs for age (10-year increments): OR (50/40) = 1.271; OR (70/60) = 1.414; OR (90/80) = 1.573. ORs for SBP (10-mmHg decreases): OR (110/120) = 1.118; OR (90/100) = 1.219; OR (70/80) = 1.497. Approximated ORs are provided for illustrative purposes only and were not used for making predictions.
Based on the logistic regression model, the probability of critical COVID-19 illness could be calculated as:
Probability (%) = 100/(1+exp(–z)), where z = –4.665 + 2.663·((age/100)2) + 0.164·(dependency) + 0.316·(cardiovascular disease) + 0.586·(chronic kidney disease) + 0.504·(dyspnoea) + 0.844·(1/(SBP/100)2) + 0.911·(tachypnoea) + 1.200·(SpO2 ≤93% or oxygen requirement) + 0.681·(confusion).
All predictors were coded as binary variables (1 when present and 0 when absent) except for age (years) and systolic blood pressure (SBP, mmHg). We also developed an online calculator based on this model (Supplementary Material Fig. S2), accessible at https://www.evidencio.com/models/show/2344.
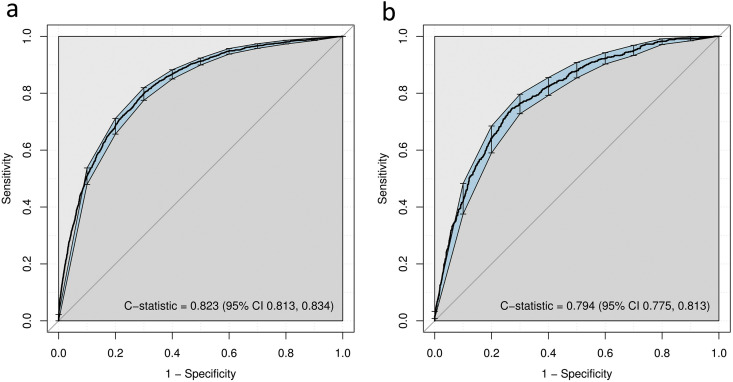
In the development cohort, the PRIORITY model had an R2 of 0.347 and a Brier score of 0.138. The apparent C-statistic was 0.823 (95%CI 0.813, 0.834) (Fig. 1 a). After bootstrap internal validation, optimism-corrected C-statistic was 0.821 (95%CI 0.810, 0.832). The model showed good calibration across the range of predicted probabilities within the development cohort (calibration slope 0.996, 95%CI 0.989, 0.999; Supplementary Material Fig. S3a).
Fig. 1.
Discriminatory ability of the PRIORITY model in (a) the development and (b) the validation cohorts. Discriminative ability was assessed using the C-statistic, as the area under the receiver operating characteristic curve, with 95% confidence intervals (CIs) computed with 1000 bootstrap replicates.
External validation
The validation cohort included 2583 patients, of whom 698 (27.0%) presented critical illness: 200 (7.7%) were admitted to the ICU and 594 (23.0%) died. The mean age was 69.5 ± 16.0 years, 54.8% (1415/2580) were male (Table 1). The PRIORITY model showed good discrimination for critical illness within the validation cohort (C-statistic 0.794, 95%CI 0.775, 0.813) (Fig. 1b), and a calibration slope of 0.875, 95%CI 0.790, 0.960 (Supplementary Material Fig. S3b).
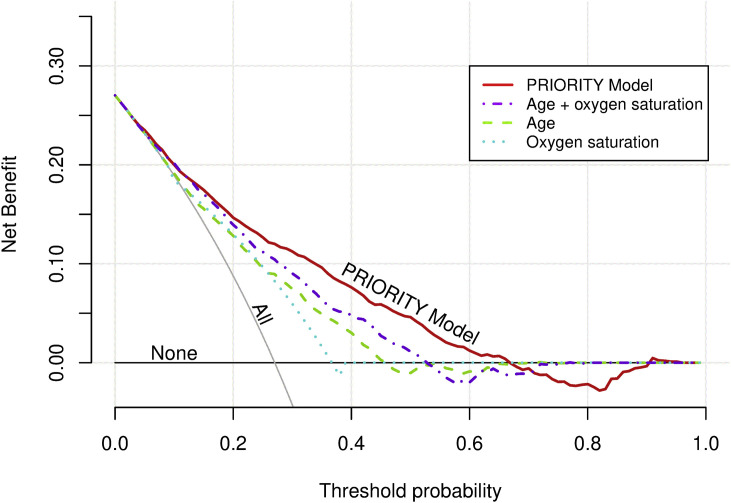
Our model compared well against the risk stratification based on univariate models including age (C-statistic 0.707, 95%CI 0.686, 0.729) or SpO2 ≤93%/oxygen requirement at admission (C-statistic 0.652, 95%CI 0.635, 0.670) as sole predictors. Likewise, the PRIORITY model had better discriminative ability than the model including both age and SpO2 ≤93%/oxygen supply (C-statistic 0.751, 95%CI 0.731, 0.771). Additionally, decision curve analysis showed that the PRIORITY model had higher net benefit across a wide range of threshold probabilities for developing critical illness compared to risk stratification using age and/or SpO2 ≤93%/oxygen supply (Fig. 2 ).
Fig. 2.
Decision curve analysis within the validation cohort. Clinical usefulness of the PRIORITY model compared to risk stratification based on oxygen saturation (binary: SpO2 ≤93% or oxygen requirement) and/or age (quadratic term). The x-axis represents the whole range of decision threshold probabilities for critical illness (pt) and the y-axis represents the net benefit (NB). NB calculated as: True positives/N – (false positives/N)∗(pt/(1–pt)), with N total sample size.
Sensitivity analysis
We carried out a complete-case analysis selecting as development cohort the 5513 patients with complete data on the 21 potential predictors and the outcome. The resulting model had the same predictors as the PRIORITY model with apparent C-statistic 0.813 (95%CI 0.800, 0.826) and calibration slope 0.993 (95%CI 0.986, 0.997). Next, we fitted a new model with no restriction in the maximum number of variables, resulting in a model which added sex, diabetes mellitus, malignancy, immunocompromised status, pulmonary rales, and heart rate cubed to the predictors in the PRIORITY model. C-statistic was 0.831 (95%CI 0.821, 0.842) and slope 0.990 (95%CI 0.986, 0.996). Likewise, we fitted an alternative model using linear continuous predictors instead of non-linear terms, which included sex but excluded the systolic blood pressure. C-statistic was 0.823 (95%CI 0.812, 0.833) and slope 0.994 (95%CI 0.988, 0.999).
Discussion
We developed and validated a new clinical risk model to predict COVID-19 critical illness based on nine simple clinical features easily available on initial assessment, which would be useful in resource-limited or out-of-hospital settings without access to other complementary tests. The model was well calibrated, had good discrimination, and performed robustly in an external validation cohort. Moreover, it showed a potential clinical benefit in a variety of scenarios covering different healthcare situations over a range of threshold probabilities for critical illness. The web-based calculator can facilitate its immediate application for frontline clinicians.
Previously, an external validation of 22 prognostic models showed that none of the multivariate models offered incremental value for patient stratification compared to oxygen saturation or age [15]. In this regard, the PRIORITY model showed higher discriminative ability and net benefit than age and/or oxygen saturation. Additionally, despite its simplicity, our model had a performance similar to previously published prognostic tools including laboratory and imaging tests [[9], [10], [11], [12], [13], [14], [15], [16]].
