Dear editor,
Coronavirus disease-19 (COVID-19), which was caused by Severe Acute Respiratory Syndrome-related Coronavirus 2 (SARS-CoV-2), has been a global pandemic. The first key step of SARS-CoV-2 entry is attachment and binding to cell receptor angiotensin-converting enzyme 2 (ACE2) by spike protein (S).1 Integrin family consists of 18 α and 8 β subunits, which form tens of transmembrane heterodimers. It has been determined that many integrins associated with other proteins by recognizing RGD or KGD peptide. Previously we proposed an inhibitory role for integrin in the receptor targeting of SARS-CoV-2.2 Both S protein of SARS-CoV-2 and ACE2 have an RGD in their sequences, with ACE2 possessing a KGD.2 Based on the integrin-binding motif in S sequence, other researchers hypothesized that integrin could be an alternative receptor for SARS-CoV-2.3 However, the real function of integrin in SARS-CoV-2 entry is not determined experimentally.
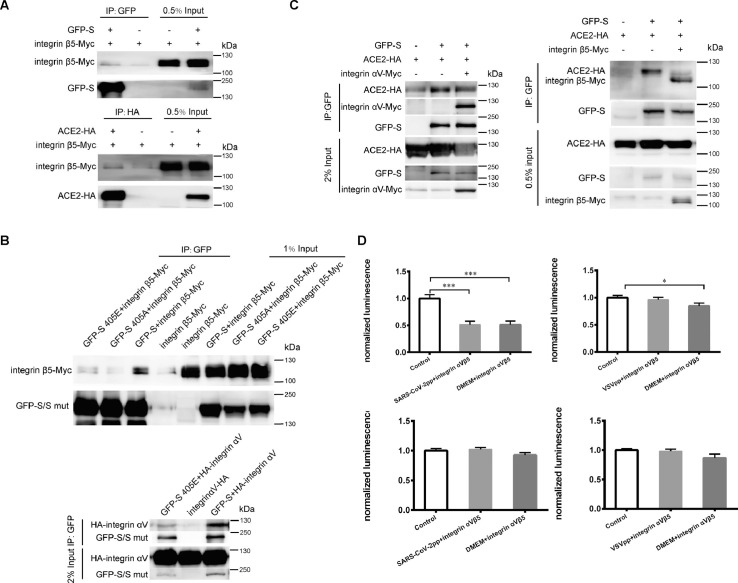
To verify the interaction of integrin with SARS-CoV-2 S and ACE2, we over-expressed these proteins in 293T cells and performed co-immunoprecipitation (co-IP). Integrin β5 was found to interact with S and ACE2 (Fig. 1 A). The association of integrin αV with S and ACE2 was also confirmed (Fig. S1). These results indicated that integrin can interact with the S protein of SARS-CoV-2 and ACE2, respectively. Because RGD/KGD residues are classical integrin-binding motifs, we went on to determine the dependence of these motifs in S-integrin and ACE2-integrin interactions. We constructed RGE and RGA mutants of S. Co-IP showed that either RGE or RGA bound with integrin β5 less (Fig. 1B), suggesting that RGD motif was important for integrin binding. S 405E mutation significantly suppressed integrin αV binding (Fig. 1B). The KGD motif in ACE2 was mutated to KGE and KGA. The amount of mutant ACE2 co-immunoprecipitated with integrin αV and β5 was lower than wild type ACE2 (Fig. S2), indicating that KGD residues was involved in ACE2-integrin interaction.
Fig. 1.
Integrin inhibited SARS-CoV-2 pseudovirus infection through inhibition of S-ACE2 binding. (A) Integrin β5 interacted with SARS-CoV-2 S and ACE2. Top, Myc-tagged integrin β5 with or without GFP-tagged S was transfected into 293T cells. Cell lysates were immunoprecipitated using anti-GFP antibody. Bottom, integrin β5-Myc was expressed in 293T cells with or without ACE2-HA. Cell lysates were precipitated with anti-HA antibody. (B) RGD motif in S protein was important for its binding with integrin β5 and integrin αV. Top, Wild type S and two single-amino acid mutants (405E, 405A) were expressed in 293T cells receptively with integrin β5-Myc. Bottom, wild type S or S 405E mutant was expressed in 293T cells. Lysates was immunoprecipitated by GFP. (C) SARS-CoV-2 S and ACE2 interaction was inhibited by integrin αV and integrin β5. Left, GFP-tagged full-length S and HA-tagged ACE2 were co-expressed in 293T cells with or without Myc-tagged integrin αV. Right, GFP-S and ACE2-HA were transfected with or without integrin β5-Myc. Cell lysates were immunoprecipitated by GFP antibody. (D) Integrin αVβ5 protein inhibited SARS-CoV-2 pseudovirus infection. 293T cells were transiently transfected with ACE2-expressing plasmid. Left top: SARS-CoV-2 pseudovirus and ACE2-expressing cells were separately treated with recombinant human integrin αVβ5 protein. Then the 293T cells were infected by SARS-CoV-2 pseudovirus for 48 h and pseudovirus entry was indicated with luciferase activity. Left down: Cell viabilities were measured. Right top: ACE2-expressing 293T cells and VSVpp were pretreated with integrin αVβ5 protein. Then the cells were infected with integrin αVβ5 conditioned or unconditioned VSVpp. Luciferase activity was measured after 48 h. Right down: Cell viability under two conditions were not affected.
Having confirmed the interaction of integrin with S and ACE2, we proposed that integrin potentially interrupted receptor targeting of SARS-CoV-2 S. To test our hypothesis, we transfected plasmids expressing ACE2 and S into 293T cells in the presence or absence of integrin. GFP-tagged S co-precipitated less ACE2 in the presence of integrin αV (Fig. 1C). Co-IP using S, ACE2 and integrin β5 came to the same result (Fig. 1C). These results demonstrated that integrin could interfere with the binding of ACE2 and SARS-CoV-2 S protein.
The inhibitory role of integrin in S-ACE2 binding suggested that the integrin could perturb the entry of SARS-CoV-2. Taking advantage of pseudovirus system which has been successfully established in our lab, we went further to determine the role of integrin in SARS-CoV-2 entry. This SARS-CoV-2 pseudovirus particle (SARS-CoV-2pp) was packaged by four plasmids expressing firefly luciferase, gag, rev, S protein, respectively. The S is changed to envelope G protein of vesicular stomatitis virus (VSV) for constructing VSVpp. We first tested whether integrin itself supported SARS-CoV-2 entry by transfecting 293T cells with integrin αV only. In contrast to 293T cells transfected with human ACE2, 293T cells expressing integrin αV produced undetectable luciferase activity signal (Fig. S3). This result indicated that integrin was not a receptor for SARS-CoV-2 pseudovirus. Then we examined the effect of exogeneous integrin on SARS-CoV-2 entry into 293T cells expressing ACE2. Compared to unconditioned group, pretreatment of either SARS-CoV-2pp or cells with exogenous integrin αVβ5 reduced luciferase activity (Fig. 1D). However, incubating VSVpp with integrin αVβ5 did not affect VSVpp entry (Fig. 1D). Pretreating 293T cells slightly inhibited VSVpp entry. Integrin αVβ5 addition did not change cell viability in all these conditions (Fig. 1D).
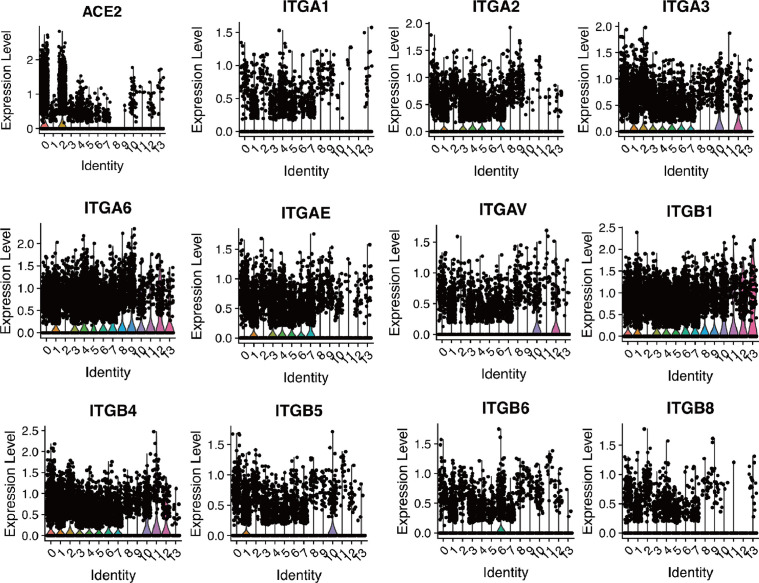
Gastrointestinal symptoms have been described in COVID-19 patients.4, 5, 6, 7 However, the incidence rate of diarrhoea in COVID-19 patients is low.8 We evaluated the expression of SARS-CoV-2 receptor ACE2 in intestinal tract by analyzing single-cell sequencing data (GSE125970) containing 14,537 human intestinal cells. 14 different cell clusters were verified in intestinal tissues. ACE2 was found mainly in intestinal epithelial progenitor cells and intestinal epithelial cells. Since integrin plays an inhibitory role in the receptor targeting of SARS-CoV-2, we examined the expression of various integrin subtypes in intestinal tract and found broad distribution of integrin A1, A2, A3, A6, AE, AV, B1, B4, B5, B6 and B8 in all 14 cell clusters (Fig. 2 ). In this case, the high level of intestinal integrin may be related to rare diarrhoea in COVID-19 patients.
Fig. 2.
Single-cell analysis of ACE2 and different integrin subtypes expression in human intestine. The expression of ACE2, integrin A1 (ITGA1), A2(ITGA2), A3(ITGA3), A6(ITGA6), AE(ITGAE), AV(ITGAV), B1(ITGB1), B4(ITGB4), B5(ITGB5), B6(ITGB6) and B8(ITGB8) in 14 cell populations were presented by violin plots. 0, small intestine enterocyte progenitor cell; 1, large intestine paneth-like cell; 2, small intestine enterocyte; 3, large intestine stem cell; 4, small intestine stem cell; 5, small intestine paneth cell; 6, large intestine enterocyte; 7, small intestine transit amplifying cell; 8, large intestine goblet cell; 9, small intestine goblet cell; 10, large intestine enterocyte progenitor cell; 11, large intestine enteroendocrine cell; 12, large intestine transit amplifying cell; 13, small intestine enteroendocrine cell.
In conclusion, our work found that both αV and β5 subtypes of integrin inhibited S-ACE2 interaction and entry of SARS-CoV-2pp by binding to S and ACE2, which largely relied on RGD/KGD motifs. ACE2 has been proven to mediate SARS-CoV-2 entry into human cells by researches on multiple levels. ACE2 was found highly expressed in digestive system by single-cell RNA sequencing, while diarrhoea was not often reported in patients infected with SARS-CoV-2. This raised a question about the existence of inhibitory factors. The integrin-binding motif in S sequence makes integrin on plasma membrane become a candidate mediator. Our research for the first time experimentally identified the association between integrin and SARS-CoV-2 S. At the same time, co-IP results showed that both two types of integrin interacted with ACE2. These results indicate that integrin could play a role in S-ACE2 binding, which may not be promoting effect but inhibitory function. To verify our hypothesis, we performed co-IP in the presence of integrin and confirmed the inhibitory role of it in S-ACE2 binding. Integrin kills two birds with one stone that neutralizing virus by binding to S, meanwhile, masking the receptor by binding to ACE2. In accordance with co-IP results, less SARS-CoV-2pp entered into 293T cells when the virus and cells were preincubated with recombinant integrin αVβ5 protein. This result not only demonstrated the inhibitory effect of integrin on SARS-CoV-2 entry, but also provided a strategy for antivirus research. Combined with our sc-RNA analysis which showed high expression levels of multiple integrin types in intestine, the low incidence of diarrhoea in COVID-19 patients may be partially explained. The function of integrin in virus-host interaction may need re-defined.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China [82072270 and 81871663], Shandong Provincial Natural Science Foundation [ZR2020QH273], and Academic Promotion Programme of Shandong First Medical University [2019LJ001]. We thank Dr. Ping Zhao and Zhaohui Qian for reagents.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2021.07.007.
Appendix. Supplementary materials
References
- 1.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luan J., Lu Y., Gao S., Zhang L. A potential inhibitory role for integrin in the receptor targeting of SARS-CoV-2. J Infect. 2020;81:318–356. doi: 10.1016/j.jinf.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigrist C.J., Bridge A., Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 2020;177 doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin X., Lian J.S., Hu J.H. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang W., Feng Z., Rao S. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 6.Lin L., Jiang X., Zhang Z. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 7.Song Y., Liu P., Shi X.L. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69:1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- 8.Mao R., