Abstract
Helicobacter pylori is the strongest risk factor for gastric adenocarcinoma. The H. pylori cancer-associated cag pathogenicity island (cag-PAI) encodes a type IV secretion system (T4SS), which translocates microbial DNA and activates TLR9; however, most cag-PAI+–infected persons do not develop cancer and cag-PAI–independent regulators of pathogenesis, including strain-specific adhesins, remain understudied. We defined the relationships between H. pylori HopQ adhesin allelic type, gastric injury, and TLR9 activation. Type I hopQ alleles were significantly associated with magnitude of injury, cag-T4SS function, and TLR9 activation. Genetic deletion of hopQ significantly decreased H. pylori-induced TLR9 activation, implicating this adhesin in H. pylori-mediated disease.
Keywords: Helicobacter pylori, gastric cancer, HopQ, TLR9, secretion systems
Helicobacter pylori incurs the highest known risk for developing gastric cancer [1]; however, only 1%–3% of infected individuals develop gastric adenocarcinoma [2]. One strain-specific H. pylori oncogenic determinant is the cag pathogenicity island (cag-PAI), which encodes a type IV secretion system (T4SS). The cag-T4SS translocates effectors, such as CagA, peptidoglycan, heptose-bisphosphate, and DNA into epithelial cells [3]. Translocated DNA subsequently activates Toll-like receptor 9 (TLR9) and H. pylori strains that confer a higher risk for gastric cancer are more potent in their ability to activate TLR9 [4]. However, the precise molecular mechanisms regulating H. pylori-dependent TLR9 activation remain incompletely defined.
Most persons colonized with cag-PAI+ strains do not develop gastric cancer, raising the hypothesis that other H. pylori constituents may also affect disease risk. The outer membrane protein HopQ, which binds human carcinoembryonic antigen-related cell adhesion molecule (CEACAM) receptors, has been reported to facilitate CagA translocation [5, 6]. H. pylori hopQ exhibits a high level of diversity and 2 genetically distinct families of hopQ alleles (type I and type II) have been previously described [7]. Type I alleles are present significantly more frequently in cagA+ versus cagA− strains [7], suggesting that HopQ may represent a microbial component that can regulate DNA translocation and TLR9 activation and play a role in disease.
METHODS
Clinical Specimens
Gastric antral biopsies were collected for culture and immunohistochemistry [8]. Patients were prospectively enrolled after written informed consent and the study was approved by the institutional review boards of Vanderbilt University and the Nashville Department of Veterans Affairs. Histologic parameters were scored from 0 to 3 as outlined by the Sydney System [8].
Bacterial Strains
H. pylori cag-PAI+ strain 26695 (which contains a single type I hopQ allele) [9], isogenic mutants, and clinical isolates were maintained on trypticase soy agar plates with 5% sheep blood (Hemostat Laboratories). Allele-specific polymerase chain reactions (PCRs) were used to type hopQ [7] and stratify strains into hopQ allelic categories based on detection of type I alleles, type II alleles, or both. A kanamycin-resistant 26695 cagE− mutant [4], which lacks functional cag-T4SS activity, and a chloramphenicol-resistant 26695 hopQ− mutant (hopQ−#1) were previously described [9]. An H. pylori 26695 hopQ complemented strain was generated by insertion of the hopQ gene into the hp0177/0178 intergenic chromosomal region of hopQ−#1. A second independent 26695 hopQ− mutant (hopQ−#2) was constructed as previously described by insertional mutagenesis [4]. Additionally, a hopQ deletion mutant derivative of strain 7.13 was constructed by inserting chloramphenicol and kanamycin resistance cassettes into the 2 hopQ loci [10]. PCR-based typing of clinical isolates was performed for hopQ alleles and cagA status (Supplementary Table 1) [7, 8]. TLR9 activation, adherence, CagA translocation, and interleukin-8 (IL-8) production assays were performed using the following multiplicities of infection (MOIs): 10:1, 25:1, 50:1, 100:1, and 200:1 (Supplementary Figure 1). Activation with minimal control activation by wild-type H. pylori for CagA translocation and IL-8 induction occurred at an MOI of 50:1 and for adherence and TLR9 activation at an MOI of 100:1.