It is worth noting that the PRIORITY model could be applied in triage, using easily measurable variables available in settings without access to laboratory or radiology tests, identifying high-risk patients for referral to hospital. This model could be useful for supporting clinical management decisions over a range of risk thresholds for critical illness which could be considered as relevant in clinical practice. The choice of thresholds will vary across different regions, according to changing epidemiological situations and availability of health resources. For example, under pandemic peak pressure or low-resource healthcare systems, policy-makers may consider a cut-off point up to 20%, a threshold that will be associated with higher reduction in unnecessary critical care admissions. However, at low risk of overwhelming the critical care capacity, a lower threshold may be considered at the expense of unnecessary referrals. We recommend objectively defining specific cut-off points considering the circumstances and the availability of health resources.
This study has several methodological strengths maximizing internal and external validity [23]. To the best of our knowledge, this is the first generalizable COVID-19 predictive model built with simple clinical information (excluding imaging and laboratory data). We developed and validated the model in a large multicentre national cohort. The methodology was rigorous, avoiding data-driven predictor selection and biases that have affected previous studies [14]. The practical application of the model was maximized by forging an agreement among an expert panel on key issues. Moreover, the model was validated in a separate cohort of patients admitted in smaller hospitals, showing transportability to a setting with a different level of healthcare [21,22].
The strengths of our findings should be interpreted in light of some limitations. First, although we carefully selected easily available clinical and demographic variables, the data were collected at the time of hospital admission, which represents an important selection bias that would require further studies in an outpatient setting. Second, it could be suggested that, taking into account the situation of healthcare pressure, data quality may be affected. In this regard, it is notable that in this study there were relatively few missing data and we used imputation to reduce their impact. Third, since the COVID-19 pandemic has demonstrated significant differences between countries and time periods, it could affect the applicability of the model to other settings. However, we considered this early pandemic period in Spain to reflect a scenario with an overwhelmed healthcare system, where our predictive model could be particularly useful. Nevertheless, further studies introducing factors such as viral strains, healthcare system actions, new treatments, or vaccination could improve the applicability of the PRIORITY model. Finally, even though we compared the net benefit of using the model with discrimination based on oxygen saturation and/or age, its real clinical usefulness would require comparison with the best existing scores or the clinician's decision.
In summary, we have developed and validated a new prediction model, called PRIORITY, to estimate the risk of critical illness in patients with COVID-19 based on nine clinical variables easily measurable in resource-limited or out-of-hospital settings. The study could provide underpinning evidence to inform decision-making in health systems under pandemic pressure.
Author contributions
MML, AVN and MF planned and conceived the study and analysed and interpreted the data. MML, AVN, MF and LM wrote the original draft of the manuscript. MRR, SLG, FAF, JLBP, JAVN, ECM, ACEA, SJFC, JLA, PMPF, AP, AMAS, ASA, BGL, JLP, JSC, PCP, GMGG, JMNC, JMCR and RGH contributed to reading and approving the final version of the manuscript. MML and AVN are joint first authors. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. LM is the guarantor.
Transparency declaration
The corresponding author affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned have been explained. The authors have no conflicts of interest to declare. No funding was received for this work.
Acknowledgements
We gratefully acknowledge all the investigators who participate in the SEMI-COVID-19 Registry (listed at Supplementary Appendix S1), especially those from Hospital Universitario Ramón y Cajal (Luis F. Abrego-Vaca, Ana Andreu-Arnanz, Octavio A. Arce-García, Marta Bajo-González, Pablo Borque-Sanz, Alberto Cózar-Llistó, Beatriz Del Hoyo-Cuenda, Alejandra Gamboa-Osorio, Isabel García-Sánchez, Óscar A. López-Cisneros, Borja Merino-Ortiz, Elisa Riera-González, Jimena Rey-García, Cristina Sánchez-Díaz, Grisell Starita-Fajardo, Cecilia Suárez-Carantoña, Svetlana Zhilina Zhilina). We especially thank our colleagues at Hospital Universitario Ramon y Cajal, IRYCIS: Nuria Bara Ledesma, Andrés González García and José Luis Calleja López (Internal Medicine Dept), Javier Zamora and Borja M. Fernandez-Felix (Clinical Biostatistics Unit), and Javier Soto Pérez-Olivares (Radiology Dept) for their valuable contributions to the review of the manuscript. The authors would also like to gratefully acknowledge Professor Khalid S. Khan, Distinguished Investigator at the University of Granada, Spain, for his support and advice on the manuscript. Finally, we also thank the SEMI-COVID-19 Registry Coordinating Center, S&H Medical Science Service, for their quality control data, logistic and administrative support.
