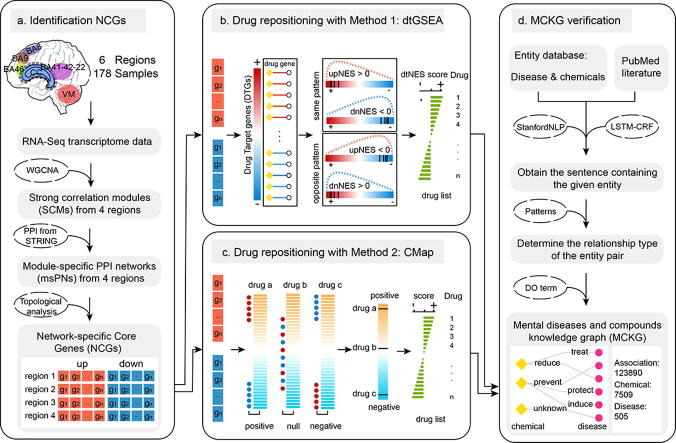
Graphical abstract
Keywords: Autism spectrum disorder, Coexpression network, Drug repositioning, Knowledge graph, Natural language processing
Abstract
Identification of exact causative genes is important for in silico drug repositioning based on drug-gene-disease relationships. However, the complex polygenic etiology of the autism spectrum disorder (ASD) is a challenge in the identification of etiological genes. The network-based core gene identification method can effectively use the interactions between genes and accurately identify the pathogenic genes of ASD. We developed a novel network-based drug repositioning framework that contains three steps: network-specific core gene (NCG) identification, potential therapeutic drug repositioning, and candidate drug validation. First, through the analysis of transcriptome data for 178 brain tissues, gene network analysis identified 365 NCGs in 18 coexpression modules that were significantly correlated with ASD. Second, we evaluated two proposed drug repositioning methods. In one novel approach (dtGSEA), we used the NCGs to probe drug-gene interaction data and identified 35 candidate drugs. In another approach, we compared NCG expression patterns with drug-induced transcriptome data from the Connectivity Map database and found 46 candidate drugs. Third, we validated the candidate drugs using an in-house mental diseases and compounds knowledge graph (MCKG) that contained 7509 compounds, 505 mental diseases, and 123,890 edges. We found a total of 42 candidate drugs that were associated with mental illness, among which 10 drugs (baclofen, sulpiride, estradiol, entinostat, everolimus, fluvoxamine, curcumin, calcitriol, metronidazole, and zinc) were postulated to be associated with ASD. This study proposes a powerful network-based drug repositioning framework and also provides candidate drugs as well as potential drug targets for the subsequent development of ASD therapeutic drugs.
1. Introduction
Autism is a highly heterogeneous and polygenic neurodevelopmental disorder that has been reported to occur in approximately 1 in 59 children [1], [2], [3]. Currently, risperidone and aripiprazole are the only approved therapeutic options for partially improving autism symptoms [4]. As the global prevalence of ASD continues to rise, there is an urgent need for developing efficient and cost-effective therapeutic options for this disease [5]. However, de novo drug discovery is an expensive and time-consuming process; it takes approximately 10 years and more than $1 billion to develop a new drug [6]. As a complex polygenic disease, ASD is not suited to the traditional drug development pattern of identifying lead compounds that show activity against single therapeutic targets. Therefore, there is a need to establish new and effective drug discovery strategies for the treatment of ASD. In silico drug repositioning is one of the attractive strategies for identifying the potential therapeutic drugs for ASD.
In silico drug repositioning is aimed at identifying new indications for existing compounds. Compared to the traditional drug discovery approaches, in silico approaches combine and analyze various forms of drug data and disease omics data to identify new potential functions of existing drugs. Since this approach has the advantage of systematically filtering multiple candidate drugs, development time and costs are significantly reduced [7], [8]. Due to the generation of large amounts of transcriptome data, transcriptome signature matching has become a promising method for in silico drug repositioning [9]. This method assumes that if a drug can reverse the gene expression pattern associated with a diseased state back to the pattern associated with the healthy state, then that drug may be a potential therapeutic option for the disease [7]. The first step in this method involves the identification of disease-associated gene expression signatures, and then, the identification of drug-induced changes in gene expression using various resources, such as Connectivity Map (CMap) [10], in order to screen candidate drugs [11]. For example, Dudley et al. used CMap to compare gene signatures to transcriptome signatures and found that the anticonvulsant, topiramate, can treat inflammatory bowel disease [12]. So et al. proposed a framework for drug repositioning that links GWASs and drug expression profiles from CMap [13]. However, ASD is heterogeneous and the molecular mechanism involved have not been established, therefore, Dudley et al.'s method, which involves using differential expression analysis to identify disease gene signatures is not suitable for ASD. Accurate identification of disease-relevant gene expression signatures is key in transcriptomic signature matching. In addition to drug transcriptome label data in CMap, there are large amounts of drug-induced gene expression data that have not been used. In So et al.'s framework, evidence for therapeutic potential of candidate drugs found by manual literature search is not comprehensive. Addressing these challenges will require a more effective and systemic approach.
The network-based analysis method provides an opportunity for accurately identifying ASD pathogenic genes, revealing molecular mechanisms, and identifying drug targets, which offer possibilities for drug repositioning. ASD is a polygenetic disease, gene interactions significantly contribute to the highly heterogeneous genetic structure and phenotypic complexity of ASD [14], [15]. Network analysis is an effective method for evaluating gene interactions that can be used to develop potential gene interaction networks with similar functions from high-dimensional transcriptome data. These genes and gene interaction pathways may represent novel drug targets for ASD [16]. In addition, network-based analysis methods can organize protein–protein interactions, drug-target interactions, and drug disease modules in form of a network to on molecular mechanisms, potential therapeutic drugs, and offer evidence related to potential therapeutic drugs [17], [18].
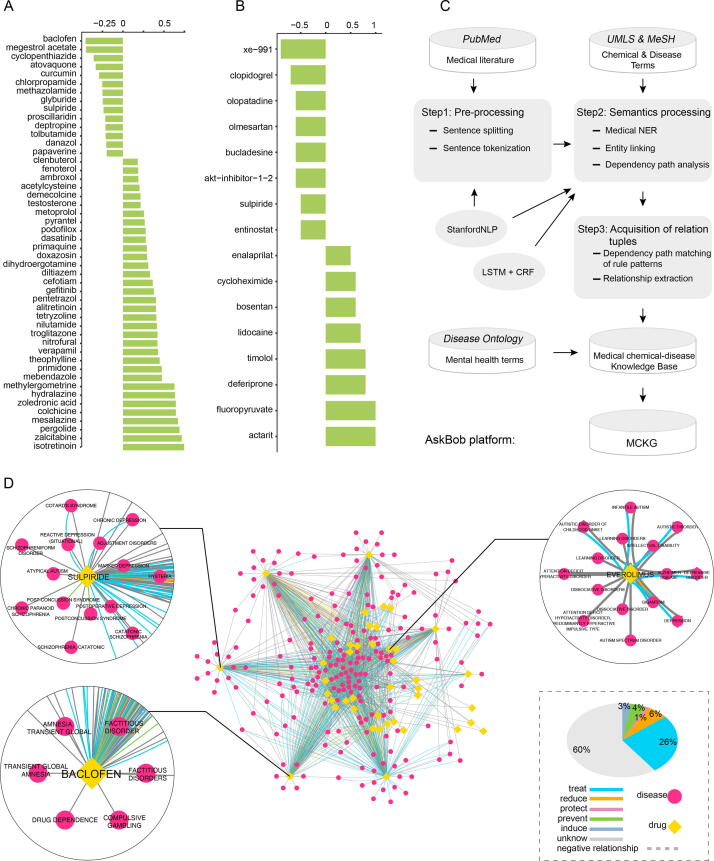
In this study, a novel network-based drug repositioning framework was developed which integrates network-specific core gene (NCG) screening from the transcriptome data of different brain regions from children with ASD, drug repositioning methods based on drug-gene networks and transcriptome signature matching, as well as evaluation of the therapeutic potential of candidate drugs based on a knowledge graph (Fig. 1). Moreover, we evaluated and compared gene expression patterns and gene regulation networks in different brain regions of children with ASD. Molecular mechanisms were further investigated by functional enrichment analysis.
Fig. 1.
Workflow for potential therapeutic drug repurposing for ASD. The workflow contains 4 main steps: a identification of NCGs based on network analysis methods; b drug repositioning with method 1: drug-target gene set enrichment analysis (dtGSEA); c drug repositioning with method 2: the Connectivity Map webtool; and d knowledge graph construction.
2. Materials and methods
2.1. Data collection and sample selection
For ASD it is easier to use brain tissue transcriptome data to identify dydregulated gene signals. To reduce technical errors, a total of 5 datasets that qualified for our study were obtained, including GSE102741 [19], GSE59288 [20], GSE51264 [21], GSE62098 [22], and the dataset developed by Parikshak et al. [16]. These 5 public datasets have the same sequencing platform and a large sample size. Transcriptome sequencing data for a total of 178 brain tissue samples were obtained from 5 datasets. The tissues used for sequencing were obtained from 6 different brain regions, including the dorsolateral prefrontal cortex (DPC, BA46), superior frontal gyrus (SFG, BA8), corpus callosum (CC), BA9, BA41-42-22, and Vermis (VM) (Table S1). All the samples had been obtained from children (age < 18 years). The control group consisted of 97 healthy individuals (24 girls and 73 boys, mean age: 10.07 years), while the ASD group consisted of 81 patients (19 girls and 62 boys, mean age: 10.21 years). There was no statistically significant difference in age between the two groups (p > 0.05).