Real-Time PCR
RNA was extracted using the RNAeasy Mini Kit (Qiagen) from log-phase H. pylori cultures. cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (ThermoFisher) and quantitative real-time PCR was performed using Power SYBR Green Master Mix (ThermoFisher) with gene-specific primers (Supplementary Table 1).
TLR9 and NOD1 Activation Assays
HEK-Blue-hTLR9 cells (TLR9+), HEK-Blue-hNOD1 cells (NOD1+), and HEK-Blue-Null1 (parental) cells were seeded in 96-well plates (Corning) in Dulbecco’s Modified Eagle’s Medium without antibiotics and challenged with agonist, H. pylori, or sterile phosphate-buffered saline for 24 hours. Supernatants were added to QUANTI-Blue solution (Invivogen) and analyzed by spectrophotometer (Bitoek) at 650 nm.
Statistical Analysis
All experiments were repeated at least 3 times. The Mann-Whitney test or student t test was used for 2 group comparisons, while 1-way analysis of variance (ANOVA) with Bonferroni correction was used for multiple group comparisons. Statistical significance was set at a P value of < .05.
RESULTS
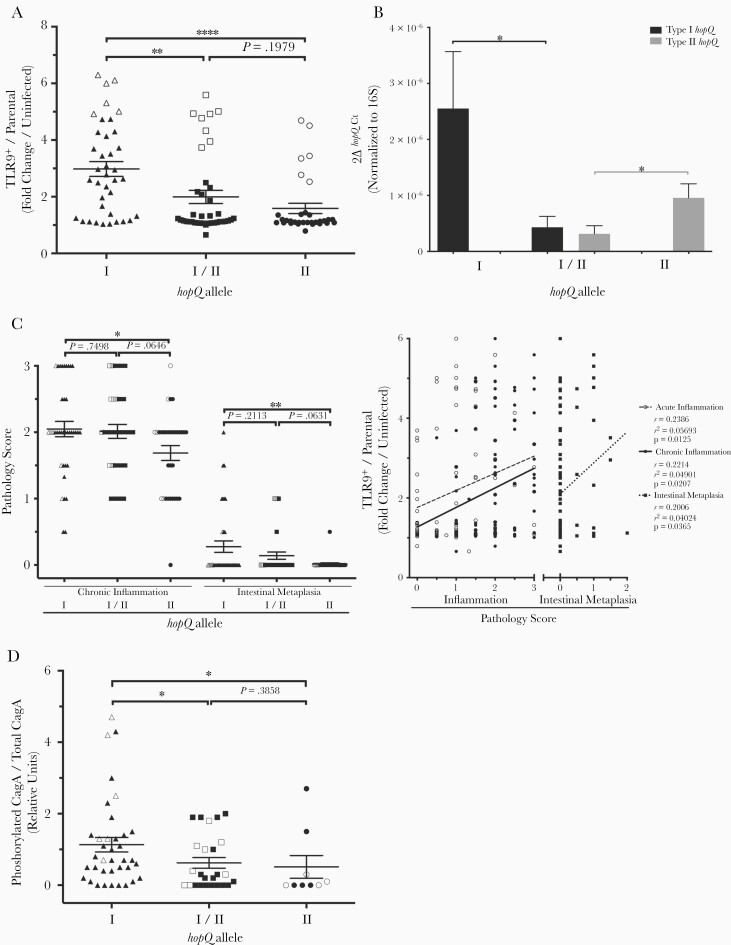
We first PCR-genotyped a cohort of clinical strains for hopQ allelic type and analyzed their ability to activate TLR9. Type I strains induced significantly higher levels of TLR9 activation compared to type I/II or type II strains (Figure 1A and Supplementary Figure 2). To determine whether gene expression may be associated with these differences, we analyzed expression of hopQ alleles in a subset of clinical strains by real-time PCR. Expression of type I hopQ was significantly greater in type I strains compared to type I/II strains while expression of type II hopQ was significantly higher in type II strains compared to type I/II strains (Figure 1B). To study potential downstream ramifications of these findings in vivo, severity of inflammation and premalignant lesions (eg, intestinal metaplasia) in biopsies obtained from the source patients was stratified by hopQ genotypes of the infecting H. pylori isolates. Severity of chronic inflammation and intestinal metaplasia were significantly increased in patients infected with type I strains compared to patients infected with type II strains (Figure 1C), while levels of acute inflammation were no different (Supplementary Figure 3). We also directly compared levels of H. pylori-induced TLR9 activation in vitro to the severity of gastric inflammation and injury induced by the same strains in vivo. Levels of TLR9 activation were significantly associated, albeit of weaker magnitude, with the intensity of chronic inflammation, intestinal metaplasia, as well as acute inflammation (Figure 1C), suggesting that the capacity of H. pylori strains to induce higher levels of TLR9 activation in vitro is related in part to the extent of damage induced by these same strains in vivo. H. pylori cagA+ type I hopQ strains also translocated significantly higher amounts of CagA per level of total CagA/strain compared to strains containing type I/II alleles or a type II allele; however, there were no significant associations between type I hopQ expression levels and pathologic outcomes or levels of CagA translocation (Figure 1D, Supplementary Figure 4, and Supplementary Figure 5). Collectively, these results indicate that H. pylori type I strains induce more intense TLR9 activation in vitro and enhanced chronic inflammation and damage in vivo. This may reflect enhanced translocation of CagA, which has been shown to induce inflammation and promote the development of intestinal metaplasia [11].