Editor: M. Paul
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.07.006.
Contributor Information
for the SEMI-COVID-19 Network:
Luis F. Abrego-Vaca, Ana Andreu-Arnanz, Octavio A. Arce-García, Marta Bajo-González, Pablo Borque-Sanz, Alberto Cózar-Llistó, Beatriz Del Hoyo-Cuenda, Alejandra Gamboa-Osorio, Isabel García-Sánchez, Óscar A. López-Cisneros, Borja Merino-Ortiz, Elisa Riera-González, Jimena Rey-García, Cristina Sánchez-Díaz, Grisell Starita-Fajardo, Cecilia Suárez-Carantoña, and Svetlana Zhilina Zhilina
Dissemination to participants and related patient and public communities
Results of this study have been made available to the public through an open access preprint posted to MedRxiv (https://doi.org/10.1101/2020.11.27.20237966). The Spanish Society of Internal Medicine (SEMI) shares the results of the studies derived from the SEMI-COVID-19 Registry through its public-facing website (https://www.fesemi.org/investigacion/proyectos/registro-semi-covid-19) and its twitter account (@Sociedad_SEMI).
Data sharing
The data that support the findings of this study are available on request from the SEMI-COVID-19 Scientific Committee and the Registry Coordinating Center.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Docherty A.B., Harrison E.M., Green C.A., Hardwick H.E., Pius R., Norman L., et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ. 2020;369:m1985. doi: 10.1136/bmj.m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med;382:1708-1720. [DOI] [PMC free article] [PubMed]
- 3.WHO . 2020. COVID-19 situation reports.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports [Google Scholar]
- 4.Ministerio De Sanidad, Consumo y Bienestar Social . 2020. Enfermedad por el coronavirus (COVID-19)https://www.mscbs.gob.es/en/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/Actualizacion_288_COVID-19.pdf (in Spanish) [Google Scholar]
- 5.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berenguer J., Ryan P., Rodríguez-Baño J., Jarrín I., Carratalà J., Pachón J., et al. Characteristics and predictors of death among 4,035 consecutively hospitalized patients with COVID-19 in Spain. Clin Microbiol Infect. 2020;26:1525–1536. doi: 10.1016/j.cmi.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fumagalli C., Rozzini R., Vannini M., Coccia F., Cesaroni G., Mazzeo F., et al. Clinical risk score to predict in-hospital mortality in COVID-19 patients: a retrospective cohort study. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-040729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang W., Liang H., Ou L., Chen B., Chen A., Li C., et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji D., Zhang D., Xu J., Chen Z., Yang T., Zhao P., et al. Prediction for progression risk in patients with COVID-19 pneumonia: the CALL Score. Clin Infect Dis. 2020;71:1393–1399. doi: 10.1093/cid/ciaa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight S.R., Ho A., Pius R., Buchan I., Carson G., Drake T.M., et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ. 2020;370:m3339. doi: 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartoletti M., Giannella M., Scudeller L., Tedeschi S., Rinaldi M., Bussini L., et al. Development and validation of a prediction model for severe respiratory failure in hospitalized patients with SARS-Cov-2 infection: a multicenter cohort study (PREDI-CO study) Clin Microbiol Infect. 2020;26:1545–1553. doi: 10.1016/j.cmi.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., et al. Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta R.K., Marks M., Samuels T., Luintel A., Rampling T., Chowdhury H., et al. Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: an observational cohort study. Eur Respir J. 2020;56:2003498. doi: 10.1183/13993003.03498-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yadaw A.S., Li Y.C., Bose S., Iyengar R., Bunyavanich S., Pandey G. Clinical features of COVID-19 mortality: development and validation of a clinical prediction model. Lancet Digit Health. 2020;2:e516–e525. doi: 10.1016/S2589-7500(20)30217-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ajayi-Obe E.K., Lodi E., Alkali A.S., Galbati M., Rooney C., Mannoni B., et al. Prognostic scores for use in African meningococcal epidemics. Bull World Health Organ. 1998;76:149–152. [PMC free article] [PubMed] [Google Scholar]
- 18.Bassat Q., Lanaspa M., Machevo S., O'Callaghan-Gordo C., Madrid L., Nhampossa T., et al. Hypoxaemia in Mozambican children <5 years of age admitted to hospital with clinical severe pneumonia: clinical features and performance of predictor models. Trop Med Int Health. 2016;21:1147-56. doi: 10.1111/tmi.12738. [DOI] [PubMed] [Google Scholar]
- 19.Lanaspa M., Valim C., Acacio S., Almendinger K., Ahmad R., Wiegand R., et al. High reliability in respiratory rate assessment in children with respiratory symptomatology in a rural area in Mozambique. J Trop Pediatr. 2014;60:93-8. doi: 10.1093/tropej/fmt081. [DOI] [PubMed] [Google Scholar]
- 20.Casas Rojo J.M., Antón Santos J.M., Nuñez-Cortés J., Lumbreras-Bermejo C., Ramos-Rincón J.M., Roy-Vallejo E., et al. Clinical characteristics of patients hospitalized with COVID-19 in Spain: results from the SEMI-COVID-19 Network. Rev Clin Esp. 2020;220:480–494. doi: 10.1016/j.rce.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Justice A.C., Covinsky K.E., Berlin J.A. Assessing the generalizability of prognostic information. Ann Intern Med. 1999;130:515–524. doi: 10.7326/0003-4819-130-6-199903160-00016. [DOI] [PubMed] [Google Scholar]
- 22.Steyerberg E.W., Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J. 2014;35:1925-31. doi: 10.1093/eurheartj/ehu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ministerio de Sanidad, Consumo y Bienestar Social . 2020. Certificates of discharge of the national health system register.https://www.mscbs.gob.es/en/estadEstudios/estadisticas/cmbd.htm CMBD. State Regulation (in Spanish) [Google Scholar]
- 24.Metlay J.P., Waterer G.W., Long A.C., Anzueto A., Brozek J., Crothers K., et al. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200:e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evangelidis, Nicole Tong A., Howell M., Teixeira-Pinto A., Elliott J.H., Azevedo L.C., et al. International survey to establish prioritized outcomes for trials in people with coronavirus disease 2019. Crit Care Med. 2020;48:1612–1621. doi: 10.1097/CCM.0000000000004584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitch K., Bernstein S., Aguilar M.D., Burnand B., LaCalle J.R., Lazaro P., et al. 1st ed. RAND Corporation; Santa Monica, CA: 2001. The RAND/UCLA appropriateness method user’s manual. [Google Scholar]
- 27.Steyerberg E.W. 2nd ed. Springer; New York: 2019. Clinical prediction models: a practical approach to development, validation and updating. [Google Scholar]
- 28.Royston P., Altman D.G. Regression using fractional polynomials of continuous covariates: parsimonious parametric modelling. Appl Stat. 1994;43:429–467. [Google Scholar]
- 29.Hastie T., Tibshirani R., Friedman J.H. 2nd ed. Springer; New York: 2009. The elements of statistical learning: data mining, inference, and prediction. [Google Scholar]
- 30.Vickers A.J., Elkin E.B. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.