Abstract
Inflammation is intimately involved at all stages of atherosclerosis and remains a substantial residual cardiovascular risk factor in optimally treated patients. The proof of concept that targeting inflammation reduces cardiovascular events in patients with a history of myocardial infarction has highlighted the urgent need to identify new immunotherapies to treat patients with atherosclerotic cardiovascular disease. Importantly, emerging data from new clinical trials show that successful immunotherapies for atherosclerosis need to be tailored to the specific immune alterations in distinct groups of patients. In this Review, we discuss how single-cell technologies — such as single-cell mass cytometry, single-cell RNA sequencing and cellular indexing of transcriptomes and epitopes by sequencing — are ideal for mapping the cellular and molecular composition of human atherosclerotic plaques and how these data can aid in the discovery of new precise immunotherapies. We also argue that single-cell data from studies in humans need to be rigorously validated in relevant experimental models, including rapidly emerging single-cell CRISPR screening technologies and mouse models of atherosclerosis. Finally, we discuss the importance of implementing single-cell immune monitoring tools in early phases of drug development to aid in the precise selection of the target patient population for data-driven translation into randomized clinical trials and the successful translation of new immunotherapies into the clinic.
Subject terms: Immunology, Cardiology, Systems biology
In this Review, Fernandez and Giannarelli discuss how single-cell technologies can advance our understanding of the cellular and molecular composition of atherosclerotic plaques and how these approaches can guide the design of new, personalized immunotherapies and immune monitoring tools for the management of patients with atherosclerotic cardiovascular disease.
Key points
Atherosclerosis is initiated by the subendothelial accumulation of lipids that trigger maladaptive, non-resolving, chronic inflammation.
Anti-inflammatory interventions have substantially reduced the risk of adverse cardiovascular events in patients with recent acute myocardial infarction.
Emerging data from clinical trials published since 2017 show that successful treatments need to be tailored to specific groups of patients.
Single-cell technologies are ideal for studying immune system dynamics and can advance our understanding of the cellular and molecular architecture of human atherosclerotic tissue to advance drug discovery.
The inclusion of single-cell immune-monitoring tools in early phases of drug testing could advance drug discovery and precision medicine in cardiovascular disease to reduce the risk of adverse cardiovascular outcomes and death in patients.
Introduction
Atherosclerosis is a chronic inflammatory disease initiated by the subendothelial accumulation of lipids that trigger maladaptive, non-resolving immune responses1–4. The causal role of the immune system and inflammation in atherosclerosis has been well established in preclinical models2,3, and inflammation remains a substantial residual risk factor for cardiovascular events in optimally treated patients5. The positive results of the CANTOS6 and COLCOT7 trials in the past 4 years proved the pathogenic role of inflammation in atherosclerosis in patients. Importantly, the negative results of the CIRT trial8, which targeted inflammation broadly with the use of low-dose methotrexate in patients with previous myocardial infarction or multivessel coronary artery disease who had either type 2 diabetes mellitus or metabolic syndrome, also indicated that not all immunotherapies are atheroprotective. Moreover, new evidence is emerging showing that a one-size-fits-all therapeutic approach is ineffective due to variation in patient responses. For instance, in the CANTOS trial6, a greater benefit with canakinumab therapy, a monoclonal antibody against IL-1β, was seen in responder patients in whom the levels of inflammation decreased 3 months after treatment initiation (measured as a decrease in high-sensitivity C-reactive protein (hsCRP) to <2 mg/l). The results of other clinical trials suggest that, to be effective, immunotherapies need to be tailored to specific groups of patients with atherosclerotic cardiovascular disease depending on their clinical status. For example, the LoDoCo2 trial9 showed the efficacy of low-dose colchicine (also tested in the COLCOT trial7) for the secondary prevention of cardiovascular events in patients with stable coronary artery disease. This effect was associated with a reduction in the circulating levels of hsCRP and of proteins involved in the activation of the NLRP3 inflammasome10,11. However, treatment with colchicine did not reduce the risk of cardiovascular death, and the incidences of death from any cause and non-cardiovascular death were higher in the colchicine-treated group than in the placebo group. Moreover, in the COPS trial12 in patients with acute coronary syndrome, colchicine treatment resulted in a higher mortality and had no benefit in reducing cardiovascular outcomes at 12 months compared with placebo.
Most research efforts so far have focused on reducing the inflammatory status of patients with atherosclerosis, an approach that is typically associated with an increased risk of infection6,7. However, new strategies that harness anti-atherosclerotic functions of the immune system, including the use of low-dose IL-2 to promote the polarization of anti-atherosclerotic regulatory T (Treg) cells, are under investigation in the ongoing LILACS13 and IVORY14 clinical trials.
Nevertheless, despite remarkable advances in the field, including the identification of novel molecular targets and the development of new experimental treatments (extensively reviewed prevously15), the clinical translation of many immunotherapies identified by single-target drug discovery has proved difficult16,17. Most studies have focused on systemic immune alterations in patients with atherosclerotic cardiovascular disease18, while our knowledge of the cellular composition of the human atherosclerotic plaque, where the plaque disruption underlying the clinical events occurs, remains limited. The importance of immune cell plasticity in atherosclerosis — with immune cells acquiring pro-atherogenic and anti-atherogenic functions across a spectrum of differentiation states that determine the fate of an atherosclerotic lesion — has long been investigated in animal models1,19. However, the delineation of the dynamic alterations and crosstalk between heterogeneous immune cell subsets in the human atherosclerotic plaque has been restricted to low-dimensional approaches that do not capture the coordinated activity of heterogeneous cells in health and disease states20.
The past decade has seen a revolutionary advance in single-cell technologies — such as single-cell mass cytometry (cytometry by time of flight; CyTOF)21,22, single-cell RNA sequencing (scRNA-seq)23 and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq)24 — that has enabled high-dimensional profiling and analysis of the proteome and transcriptome of individual cells with unprecedented resolution and throughput. Therefore, single-cell technologies are ideal for mapping human atherosclerotic plaques and have already provided a first glimpse of newly identified immune alterations that are specific to the human plaque microenvironment and are associated with clinical complications25 (Fig. 1).
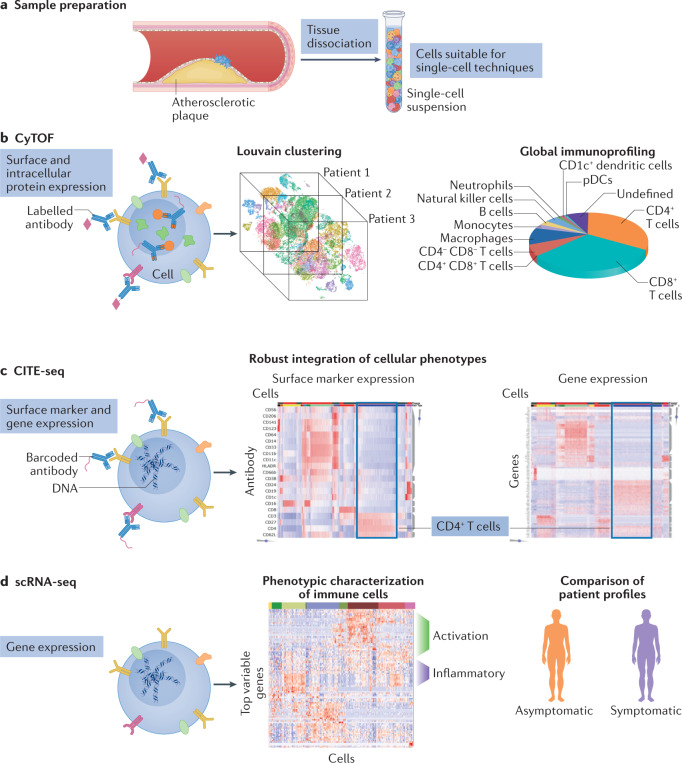
Fig. 1. Single-cell approaches to study human atherosclerosis.
a | After atherosclerotic tissue is dissociated into single cells, the sample is analysed using three approaches: cytometry by time of flight (CyTOF), cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) and single-cell RNA sequencing (scRNA-seq). b | CyTOF can be used to analyse the broad cell types and frequencies of immune cells across patients, with the use of unbiased approaches such as Louvain clustering. c | CITE-seq accurately integrates proteomics and gene expression signatures. d | scRNA-seq can be used to characterize phenotypically the immune cells from patients and to compare profiles of different cell types. pDCs, plasmacytoid dendritic cells.
In this Review, we discuss how single-cell technologies can advance our understanding of the cellular and molecular architecture of human atherosclerotic plaques to advance drug discovery. First, single-cell approaches provide the high-dimensional molecular resolution that is required to resolve heterogeneous cell populations on the basis of the molecular make-up of each cell in the complex plaque microenvironment. Second, single-cell data offer the opportunity to computationally predict cell trajectories of differentiation and infer the key cell–cell communications that contribute to specific cell functions in the atherosclerotic niche. We also argue that to identify new molecular targets successfully, future studies will need to harness single-cell studies in humans used as model systems, coupled with rigorous validation in relevant experimental models. Finally, we discuss the promise of single-cell omics studies in humans for guiding the design of new personalized immunotherapies and immune monitoring tools in patients with atherosclerotic cardiovascular disease.
Immune mechanism in atherosclerosis
Decades of intense research efforts have rigorously established fundamental steps in atherosclerotic plaque formation and progression. This knowledge is largely based on the use of well-validated animal models of atherosclerosis and state-of-the art pathology studies of human plaques1–3,26–28. Key atherosclerotic mechanisms include the immune reaction to lipid accumulation in the arterial wall. Of particular importance in atherogenesis are inflammatory macrophages and foam cell formation in plaque progression, processes that have been extensively reviewed previously1–3. Cholesterol loading of macrophages triggers the synthesis of pro-inflammatory cytokines, including the activation of the NLRP3–IL-1β pathway29,30. As the disease progresses, macrophage function deteriorates, their capacity to metabolize and clear lipids from the arterial wall becomes inefficient, and they undergo apoptosis contributing to the formation of a necrotic core. Defective efferocytosis — the clearing of apoptotic cells by macrophages, which promotes inflammation resolution — further promotes a chronic non-resolving inflammatory state that results in plaque progression31. Several research groups have provided strong evidence that restoring macrophage efferocytosis and inducing a reparative phenotype promotes plaque regression1–3. Preclinical studies (extensively reviewed previously32,33) have shown that macrophage phenotype and function are dictated by complex microenvironmental cues that promote either pro-inflammatory responses or reparative functions to promote healing and restore tissue homeostasis. Several subsets of pro-atherogenic or pro-resolving macrophages have been reported in atherosclerotic plaques in animal models and are likely to correspond to distinct functional states rather than predetermined cell subsets32,33. Similarly, foam cell formation can be either pro-atherosclerotic or reparative depending on environmental cues that regulate or impair the capacity of the macrophage to uptake, metabolize and remove cholesterol from the atherosclerotic plaque32,33. Overall, these preclinical studies highlight the fundamental role of macrophages in atherosclerosis and provide the important concept that manipulating macrophage polarization to restore their reparative functions is a promising strategy to halt plaque inflammation and promote plaque regression.