Figure 1.
Clinical Helicobacter pylori type I hopQ strains enhance Toll-like receptor 9 (TLR9) activation and are more virulent than type I/II or type II strains. The highest H. pylori responder strains for each hopQ allelic category (type I, n = 6; type I/II, n = 8; type II, n = 6) are indicated by open symbols (A); data originating from gastric tissue harvested from patients infected with these same strains (C) and levels of CagA translocation by these same strains (D) are also indicated by open symbols. A, TLR9-reporter or parental cells were challenged with clinical H. pylori strains. Data are fold change in infected TLR9+ cells/parental cells over uninfected controls. Each strain was tested in duplicate at least 3 times (type I, n = 38; type I/II, n = 39; type II, n = 32). B, Expression of either type I hopQ or type II hopQ was assessed by real-time PCR (RT-PCR) on a random selection of clinical strains (type I, n = 7; type I/II, n = 8; type II, n = 9). 2ΔhopQCt is the expression level of hopQ normalized to the reference gene 16S rRNA. C, Inflammation and intestinal metaplasia scores from patients infected with either H. pylori type I, type I/II, or type II hopQ strains. Each data point represents the score from an individual patient (type I, n = 38; type I/II, n = 39; type II, n = 32). TLR9 activation levels induced by the corresponding infecting H. pylori strains (n = 109) are shown as fold change in infected TLR9+ cells/parental cells over uninfected controls. Spearman correlation was performed to determine linear correlation. D, AGS cells were co-cultured with cagA+ clinical H. pylori strains (n = 71) at multiplicities of infection 50:1 for 4 hours. Levels of translocated CagA were quantified in cell lysates by Western blotting for phosphorylated CagA and total CagA. Data are represented as phosphorylated CagA over total CagA (type I, n = 37; type I/II, n = 25; type II n = 9). Mean ± SEM are shown for all groups. Mann-Whitney tests or student t test were used to determine statistical significance between groups *P < .05, **P < .01, ****P < .0001.