2.2. RNA sequencing data preprocessing
Raw data for the four datasets (GSE102741, GSE59288, GSE51264, GSE62098) from the GEO database were converted to “.fastq” format files using the fasterq-dump tool. We used fastqc v0.11.8 to check the sequencing quality of the data and trimmomatic-0.39 to trim and filter low-quality reads [23]. The reads were mapped to GRCh37.73 annotations using TopHat v2.1.1 [24]. Quantification of gene expression levels were performed using union exon models with HTSeq v0.11.0 [25]. Processing methods for the data in the 4 datasets were the same as those for the Parikshak et al. dataset. Based on the reprocessed quantitative file for gene expression levels, we divided the data into 6 gene expression datasets according to tissue sources. Genes expressed in at least 50% of the samples (counts of 10 or more) were retained.
2.3. Coexpression module analysis
Filtered raw read counts for all samples were normalized using the “VST” command in DESeq2 [26]. After using the ComBat function in R package SVA to correct for the batch effect [27], expression data were divided into 6 datasets based on different brain regions. WGCNA v1.67 was used to perform weighted gene coexpression network analysis on each dataset [28]. The “BlockwiseModules” function was used to construct signed coexpression modules, with each module containing at least 50 genes. Modules were summarized by their module eigengene (ME), and modules with eigengene correlations of > 0.85 were merged. For module-trait analyses, “bicor” was used to establish the correlation between gene expression and phenotypic data (ASD diagnosis). A |cor| > 0.35 and p-value < 0.05 were set as cutoff criteria for ASD-associated module screening. A positive correlation implied that a module was upregulated in the disease group compared to the control group while a negative correlation indicated downregulation. We used gene significance (GS) and module membership (MM) values to define the hub genes (GS > 0.30, MM > 0.6, p-value < 0.05). We classified the significantly associated ASD modules composed of hub genes as strong correlation modules (SCMs), which included upregulated SCMs and downregulated SCMs.
2.4. ASD-related genes and evaluation of SCMs
We constructed an ASD-related gene set, that contained differentially expressed genes (DEGs) and previously reported ASD pathogenic genes. First, we identified DEGs between ASD samples and controls in each brain region using edgeR v3.26.5 [29]. Read counts were subjected to TMM normalization. Log2 (counts per million, CPM) transformation was used for clustering to remove outlier samples. To ensure that the different signals between groups were not affected by hidden confounding factors, we used R package “SVA” to calculate two covariates and design them into the model. Using the exact test model to determine DEG in the expression data for each brain region, multiple testing correction was performed using the Benjamini-Hochberg method. Genes with FDR values < 0.05 and logFC values > 1 (upregulated) or logFC values <−1 (downregulated) were reported as significant DEGs. Second, we searched for previously reported ASD causative genes in the SFARI Gene and AutismKB 2.0 databases [30], [31]. Based on their scores, the genes were into three categories high confidence (SFARI score = 1 or AutismKB score > 16), strong candidate (SFARI score = 2 or AutismKB score > 10), and suggestive evidence (SFARI score = 3 or AutismKB score > 5) categories. Finally, hypergeometric distribution was used for enrichment analysis to establish the significance of the overlap between the ASD-related gene set and SCMs.
2.5. msPN construction and NSGs screening
Protein-protein interactions (PPIs) were downloaded from the STRING database (Version 11, http://string-db.org/), and interactions with a combined score of > 400 were selected for the construction of the PPI network. We mapped the genes in SCMs to the PPI network to obtain module-specific PPI networks (msPNs), which included upregulated and downregulated msPNs. For each msPN, we calculated the degree of each node using “degree” function in the R package igraph v1.2.2. Then, we selected nodes whose degrees were greater than the mean value of the degrees for all nodes in the network as network-specific core genes (NCGs):
where by N[m] is the node in each msPN, dm is the degree of the node, and n is the total number of nodes in each msPN. If N[m] is equal to 1, then the node is NCG. Cytoscape v3.7.1 software was used for network visualization.
2.6. Drug-target gene set enrichment analysis (dtGSEA)
We downloaded pharmaco-transcriptomics data from the DrugBank database (https://www.drugbank.ca/) and drug-gene interactions from Drug Targetor v1.21 (http://drugtargetor.com/), which contains information on up-/downregulated genes due to the metabolism of pharmaceutical compounds [32], [33]. We combined drug-target interactions from the two databases and deleted the duplicates. Then, target genes for each drug were mapped to the PPI network to construct drug-specific PPI networks (dsPNs). Genes in each dsPN network were postulated to be drug target genes (DTGs). The ENSEMBL peptide ID and gene name were converted to an Entrez Gene ID using biomaRt.
Drug-target normalized enrichment scores (dtNES), were determined in four steps. First, for each dsPN, we calculated the degree of each node using “degree” function in R pacakage igraph v1.2.2. Then, based on the type of drug-gene interaction, we divided the DTGs into drug-upregulated and drug-downregulated gene sets. Node degrees of the drug-upregulated genes were positive while those of drug-downregulated genes were negative. The DTGs were ranked in a descending order of node degrees. A ranking list of gene importance was generated for each dsPN. The most a gene was top- and bottom-ranked, the more important it was in the network. The ranked gene list was further used for GSEA analysis. Second, based on the msPN type, NCGs were separated into up- and downregulated gene sets and simultaneously scanned with the ranked lists of all dsPNs through gene set enrichment analysis (GSEA) [34]. For each dsPN, NCGs and DTGs overlapped by at least two genes. Then GSEA pre-ranked list analysis was applied using 1000 permutations to obtain significant dsPNs (FDR < 0.25). Third, based on the results of GSEA, the drug action type was divided into two patterns. The phenomenon in which upregulated NCGs were enriched in the upregulated DTG set (upNES > 0) while downregulated NCGs were enriched in the downregulated DTG set (dnNES < 0) was defined as the “same” pattern. The phenomenon in which the upregulated NCGs were enriched in the downregulated DTG set (upNES < 0) and the downregulated NCGs were enriched in the upregulated DTG set (dnNES > 0) was defined as the “opposite” pattern. Fourth, the maximum absolute NESs against up- and downregulated NCGs were defined as drug-target normalized enrichment scores (dtNESs) for overlap, as follows: dtNES = max(abs(upNES), abs(dnNES)). The dtNES exhibiting the same pattern were positive, while scores of the opposite pattern were negative. Drugs corresponding to a dsPN with a negative dtNES may reverse the regulatory direction of the gene network. When DTG sets overlapped with either up- or downregulated NCG genes but not both, the direction and score for the overlap were determined using the NES of the more significant set. Results for up- and downregulated NCGs that were simultaneously enriched in the up- or downregulated DTG sets were discarded. For simplicity, we referred to this approach for drug repositioning as the drug-target gene set enrichment analysis (dtGSEA).
2.7. Drug repositioning based on Connectivity Map
Based on the principle of matching transcriptome signatures, we used the “Query” web tool in CMap (https://clue.io/) to analyze NCGs in the up- and downregulated msPNs to identify drugs with therapeutic repositioning potential in ASD [35]. We considered CMap connectivity scores (tau values) of + 95 or higher, and −95 or lower to be strong scores for identifying potential compounds that affect the disease state. The most negative connections were considered to be perturbations that elicit transcriptional effects opposite of the disease state; these compounds are potential therapeutic drugs. Similarly, the most positive connections were considered to be perturbations with similar effects.
2.8. Construction MCKG
The mental diseases and compounds knowledge graph (MCKG), a comprehensive biological knowledge graph that associates compounds and mental diseases, was constructed based on the AskBob drug discovery platform (http://lwj1.ens.yun.pingan.com:5001/). AskBob is a drug discovery commercial platform that was designed and developed by PingAn HealthCare Technology. First, we used the StanfordNLP tool to preprocess all PubMed documents published before 2019. The preprocessing involved splitting of the documents into sentences, sentence tokenization, and part-of-speech tagging. Second, we applied named entity recognition (NER) based on the LSTM-CRF model to every sentence for medical entity mention detection [36]. Sentences that did not contain chemical or disease entities were discarded. The objective of entity linking was to link each mentioned entity to a dictionary of chemical and disease medical entities as defined by the Unified Medical Language System (UMLS) (www.nlm.nih.gov/research/umls) and Medical Subject Headings (MeSH) (http://www.ncbi.nlm.nih.gov/mesh). In case the entity could not be linked to the dictionary, it was discarded. Dependency analysis was performed using the StanfordNLP tool to obtain the dependency path of a given chemical and disease entity pair in a sentence [37]. Third, relation type of the given entity pair in the sentence was identified by matching the dependency path and high-confidence rule patterns as evaluated by experts. In case the rule pattern could be matched, a relation tuple containing the chemical, relation, and disease was obtained. The relation tuples were stored in the medical knowledge database. Finally, we used all DO terms under the disease of mental health term (DOID: 150) in the Disease Ontology (DO, http://www.disease-ontology.org) database to extract mental disease-related relational tuples from the medical knowledge base [38]. Finally, we constructed the MCKG and visualized it in the AskBob platform.