Dysregulated innate immune responses within the plaque microenvironment are tied to equally complex alterations of the adaptive immune system. T cell subsets in progressing and regressing atherosclerotic plaques are highly heterogeneous and have a dynamic spectrum of pro-atherogenic and anti-atherogenic functions3,34. CD4+ T helper 1 (TH1) cells have pro-atherogenic functions, whereas Treg cells canonically have atheroprotective roles, although Treg cell subsets with detrimental effects in atherosclerosis have been described34. Other CD4+ T cell subsets including TH2 cells and TH17 cells are present in the plaque microenvironment but their role in atherosclerosis is still controversial and mechanistically not fully understood34–38. In addition to macrophages and T cells, other innate and adaptive immune cells contribute to the pathogenesis of atherosclerosis, including neutrophils, natural killer (NK) T cells and B cells, and less clearly NK cells, and the role of these cells in atherosclerosis has been extensively reviewed19,39–43.
These advances in the field clearly show that the immune system has a crucial role in atherosclerosis and that targeting specific immune mechanisms of plaque progression and regression is a successful strategy to pursue in experimental models. However, important challenges remain in resolving the complex cellular and molecular architecture of atherosclerotic plaques and in translating the findings of animal studies to humans.
Animal models of atherosclerosis offer tremendous capabilities in terms of genetic manipulation of the disease with molecular and cellular specificity and have undoubtedly enabled remarkable advances in the field44. For example, mice and humans show a strong concordance of risk factors for atherosclerosis and a substantial overlap between mouse and human genes associated with atherosclerosis identified in genome-wide association studies (GWAS). However, fundamental differences between experimental and human atherosclerosis exist. Mouse models of atherosclerosis are created using genetically modified mice in which atherosclerotic plaques often develop under the influence of toxic levels of circulating lipids over a short period of time. Other translational challenges include the lack of genetic diversity in most experimental mouse models, because they are largely limited to one genetic background (C57BL/6). Moreover, differences in the regulation of gene expression in adaptive immune cells and of genes related to metabolic pathways include the presence of non-conserved enhancers and long non-coding RNAs across species44. Finally, the lack of microbial experience of laboratory mice might limit the adequate modelling of immunological events relevant to humans45.
The natural course of atherosclerotic plaque development shows other crucial differences across species. First, human plaques develop over decades under the multifactorial influence of high plasma cholesterol levels and cardiometabolic and environmental risk factors acting on heterogeneous genetic backgrounds. Seminal work has documented that human atherosclerotic plaques can progress to a vulnerable state, characterized by a large necrotic core, a thin fibrous cap, an inflammatory phenotype and intraplaque haemorrhage27,28,46–49. Vulnerable plaques are prone to spontaneous rupture or erosion, events exclusive to humans that can trigger occlusive atherothrombosis and consequent ischaemic clinical events such as stroke or myocardial infarction. Second, emerging evidence supports a nonlinear evolution of human atherosclerosis, because plaque disruption (rupture or erosion) does not always lead to clinical events. When formation of a thrombus is prevented, plaque healing promotes the repair of the damaged vasculature and stabilization of the atherosclerotic plaque50. The clinical significance of plaque healing is still debated but it is increasingly suggested that the reiteration of clinically silent plaque disruption coupled to impaired healing could contribute to the development of vulnerable plaques and to future ischaemic cardiovascular events resulting from plaque distruption50.
Therefore, the natural history of human atherosclerotic disease involves pivotal processes that are common to those in mice, and others that are unique to humans and not fully recapitulated in existing experimental models. Owing to the intrinsic complexity and tissue specialization of the immune system, the precise involvement of specific immune cell populations and subsets and the specific cell–cell communications occurring in the atherosclerotic niche in mice and humans and the differences across species remain to be fully understood. Single-cell multiomics analyses hold the potential to transform the way that we characterize immune processes within human and experimental atherosclerotic lesions and to advance our capacity to identify new cell targets, pathways and immune crosstalk to harness the power of the immune system and develop new and precise immunotherapies for atherosclerotic cardiovascular disease. Immune mapping of experimental atherosclerosis will help improve the selection of relevant mouse models for the mechanistic validation of the identified therapeutic targets and signalling pathways and of new immunotherapies.
Single-cell proteomics
The capacity to study individual cells within tissues has been around for decades51. Conventional immunohistochemistry and conventional flow cytometry are robust and validated single-cell methods that rely on the use of light and fluorescence to investigate individual cells. However, multiparameter detection of heterogeneous cells including immune cells infiltrating human atherosclerotic plaques has long been limited to a few cell types or subsets owing to the spectral overlap of fluorescent probes52. Since the first introduction of advanced spectral flow cytometry detection in 2005, advances and commercialization of spectral flow cytometry technologies have enabled a more efficient spectrum capture, pushing the limits to more than 30 colours53. Spectral flow cytometry can detect the shape, and not the peak as in conventional flow cytometry, of emission spectra along a range of continuous wavelengths. Therefore, spectral flow cytometry can discriminate different fluorochromes with similar emission peaks and allows multiparameter analysis without compensation requirements. These remarkable advances now allow in-depth analyses of heterogeneous cell populations. However, some disadvantages remain for specific analyses. These disadvantages include limitation in barcoding capacity and, when analysing complex tissue samples, intrinsic tissue background autofluorescence that can limit the detection of low-expression proteins54–56. Sources of autofluorescence in atherosclerotic plaques include cellular and extracellular components. Cellular autofluorescence can dynamically reflect the metabolic, differentiation and survival state of cells57. Macrophage autofluorescence can vary based on the differentiation state of the cells58, and foam cells expressing inducible nitric oxide synthase can produce autofluorescent (per)oxidized lipids (ceroid)59. Furthermore, changes in the levels of NADPH and FAD autofluorescence reflect T cell activation and subtype60. Collagen, elastin and oxidized LDL components of the lipid core are other potential sources of extracellular autofluorescence in atherosclerotic plaques that can interfere with sample analysis61,62. Specific to human advanced atherosclerotic plaques, near-infrared autofluorescence derived from intraplaque haemorrhage and haem degradation can variably affect samples from plaques either at high risk of rupture or that have ruptured63.
Mass cytometry (CyTOF)21 overcomes these limitations by using probes labelled with elemental isotopes, with a detection overlap among heavy metal isotopes generally <2%64. Therefore, CyTOF allows high parameterization and the quantitative detection of 40–50 parameters in a single tube and is highly suitable for mapping circulating immune cells as well as the cellular composition of human atherosclerotic plaques. The specific details of basic CyTOF workflows and technical performance have been elegantly described previoulsy21,65,66. A large number of antibodies conjugated to heavy metals are now commercially available, and antibody panels can be tailor-made using commercially available heavy metal conjugation kits67. Indeed, with 135 CyTOF detection channels now available, the current limitation to ~40 markers is due to the availability of pure and stable reporter metals and the chemistry to conjugate them68. To overcome this constraint, atomic mass tags are continually being developed to expand parameter detection and multiplexing capacity69. The high parameterization capacity of CyTOF also enables many samples to be multiplexed together with the use of specific barcoding schemes70,71 to reduce batch effects and remove staining variation across samples72. Moreover, CyTOF can reproducibly detect cell subsets in samples with as few as 10,000 cells, thereby making the technique ideal for use with limited specimens from patients73. Multiplexing capabilities can also protect against sample loss during the antibody staining process. Therefore, CyTOF allows the gathering of maximal information from limited clinical tissue samples.
High-parameter CyTOF data can be analysed either with traditional gating methods on the basis of previous knowledge or with unbiased approaches that enable the discovery of new cell clusters and functional states. The web-based platform Cytobank incorporates several computational methods to analyse CyTOF data, including traditional gating, viSNE, FlowSOM, SPADE and CITRUS74. Cytobank tools are user-friendly and well-suited to use by many non-computational bench scientists. Other flow cytometry software, such as FlowJo and FCS Express, have implemented plugins to allow the visualization and analysis of CyTOF data. Kimball and colleagues have published a practical guide to analysing and visualizing mass cytometry data, with an overview on the use of algorithms such as viSNE, PhenoGraph, X-shift, SPADE and CITRUS, and detailing the insights garnered by each platform75. A systematic comparison of seven unsupervised methods and two semi-supervised methods tested on six mass cytometry datasets clearly showed the advantages of each method, including the resolving capabilities and precision or coherence of the different algorithms and a decision pipeline for choosing the most appropriate one76. New tools and algorithms for fast and automated unbiased clustering from high-dimensional data are continually being developed77, with one of the goals being the discovery of new cell subsets or functional states78,79.
CyTOF analyses provide novel clinical insights
CyTOF can detect both surface and intracellular markers, making this technique ideal for phosphoproteomics studies of heterogeneous cell populations as well as pharmacodynamics and toxicity studies80,81. Single-cell phospho-signatures of activated monocyte–platelet aggregates in patients with sickle cell disease have been described82. In another clinical study, the activation of STAT3, CREB and NF-κB signalling in monocyte subsets correlated with functional impairment and pain in patients after surgical hip replacement83. CyTOF has also been used to profile differential immune responses to drugs in human haematopoietic cells22. In cancer, CyTOF analyses have contributed to the identification of circulating immune correlates predictive of overall patient survival with new cancer treatments84–86 and of tumour-specific innate and adaptive immune cell alterations in early lung adenocarcinoma lesions87.
Overall, CyTOF is increasingly being used in large-scale clinical immune monitoring and precision medicine studies83. More than 100 clinical trials registered in clinicaltrials.gov use CyTOF as the main immune-monitoring tool. The studies range from phase I to phase IV clinical trials across several disciplines, including immunology, immuno-oncology, vaccine development, autoimmunity, surgery, biomarker discovery, infectious disease and, more recently, COVID-19 (ref.88). The analysed tissues include peripheral blood, tissue biopsies and bone marrow samples, depending on the type of disease and availability of tissue sampling.
The increasing use of CyTOF in clinical research requires standardized workflows and protocols for the acquisition and analysis of large-scale, multicentre, immune-monitoring clinical studies. Batch-correcting algorithms89 and reproducible data generation are the basis for reliable and robust clinical analyses. For this objective, a method that uses a lyophilized core antibody panel to streamline blood sample processing has been implemented to reduce technical variation and standardize operations90 and includes quality control by removing events caused by loss of stability and compensating for signal spillover resulting from isotopic impurities or oxide formation91. New, streamlined CyTOF workflows that are based on a simple method to cryopreserve samples at the sample collection site and analysis at a central core facility have been implemented to facilitate multicentre studies, including in patients with COVID-19 (refs92,93). Overall, immune monitoring by CyTOF is robust and reproducible for clinical sample types across different institutions94.
CyTOF analyses uncover new immune perturbations in atherosclerosis
In cardiovascular research, CyTOF has been instrumental in developing precise new gating strategies for improved flow cytometry analysis of circulating monocytes95 and in disentangling their heterogeneity in healthy individuals and in patients with coronary artery disease96. In particular, the latter study identified eight clusters of circulating monocytes, including three classic subsets and four non-classic subsets96, challenging the commonly accepted understanding that human monocyte populations are limited to three subsets. The presence of the new non-classic subset of Slan+CXCR6+ monocytes, with the potential to migrate into atherosclerotic plaques, positively correlated with coronary artery disease severity.
CyTOF studies have also contributed to resolving the immune cell composition of atherosclerotic plaques in humans and mice. Cole and colleagues provided one of the first comprehensive, single-cell, immune proteomic mapping studies of the atherosclerotic aorta of Apoe−/− mice fed a chow or a high-fat diet; plaque macrophages comprised ~50% of the aortic CD45+ cells97, a proportion that was similar to that observed by Cochain and colleagues in a scRNA-seq study in Ldlr−/− mice98. In Apoe−/− mice, a high-fat diet had a profound effect on the myeloid cell repertoire present in atherosclerotic plaques, by inducing more inflammatory monocyte–macrophage subsets and fewer CD206+CD169+ macrophages and type 2 conventional dendritic cells97. Winkels and colleagues used CyTOF to validate the 11 heterogeneous leukocyte populations identified with scRNA-seq of aortas from Apoe−/− mice fed a Western or chow diet and to compare the CyTOF data in mice with CyTOF data from a human atherosclerotic lesion99.
Our group used CyTOF to provide a first immune profile dataset of human atherosclerosis in patients with carotid atherosclerosis25. In a first cohort of 15 patients, unbiased Louvain clustering of immune cells in the atherosclerotic plaque and blood from the same patient revealed that T cells were a major and heterogeneous cell type among all CD45+ cells infiltrating atherosclerotic plaques. The analysis of a second cohort of 27 patients confirmed these findings and identified a remarkable heterogeneity of T cells, that comprised 25 distinct metaclusters across blood and atherosclerotic plaques. Four T cell subsets were enriched in atherosclerotic lesions and expressed tissue-residency markers, were more activated and differentiated, and expressed higher levels of PD1 compared with corresponding metaclusters in blood25. Atherosclerotic plaques from patients with stroke were enriched in a specific subset of CD4+ effector memory T cells compared with atherosclerotic plaques from patients without stroke25. Overall, these results uncovered a new role for T cells, which accounted for ~65% of all immune cells in atherosclerotic plaques, and confirmed the importance of macrophages, which corresponded to ~13% (3–28%) of all CD45+ cells in the plaques25.
Macrophages are considered to be the main cell type in atherosclerotic plaques on the basis of traditional pathology studies and, more recently, cell-type deconvolution methods used to infer the cellular composition of atherosclerotic plaques from bulk RNA-seq of mouse and human atherosclerosis99. The discrepancy with human single-cell analysis data could simply be due to a technical artefact owing to low efficiency of macrophage retrieval during tissue digestion. However, other explanations could include the heterogeneous distribution of immune cells within plaques, the histological quantification method adopted and, for computational prediction methods of tissue cell composition, the lack of tissue-specific input of ‘signature matrices’ that reflect the exact molecular adaptation of immune cells residing in atherosclerotic plaques. Previous histological analysis of human atherosclerotic plaques revealed remarkable differences in the spatial distribution of plaque macrophages, with higher frequencies in the necrotic core (~60%) than in the adjacent intima (~8.6%), the shoulder of the plaque (~18.2%) or the fibrotic cap (~23.9%)100. The average macrophage frequency, excluding the necrotic core, was ~16.9%, which is consistent with that identified by CyTOF and scRNA-seq in human carotid plaques25,101. Indeed, apoptotic and necrotic cells, largely macrophages present in the necrotic core46, would not be captured by single-cell analyses. Other studies based their findings on the quantification of stained areas instead of number of cells49,102. Given that T cells are much smaller (5–7 μm) than macrophages and foam cells (20–30 μm), a lower number of macrophages and a higher number of T cells would occupy the same area. Moreover, when macrophage content is measured as the percentage of stained areas, the measured area might include apoptotic or dead macrophages, which could potentially lead to overestimation of the frequency and number of macrophages compared with single-cell methods such as CyTOF and scRNA-seq, which analyse live cells. A scRNA-seq analysis published in 2020 of all immune and non-immune cells from carotid atherosclerotic plaques from 18 patients confirmed that T cells (~52.4%) are the major cell type in human atherosclerotic plaques, followed by myeloid cells (~18.5%)101. In this study, data were validated by histological analysis of a subset of matched samples, indicating that dissociation methods did not alter the immune cell composition of plaques by overestimating the proportion of T cells. The application of high-dimensional multiomics analysis in situ (Table 1), systematically performed on serial sections of different parts of the atherosclerotic plaque and combined with automated image segmentation processing, will help determine accurately the immune cell composition in intact tissue and the spatial relationships between cells in human atherosclerotic plaques. In situ approaches will offer complementary information to that obtained from global, high-throughput analysis of whole tissue, and will thereby offer new opportunities for the identification of novel molecular targets for drug development (Fig. 2).
Table 1.
Single-cell technologies to study in situ complex tissue architecture for drug target discovery
| Technology | Approach | Tissue | Advantages | Disadvantages | Resolution | Platform | Ref. |
|---|---|---|---|---|---|---|---|
| Proteomics | |||||||
| Co-detection by indexing (CODEX) | Tissues are stained with oligo-conjugated antibodies and are specifically detected by reporters that are imaged in cycles with the use of a standard microscope | FFPE; fresh frozen tissue |
Reagents available for custom conjugating of antibodies Non-destructive staining procedure that allows morphological analysis after image acquisition |
Multiple rounds of imaging with long acquisition time Limited commercially available antibody panels |
Subcellular | Akoya | 145 |
| Multiplexed ion beam imaging (MIBI) | Tissues are stained with antibodies conjugated to heavy metals and are imaged using a specialized mass cytometer | FFPE; fresh frozen tissue | – | Long acquisition time | Subcellular | IonPath | 146 |
| Imaging mass cytometry (IMC) | Tissues are stained with antibodies conjugated to heavy metals and imaged using an atmospheric laser ablation chamber interfaced to a mass cytometer | FFPE; fresh frozen tissue | Large selection (>100) of validated antibodies | – | Subcellular | Fluidigm | 147 |
| Transcriptomics | |||||||
| Spatial transcriptomics |
Tissues are attached to slides that contain barcoded probes capable of capturing RNA from the permeabilized sample cDNA synthesis occurs on the slide and is subsequently used for sequencing |
FFPE; fresh frozen tissue | – | – | 50–100 μm | Visium 10× Genomics | 148 |
| Multiplexed error-robust fluorescence in situ hybridization (MERFISH) | Uses combinational labelling with sequential imaging and error-robust barcoding of individual mRNAs | Fresh frozen tissue |
High-throughput, single-cell resolution with up to 1 cm2 of tissue imaged per single run High multiplexing power that measures thousands of transcripts Subcellular localization of transcripts Can detect low-expression genes with single-molecule sensitivity |
Molecular crowding of signal and possible spatial overlapping of signal Lengthy imaging workflow Depends on a finite number of hybridized probes to known mRNA sequences |
Subcellular | Vizgen | 149 |
| Slide-seq | A monolayer of DNA-barcoded beads placed on a slide are set to capture the RNA from tissue placed onto that slide | Fresh frozen tissue | – |
Typically detects a low number of genes Incompatible with FFPE |
Cellular (~10 μm) | NR | 150 |
| RNAscope |
Novel in situ hybridization assay for detection of target RNAs within intact cells or tissue Based on proprietary probe design (‘ZZ’ oligonucleotide probe pairs) to amplify target-specific signals but not background noise from non-specific hybridization |
FFPE; fresh frozen tissue |
Low background noise with a single RNA molecule level of sensitivity within intact cells The probe design (based on short target regions) allows successful hybridization of partially degraded RNA (degraded-sample compatible) Suitable when the target cannot be detected by antibodies (commercial antibodies unavailable, targets are low abundance, extracellular targets) |
Typically detects a low number of targeted transcripts | Cellular | Bio-Techne | 151 |
| Proteomics and transcriptomics | |||||||
| Digital spatial profiling |
The tissue slide is stained with fluorescence labelled reagents to select a region of interest Tissues are also stained with a panel of proteins or RNA targets of interest by using specialized UV-cleavable oligo-barcodes attached to either a target complementary sequence (transcriptomics) or a target antibody (proteomics) The oligos are cleaved from the region of interest and counted for digital quantification |
FFPE; fresh frozen tissue |
Non-destructive staining procedure for tissues Quantitative expression data |
Limited markers (three) for visualization | Regions of interest comprising many cells | NanoString | 152 |
| Sequential fluorescence in situ hybridization (SeqFISH) | Sequential rounds of fluorescent in situ hybridization and imaging | Fresh frozen tissue |
Multiplexing (>10,000 molecules) Multiomics capability No quantification bias caused by the use of reverse transcription Can detect low copy number mRNAs that are undetectable using scRNA-seq or in situ hybridization |
Molecular crowding of signal and spatial overlapping of signal | Subcellular | NR | 153 |
| Deterministic barcoding in tissue for spatial omics sequencing (DBiT-seq) | Microfluidic barcoding of mRNAs and proteins in tissues on slides is followed by high-throughput sequencing | FFPE | Microfluidic device that requires little microfluidics expertise | Limited resolution, might not ensure single-cell readouts | Cellular (~10 μm) | NR | 154 |
FFPE, formalin-fixed paraffin-embedded; NR, not reported; scRNA-seq, single-cell RNA sequencing.
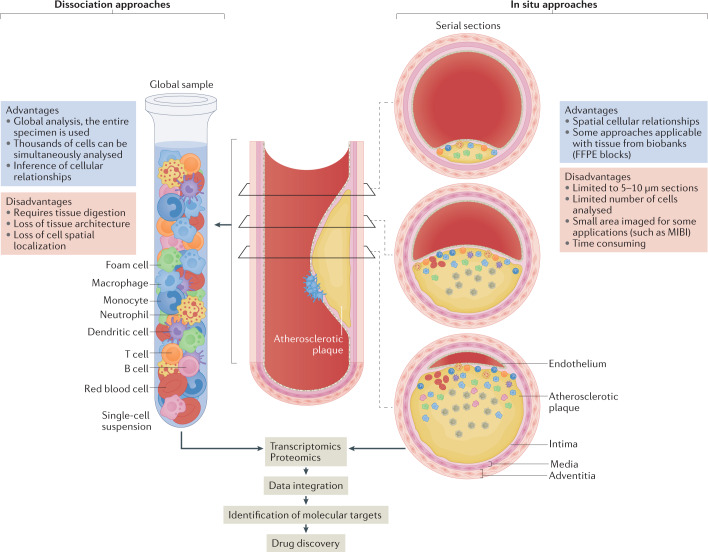
Fig. 2. Single-cell approaches to study atherosclerotic tissue.
Tissue dissociation methods and in situ approaches are complementary systems to understand cell properties at the single-cell level. Several methods exist for each approach, and both approaches can be used for proteomics and transcriptomics analyses. The integration of the resulting data can be useful for the identification of molecular targets for disease therapies and subsequent drug discovery. FFPE, formalin-fixed paraffin-embedded; MIBI, multiplexed ion beam imaging.
Single-cell transcriptomics
Together with substantial advances in single-cell proteomics (such as CyTOF), scRNA-seq offers increased sensitivity and granularity in identifying molecular dysregulation of heterogeneous cells in tissues. Gene expression analysis and systems genetics have revolutionized our understanding of basic biological processes of common complex diseases103. Most systems genetics studies have been based on bulk RNA-seq that averages transcriptional expression across cells and cannot provide detail on the transcriptional alterations of individual cells from heterogeneous cell pools composing the tissue. scRNA-seq analysis reveals transcriptional differences not only within populations of heterogeneous cells but also among cells with homogeneous proteomes based on canonical markers104. Therefore, single-cell technologies are ideal for uncovering new alterations of the immune system, a complex hierarchical set of molecular and cellular networks that contribute to atherosclerotic cardiovascular disease and clinical outcomes. The identification of the complex cell type composition and functional states of cells in disease-relevant tissues, such as atherosclerotic plaques, is crucial for the identification of the cellular contribution to disease and of new druggable cellular targets.
In dissociative scRNA-seq techniques, individual cells are isolated and lysed, and RNA is captured before being converted to cDNA by reverse transcription. cDNA is then amplified to prepare sequencing libraries23. Depending on the experimental needs (for example, number of cells to be analysed, throughput requirements or cost), cell-capture methods differ between distinct techniques. A useful framework for benchmarking the most suitable technique has been comprehensively discussed previously105–107. Some methods, including SMART-seq2 and MARS-seq, are based on florescence activated cell sorting (FACS) to isolate cells of interest106. Although upstream FACS alleviates the constraints of cell size, shape and homogeneity, and total cell number, this technique also subjects cells to mechanical stress, which induces oxidative stress in the cells and might affect their molecular state108,109. Alternatively, cells can either be isolated into microwells using a microfluidics system (such as Fluidigm C1 HT) or encapsulated into individual droplets (with Drop-seq or 10× Chromium technology)110. The development of droplet capture techniques has revolutionized scRNA-seq to enable the transcriptional detection from thousands of cells and the identification of rare cells, and has minimized the mechanical stress to the cells108,109. Other systems rely on single-cell isolation with magnetic beads in microwells, where individual cells are lysed and the beads are retrieved (BD Rhapsody) for targeted gene expression analysis. This method and other scRNA-seq methods have been discussed previously111,112.
scRNAseq data are often analysed with the use of established pipelines, such as Seurat113, SCANPY114, Clustergrammer2 and other tools (which have been comprehensively reviewed previously115 and are listed in the publicly available database scRNA-tools), that act as frameworks to enable data analysis. Specific tools also allow the inference of biological processes of interest from scRNA-seq data, including developmental, differentiation and cell cycle processes116,117. Luecken and Theis established a best practices tutorial that describes the processes of data analysis from start to finish118. Systems genetics of bulk RNA-seq data has shown that the pathophysiology of common complex disease is better explained by gene regulatory networks103,119–123, a set of genes that interact with each other to control a specific biological function. Several algorithms are being developed to infer gene regulatory networks from single-cell data. However, these analyses and the rigorous identification of gene regulatory networks are currently hindered by high cell heterogeneity, cell–cell variation in sequencing depth and the data sparsity of scRNA-seq datasets owing to the high rate of dropout events124,125. To facilitate a rigorous and reproducible evaluation of inference methods of gene regulatory networks and to provide recommendations to the end users, Pratapa and colleagues have developed an evaluation framework called BEELINE126. BEELINE comprehensively evaluates the accuracy, robustness and efficacy of several algorithms to identify gene regulatory networks and help avoid pitfalls in single-cell transcriptomic data analysis.
scRNA-seq analysis also offers the unique opportunity to infer crucial cell–cell communications that coordinate the function of immune cells within heterogeneous tissues127. This approach has enabled the identification of specific ligand–receptor interactions associated with cell development and disease states, including clinical cardiovascular outcomes in patients with carotid atherosclerosis25.
Despite the substantial advances in the use of high-throughput scRNA-seq to study heterogeneous cell populations, the findings obtained with this approach cannot be matched with 100% accuracy with traditional immune cell classifications that are based on the expression of canonical markers. In 2017, Stoeckius and colleagues introduced the method of CITE-seq, which is based on the use of oligonucleotide-labelled antibodies to integrate cellular protein and transcriptome measurements in a single cell with high throughput24. This multiomic approach offers unique capabilities to study the immune system with the use of both data-driven and unbiased methods of analysis. CITE-seq is compatible with existing scRNA-seq approaches and can achieve a parallel characterization of immune cell populations on the basis of the expression of canonical and other surface protein markers and unbiased transcriptomics analyses. Combined with the Cell Hashing method128 for multiplex and doublet detection, CITE-seq has remarkably expanded the immune-profiling capabilities. The method has been applied to ECCITE-seq, which enables the detection of six modalities (transcriptome, T cell receptor (αβ TCR and γδ TCR), surface protein, sample identity by hashtags, and CRISPR single guide RNA) in a single experiment129.
scRNA-seq and CITE-seq as new tools to study experimental and human atherosclerosis
scRNA-seq technologies have been used to profile the immune cell landscape of atherosclerotic vessels from Apoe−/− mice and Ldlr−/− mice and atherosclerotic plaques from humans (Table 2). Collectively, these results provide a new understanding of the immune cell landscape in mouse atherosclerotic tissue. Winkels and colleagues used scRNA-seq followed by an unsupervised clustering algorithm and found a greater diversity of leukocytes in aortic atherosclerotic tissue from Apoe−/− mice and Ldlr−/− mice than in tissue from wild-type mice, and validated these findings with the use of CyTOF99. Using scRNA-seq, Cochain and colleagues identified three distinct macrophage populations in aortic atherosclerotic tissue from Apoe−/− mice and Ldlr−/− mice98. One subset of resident-like macrophages was found in both atherosclerotic mice and healthy mice, whereas two subsets of inflammatory and Trem2high macrophages were present exclusively in atherosclerotic tissue and had a transcriptional profile similar to that of intimal foamy macrophages described by Kim and colleagues130. Lin and colleagues further characterized the dynamic changes of macrophages in the mouse aortic arch with the use of validated models of atherosclerotic plaque progression and regression131. Specifically, they identified populations of inflammatory and Trem2high macrophages, analogous to those subsets observed in atherosclerotic tissue by Cochain and colleagues98. Furthermore, combining scRNA-seq with genetic fate mapping, Lin and colleagues resolved the diversity of macrophage transcriptional states and identified a cluster of proliferating monocytes that could contribute to overall macrophage accumulation in atherosclerotic lesions131, consistent with previous observations that local proliferation is a main source of plaque macrophages132. A large meta-analysis of nine scRNA-seq and two CyTOF studies that focused on the leukocyte diversity of mouse atherosclerosis has provided new insights into experimental atherosclerosis and confirmed known biology of the disease133. The researchers used Harmony134 to integrate nine scRNA-seq datasets and analysed a total of 15,288 cells and identified 17 cell clusters that encompassed all major immune cells. This analysis showed that macrophages are the most abundant cell type in the atherosclerotic aorta of mice and included Trem2+ foamy macrophages that were not inflammatory and tissue-resident macrophages expressing Pf4. A substantial proportion of all foam cells were predicted to be derived from vascular smooth muscle cells133. This result is consistent with previous observations in both mice and humans suggesting that vascular smooth muscle cells contribute to a considerable number of lipid-rich macrophage-like cells in atherosclerotic plaques135–139. The plasticity of vascular smooth muscle cells in atherosclerosis has been extensively reviewed previously140.
Table 2.
Single-cell studies of human and mouse atherosclerotic plaques
| Sample and model | Cell type focus | Analysis (platform) | Ref. |
|---|---|---|---|
| Studies in mice | |||
| Atherosclerotic aorta from Apoe−/− or Ldlr−/− mice | Immune cells | CyTOF; scRNA-seq (10× Genomics) | 99 |
| Atherosclerotic aorta from Cx3cr1CreERT2-EYFP/+Rosa26tdTomato/+ mice injected with AAV-mPCSK9 | Macrophages | scRNA-seq (10× Genomics) | 131 |
| Normal aorta and atherosclerotic aorta from Apoe−/− or Ldlr−/− mice | CD45+ immune cells, macrophages | scRNA-seq (10× Genomics) | 98 |
| Normal aorta and atherosclerotic aorta from Apoe−/− mice | Vascular smooth muscle cells | scRNA-seq (Fluidigm C1, Smart-Seq2, 10× Genomics) | 155 |
| Atherosclerotic aortas from Apoe−/− or Ldlr−/− mice | CD45+ immune cells, macrophages | FACS combined with scRNA-seq (10× Genomics) | 130 |
| Aortas from Apoe−/− mice fed a chow or a high-fat diet | CD45+ immune cells, macrophages | CyTOF | 97 |
| Aortas from Apoe−/− mice treated with pro-efferocytosis nanoparticles | CD45+ immune cells, macrophages | scRNA-seq (10× Genomics) | 156 |
| Aortas from Apoe−/− mice fed a chow or a Western diet | CD4+ T cells | scRNA-seq (10× Genomics) | 157 |
| Studies in humans | |||
| Carotid atherosclerotic plaque and peripheral blood mononuclear cells | CD45+ immune cells, macrophages, T cells | CD45+ bead-based enrichment combined with CyTOF, CITE-seq and scRNA-seq (10× Genomics) | 25 |
| Carotid atherosclerotic plaque | Immune cells, vascular smooth muscle cells, endothelial cells | FACS combined with scRNA-seq (CEL-seq2) and scATAC-seq | 101 |
| Studies in humans and mice | |||
| Human atherosclerotic coronary arteries and mouse atherosclerotic aorta | Immune cells, vascular smooth muscle cells | FACS combined with CITE-seq and scRNA-seq (10× Genomics) | 142 |
AAV, adeno-associated virus; CITE-seq, cellular indexing of transcriptomes and epitopes by sequencing; CyTOF, cytometry by time of flight; FACS, fluorescence-activated cell sorting; scATAC-seq, single-cell assay for transposase-accessible chromatin using sequencing; scRNA-seq, single-cell RNA sequencing.
scRNA-seq analysis of human atherosclerotic plaques has been so far limited to a few studies. Our group used scRNA-seq and CITE-seq (10× Genomics) to provide the first single-cell, immune cell mapping of human carotid artery atherosclerotic plaques that uncovered new innate and adaptive immune dysregulation associated with clinical cerebrovascular events25. Overall, plaque CD4+ T cells from patients with recent stroke were characterized by signalling involved in T cell migration, activation and differentiation. Plaque CD8+ T cells were activated in both patients with carotid artery plaques with recent stroke and patients with carotid artery plaques without recent stroke. However, in patients with stroke, CD8+ T cell activation coexisted with exhaustion signalling, suggesting the initiation of exhaustion reprogramming, possibly caused by chronic, non-resolving inflammation. Plaque macrophages comprised highly heterogeneous subsets, including activated and inflammatory cells and foam cells25, reminiscent of the heterogeneity found in experimental atherosclerosis97–99,131. In patients with stroke, macrophages expressed sets of genes associated with plaque instability (such as CCL5 and genes encoding granzymes), and included a small subset of macrophages expressing genes related to iron metabolism25, resembling the pro-atherogenic, iron-metabolizing macrophages described by Guo and colleagues141. Ligand–receptor interaction analyses of carotid artery plaques from patients with recent stroke and patients without a recent stroke identified top candidate interactions between plaque macrophages and T cells, suggesting highly specialized cell crosstalk25.
A subsequent scRNA-seq study of all plaque cells confirmed all the identified major immune cell subsets in the plaques, including endothelial cells, vascular smooth muscle cells, mast cells, B cells, myeloid cells and T cells101. This analysis identified subsets of endothelial cells expressing genes indicative of angiogenic capacity and endothelial-to-mesenchymal transition. Plaque T cells had transcriptional profiles suggestive of a transition from a cytotoxic to a quiescent phenotype. The two myeloid cell subsets identified included pro-inflammatory macrophages and a TREM2high foam cell-like population with a fibrotic phenotype. Vascular smooth muscle cells expressed a synthetic transcriptional profile101. In this study, the investigators also used single-cell ATAC-seq (assay for transposase-accessible chromatin using sequencing) and cell–cell interaction analysis to identify candidate transcription factors involved in gene regulation and putative intercellular communications involved in atherosclerotic plaque inflammation. Using scRNA-seq of human and mouse atherosclerotic plaques, Wirka and colleagues identified the transcription factor TCF21 as an important factor governing the atheroprotective transition of vascular smooth muscle cells to fibroblast-like cells, termed fibromyocytes142. Specifically, deletion of Tcf21 in Apoe−/− mice significantly reduced the number of vascular smooth muscle cell-derived fibromyocytes and resulted in a thinner fibrous cap. The researchers identified analogous populations of fibromyocytes in human atherosclerotic plaques142. The relevance of these findings in humans was further supported by the analysis of large GWAS datasets and a cis-eQTL (expression quantitative trait loci) dataset, showing that individuals with lower TCF21 expression levels resulting from genetic variants in this gene had an increased risk of cardiovascular events.
Collectively, these studies provide a first step towards the ambitious goal of building a comprehensive single-cell atlas of human and experimental atherosclerosis that will provide a robust framework to explore the immune system in the diseased vasculature. Moreover, these studies provide robust evidence that single-cell transcriptional analysis is a powerful approach to resolving complex cellular and molecular alterations and to identifying new molecular targets that will aid the development of new, precise immunotherapies for patients with atherosclerotic cardiovascular disease.
Immune profiling for precision medicine
Most research efforts in drug discovery and repurposing of existing drugs for use in patients with atherosclerosis so far have focused on targeting systemic inflammation. However, emerging single-cell data show specific tissue specialization of innate and adaptive immune cells in plaque versus blood in the same patient. Identifying new, precise treatments that restore the function of immune cellular and molecular networks specifically acting in blood and/or in atherosclerotic lesions in the same patient is an innovative strategy to reduce both systemic and plaque inflammation and ultimately treat cardiovascular disease (Fig. 3). Existing single-cell studies have already enabled new discoveries of novel inflammatory mechanisms in human and mouse atherosclerosis25,97–99,101,131,133 through the identification of new immune cell subsets or functional states and putative intercellular communications on the basis of the expression of receptors, co-receptors and ligands, and differences in patients with distinct clinical status25,101. The integration of human and mouse single-cell datasets could help achieve a dual goal by providing information on the relevance in humans of mechanisms identified in experimental models and on the most suitable experimental model to study in vivo the shared mechanisms across species.
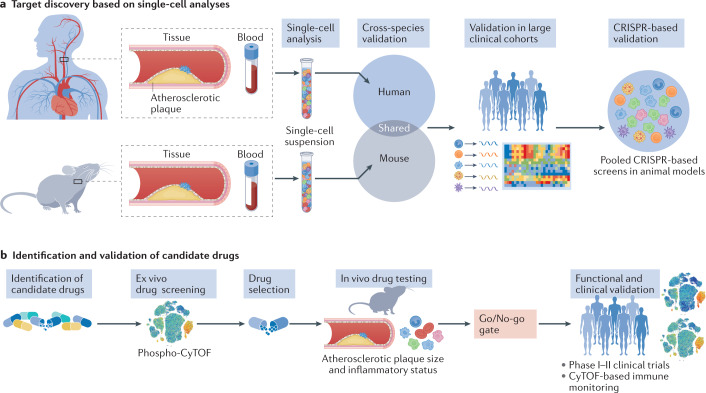
Fig. 3. Integration of single-cell methods for the discovery and validation of drug targets.
a | Single-cell studies in humans and mice provide information about the disease. Whereas studies in humans define the actual disease state, mechanistic studies in mice can aid in the understanding of how perturbations affect the disease. Integration and cross-species validation of these studies can be used to identify novel molecular targets. Understanding these molecular pathways in large clinical cohorts can be used as validation and then secondarily validated in animal models with the use of pooled CRISPR screening. b | When new targets are identified and validated, candidate drugs can be assessed for their specific effect in modulating these pathways. One evaluation method is phosphoproteomics with cytometry by time of flight (phospho-CyTOF). In vivo testing in animals can be used to investigate further the efficacy of the drug and to evaluate the go/no-go decisions to enter clinical phases of drug development. The adoption of immune monitoring in the early phases of clinical trials can provide crucial information on patient selection and efficacy for the design of future end point-driven clinical trials.
The biological and clinical translation of single-cell findings from both human and mouse datasets into drug development is a long-term goal that requires several additional steps. These steps include the validation of cell types and target genes in larger bulk RNA-seq clinical datasets, for example with the use of deconvolution approaches143, to establish potential clinical associations. The next steps required include functional validation in relevant model systems to provide functional annotations of identified cell clusters and genes, for example with the use of fate mapping (as described by Lin and colleagues to study macrophages in progression and regression models of atherosclerosis131) or cell-specific deletion of candidate genes in mice. To complement these approaches, high-dimensional multiplexing strategies have enabled the detection of gene function in response to in vitro perturbations with the use of innovative single-cell CRISPR screen technology. Several approaches have been developed to allow the simultaneous examination of the effect of several genes or proteins on the phenotype and function of individual cells (Box 1). Rigorous single-cell CRISPR screens to determine the immune functions of candidate genes and proteins requires further experimental dissection of immune responses in vivo and is crucial to understanding fully the computationally predicted biological functions and to identifying new molecular targets for future drug development. New methods for single-cell CRISPR screenings that are based on protein barcodes suitable for complementary in vitro and in vivo studies are emerging, and the future implementation of this method to screen for the function of new target proteins could aid in the identification of new drug targets144.
When a candidate target is identified and new drugs developed, ex vivo phospho-CyTOF drug screenings can provide a suitable platform to determine the immune response to drugs in heterogeneous samples of immune cells22 and to select top candidate drugs to be tested for efficacy in mouse models of atherosclerosis. These steps will provide a robust framework before moving into early-phase clinical trials that use scalable, single-cell technologies such as CyTOF (Fig. 3b). Immune monitoring implemented in phase I and phase IIa studies to analyse peripheral blood and extravascular tissue samples, when available based on the standard-of-care, could provide specific information on immune responses to drugs that could inform and guide phase IIb and phase III studies with regard to patient variability in drug responses and the effect of comorbidities and comedications. These studies could provide evidence-based information on the best strategies for patient selection for future large, randomized, outcome-based clinical studies, which are the gold-standard evidence to approve new therapeutics for clinical use. Indeed, these trials are large and expensive, and the failure of a large clinical trial imposes huge costs on the biomedical enterprise. Therefore, early-stage incorporation of an integrative approach to mechanistic data in experimental models and humans could aid in the selection of precise treatments for specific groups of patients who would derive the most benefit.
Box 1 Single-cell CRISPR technologies for drug target functional validation.
Single-cell CRISPR screenings are an important and innovative tool that can contribute to the identification of gene functions in individual cells. Single-cell CRISPR screenings can be easily implemented in laboratory cell lines or in human primary cells. Single-cell CRISPR screenings can provide a direct correlation between the effect of specific guide RNAs and the phenotypic data from heterogeneous cell populations. Several single-cell CRISPR strategies exist and have similar capabilities (see the table). Pooled CRISPR libraries of guide RNAs are used to genetically modify cells that are subsequently used for single-cell approaches that can resolve individual cells from heterogeneous cell populations. Several approaches can be used to incorporate single guide RNA into cells, including transfection, electroporation and viral transduction. The selection of the most suitable method depends on the cell type, owing to differences in the inherent resistance of the cells to incorporating these constructs.
| Screening | Technology | Description | Advantages and disadvantages | Ref. |
|---|---|---|---|---|
| Proteomics | Procode |
Barcoding system that leverages the use of protein tags to enable the multiplexing of >100 unique samples Can be applied to CRISPR screens with the use of high-dimensional methods, such as CyTOF, to characterize knockout constructs en masse |
Can also be used to evaluate overexpression cDNA constructs | 144 |
| Transcriptomics | Perturb-seq | Pooled single guide RNA libraries are transduced in cells of choice and used in conjunction with scRNA-seq |
Combines scRNA-seq and CRISPR-based perturbations to perform many assays in a pool Limited by reliance on indirect indexing of single guide RNAs |
158 |
| Direct capture Perturb-seq | Expression from single guide RNAs is sequenced alongside transcriptomic measurements | Targets individual genes with multiple single guide RNAs per cell; allows scRNA-seq experiments | 159 | |
| Mosaic single-cell analysis by indexed CRISPR sequencing (Mosaic-seq) | Uses a CRISPR barcoding system in combination with the measurement of single cell gene expression to readout both the phenotypic perturbations and the barcode of the specific single guide RNA | High-throughput endogenous interrogation of enhancers evaluated in single cells | 160 | |
| CRISPR droplet sequencing (CROP-seq) | A guide RNA serves as the barcode |
Enables pooled CRISPR screens with single-cell transcriptome resolution Overcomes the problem of lentiviral template switching by using CROP-seq lentiviral constructs |
161 | |
| Chromatin status | Perturb-ATAC | Combines CRISPR screening with scATAC-seq to measure the effect of CRISPR perturbations on chromatin status in single cells | NR | 162 |
CROP-seq, CRISPR droplet sequencing; CyTOF, cytometry by time of flight; NR, not reported; scATAC-seq, single-cell assay for transposase-accessible chromatin using sequencing; scRNA-seq, single-cell RNA sequencing.
Conclusions
Single-cell technologies have the potential to advance our basic knowledge of the complex immune mechanisms underlying atherosclerosis directly in humans. The analysis of experimental atherosclerosis with the use of the same single-cell tools offers the unique opportunity to build an immune atlas of experimental and human disease that will aid in the validation of human mechanisms in relevant mouse models. Paired with cell-based, pooled single-cell CRISPR screens to identify the specific function of several genes and proteins on heterogeneous immune cells, single-cell studies offer new opportunities for molecular-targeted drug development. The growing adoption of immune monitoring tools such as CyTOF in ongoing clinical trials shows the importance of how immune cells respond to new drugs and highlights that a similar approach might aid in the discovery of new, precise immunotherapies for patients with cardiovascular disease. The inclusion of immune monitoring in early phases of drug testing could advance drug discovery and precision medicine in cardiovascular disease to reduce adverse cardiovascular outcomes and death.
Acknowledgements
D.M.F. was supported by grant NIH T32HL007824. C.G. acknowledges research support from NIH-NHLBI (R01 HL153712-01), NIH-NCATS UH3TR002067, AHA (20SFRN35210252) and CZI (NFL-2020-218415).
Glossary
- Phosphoproteomics
A proteomic analysis to identify and characterize protein phosphorylation, a post-translational modification that regulates cellular processes.
- Louvain clustering
Graph-based community detection algorithm that is used to infer clusters of cells from single-cell data.
- Cell-type deconvolution methods
Computational methods used to infer cell type proportions from bulk transcriptomics data.
- Image segmentation
Image processing algorithm for the automatic identification of cells in intact tissue.
- Data sparsity
Refers to a matrix of numbers that includes many zeros. In single-cell analysis, data sparsity is due to dropout events.
- Dropout events
Missing values as a result of extremely low RNA input in the sequencing experiments or the stochastic nature of the gene expression pattern at the single-cell level. As a consequence, a gene can be observed at a moderate-to-high expression level in one cell but might be undetected in another cell.
- Single guide RNA
A single RNA molecule that contains both the custom-designed short CRISPR RNA (a sequence of 17–20 nucleotides complementary to the target DNA) and a trans-activating CRISPR RNA sequence that serves as a binding scaffold for the Cas nuclease. Single guide RNAs are used for CRISPR–Cas9 technology. The use of single guide RNA allows multimodal readout of gene perturbations at the single-cell level.
- Protein barcodes
Vector barcoding system that operates at the protein level and is based on sequences of linear epitopes that are detectable by antibodies.
Author contributions
Both authors researched data for the article, discussed its content and wrote the manuscript. C.G. reviewed and edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Cardiology thanks A. Zernecke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 2013;13:709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145:341–355. doi: 10.1016/j.cell.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabas I, Lichtman AH. Monocyte-macrophages and T cells in atherosclerosis. Immunity. 2017;47:621–634. doi: 10.1016/j.immuni.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross R. Atherosclerosis–an inflammatory disease. N. Engl. J. Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 5.Pradhan AD, Aday AW, Rose LM, Ridker PM. Residual inflammatory risk on treatment with PCSK9 inhibition and statin therapy. Circulation. 2018;138:141–149. doi: 10.1161/CIRCULATIONAHA.118.034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridker PM, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 2017;377:1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 7.Tardif JC, et al. Efficacy and safety of low-dose colchicine after myocardial infarction. N. Engl. J. Med. 2019;381:2497–2505. doi: 10.1056/NEJMoa1912388. [DOI] [PubMed] [Google Scholar]
- 8.Ridker PM, et al. Low-dose methotrexate for the prevention of atherosclerotic events. N. Engl. J. Med. 2019;380:752–762. doi: 10.1056/NEJMoa1809798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nidorf SM, et al. Colchicine in patients with chronic coronary disease. N. Engl. J. Med. 2020;383:1838–1847. doi: 10.1056/NEJMoa2021372. [DOI] [PubMed] [Google Scholar]
- 10.Fiolet ATL, et al. Short-term effect of low-dose colchicine on inflammatory biomarkers, lipids, blood count and renal function in chronic coronary artery disease and elevated high-sensitivity C-reactive protein. PLoS ONE. 2020;15:e0237665. doi: 10.1371/journal.pone.0237665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opstal TSJ, et al. Colchicine attenuates inflammation beyond the inflammasome in chronic coronary artery disease: a LoDoCo2 proteomic substudy. Circulation. 2020;142:1996–1998. doi: 10.1161/CIRCULATIONAHA.120.050560. [DOI] [PubMed] [Google Scholar]
- 12.Tong DC, et al. Colchicine in patients with acute coronary syndrome: the Australian COPS randomized clinical trial. Circulation. 2020;142:1890–1900. doi: 10.1161/CIRCULATIONAHA.120.050771. [DOI] [PubMed] [Google Scholar]
- 13.Zhao TX, et al. Low-dose interleukin-2 in patients with stable ischaemic heart disease and acute coronary syndromes (LILACS): protocol and study rationale for a randomised, double-blind, placebo-controlled, phase I/II clinical trial. BMJ Open. 2018;8:e022452. doi: 10.1136/bmjopen-2018-022452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04241601 (2020)
- 15.Zhao TX, Mallat Z. Targeting the immune system in atherosclerosis: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2019;73:1691–1706. doi: 10.1016/j.jacc.2018.12.083. [DOI] [PubMed] [Google Scholar]
- 16.Charo IF, Taub R. Anti-inflammatory therapeutics for the treatment of atherosclerosis. Nat. Rev. Drug Discov. 2011;10:365–376. doi: 10.1038/nrd3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhury RP, et al. Arterial effects of canakinumab in patients with atherosclerosis and type 2 diabetes or glucose intolerance. J. Am. Coll. Cardiol. 2016;68:1769–1780. doi: 10.1016/j.jacc.2016.07.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridker PM. From C-reactive protein to interleukin-6 to Interleukin-1: moving upstream to identify novel targets for atheroprotection. Circ. Res. 2016;118:145–156. doi: 10.1161/CIRCRESAHA.115.306656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf D, Ley K. Immunity and inflammation in atherosclerosis. Circ. Res. 2019;124:315–327. doi: 10.1161/CIRCRESAHA.118.313591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358:58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 22.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen TK, Baryawno N. Introduction to single-cell RNA sequencing. Curr. Protoc. Mol. Biol. 2018;122:e57. doi: 10.1002/cpmb.57. [DOI] [PubMed] [Google Scholar]
- 24.Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez DM, et al. Single-cell immune landscape of human atherosclerotic plaques. Nat. Med. 2019;25:1576–1588. doi: 10.1038/s41591-019-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witztum JL, Lichtman AH. The influence of innate and adaptive immune responses on atherosclerosis. Annu. Rev. Pathol. 2014;9:73–102. doi: 10.1146/annurev-pathol-020712-163936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burke AP, Taylor A, Farb A, Malcom GT, Virmani R. Coronary calcification: insights from sudden coronary death victims. Z. Kardiol. 2000;89:49–53. doi: 10.1007/s003920070099. [DOI] [PubMed] [Google Scholar]
- 28.Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM. Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 2000;20:1262–1275. doi: 10.1161/01.atv.20.5.1262. [DOI] [PubMed] [Google Scholar]
- 29.Sheedy FJ, et al. CD36 coordinates NLRP3 inflammasome activation by facilitating intracellular nucleation of soluble ligands into particulate ligands in sterile inflammation. Nat. Immunol. 2013;14:812–820. doi: 10.1038/ni.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajamaki K, et al. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PLoS ONE. 2010;5:e11765. doi: 10.1371/journal.pone.0011765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doran AC, Yurdagul A, Jr., Tabas I. Efferocytosis in health and disease. Nat. Rev. Immunol. 2020;20:254–267. doi: 10.1038/s41577-019-0240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabas I, Bornfeldt KE. Macrophage phenotype and function in different stages of atherosclerosis. Circ. Res. 2016;118:653–667. doi: 10.1161/CIRCRESAHA.115.306256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koelwyn GJ, Corr EM, Erbay E, Moore KJ. Regulation of macrophage immunometabolism in atherosclerosis. Nat. Immunol. 2018;19:526–537. doi: 10.1038/s41590-018-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saigusa R, Winkels H, Ley K. T cell subsets and functions in atherosclerosis. Nat. Rev. Cardiol. 2020;17:387–401. doi: 10.1038/s41569-020-0352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taleb S, et al. Loss of SOCS3 expression in T cells reveals a regulatory role for interleukin-17 in atherosclerosis. J. Exp. Med. 2009;206:2067–2077. doi: 10.1084/jem.20090545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon T, et al. Circulating levels of interleukin-17 and cardiovascular outcomes in patients with acute myocardial infarction. Eur. Heart J. 2013;34:570–577. doi: 10.1093/eurheartj/ehs263. [DOI] [PubMed] [Google Scholar]
- 37.Poizeau F, et al. Association between early severe cardiovascular events and the initiation of treatment with the anti-interleukin 12/23p40 antibody ustekinumab. JAMA Dermatol. 2020;156:1208–1215. doi: 10.1001/jamadermatol.2020.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim H, et al. Proatherogenic conditions promote autoimmune T helper 17 cell responses in vivo. Immunity. 2014;40:153–165. doi: 10.1016/j.immuni.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sage AP, Tsiantoulas D, Binder CJ, Mallat Z. The role of B cells in atherosclerosis. Nat. Rev. Cardiol. 2019;16:180–196. doi: 10.1038/s41569-018-0106-9. [DOI] [PubMed] [Google Scholar]
- 40.Getz GS, Reardon CA. Natural killer T cells in atherosclerosis. Nat. Rev. Cardiol. 2017;14:304–314. doi: 10.1038/nrcardio.2017.2. [DOI] [PubMed] [Google Scholar]
- 41.Winkels H, Ley K. Natural killer cells at ease: atherosclerosis is not affected by genetic depletion or hyperactivation of natural killer cells. Circ. Res. 2018;122:6–7. doi: 10.1161/CIRCRESAHA.117.312289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silvestre-Roig C, Braster Q, Ortega-Gomez A, Soehnlein O. Neutrophils as regulators of cardiovascular inflammation. Nat. Rev. Cardiol. 2020;17:327–340. doi: 10.1038/s41569-019-0326-7. [DOI] [PubMed] [Google Scholar]
- 43.Nour-Eldine W, et al. Genetic depletion or hyperresponsiveness of natural killer cells do not affect atherosclerosis development. Circ. Res. 2018;122:47–57. doi: 10.1161/CIRCRESAHA.117.311743. [DOI] [PubMed] [Google Scholar]
- 44.von Scheidt M, et al. Applications and limitations of mouse models for understanding human atherosclerosis. Cell Metab. 2017;25:248–261. doi: 10.1016/j.cmet.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abolins S, et al. The comparative immunology of wild and laboratory mice, Mus musculus domesticus. Nat. Commun. 2017;8:14811. doi: 10.1038/ncomms14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolodgie FD, et al. Localization of apoptotic macrophages at the site of plaque rupture in sudden coronary death. Am. J. Pathol. 2000;157:1259–1268. doi: 10.1016/S0002-9440(10)64641-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor AJ, et al. A comparison of the Framingham risk index, coronary artery calcification, and culprit plaque morphology in sudden cardiac death. Circulation. 2000;101:1243–1248. doi: 10.1161/01.cir.101.11.1243. [DOI] [PubMed] [Google Scholar]
- 48.Virmani R, et al. Atherosclerotic plaque progression and vulnerability to rupture: angiogenesis as a source of intraplaque hemorrhage. Arterioscler. Thromb. Vasc. Biol. 2005;25:2054–2061. doi: 10.1161/01.ATV.0000178991.71605.18. [DOI] [PubMed] [Google Scholar]
- 49.Yahagi K, et al. Pathology of human coronary and carotid artery atherosclerosis and vascular calcification in diabetes mellitus. Arterioscler. Thromb. Vasc. Biol. 2017;37:191–204. doi: 10.1161/ATVBAHA.116.306256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vergallo R, Crea F. Atherosclerotic plaque healing. N. Engl. J. Med. 2020;383:846–857. doi: 10.1056/NEJMra2000317. [DOI] [PubMed] [Google Scholar]
- 51.Melamed MR. A brief history of flow cytometry and sorting. Methods Cell Biol. 2001;63:3–17. doi: 10.1016/s0091-679x(01)63005-x. [DOI] [PubMed] [Google Scholar]
- 52.Bonanno E, Mauriello A, Partenzi A, Anemona L, Spagnoli LG. Flow cytometry analysis of atherosclerotic plaque cells from human carotids: a validation study. Cytometry. 2000;39:158–165. doi: 10.1002/(sici)1097-0320(20000201)39:2<158::aid-cyto9>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 53.Feher K, et al. Multispectral flow cytometry: the consequences of increased light collection. Cytometry A. 2016;89:681–689. doi: 10.1002/cyto.a.22888. [DOI] [PubMed] [Google Scholar]
- 54.Parks DR. Multispectral flow cytometry: unaddressed issues and recommendations for improvement. Cytometry A. 2020;97:1276–1277. doi: 10.1002/cyto.a.24166. [DOI] [PubMed] [Google Scholar]
- 55.Mazza EMC, et al. Background fluorescence and spreading error are major contributors of variability in high-dimensional flow cytometry data visualization by t-distributed stochastic neighboring embedding. Cytometry A. 2018;93:785–792. doi: 10.1002/cyto.a.23566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nettey L, Giles AJ, Chattopadhyay PK. OMIP-050: A 28-color/30-parameter fluorescence flow cytometry panel to enumerate and characterize cells expressing a wide array of immune checkpoint molecules. Cytometry A. 2018;93:1094–1096. doi: 10.1002/cyto.a.23608. [DOI] [PubMed] [Google Scholar]
- 57.Surre J, et al. Strong increase in the autofluorescence of cells signals struggle for survival. Sci. Rep. 2018;8:12088. doi: 10.1038/s41598-018-30623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Njoroge JM, et al. Characterization of viable autofluorescent macrophages among cultured peripheral blood mononuclear cells. Cytometry. 2001;44:38–44. doi: 10.1002/1097-0320(20010501)44:1<38::aid-cyto1080>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 59.Kockx MM, et al. Phagocytosis and macrophage activation associated with hemorrhagic microvessels in human atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2003;23:440–446. doi: 10.1161/01.ATV.0000057807.28754.7F. [DOI] [PubMed] [Google Scholar]
- 60.Walsh AJ, et al. Classification of T-cell activation via autofluorescence lifetime imaging. Nat. Biomed. Eng. 2021;5:77–88. doi: 10.1038/s41551-020-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laifer LI, et al. Biochemical basis for the difference between normal and atherosclerotic arterial fluorescence. Circulation. 1989;80:1893–1901. doi: 10.1161/01.cir.80.6.1893. [DOI] [PubMed] [Google Scholar]
- 62.Lee MW, et al. Comprehensive intravascular imaging of atherosclerotic plaque in vivo using optical coherence tomography and fluorescence lifetime imaging. Sci. Rep. 2018;8:14561. doi: 10.1038/s41598-018-32951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Htun NM, et al. Near-infrared autofluorescence induced by intraplaque hemorrhage and heme degradation as marker for high-risk atherosclerotic plaques. Nat. Commun. 2017;8:75. doi: 10.1038/s41467-017-00138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bjornson ZB, Nolan GP, Fantl WJ. Single-cell mass cytometry for analysis of immune system functional states. Curr. Opin. Immunol. 2013;25:484–494. doi: 10.1016/j.coi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hartmann FJ, et al. Scalable conjugation and characterization of immunoglobulins with stable mass isotope reporters for single-cell mass cytometry analysis. Methods Mol. Biol. 2019;1989:55–81. doi: 10.1007/978-1-4939-9454-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schulz AR, Stanislawiak S, Baumgart S, Grutzkau A, Mei HE. Silver nanoparticles for the detection of cell surface antigens in mass cytometry. Cytometry A. 2017;91:25–33. doi: 10.1002/cyto.a.22904. [DOI] [PubMed] [Google Scholar]
- 69.Han G, et al. Atomic mass tag of bismuth-209 for increasing the immunoassay multiplexing capacity of mass cytometry. Cytometry A. 2017;91:1150–1163. doi: 10.1002/cyto.a.23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartmann FJ, Simonds EF, Bendall SC. A universal live cell barcoding-platform for multiplexed human single cell analysis. Sci. Rep. 2018;8:10770. doi: 10.1038/s41598-018-28791-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai L, Ong R, Li J, Albani SA. CD45-based barcoding approach to multiplex mass-cytometry (CyTOF) Cytometry A. 2015;87:369–374. doi: 10.1002/cyto.a.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zunder ER, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yao Y, et al. CyTOF supports efficient detection of immune cell subsets from small samples. J. Immunol. Methods. 2014;415:1–5. doi: 10.1016/j.jim.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 2010 doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kimball AK, et al. A beginner’s guide to analyzing and visualizing mass cytometry data. J. Immunol. 2018;200:3–22. doi: 10.4049/jimmunol.1701494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu X, et al. A comparison framework and guideline of clustering methods for mass cytometry data. Genome Biol. 2019;20:297. doi: 10.1186/s13059-019-1917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meehan S, et al. Automated subset identification and characterization pipeline for multidimensional flow and mass cytometry data clustering and visualization. Commun. Biol. 2019;2:229. doi: 10.1038/s42003-019-0467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palit S, Heuser C, de Almeida GP, Theis FJ, Zielinski CE. Meeting the challenges of high-dimensional single-cell data analysis in immunology. Front. Immunol. 2019;10:1515. doi: 10.3389/fimmu.2019.01515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weber LM, Robinson MD. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytometry A. 2016;89:1084–1096. doi: 10.1002/cyto.a.23030. [DOI] [PubMed] [Google Scholar]
- 80.Morris MK, Chi A, Melas IN, Alexopoulos LG. Phosphoproteomics in drug discovery. Drug Discov. Today. 2014;19:425–432. doi: 10.1016/j.drudis.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 81.Nassar AF, Ogura H, Wisnewski AV. Impact of recent innovations in the use of mass cytometry in support of drug development. Drug Discov. Today. 2015;20:1169–1175. doi: 10.1016/j.drudis.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Glassberg J, et al. Application of phospho-CyTOF to characterize immune activation in patients with sickle cell disease in an ex vivo model of thrombosis. J. Immunol. Methods. 2018;453:11–19. doi: 10.1016/j.jim.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gaudilliere B, et al. Clinical recovery from surgery correlates with single-cell immune signatures. Sci. Transl. Med. 2014;6:255ra131. doi: 10.1126/scitranslmed.3009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nair N, et al. Single-cell immune competency signatures associate with survival in phase II GVAX and CRS-207 randomized studies in patients with metastatic pancreatic cancer. Cancer Immunol. Res. 2020;8:609–617. doi: 10.1158/2326-6066.CIR-19-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang AC, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deng M, et al. Apatinib exhibits anti-leukemia activity in preclinical models of acute lymphoblastic leukemia. J. Transl. Med. 2018;16:47. doi: 10.1186/s12967-018-1421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lavin Y, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fluidigm. Use of CyTOF technology in clinical research trials. https://www.fluidigm.com/binaries/content/documents/fluidigm/marketing/mass-cytometry-clinical-trials-list-public_202101/mass-cytometry-clinical-trials-list-public_202101/fluidigm%3Afile (2021).
- 89.Trussart M, et al. Removing unwanted variation with CytofRUV to integrate multiple CyTOF datasets. eLife. 2020;9:e59630. doi: 10.7554/eLife.59630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amir ED, et al. Development of a comprehensive antibody staining database using a standardized analytics pipeline. Front. Immunol. 2019;10:1315. doi: 10.3389/fimmu.2019.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chevrier S, et al. Compensation of signal spillover in suspension and imaging mass cytometry. Cell Syst. 2018;6:612–620.e5. doi: 10.1016/j.cels.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Geanon D, et al. A streamlined CyTOF workflow to facilitate standardized multi-site immune profiling of COVID-19 patients. Preprint at. medRxiv. 2020 doi: 10.1101/2020.06.26.20141341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sumatoh HR, Teng KW, Cheng Y, Newell EW. Optimization of mass cytometry sample cryopreservation after staining. Cytometry A. 2017;91:48–61. doi: 10.1002/cyto.a.23014. [DOI] [PubMed] [Google Scholar]
- 94.Hartmann FJ, et al. Comprehensive immune monitoring of clinical trials to advance human immunotherapy. Cell Rep. 2019;28:819–831.e4. doi: 10.1016/j.celrep.2019.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thomas GD, et al. Human blood monocyte subsets: a new gating strategy defined using cell surface markers identified by mass cytometry. Arterioscler. Thromb. Vasc. Biol. 2017;37:1548–1558. doi: 10.1161/ATVBAHA.117.309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hamers AAJ, et al. Human monocyte heterogeneity as revealed by high-dimensional mass cytometry. Arterioscler. Thromb. Vasc. Biol. 2019;39:25–36. doi: 10.1161/ATVBAHA.118.311022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cole JE, et al. Immune cell census in murine atherosclerosis: cytometry by time of flight illuminates vascular myeloid cell diversity. Cardiovasc. Res. 2018;114:1360–1371. doi: 10.1093/cvr/cvy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cochain C, et al. Single-cell RNA-Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ. Res. 2018;122:1661–1674. doi: 10.1161/CIRCRESAHA.117.312509. [DOI] [PubMed] [Google Scholar]
- 99.Winkels H, et al. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ. Res. 2018;122:1675–1688. doi: 10.1161/CIRCRESAHA.117.312513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jonasson L, Holm J, Skalli O, Bondjers G, Hansson GK. Regional accumulations of T cells, macrophages, and smooth muscle cells in the human atherosclerotic plaque. Arteriosclerosis. 1986;6:131–138. doi: 10.1161/01.atv.6.2.131. [DOI] [PubMed] [Google Scholar]
- 101.Depuydt MA, et al. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circ. Res. 2020;127:1437–1455. doi: 10.1161/CIRCRESAHA.120.316770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burke AP, et al. Morphologic findings of coronary atherosclerotic plaques in diabetics: a postmortem study. Arterioscler. Thromb. Vasc. Biol. 2004;24:1266–1271. doi: 10.1161/01.ATV.0000131783.74034.97. [DOI] [PubMed] [Google Scholar]
- 103.Bjorkegren JL, Kovacic JC, Dudley JT, Schadt EE. Genome-wide significant loci: how important are they? Systems genetics to understand heritability of coronary artery disease and other common complex disorders. J. Am. Coll. Cardiol. 2015;65:830–845. doi: 10.1016/j.jacc.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Choi YH, Kim JK. Dissecting cellular heterogeneity using single-cell RNA sequencing. Mol. Cell. 2019;42:189–199. doi: 10.14348/molcells.2019.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.See P, Lum J, Chen J, Ginhoux F. A single-cell sequencing guide for immunologists. Front. Immunol. 2018;9:2425. doi: 10.3389/fimmu.2018.02425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ziegenhain C, et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65:631–643.e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 108.Binek A, et al. Flow cytometry has a significant impact on the cellular metabolome. J. Proteome Res. 2019;18:169–181. doi: 10.1021/acs.jproteome.8b00472. [DOI] [PubMed] [Google Scholar]
- 109.Llufrio EM, Wang L, Naser FJ, Patti GJ. Sorting cells alters their redox state and cellular metabolome. Redox Biol. 2018;16:381–387. doi: 10.1016/j.redox.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nguyen QH, Pervolarakis N, Nee K, Kessenbrock K. Experimental considerations for single-cell RNA sequencing approaches. Front. Cell Dev. Biol. 2018;6:108. doi: 10.3389/fcell.2018.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Valihrach L, Androvic P, Kubista M. Platforms for single-cell collection and analysis. Int. J. Mol. Sci. 2018;19:807. doi: 10.3390/ijms19030807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gao C, Zhang M, Chen L. The comparison of two single-cell sequencing platforms: BD Rhapsody and 10x Genomics Chromium. Curr. Genomics. 2020;21:602–609. doi: 10.2174/1389202921999200625220812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zappia L, Phipson B, Oshlack A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput. Biol. 2018;14:e1006245. doi: 10.1371/journal.pcbi.1006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 120.Schadt EE, Bjorkegren JL. NEW: network-enabled wisdom in biology, medicine, and health care. Sci. Transl. Med. 2012;4:115rv111. doi: 10.1126/scitranslmed.3002132. [DOI] [PubMed] [Google Scholar]
- 121.Foroughi Asl H, et al. Expression quantitative trait loci acting across multiple tissues are enriched in inherited risk for coronary artery disease. Circ. Cardiovasc. Genet. 2015;8:305–315. doi: 10.1161/CIRCGENETICS.114.000640. [DOI] [PubMed] [Google Scholar]
- 122.Franzen O, et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–830. doi: 10.1126/science.aad6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Talukdar HA, et al. Cross-tissue regulatory gene networks in coronary artery disease. Cell Syst. 2016;2:196–208. doi: 10.1016/j.cels.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat. Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 2016;34:1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pratapa A, Jalihal AP, Law JN, Bharadwaj A, Murali TM. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods. 2020;17:147–154. doi: 10.1038/s41592-019-0690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell-cell interactions and communication from gene expression. Nat. Rev. Genet. 2021;22:71–88. doi: 10.1038/s41576-020-00292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stoeckius M, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mimitou EP, et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods. 2019;16:409–412. doi: 10.1038/s41592-019-0392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim K, et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ. Res. 2018;123:1127–1142. doi: 10.1161/CIRCRESAHA.118.312804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lin JD, et al. Single-cell analysis of fate-mapped macrophages reveals heterogeneity, including stem-like properties, during atherosclerosis progression and regression. JCI Insight. 2019;4:e124574. doi: 10.1172/jci.insight.124574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Robbins CS, et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 2013;19:1166–1172. doi: 10.1038/nm.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zernecke A, et al. Meta-analysis of leukocyte diversity in atherosclerotic mouse aortas. Circ. Res. 2020;127:402–426. doi: 10.1161/CIRCRESAHA.120.316903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Allahverdian S, Chehroudi AC, McManus BM, Abraham T, Francis GA. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation. 2014;129:1551–1559. doi: 10.1161/CIRCULATIONAHA.113.005015. [DOI] [PubMed] [Google Scholar]
- 136.Wang Y, et al. Smooth muscle cells contribute the majority of foam cells in ApoE (apolipoprotein E)-deficient mouse atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2019;39:876–887. doi: 10.1161/ATVBAHA.119.312434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shankman LS, et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat. Med. 2015;21:628–637. doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rong JX, Shapiro M, Trogan E, Fisher EA. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc. Natl Acad. Sci. USA. 2003;100:13531–13536. doi: 10.1073/pnas.1735526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vengrenyuk Y, et al. Cholesterol loading reprograms the microRNA-143/145-myocardin axis to convert aortic smooth muscle cells to a dysfunctional macrophage-like phenotype. Arterioscler. Thromb. Vasc. Biol. 2015;35:535–546. doi: 10.1161/ATVBAHA.114.304029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Basatemur GL, Jorgensen HF, Clarke MCH, Bennett MR, Mallat Z. Vascular smooth muscle cells in atherosclerosis. Nat. Rev. Cardiol. 2019;16:727–744. doi: 10.1038/s41569-019-0227-9. [DOI] [PubMed] [Google Scholar]
- 141.Guo L, et al. CD163+ macrophages promote angiogenesis and vascular permeability accompanied by inflammation in atherosclerosis. J. Clin. Invest. 2018;128:1106–1124. doi: 10.1172/JCI93025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wirka RC, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 2019;25:1280–1289. doi: 10.1038/s41591-019-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wroblewska A, et al. Protein barcodes enable high-dimensional single-cell CRISPR screens. Cell. 2018;175:1141–1155.e16. doi: 10.1016/j.cell.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Goltsev Y, et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–981.e15. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Angelo M, et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 148.Stahl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 149.Xia C, Fan J, Emanuel G, Hao J, Zhuang X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl Acad. Sci. USA. 2019;116:19490–19499. doi: 10.1073/pnas.1912459116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rodriques SG, et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr. Protoc. Mol. Biol. 2011 doi: 10.1002/0471142727.mb25b10s94. [DOI] [PubMed] [Google Scholar]
- 153.Coskun AF, Cai L. Dense transcript profiling in single cells by image correlation decoding. Nat. Methods. 2016;13:657–660. doi: 10.1038/nmeth.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu Y, et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell. 2020;183:1665–1681.e18. doi: 10.1016/j.cell.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dobnikar L, et al. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat. Commun. 2018;9:4567. doi: 10.1038/s41467-018-06891-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Flores AM, et al. Pro-efferocytic nanoparticles are specifically taken up by lesional macrophages and prevent atherosclerosis. Nat. Nanotechnol. 2020;15:154–161. doi: 10.1038/s41565-019-0619-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wolf D, et al. Pathogenic autoimmunity in atherosclerosis evolves from initially protective apolipoprotein B100-reactive CD4(+) T-regulatory cells. Circulation. 2020;142:1279–1293. doi: 10.1161/CIRCULATIONAHA.119.042863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dixit A, et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Replogle JM, et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol. 2020;38:954–961. doi: 10.1038/s41587-020-0470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xie S, Duan J, Li B, Zhou P, Hon GC. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell. 2017;66:285–299.e5. doi: 10.1016/j.molcel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 161.Datlinger P, et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rubin AJ, et al. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell. 2019;176:361–376.e17. doi: 10.1016/j.cell.2018.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]