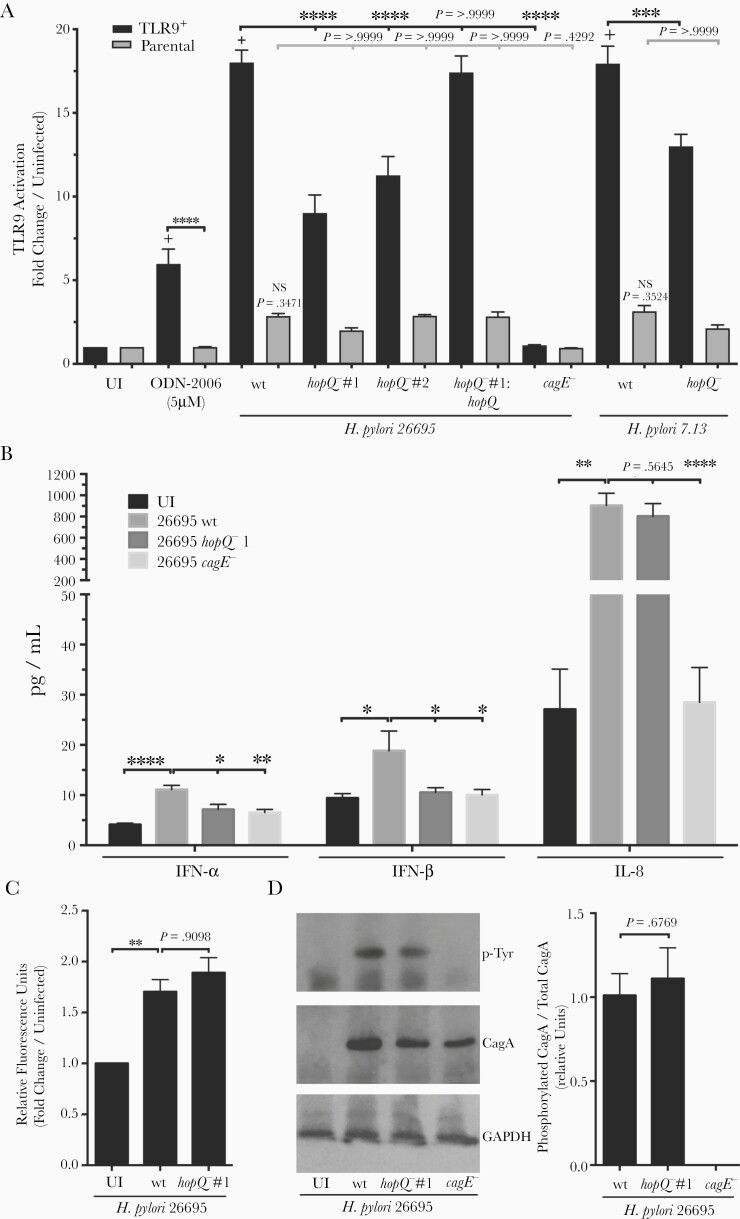
We next more definitively defined the role of HopQ in TLR9 activation by genetically inactivating hopQ. H. pylori wild-type strain 26695 robustly induced TLR9 activation when compared to uninfected cells, while a cagE− mutant minimally activated TLR9. TLR9 activation was significantly diminished following co-culture with the hopQ− mutant compared to H. pylori wild-type 26695-infected cells, and complementation fully restored levels of TLR9 activation to levels induced by wild-type strain 26695 (Figure 2A). To more strongly implicate hopQ allele status in TLR9 activation, we generated a second independent isogenic hopQ− mutant in the H. pylori 26695 strain background as well as a double hopQ mutant in H. pylori strain 7.13, which contains 2 identical copies of type I hopQ. Levels of TLR9 activation induced by 26695 hopQ−#2 and the 7.13 hopQ− mutant were significantly reduced compared to levels induced by the wild-type strains (Figure 2A). Of note, levels of activation in parental cells infected by either of the wild-type strains were higher than in uninfected parental cells although this was not statistically significant, which may represent residual cag-T4SS-dependent but HopQ-independent activation of nuclear factor-κB (NF-κB). Downstream signaling effectors activated by TLR9 include type I interferons (IFN-α and IFN-β). To investigate consequences of TLR9 activation, we co-cultured wild-type H. pylori strain 26695 and the 26695 isogenic hopQ− mutant with AGS cells and quantified IFN-α and IFN-β production. Similar to TLR9, wild-type H. pylori strain 26695 induced significantly higher levels of type I interferons compared to the 26695 hopQ− mutant (Figure 2B).
Figure 2.
Deletion of hopQ significantly decreases TLR9 activation independent of cellular adhesion and cag-T4SS function. A, TLR9-reporter or parental cells were challenged with TLR9 agonist ODN-2006, Helicobacter pylori wild-type cag-PAI+ strain 26695, wild-type cag-PAI+ strain 7.13, respective hopQ− or cagE− isogenic mutant strains, or a complemented 26695 hopQ mutant. Samples were tested in duplicate at least 3 times and data are fold change in infected over uninfected controls. B, Levels of IFN-α, IFN-β, and IL-8 were determined via ELISA in H. pylori:AGS cell supernatants. In each experiment, strains were tested at least 3 times and mean ± SEM are shown. C, Fluorescently labeled H. pylori wild-type strain 26695 or a 26695 hopQ− isogenic mutant were co-cultured with AGS cells for 4 hours and analyzed for fluorescence. Strains were tested in duplicate and data are fold change of infected over uninfected control. D, CagA translocation was determined by quantifying levels of phospho-CagA in AGS cell lysates during H. pylori co-culture by Western blotting. Representative Western blots and densitometric analysis normalizing levels of phosphorylated CagA to total CagA from 3 replicates are shown. GAPDH served as a loading control. ANOVA with Bonferroni correction or student t test was used to determine statistical significance between groups. *P < .05, **P < .01, ***P < .001, ****P < .0001; + P < .00001 compared to uninfected TLR9+ cells; NS, nonsignificant compared to uninfected parental cells. Abbreviations: ANOVA, analysis of variance; ELISA, enzyme-linked immunosorbent assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IFN, interferon; IL-8, interleukin-8; TLR9, Toll-like receptor 9; UI, uninfected; wt, wild type.
Because HopQ is a bacterial adhesin, we determined whether reductions in TLR9 activation induced by the hopQ− mutant were dependent upon decreased adherence. No differences in binding to AGS cells were identified between the wild-type strain 26695 and the hopQ− isogenic mutant (Figure 2C). Soft agar motility assays demonstrated that H. pylori parental strain 26695 exhibited similar motility to other H. pylori strains included in this study (data not shown). Type I hopQ alleles are in linkage disequilibrium with the cag-T4SS [7] and specific HopQ-CEACAM interactions have been reported to be required for translocation of CagA into epithelial cells [5, 6]. Thus, to discern whether reductions in TLR9 activation were due to inactivation of hopQ per se and not due to concomitant loss of cag-T4SS function, we analyzed cag-T4SS-associated phenotypes in the hopQ− mutant. Both wild-type strains 26695 and 7.13 and their respective hopQ− or complemented hopQ mutants were similar in their ability to translocate CagA, as determined by levels of phosphorylated CagA per level of total CagA/strain, while, as expected, the cagE− mutant failed to translocate CagA (Figure 2D and Supplementary Figure 6). Furthermore, no significant differences in levels of IL-8 production were observed between 26695 and 7.13 wild type, hopQ−, and hopQ complemented mutant-infected samples (Figure 2B and Supplementary Figure 6). There were also no differences between wild-type H. pylori strain 26695 and the hopQ− mutant in the ability to activate an independent cag-T4SS−dependent effector, NOD1 (Supplementary Figure 7).
DISCUSSION
We identified a strain-variable cag-PAI–independent H. pylori component, HopQ, that is associated with TLR9 activation and is linked to carcinogenic potential. There are several potential mechanisms that may underpin these observations. Structural analyses comparing type I HopQ to type II HopQ proteins have revealed a differential ability to bind specific CEACAMs [12]. Type I HopQ harbors a higher affinity for human CEACAM1 versus CEACAM6, raising the possibility that HopQ-CEACAM1 interactions are necessary for translocation of microbial DNA and TLR9 activation. Of interest, H. pylori cagA+ strains induce higher levels of CEACAM expression than cagA− strains, and TLR9-regulated transcription factors such as NF-κB and activator protein 1 (AP-1) are linked to H. pylori infection and CEACAM regulation [5]. Although further studies linking discrete cell signaling cascades to specific HopQ alleles and CEACAMs will be required, H. pylori has likely evolved to harbor different alleles of hopQ that may confer selective binding and molecular signaling capacities.
Our current results also indicate that cag-T4SS function in H. pylori strains 26695 and 7.13 is not dependent on HopQ. Some, but not all, previous studies have demonstrated a more direct role for HopQ in cag-PAI functions [5, 6]. However, these studies utilized independent H. pylori strains in different cell models under different conditions compared to our current study, which may account for the varying results.
We recognize that not all of the in vitro and in vivo data are fully aligned and speculate that the lack of absolute concordance represents fundamental differences that exist between the reductionist in vitro TLR9 activation assay and the in vivo milieu colonized by H. pylori. For example, varying expression levels of CEACAM proteins may be present within different patient samples, which may alter HopQ function. Further, other microbial constituents and host signaling pathways, such as peptidoglycan and NOD1 as well as heptose-bisphosphate and NF-κB, have been shown to affect inflammation in vivo [3]. The complexity of such interactions is heightened when comparing results from human tissue to rodent tissue. In humans, genetic polymorphisms within TLR9 have been linked to H. pylori persistence [13]. We previously demonstrated that H. pylori strains harvested from persons at increased risk for gastric cancer activated TLR9 more robustly than strains isolated from patients residing in a low-risk cancer region [4]. These data are consistent with the current results and with data from Qin et al demonstrating that H. pylori and H. pylori DNA induce TLR9-dependent proliferation, migration, and invasion of human gastric epithelial cells [14]. However, our group also reported enhanced inflammation in H. pylori-infected tlr9−/− mice when compared to H. pylori-infected wild-type mice [15]. We speculate that this discordance may be related to the duration of infection (years in humans, weeks in mice) as well as inherent differences in host responses to microbial pathogens across species, which for TLR9 carries increased complexity because activation of TLR9 can lead to either pro- or anti-inflammatory responses in vivo depending on cellular context.
Collectively, the current results aid in delineating the route by which microbial DNA is delivered to host cells, and also may reveal the impact that DNA translocation has on carcinogenesis in vivo.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the National Institutes of Health (grant numbers T32AI112541-02 to S. D. R. D; R01CA077955, R01DK058587, P01CA116087, and P30DK058404 to R. M. P.; and R01AI118932 to T. L. C.); and the Department of Veterans Affairs (grant number I01BX004447 to T. L. C.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Amieva M, Peek RM Jr. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology 2016; 150:64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wroblewski LE, Peek RM Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev 2010; 23:713–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Backert S, Haas R, Gerhard M, Naumann M. The Helicobacter pylori type IV secretion system encoded by the cag pathogenicity island: architecture, function, and signaling. Curr Top Microbiol Immunol 2017; 413:187–220. [DOI] [PubMed] [Google Scholar]
- 4. Varga MG, Shaffer CL, Sierra JC, et al. Pathogenic Helicobacter pylori strains translocate DNA and activate TLR9 via the cancer-associated cag type IV secretion system. Oncogene 2016; 35:6262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Javaheri A, Kruse T, Moonens K, et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat Microbiol 2016; 2:16189. [DOI] [PubMed] [Google Scholar]
- 6. Zhao Q, Busch B, Jiménez-Soto LF, et al. Integrin but not CEACAM receptors are dispensable for Helicobacter pylori CagA translocation. PLoS Pathog 2018; 14:e1007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao P, Cover TL. Two different families of hopQ alleles in Helicobacter pylori. J Clin Microbiol 2002; 40:4504–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peek RM Jr, van Doorn LJ, Donahue JP, et al. Quantitative detection of Helicobacter pylori gene expression in vivo and relationship to gastric pathology. Infect Immun 2000; 68:5488–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loh JT, Torres VJ, Algood HM, McClain MS, Cover TL. Helicobacter pylori HopQ outer membrane protein attenuates bacterial adherence to gastric epithelial cells. FEMS Microbiol Lett 2008; 289:53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loh JT, Beckett AC, Scholz MB, Cover TL. High-salt conditions alter transcription of Helicobacter pylori genes encoding outer membrane proteins. Infect Immun 2018; 86:e00626-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yokoyama K, Higashi H, Ishikawa S, et al. Functional antagonism between Helicobacter pylori CagA and vacuolating toxin VacA in control of the NFAT signaling pathway in gastric epithelial cells. Proc Natl Acad Sci U S A 2005; 102:9661–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moonens K, Hamway Y, Neddermann M, et al. Helicobacter pylori adhesin HopQ disrupts trans dimerization in human CEACAMs. EMBO J 2018; 37:e98665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loganathan R, Nazeer M, Goda V, et al. Genetic variants of TLR4 and TLR9 are risk factors for chronic Helicobacter pylori infection in South Indian Tamils. Hum Immunol 2017; 78:216–20. [DOI] [PubMed] [Google Scholar]
- 14. Qin XR, Wu J, Yao XY, Huang J, Wang XY. Helicobacter pylori DNA promotes cellular proliferation, migration, and invasion of gastric cancer by activating toll-like receptor 9. Saudi J Gastroenterol 2019; 25:181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Varga MG, Piazuelo MB, Romero-Gallo J, et al. TLR9 activation suppresses inflammation in response to Helicobacter pylori infection. Am J Physiol Gastrointest Liver Physiol 2016; 311:G852–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.