2.9. Toxicogenomic data analysis
We downloaded rat in vivo toxicogenomic data from DrugMatrix and Open TG-GATEs databases [39]. The DrugMatrix database includes two datasets: GSE59927 and GSE57822 [40], which can be downloaded from the GEO database. The rat GeneChip array data, which was downloaded from the Open TG-GATEs database, includes two subsets: rat in vivo liver single dose, and rat in vivo liver repeat dose. Microarray data were downloaded from ftp://ftp.biosciencedbc.jp/archive/open-tggates/LATEST/. Repeat probes were combined with the maximum value and annotated using R package annotate v.1.52.1 to obtain gene symbols. Entrez gene ID and gene symbol were mapped using the R biomaRt package for both human and rat. Limma V3.30.13 was used to obtain differentially expressed genes [41]. The threshold for DEGs was set as |logFC| > 0 and p < 0.05. The DEGs for each compound under different conditions (time, dose, and tissue) were sorted in a descending order of logFC to generate a ranked list. Then, the GSEA algorithm was used to determine reversal of the expression profile of NCGs in the 4 rat in vivo data subsets. The principle was the same as that of dtGSEA.
2.10. Functional enrichment analysis
Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomics (KEGG) pathways enrichment analyses were performed using clusterProfiler v3.12.0 R package [42]. In case large numbers of comparisons were performed, the Benjamini-Hochberg method was used to adjust raw p-values. GO terms with q values < 0.05 and KEGG pathways with q-values < 0.05 were considered significantly enriched. The list of abbreviations is provided in Box 1.
Box 1. List of abbreviations.
3. Results
3.1. ASD brain region-specific gene coexpression module dysregulation
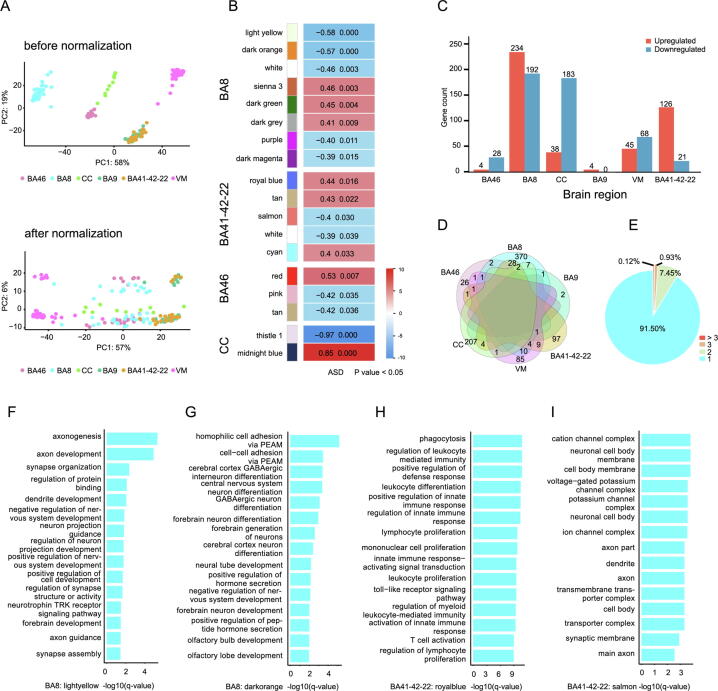
Raw data were analyzed and processed via a unified data processing flow to eliminate bias (Fig. S1A). Fig. 2A shows changes in the PCA plot before and after batch effect adjustment. After quality control and filtering of the gene expression data, we obtained a total of 20,119 genes. A total of 18 SCMs were identified in 4 brain regions, BA46, BA8, CC, and BA41-42–22(Fig. 2B, Fig. S2E). No SCMs were identified in the BA9 and VM brain regions. Among these SCMs, 8 modules (upregulated: 3, downregulated: 5) were in BA8; 5 modules (upregulated: 3, downregulated: 2) were in BA41-42–22; 3 modules (upregulated: 1, downregulated: 2) were in BA46, while 2 modules (upregulated: 1, downregulated: 1) were in CC (Fig. 2B). More infiormation on the SCMs is provided in Table S2.
Fig. 2.
Coexpression network analysis and functional enrichment network for the identified genes. a PCA plot before batch effect removal based on gene expression and after ComBat batch effect removal. Dots with different colors correspond to different brain regions. b Relationships of MEs (module eigengenes) and ASD in the 4 brain region. Each row in the table corresponds to a module. The numbers in the table report the correlations of the corresponding MEs and traits (P < 0.05, modified Bonferroni test). The table is color-coded by correlation according to the color legend. c Bar plot representation of differential gene expression in different brain regions. d Venn diagrams of differentially expressed genes. e The percentages of all differentially expressed genes distributed in the brain regions are shown in the pie chart. f GO term enrichment analysis of genes in the light yellow SCM of BA8. g GO term enrichment analysis of genes in the dark orange SCM of BA8. h GO term enrichment analysis of genes in the royal blue SCM of BA41-42-22. i GO term enrichment analysis of genes in the salmon SCM of BA41-42-22. The x-axis shows the -log (q-value), and the y-axis shows the GO terms. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The ASD-related gene set contained a total of 2,475 genes, including 773 differentially expressed genes (DEGs)and 1,805 previously reported ASD causative genes (Table S3, see Methods). A total of 426 DEGs, including 234 upregulated and 192 downregulated genes were found in BA8. BA8 was found to have the highest number of DEGs, followed by CC (221 DEGs), BA41-42–22 (147 DEGs), VM (113 DEGs), BA46 (32 DEGs), and BA9 (4 DEGs) (Fig. 2C and Fig. S1B). Among the DEGs, there was no gene that was differentially expressed in all regions (Fig. 2D). About 91.50% of the genes were dysregulated in only one region while 8.5% of the genes were deregulated in more than two regions (Fig. 2E, Table S4). GO enrichment analysis revealed that downregulated DEGs in BA8, BA41-42–22, and CC were enriched in nervous system development, synaptic vesicle, and neurotransmitter transmission terms (Fig. S1C-E). Upregulated DEGs in BA8, BA41-42–22, and VM were enriched in immune system process term. No significant functional enrichment results were found for the DEGs in BA46 and BA9. More findings are shown in Table S5. Among the 1805 previously reported ASD causative genes, there were 318 high-confidence genes, 674 strong candidate genes, and 813 genes with suggestive evidence. Hypergeometric distribution testing revealed significant enrichments of ASD-related genes among 9 SCMs (p < 0.05, Table 1), which were mainly distributed in BA8 and BA41-42–22 (Fig. S2E). The detailed enrichment results of the genes in the SCMs are given in Table S2.
Table 1.
Hypergeometric distribution testing results of 18 SCMs.
| Region | Module | SCMsize | DEGs | Causative gene | P-value |
|---|---|---|---|---|---|
| BA46 | red | 169 | – | 10 | 0.94 |
| pink | 70 | 1 | 3 | 0.88 | |
| tan | 63 | 2 | 6 | 0.20 | |
| BA8 | lightyellow | 223 | 43 | 38 | 5.11E-25 |
| darkorange | 174 | 32 | 25 | 4.90E-14 | |
| white | 116 | 3 | 20 | 5.85E-04 | |
| sienna3 | 76 | 12 | 7 | 1.04E-04 | |
| darkgreen | 130 | 1 | 5 | 0.98 | |
| darkgrey | 105 | 6 | 3 | 0.61 | |
| purple | 157 | 4 | 33 | 3.00E-06 | |
| darkmagenta | 62 | 3 | 21 | 1.37E-09 | |
| BA41-42–22 | royalblue | 71 | 14 | 9 | 1.34E-07 |
| tan | 125 | 4 | 15 | 3.02E-02 | |
| salmon | 109 | – | 21 | 6.22E-04 | |
| cyan | 74 | 1 | 2 | 0.97 | |
| white | 46 | – | 8 | 0.05 | |
| CC | thistle1 | 109 | 5 | 6 | 0.39 |
| midnightblue | 118 | 2 | 3 | 0.99 |
To elucidate on the biological functions of SCMs, we performed GO analyses with genes in each SCM. The downregulated modules in BA46, BA8, and BA41-42–22 were associated with synaptic function and nervous system development. For example, the lightyellow (Fig. 2F), darkorange (Fig. 2G), and darkmagenta (Fig. S2A) modules in BA8 were enriched in axon development, neuron differentiation, and cell–cell adhesion. Salmon SCM of BA41-42–22 was enriched in synaptic and axonal functions (Fig. 2I); moreover, the upregulated royalblue module in BA41-42–22 was enriched in immune system functions (Fig. 2H). The upregulated gene in the darkgrey module in BA8 was enriched in mitochondrial respiratory electron transport chain and mitochondrial ATP synthesiscoupled electron transport functions (Fig. S2B). The downregulated white module in BA41-42–22 was associated with sodium ion transport and ion transmembrane transport (Fig. S2C). The pink module in BA46 was enrichment in mitochondrial functions (Fig. S2D). Enriched GO terms for these SCMs are shown in Table S6.
3.2. NCGs are associated with ASD
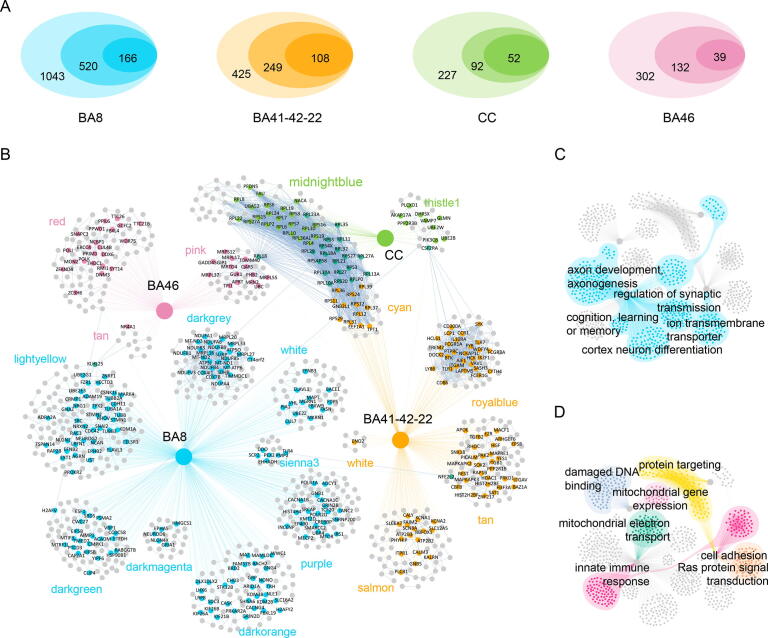
Pathways and hub proteins within PPI networks are important drug targets. Therefore, precise selection of target genes involved in molecular mechanisms of ASD is critical for drug repositioning. The combined use of gene coexpression data and protein interaction data improves the confidence of the corresponding PPI network, thereby enabling a high-confidence ASD-related molecular network to be obtained. The strong correlation between network topology and biological function is associated with the presence of proteins with high node degrees in the network that may exert a strong influence on network function through multiple interactions. In this study, we constructed a pipeline that uses transcriptome data and PPI networks to screen network-specific core genes (NCGs) for drug repositioning (Fig. 1A). Screening procedures for NCGs are shown in Methods Section 2.5.
Based on region-specific SCMs, we constructed 18 msPNs, which contained a total of 993 protein-coding genes (Fig. 3A, Fig. 3B). After calculating the degrees of the network nodes, a total of 365 NCGs (18.28% of the total number of coexpression module genes) were screened out of the 993 genes (Fig. 3A). There were 166 (31.92% of 520), 108 (43.37% of 249), 52 (56.52% of 92), and 39 (29.55% of 132) NCGs, for the BA41-42–22–, BA8-, CC–, and BA46-msPN genes, respectively (Fig. 3A, Fig. 3B). A detailed msPN protein-coding gene list and NCGs can be found in Table S2. Functional enrichment analysis of the msPNs revealed that functions for each coexpression module were not only retained, but were also more focused than those of the larger group. For example, for the lightyellow msPN in BA8, GO enrichment analysis showed that the genes were involved in regulation of axonogenesis and axon development (Fig. 3C). The darkorange msPN network of BA8 was enriched in cerebral cortex neuron differentiation (Fig. 3C). The royalblue msPN in BA41-42–22 was enriched with phagocytosis and immune response-related genes (Fig. 3D). The darkgrey msPN network of BA8 was enriched with mitochondrial electron transport function genes(Fig. 3D) (Table S7).
Fig. 3.
Module-specific PPI network and functional analysis. a Venn diagram. In the Venn diagram, the largest ellipse represents the number of genes in the SCM, the medium ellipse represents the number of genes in the module-specific PPI network (msPN), and the smallest ellipse represents the number of NCGs. b msPNs and NCGs. The named nodes represent NCGs from the msPN, the orange nodes represent NCGs from BA41-42–22, the deep sky blue nodes represent NCGs from BA8, the light green nodes represent NCGs from CC, the pink nodes represent NCGs from BA46, and the dark cyan nodes represent the intersections of NCGs from different brain regions. The gray nodes are proteins in the msPN, and the gray edges represent protein interactions in the msPN. c Functional enrichment revealed that the genes in the msPNs are related to the nervous system. d Functional enrichment revealed that the genes in the msPNs are related to other functions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Repositioning candidates as identified by dtGSEA and CMap approaches
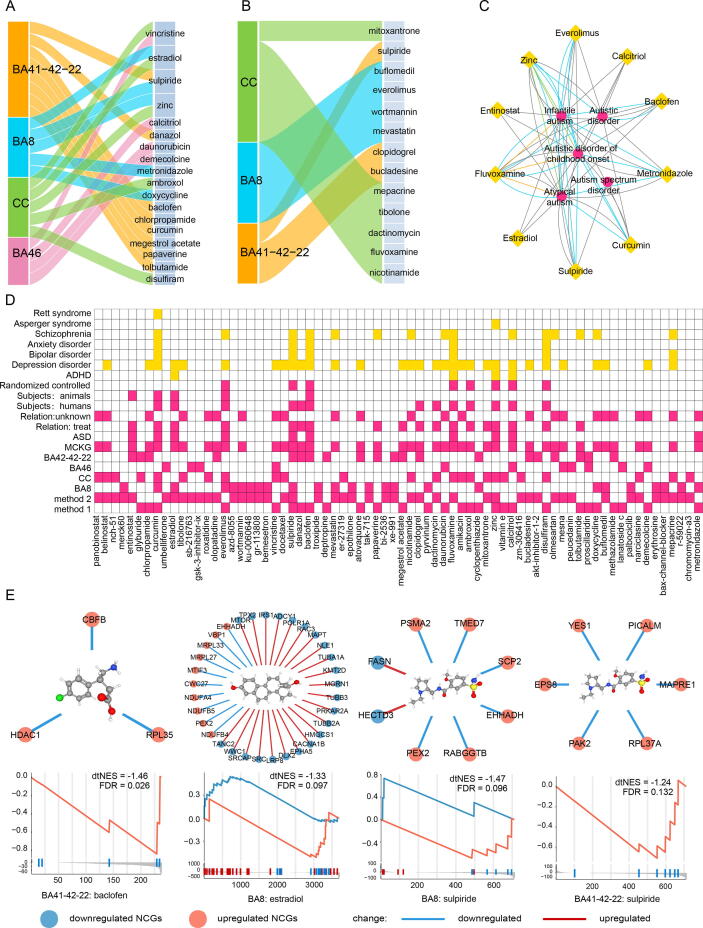
Through dtGSEA, dsPNs with NCGs were scanned to reposition drugs (Fig. 1B) while through the CMap database, drugs were repositioned (Fig. 1C, see Methods for the detailed procedures). Based on 365 NCGs from 4 brain regions, a total of 72 compounds (without duplication) that may have therapeutic effects were screened using these two methods (Table S8).
Through the dtGSEA approach, we obtained a total of 1981 dsPNs and 18,876 drug target genes (DTGs). These dsPNs contained a total of 189,899 drug-gene interactions, among which 94,581 were upregulation interactions while 95,318 were downregulation interactions. To determine the extent to which a compound targeted the NCGs, we defined dtNES with a plus or minus sign through GSEA. A negative dtNES indicated that a drug could reverse the expression of NCGs in msPNs. We obtained a total of 35 drugs (BA41-42–22: 14 drugs, BA8: 7 drugs, CC: 8 drugs, BA46: 6 drugs) with negative dtNESs, among which 7 drugs (sulpiride, estradiol, proscillaridin, vincristine, cyclopenthiazide, zinc, and glyburide) were identified in >2 brain regions (Table S9). For example, baclofen (dtNES = −1.46, FDR = 0.026), which was found in BA41-42–22, ranked first among 14 drugs (Fig. 4A). Similar findings are shown in Fig. S3. Sulpiride, identified in BA8 (dtNES = −1.47, FDR = 0.096), was also found in BA41-42–22 (dtNES = −1.24, FDR = 0.132) (Fig. S3A), while estradiol was found in both BA8 (dtNES = −1.33, FDR = 0.097) and CC (dtNES = −1.70, FDR = 0.024) (Fig. S3C).
Fig. 4.
Drug repositioning results and MCKG. a Significant drug-target normalized enrichment score (dtNES) for ASD from BA41-42–22. The x-axis shows the positive dtNES −1 and the negative dtNES + 1, and the y-axis shows the drug name. b Significant CMap connectivity score for ASD from BA41-42–22. The x-axis is the score ((positive score − 95)/10 and (negative score + 95)/10) and the y-axis is the name of the drug. The horizontal bars indicate the computationally predicted therapeutic scores for the drugs based on comparison of the gene expression signatures of the drugs with the NCGs. A negative score indicates that a drug exhibits an expression pattern that is oppositional to the disease; such, drugs are potential therapeutic drugs. c Overview of the knowledge graph and processing pipeline. d Forty-two drug candidates and the mental illness association network from MCKG. Yellow nodes represent drugs, magenta nodes represent mental illnesses, and differently colored edges indicate different relationship types. The pie charts show the percentages of relationship types. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Up- and downregulated region-specific NCGs were entered into the CMap database, and a total of 46 compounds (BA41-42–22: 8, BA8: 18, CC: 15, BA46: 5) with potential therapeutic effects were obtained, among which XE-991 was identified in both BA8 and BA41-42–22 (Table S9). These compounds included FDA-approved drugs, including sulpiride (score = −95) and entinostat (score = −95),which were found in BA41-42–22 (Fig. 4B); epothilone (score = −97) and everolimus (score = −97), which were found in BA8 (Fig. S4A); and fluvoxamine (score = −95), which was found in CC (Fig. S4C). In addition, we identified some new chemicals, including XE-991 (BA41-42–22: score = −99, BA8: score = −99), BI-2536 (BA8: score = −98), SB-216763 (BA46: score = −99), and ER-27319 (CC: score = −97).
3.4. Evaluation therapeutic potentials of drug candidates
To assess the therapeutic potentials of candidate drugs identified through drug repositioning, we constructed MCKG (Fig. 1D, Fig. 4C, see Methods). The MCKG included 2 types of entities (compounds and mental diseases), 6 types of relationships (treat, reduce, protect, prevent, induce, and unknown), 8014 nodes (7509 compounds, 505 mental diseases), and 123,890 edges. Detailed MCKG data are available on the AskBob platform (http://lwj1.ens.yun.pingan.com:5001/).
We mapped a total of 72 potentially therapeutic compounds to MCKG, amoung which 42 were covered by MCKG (Table S8), that were associated with 206 mental disease nodes and1370 edges (Fig. 4D, Table S10). Among the 37% of the compound-disease relationships that were annotated as non-induced, there were 30 drugs, accounting for 71.43% of the total number of covered drugs. They included 18 drugs (e.g., sulpiride, estradiol, baclofen, and curcumin) identified by the dtGSEA method (Fig. 5A) and 13 drugs (e.g., everolimus and fluvoxamine) identified by the CMap method (Fig. 5B). Notably, sulpiride was identified by both methods. The remaining 30 drugs that were not annotated by MCKG were mostly compounds obtained through the CMap database. In MCKG, mental diseases associated with these drugs included ASD (autistic disorder of childhood, autistic disorder, infantile autism), depressive disorder, anxiety disorder, and schizophrenia. Verification of the MCKG revealed that 10 (baclofen, calcitriol, curcumin, entinostat, estradiol, everolimus, fluvoxamine, metronidazole, sulpiride, and zinc) of the 42 drugs were associated with ASD (Fig. 5C). Among them, the therapeutic effects of everolimus (https://clinicaltrials.gov/: NCT01929642, NCT01730209) and fluvoxamine (NCT00655174) on autism have been evaluated in registered clinical trials. The remaining32 drugs were annotated to have therapeutic, protective, and reducing effects on other mental illnesses. The therapeutic effects of these drugs on autism merit further studies. Fig. 5D shows source brain regions and methods for the 72 drugs, levels of evidence associated with ASD or several mental diseases and other characteristics, such as research subjects (e.g., humans or animals) and study type (e.g., clinical trials, randomized controlled trials). More information can be found in MCKG and the expanded literature links in the database. Analysis of the mechanisms of action (MOAs) of the 72 drugs revealed that the HDAC inhibitor, estrogen receptor agonist, glycogen synthase kinase inhibitor, and mTOR inhibitor categories had more drugs, including entinostat, estradiol, SB-216763, and everolimus (Table S8). We also investigated some of the drugs identified by dtGSEA, including sulpiride, estradiol, and baclofen, and the target genes of these drugs (Fig. 5E), The target genes for sulpiride included PEX2, EHHADH and SCP2. Estradiol was found to reverse the expression patterns of some mitochondrial genes (e.g., MRPL33, MRPL27, NDUFA4, NDUFB5) and ASD-related genes (e.g., MTOR, TUBB3). Baclofen’s target genes included HDAC1, RPL35, and CBFB. More drug enrichment and target gene information is shown in Fig. S5. We also useD the rat in vivo toxicogenomic data in the DrugMatrix database and the Open TG-GATEs database to further validate the drug candidates. We found experimental data for 23 of the 72 compounds in the two databases. A total of 17 drugs (sulpiride and estradiol among others) were found to reverse the expression of NCGs (Table S11). For example, at different doses and time, Papaverine reversed the expression of some NCGs in BA41-42–22 (Fig. S7A). Sulpiride was shown to reverse the expression of NCGs in BA41-42–22, BA8, and gene expressions in 365 NCGs after 9 h of middle dose administration in rats (Fig. S7B). In addition, 7 of the 72 drugs (sulpiride and estradiol among others; Fig. S7C) were reported in the study by So et al.
Fig. 5.
Evaluation of drug candidates. a Sankey diagram of potential drugs identified by the dtGSEA method in the MCKG. b Sankey diagram of potential drugs identified by the CMap method in the MCKG. The first column (left) shows the brain regions. The second column (right) shows the potential drugs. c Ten drug candidates related to ASD in the MCKG. The yellow nodes represent drugs, and the magenta nodes represent mental illness. d Comprehensive information heatmap of 72 drug candidates. Each column represents a candidate drug, and each row represents an attribute of the candidate drug (for example, the method, brain region, whether or not it is from the MCKG, whether or not it can treat ASD, the type of study, the subject, etc.). Yellow indicates whether the drug candidate is related to other mental illnesses in the MCKG. e Target NCGs and GSEA results of drug candidates identified by the dtGSEA method. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Applicability of the pipeline in other psychiatric disorders
To verify the applicability of the pipeline for other diseases, we performed additional experiments. We used our pipeline for comprehensive analysis of a dataset from the study by Vargas et al. (data can be downloaded from GSE5281) [43], [44]. From the transcriptome data of 6 brain regions, we obtained a total of 75 candidate compounds, among which 31 drugs had evidence for the treatment of Alzheimer’s disease (Table S12). Although the 75 compounds do not include 6 drugs, our pipeline achieved a more comprehensive and accurate result.
4. Discussion
In this study, we constructed a drug repositioning framework (Fig. 1) that is different from previous studies that integrated DEGs, PPI, and CMap data to identify drug targets and candidate drugs through network analysis. Our pipeline take into account the characteristics of polygenic, heterogeneity, and dysfunctional gene network of ASD. It integrates multiple easily available data sets, using a combination method of coexpression network, PPI network and topological analysis to accurately identify key pathogenic genes. By scanning drug-induced gene expression data, we obtained an accurate and comprehensive drug candidate list, with an increasing likelihood of subsequent experimental studies success. Using this framework, we systematically analyzed functional characteristics of transcriptomes of different brain regions and repositioned potential therapeutic drugs. The MCKG provides further evidence for the effectiveness of our drug repositioning approach.
Among the 6 selected brain regions, 4 brain regions had significant gene expression level imbalances, especially BA8 and BA41-42–22. BA8 is associated with cognitive and memory functions [45], while BA41-42–22 is associated with various cognitive functions, including semantics, memory, and auditory cognition [46]. Among the downregulated DEGs, SCMs and msPNs that we identified from these 2 regions, they were highly enriched in the neuron, dendrite, and axon development terms, implying that synaptic dysfunction and abnormal neuronal homeostasis are common underlying molecular mechanisms of ASD. Upregulated SCMs in BA41-42–22 were enriched in immune processes, implying that alterations in gene pathways associated with immune function can have effects on brain development and cognition. Indeed, the central role of immune dysregulation in ASD has been reported [47]. In a study by Parikshaket al., BA9 showed dysregulation in people aged 2–67 years, however, we did not detect significant differences between diseased and healthy groups in children aged 2–18 years [16]. We only found 4 upregulated genes (SNORD14E, SLC10A1, NPAS4, and IFI6) in BA9. These results contrast to the 551 DEGs found in the BA9 brain region in Parikshak et al.’s study. The absence of significant changes in the BA9 and VM regions does not mean that children with autism do not have dysregulated genes in these two regions. We observed large differences in gene expression levels among these 6 regions. Notably, the NPAS4 gene was the only one that was upregulated in all four brain areas. NPAS4 can regulate the formation of inhibitory synapses on excitatory neurons, and its dysregulation in the brain can regulate depression, anxiety, ASD, and cognitive disorders [48]. In addition, we identified an SCM associated with mitochondrial function in both BA46 and BA8. Interestingly, a reverse regulation pattern was observed for the two SCMs: an upregulated mitochondrial-related SCM in BA46 that included mitochondrial ribosomal proteins (MRPS12, MRPL17, MRPL37, MRPL55) and a downregulated SCM in BA8 that included NADH dehydrogenase genes (NDUFA1, NDUFA4, NDUFA6, NDUFB1, NDUFB3, NDUFB4 and NDUFB5) as well as cytochrome c oxidase genes (COX7B, COX14, COX4I1). Mitochondria have been implicated in various aspects of neural development, including synaptogenesis, synaptic plasticity, neuronal differentiation, and neurotransmission. Studies have also found mitochondrial dysfunction in ASD patients [49], [50]. Abnormalities in the mitochondrial electron transport chain (ETC) complex levels and deficiencies of enzymes in the mitochondrial respiratory chain cause of brain dysfunction in children with ASD [51]. Mitochondrial complex I (NADH dehydrogenase) and complex IV (cytochrome c oxidase [COX]) are central to oxidative phosphorylation, ETC, and ATP production in eukaryotes [52]. Complex I levels in the frontal cortex were found to be suppressed in children with autism [53]. Anitha et al. reported downregulated expressions of mitochondrial complex I and complex IV genes in autistic brains [54]. Our findings in BA8 are consistent with the findings of these studies. However, it has not been established why mitochondrial SCM in BA46are upregulated. The SCMs with different functions in different brain regions elucidate on the molecular mechanisms of brain dysfunction in children with ASD. Genes and pathways in these SCMs are potential effective targets for ASD treatment and prevention.
In recent years, PPI networks and network-based methods have been used for drug target discovery and drug repositioning studies [55], [56]. msPNs constructed by integrating gene expression data and PPI data are more reliable than those integrating gene expression data or PPI data alone. Usually, networks have redundancy, therefore, identifying key genes and proteins is crucial for drug repurposing. In addition, the correlation between network topology and function determines that key genes in the network can have significant effects on network function through multiple interactions. Protein products of genes that are affected by drugs often lie at the center of important functional PPI networks [57]. If a drug can affect the expression of key genes, then tit can have a significant impact on the entire network, in turn altering the abnormal expression of multiple genes and reversing the disease state. Based on this idea, a total of 365 NCGs were selected from 18 msPNs, and the drugs screened with these genes were considered the most likely to affect the network and thus correct the disease state. Based on the two drug repositioning methods, we identified a total of 72 potential therapeutic compounds using the dtGSEA and CMAP database, a subset of which have been studied or shown to reduce and treat ASD.
ASD is a persistent neurodevelopmental disorder. It has overlapping clinical features and a common molecular mechanism with many mental disorders, including schizophrenia [58], ADHD [59], and bipolar disorder [60]. Drugs with therapeutic effects on the symptoms of mental illnesses are more likely to be useful for treating ASD. Therefore, we constructed MCKG and used it to evaluate the identified drugs. We found that 10 of the 72 drugs were directly linked to autism. They are baclofen, sulpiride, estradiol, entinostat, everolimus, fluvoxamine, curcumin, calcitriol, metronidazole, and zinc. Baclofen is a gamma-aminobutyric acid (GABA) agonist that is used for relieving pain and muscle spasms [61] and has been shown to havea therapeutic effect on autism. A recent randomized double-blind placebo-controlled trial revealed that baclofen improves hyperactivity symptoms in children with ASD [62]. Sulpiride is a dopamine D2-receptor antagonist with the potential for treating schizophrenia and autism [63], [64]. In our study, both drug repositioning methods indicated that sulpiride may have a therapeutic effect on ASD. We found that sulpiride reversed the changes in fatty acid metabolism-related genes (PEX2, EHHADH, SCP2) and genes involved in the process of cell adhesion molecule binding (PAK2, MAPR1, PICALM, and YES1). Abnormalities in fatty acid metabolism have been associated with neurodevelopmental disorders. Fatty acids levels have been shown to be dysregulated in autistic children [65]. In addition, fatty acids exert a stimulatory action on the dopamine D2 receptor, implying a potential mechanism for sulpiride treatment of autism [66]. Besides, abnormal cell adhesion molecules and pathways are important in ASD pathogenesis [67]. Estradiol (E2), a steroid sex hormone has been shown to exhibit antioxidant effects, neuroprotective roles, effects on brain cognition and memory, and the ability to alleviate symptoms of several neurological diseases, including Parkinson’s and Alzheimer’s diseases [68], [69], [70]. In addition, E2 has been shown to decrease repetitive behaviors and ameliorate ASD-associated social behavioral deficits [71]. Our findings suggest that E2 can reverse alterations in the expression of mitochondrial genes (MRPL33, MRPL27, NDUFA4, NDUFB5, NDUFB4) and in a large number of genes encoding ribosomal proteins (RPL and RPS genes). E2 has been shown to diminish complex I or complex II mediated phosphorylated respiration, thereby, protecting against mitochondrial dysfunction [72]. Additionally, E2 can cause alteration in ribosomal RNA levels within brain regions. Ribosomal proteins such as RPL10 are susceptibility genes for ASD [73]. Our results (Fig. 3B, midnightblue msPN) and those of many other studies have revealed that RPL and RPS genes are dysregulated in patients with ASD [74], [75]. Therefore, we postulate that E2 exerts a therapeutic effect on ASD by improving mitochondrial dysfunction and regulating ribosomal protein genes. This should be further investigated. Entinostat is an HDAC inhibitor that has been found to normalize abnormal epigenetic regulation of genes, thereby improving synaptic and social interaction defects associated with ASD in shank3-deficient mice with an autism-like phenotype [76]. Everolimus is used to control seizures, improve autism and depressive symptoms, and treat epilepsy and ASD in tuberous sclerosis complex [77], [78]. It has been shown that fluvoxamine can treat autistic symptoms in adults, and it can also significantly improve some clinical symptoms of autism in children, such as problems with eye contact and language use [79], [80]. Curcumin reduces autistic symptoms by suppressing mitochondrial dysfunction and oxidative nitrosative stress [81].
In general, top-ranked drugs have a greater capacity when compared to other drugs inreversing gene expression patterns in order to achieve the purpose of treating diseases. In addition to the above-mentioned drugs (baclofen and sulpiride), XE-991, SB-216763 and mepacrine were ranked highly by the two methods used to identify drugs from 4 brain regions. XE-991 and SB-216763 are small molecules discovered in the CMap database. These drugs have a strong potential for treating ASD. XE-991 is an M−current inhibitor, its inhibition may improve cognitive abilities. XE-991 has been shown to exhibit neuroprotective effects, enhance memory and learning in mice, and reverse cognitive impairment associated with neurodegeneration [82], [83]. SB-216763 is a glycogen synthase kinase inhibitor that can improve cognitive impairment by inhibiting glycogen synthase kinase-3β (GSK3β). GSK-3plays a central role in cognitive dysfunction. Fortress et al. reported that SB-216763 reduces memory as well as learning deficits and alleviates saikosaponins-induced cognitive deficits [84]. Mepacrine, a phospholipase A2(PLA2) inhibitor (quinacrine), has been used as an antimalarial drug and as an antibiotic. PLA2 reduction was shown to ameliorate cognitive deficits in a mouse model [85], while elevations in phospholipase A2 concentrations were found to lead to oxidative stress and neuroinflammation, which are causes of ASD [86]. These results findings that our pipeline is efficient at identifying ASD therapeutic candidates for drug repositioning.
Based on gene networks, the developed drug repositioning pipeline can also be used to explore the molecular mechanisms of ASD. Based on a network created to achieve the purpose of drug repositioning for ASD, drugs with therapeutic potential can be obtained and evaluated in the context of the present knowledge. To ensure reliability of drug identification, we accurately identified disease gene network as well as hub genes and verified drug functions using the knowledge graph. In addition, two drug repositioning methods were used to comprehensively scan the effects of existing drugs on gene expression patterns from different perspectives.
Our study has the following limitations. First, the dtGSEA method relies on known drug-gene pairs. This information is far from complete, which may cause some promising candidates to be missed. Besides, if a drug has too few target genes, this method will not be able to identify the drug. However, the drug repositioning method based on the CMap database makes up for these shortcomings. Second, although we found that these drugs may treat ASD, our results are preliminary, and further experimental validation is required. Nevertheless, our proposed drug repositioning pipeline has significant implications. Given the lack of effective therapeutic drugs for ASD, our proposed method can achieve large-scale screening of existing drugs, providing valuable clues to help researchers in drug development and drug screening. Third, we did not explore orther brain regions and may, therefore, have missed some transcriptomic characteristics. However, since we identified relevant ASD pathogenic mechanisms, these missing brain regions do not affect the validity and effectiveness of our entire pipeline. Due to these limitations, we recommend that before our drug repositioning pipeline is used for further research, drug characteristics (for example, their ability to penetrate the blood–brain barrier) and relevant evidence should be carefully investigated.
5. Conclusion
We have designed and built a drug repositioning pipeline that integrates pathogenic gene screening, drug repositioning, and drug verification. Our proposed workflow enables the analysis of molecular mechanisms associated with ASD and the identification of multiple candidate drugs with potential therapeutic effects, thereby providing clues for further studies on ASD treatment. In addition, this pipeline, developed based on network analysis, can be applicable to other complex psychiatric disorders and polygenic diseases.
6. Author’s contributors
HG, YN are the co-first authors and performed majority of the analyses. HG, XYM, DTL, ST, QSH, SH, and GJL processed and analyzed the transcriptome data. YN, SZ, and HG constructed the knowledge graph. HG and XYM processed and analyzed the drug data. HG, YN, HYL, LL, and YPT provided concept of the study and reviewed the overall project. HG, LL, and HYL wrote and revised the manuscript. HG and HYL designed and supervised the entire project. The author(s) read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank the Guangzhou Women and Children's Medical Center for providing funding support. We thank all investigators who provided data for this study as well as many thanks to all the participants. We thank Ping An Medical Technology for the knowledge graph construction platform.
Availability of data
RNA-seq datasets are available at the Gene Expression Omnibus (GSE102741, GSE59288, GSE51264, GSE62098), and datasets from Parikshak et al.. PPI were downloaded from the STRING database (Version 11, http://string-db.org/). The pharmaco-transcriptomics association data download from the DrugBank (https://www.drugbank.ca/) and drug-gene interaction from Drug Targetor v1.21 (http://drugtargetor.com/).
Funding source
This work was supported by the Guangdong Key Project in “Development of new tools for diagnosis and treatment of Autism ” (2018B030335001). This research was partially funded by National Natural Science Foundation of China [No. 61772375, 18ZDA325]; Hubei Provincial Natural Science Foundation of China [No. 2019CFA025]; National Key R&D Program of China (2019YFC010167); and Independent Research Project of School of Information Management Wuhan University (No: 413100032). This work was supported by the Guangzhou Institute of Pediatrics/Guangzhou Women and Children’s Medical Center (YIP-2019-026).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.06.046.
Contributor Information
Long Lu, Email: bioinfo@gmail.com.
Huiying Liang, Email: lianghuiying@hotmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jeste S.S., Geschwind D.H. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10(2):74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ecker C., Bookheimer S.Y., Murphy D.G.M. Neuroimaging in autism spectrum disorder: brain structure and function across the lifespan. Lancet Neurol. 2015;14(11):1121–1134. doi: 10.1016/S1474-4422(15)00050-2. [DOI] [PubMed] [Google Scholar]
- 3.Jon Baio E, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z: Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years — Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. Morbidity and Mortality Weekly Report (MMWR) 2018. [DOI] [PMC free article] [PubMed]
- 4.Ji N.Y., Findling R.L. An update on pharmacotherapy for autism spectrum disorder in children and adolescents. Curr Opin Psychiatr. 2015;28(2):91–101. doi: 10.1097/YCO.0000000000000132. [DOI] [PubMed] [Google Scholar]
- 5.Mazzone L., Giovagnoli G., Siracusano M., Postorino V., Curatolo P. Drug treatments for core symptoms of autism spectrum disorder: unmet needs and future directions. J Pediatr Neurol. 2017;15(03):134–142. [Google Scholar]
- 6.Xue H., Li J., Xie H., Wang Y. Review of drug repositioning approaches and resources. Int J Biol Sci. 2018;14(10):1232–1244. doi: 10.7150/ijbs.24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 8.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 9.Hodos R.A., Kidd B.A., Shameer K., Readhead B.P., Dudley J.T. In silico methods for drug repurposing and pharmacology. Wiley Interdip Rev Syst Biol Med. 2016;8(3):186–210. doi: 10.1002/wsbm.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007;7(1):54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 11.Koudijs K.K.M., Terwisscha van Scheltinga A.G.T., Böhringer S., Schimmel K.J.M., Guchelaar H.-J. Transcriptome signature reversion as a method to reposition drugs against cancer for precision oncology. Cancer J. 2019;25(2):116–120. doi: 10.1097/PPO.0000000000000370. [DOI] [PubMed] [Google Scholar]
- 12.Dudley J.T., Sirota M., Shenoy M., Pai R.K., Roedder S., Chiang A.P. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3(96) doi: 10.1126/scitranslmed.3002648. 96ra76-96ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.So H.-C., Chau C.-L., Chiu W.-T., Ho K.-S., Lo C.-P., Yim S.-Y. Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nat Neurosci. 2017;20(10):1342–1349. doi: 10.1038/nn.4618. [DOI] [PubMed] [Google Scholar]
- 14.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nature Rev Neuroence. 2015;16(9):551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 15.Noh H.J., Ponting C.P., Boulding H.C., Meader S., Betancur C., Buxbaum J.D. Network topologies and convergent aetiologies arising from deletions and duplications observed in individuals with autism. PLoS Genet. 2013;9(6):e1003523. doi: 10.1371/journal.pgen.1003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parikshak N.N., Swarup V., Belgard T.G., Irimia M., Ramaswami G., Gandal M.J. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540(7633):423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barabási A.-L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S., Lee D.G., Shin H. Network mirroring for drug repositioning. BMC Med Inf Decis Making. 2017;17(1 Supplement):55. doi: 10.1186/s12911-017-0449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright C., Shin J.H., Rajpurohit A., Deep-Soboslay A., Collado-Torres L., Brandon N.J. Altered expression of histamine signaling genes in autism spectrum disorder. Transl Psychiat. 2017;7(5) doi: 10.1038/tp.2017.87. e1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X., Han D., Somel M., Jiang X.i., Hu H., Guijarro P. Disruption of an evolutionarily novel synaptic expression pattern in autism. PLoS Biol. 2016;14(9):e1002558. doi: 10.1371/journal.pbio.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Z., Bammann H., Han D., Xie G., Khaitovich P. Conserved expression of lincRNA during human and macaque prefrontal cortex development and maturation. Palliative Med. 2014;20(7):1103–1111. doi: 10.1261/rna.043075.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Shi M, Ma Z, Zhao S, Euskirchen G, Ziskin J, Urban A, Hallmayer J, Snyder M: Integrated systems analysis reveals a molecular network underlying autism spectrum disorders. Mol Syst Biol 2014. [DOI] [PMC free article] [PubMed]
- 23.Anthony, M, Bolger, Marc, Lohse, Bjoern, Usadel: Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 2014. [DOI] [PMC free article] [PubMed]
- 24.Trapnell C, Pachter L, Salzberg SL: TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009. [DOI] [PMC free article] [PubMed]
- 25.Anders S., Pyl T.P., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12) doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffrey T, Leek W, Evan, Johnson, Hilary S, Parker, Andrew: The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (Oxford, England) 2012. [DOI] [PMC free article] [PubMed]
- 28.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9(1) doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Biogeoences. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abrahams B.S., Arking D.E., Campbell D.B., Mefford H.C., Morrow E.M., Weiss L.A. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4(1):36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu LM, Li JR, Huang Y, Zhao M, Tang X, Wei L. AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res 2012, 40(Database issue):D1016-D1022. [DOI] [PMC free article] [PubMed]
- 32.Wishart D.S. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucl Acids Res. 2006;34(90001):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A GH, Christopher H, Gerome B. Drug Targetor: a web interface to investigate the human druggome for over 500 phenotypes. Bioinformatics 2019, 14(35):2515-2517. [DOI] [PMC free article] [PubMed]
- 34.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci - PNAS. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171(6):1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lample G., Ballesteros M., Subramanian S., Kawakami K., Dyer C. Neural Architec Named Entity Recogn. 2016 [Google Scholar]
- 37.Qi P, Dozat T, Zhang Y, Manning CD. Universal Dependency Parsing from Scratch. 2019.
- 38.Bello S.M., Shimoyama M., Mitraka E., Laulederkind S.J.F., Smith C.L., Eppig J.T. Disease Ontology: improving and unifying disease annotations across species. Dis Model Mech. 2018;11(3) doi: 10.1242/dmm.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshinobu I, Noriyuki N, Tomoya Y, Atsushi O, Yasuo O, Tetsuro U, Hiroshi Y. Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res 2015(D1):D921. [DOI] [PMC free article] [PubMed]
- 40.Gusenleitner D., Auerbach S.S., Melia T., Gómez H.F., Sherr D.H., Monti S. Genomic models of short-term exposure accurately predict long-term chemical carcinogenicity and identify putative mechanisms of action. PLoS One. 2014;9(7):e102579. doi: 10.1371/journal.pone.0102579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smyth GK, Ritchie M, Thorne N, Wettenhall J, Shi W. limma: Linear Models for Microarray Data. Bioinformatics & Computational Biology Solutions Using R & Bioconductor 2010.
- 42.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vargas D.M., De B., Zimmer E.R., Klamt F. Alzheimer's disease master regulators analysis: search for potential molecular targets and drug repositioning candidates. Alzheimers Res Ther. 2018;10(1):59. doi: 10.1186/s13195-018-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang W.S., Dunckley T., Beach T.G., Grover A., Mastroeni D., Walker D.G. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28(3):311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boisgueheneuc F.d., Levy R., Volle E., Seassau M., Duffau H., Kinkingnehun S. Functions of the left superior frontal gyrus in humans: a lesion study. Brain. 2006;129(12):3315–3328. doi: 10.1093/brain/awl244. [DOI] [PubMed] [Google Scholar]
- 46.Jackson R.L., Bajada C.J., Rice G.E., Cloutman L.L., Lambon Ralph M.A. An emergent functional parcellation of the temporal cortex. Neuroimage. 2018;170:385–399. doi: 10.1016/j.neuroimage.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 47.Estes M.L., McAllister A.K. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci. 2015;16(8):469–486. doi: 10.1038/nrn3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaehne E.J., Klarić T.S., Koblar S.A., Baune B.T., Lewis M.D. Effects of Npas4 deficiency on anxiety, depression-like, cognition and sociability behaviour. Behav Brain Res. 2015;281:276–282. doi: 10.1016/j.bbr.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 49.Hollis F., Kanellopoulos A.K., Bagni C. Mitochondrial dysfunction in Autism Spectrum Disorder: clinical features and perspectives. Curr Opin Neurobiol. 2017;45:178–187. doi: 10.1016/j.conb.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 50.Giulivi C., Zhang Y.-F., Omanska-Klusek A., Ross-Inta C., Wong S., Hertz-Picciotto I. Mitochondrial dysfunction in autism. JAMA, J Am Med Assoc. 2010;304(21):2389. doi: 10.1001/jama.2010.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guevara-Campos J., González-Guevara L., Puig-Alcaraz C., Cauli O. Autism spectrum disorders associated to a deficiency of the enzymes of the mitochondrial respiratory chain. Metab Brain Dis. 2013;28(4):605–612. doi: 10.1007/s11011-013-9419-x. [DOI] [PubMed] [Google Scholar]
- 52.Bharath SM. M. Post-Translational Oxidative Modifications of Mitochondrial Complex I (NADH: Ubiquinone Oxidoreductase): Implications for Pathogenesis and Therapeutics in Human Diseases. J Alzheimers Dis 2017:1-18. [DOI] [PubMed]
- 53.Chauhan A, Gu F, Essa MM, Wegiel J, Kaur K, Brown WT, Chauhan V: Brain region-specific deficit in mitochondrial electron transport chain complexes in children with autism. J Neurochem 2011, 117(2):209-220. [DOI] [PMC free article] [PubMed]
- 54.Anitha A., Nakamura K., Thanseem I., Matsuzaki H., Miyachi T., Tsujii M. Downregulation of the expression of mitochondrial electron transport complex genes in autism brains. Brain Pathol. 2013;23(3):294–302. doi: 10.1111/bpa.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tripathi, Lokesh P, Prathipati, Philip, Mizuguchi, Kenji, Murakami, Yoichi: Network analysis and in silico prediction of protein-protein interactions with applications in drug discovery. Curr Opin Struc Biol 2017. [DOI] [PubMed]
- 56.Zhu H., Chen L., George P.D.C., Chakraborty C. Evaluating Protein-protein Interaction (PPI) Networks for Diseases Pathway, Target Discovery, and Drug-design Using ‘In silico Pharmacology‘. Curr Protein Pept Sci. 2014;15(6) doi: 10.2174/1389203715666140724090153. [DOI] [PubMed] [Google Scholar]
- 57.Kotlyar M., Fortney K., Jurisica I. Network-based characterization of drug-regulated genes, drug targets, and toxicity. Methods. 2012;57(4):499–507. doi: 10.1016/j.ymeth.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 58.Domínguez-Iturza N., Lo A.C., Shah D., Armendáriz M., Vannelli A., Mercaldo V. The autism- and schizophrenia-associated protein CYFIP1 regulates bilateral brain connectivity and behaviour. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-11203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naaijen J., Bralten J., Poelmans G., Glennon J.C., Franke B., Buitelaar J.K., Ebstein R P., Gill M. Glutamatergic and GABAergic gene sets in attention-deficit/hyperactivity disorder: association to overlapping traits in ADHD and autism. Transl Psychiat. 2017;7(1) doi: 10.1038/tp.2016.273. e999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guan J., Cai J.J., Ji G., Sham P.C. Commonality in dysregulated expression of gene sets in cortical brains of individuals with autism, schizophrenia, and bipolar disorder. Transl Psychiat. 2019;9(1) doi: 10.1038/s41398-019-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Penn R. Intrathecal baclofen for severe spinal spasticity Lancet 326(8447) 1985 125–127. [DOI] [PubMed]
- 62.Mahdavinasab SM, Saghazadeh A, Motamed-Gorji N, Vaseghi S, Mohammadi MR, Alichani R, Akhondzadeh S: Baclofen as an adjuvant therapy for autism: a randomized, double-blind, placebo-controlled trial. Eur Child Adoles Psy 2019. [DOI] [PubMed]
- 63.Wang J., Omori I.M., Fenton M., Soares B. Sulpiride augmentation for schizophrenia. Schizophrenia Bull. 2010;36(2):229–230. doi: 10.1093/schbul/sbp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott D.W., Eames P. Use of sulpiride in a case of atypical autism. J Autism dev disord. 1988;18(1):144–146. doi: 10.1007/BF02211825. [DOI] [PubMed] [Google Scholar]
- 65.Jory J. Abnormal fatty acids in Canadian children with autism. Nutrition. 2016;32(4):474–477. doi: 10.1016/j.nut.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 66.Ohara K. The n-3 polyunsaturated fatty acid/dopamine hypothesis of schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31(2):469–474. doi: 10.1016/j.pnpbp.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 67.Betancur C., Sakurai T., Buxbaum J.D. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 2009;32(7):402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zsido R.G., Heinrich M., Slavich G.M., Beyer F., Kharabian Masouleh S., Kratzsch J. Association of estradiol and visceral fat with structural brain networks and memory performance in adults. JAMA Network Open. 2019;2(6):e196126. doi: 10.1001/jamanetworkopen.2019.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dongfang S., Xiaoyan T., Binbin Z., Rongrong, Song Mechanistic evaluation of neuroprotective effect of estradiol on rotenone and 6-OHDA induced Parkinson's disease. Pharmacol Rep Pr. 2017 doi: 10.1016/j.pharep.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 70.Wenhao, Yan, Jun, Wu, Bo, Song, Qiang, Luo, Yuming, Xu: Treatment with a brain-selective prodrug of 17β-estradiol improves cognitive function in Alzheimer's disease mice by regulating klf5-NF-κB pathway. Naunyn Schmiedebergs Archives of Pharmacology 2019. [DOI] [PMC free article] [PubMed] [Retracted]
- 71.Filice F, Lauber E, V Rckel KJ, W Hr M, Schwaller B: 17-β estradiol increases parvalbumin levels in Pvalb heterozygous mice and attenuates behavioral phenotypes with relevance to autism core symptoms. Mol Autism 2018, 9(1):15. [DOI] [PMC free article] [PubMed]
- 72.Thiede A., Gellerich F., Schönfeld P., Siemen D. Complex effects of 17β-estradiol on mitochondrial function. BBA. 2012;1817 doi: 10.1016/j.bbabio.2012.02.034. [DOI] [PubMed] [Google Scholar]
- 73.Jones K.J., Chikaraishi D.M., Harrington C.A., Mcewen B.S., Pfaff D.W. In situ hybridization detection of estradiol-induced changes in ribosomal RNA levels in rat brain. Mol Brain Res. 1986;1(2):145–152. doi: 10.1016/0169-328x(86)90006-9. [DOI] [PubMed] [Google Scholar]
- 74.Lombardo M.V., Courchesne E., Lewis N.E., Pramparo T. Hierarchical cortical transcriptome disorganization in autism. Mol Autism. 2017;8(1) doi: 10.1186/s13229-017-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Griesi-Oliveira K., Fogo M.S., Pinto B.G.G., Alves A.Y., Passos-Bueno M.R. Transcriptome of iPSC-derived neuronal cells reveals a module of co-expressed genes consistently associated with autism spectrum disorder. Mol Psychiatr. 2020;21:1–17. doi: 10.1038/s41380-020-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaijie Q., Luye M., Emmanuel D., Lara J., Liu A., Yan Histone deacetylase inhibitor MS-275 restores social and synaptic function in a Shank3-deficient mouse model of autism. Neuropsychopharmacology. 2018 doi: 10.1038/s41386-018-0073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kilincaslan A., Kok B.E., Tekturk P., Yalcinkaya C., Ozkara C., Yapici Z. Beneficial effects of everolimus on autism and attention-deficit/hyperactivity disorder symptoms in a group of patients with tuberous sclerosis complex. J Child Adol Psychop. 2016:2016–2100. doi: 10.1089/cap.2016.0100. [DOI] [PubMed] [Google Scholar]
- 78.Mm A., Hi B., Ks C., Hyd E., Ys F., Ma G. Everolimus for epilepsy and autism spectrum disorder in tuberous sclerosis complex: EXIST-3 substudy in Japan. Brain Dev. 2019;41(1):1–10. doi: 10.1016/j.braindev.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 79.Mcdougle C.J., Naylor S.T., Cohen D.J., Volkmar F.R., Heninger G.R., Price L.H. A double-blind, placebo-controlled study of fluvoxamine in adults with autistic disorder. Arch Gen Psychiatry. 1996;53(11):1001. doi: 10.1001/archpsyc.1996.01830110037005. [DOI] [PubMed] [Google Scholar]
- 80.Fukuda T., Sugie H., Ito M., Sugie Y. Clinical evaluation of treatment with fluvoxamine, a selective serotonin reuptake inhibitor in children with autistic disorder. No to Hattatsu Brain Dev. 2001;33(4):314–318. [PubMed] [Google Scholar]
- 81.Bhandari R., Kuhad A. Neuropsychopharmacotherapeutic efficacy of curcumin in experimental paradigm of autism spectrum disorders. Life Sci. 2015 doi: 10.1016/j.lfs.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 82.Liu H., Jia L., Chen X., Shi L., Xie J. The Kv7/KCNQ channel blocker XE991 protects nigral dopaminergic neurons in the 6-hydroxydopamine rat model of Parkinson's disease. Brain Res Bull. 2018;137:132–139. doi: 10.1016/j.brainresbull.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 83.Fontán-Lozano Á., Suárez-Pereira I., Delgado-García J.M., Carrión Á.M. The M-current inhibitor XE991 decreases the stimulation threshold for long-term synaptic plasticity in healthy mice and in models of cognitive disease. Hippocampus. 2011;21(1):22–32. doi: 10.1002/hipo.20717. [DOI] [PubMed] [Google Scholar]
- 84.Fortress A.M., Avcu P., Wagner A.K., Dixon C.E., Pang K.C.H. Experimental traumatic brain injury results in estrous cycle disruption, neurobehavioral deficits, and impaired GSK3β/β-catenin signaling in females. Exp Neurol. 2019;315:42–51. doi: 10.1016/j.expneurol.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 85.Sanchez-Mejia R.O., Newman J.W., Toh S., Yu G., Zhou Y., Halabisky B. Phospholipase A2 reduction ameliorates cognitive deficits in a mouse model of Alzheimer's disease. Nat Neurosci. 2008;11(11):1311–1318. doi: 10.1038/nn.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qasem H., Alayadhi L., Dera H.A. Increase of cytosolic phospholipase A2 as hydrolytic enzyme of phospholipids and autism cognitive, social and sensory dysfunction severity. Lipids Health Dis. 2017;16(1):117. doi: 10.1186/s12944-016-0391-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets are available at the Gene Expression Omnibus (GSE102741, GSE59288, GSE51264, GSE62098), and datasets from Parikshak et al.. PPI were downloaded from the STRING database (Version 11, http://string-db.org/). The pharmaco-transcriptomics association data download from the DrugBank (https://www.drugbank.ca/) and drug-gene interaction from Drug Targetor v1.21 (http://drugtargetor.com/).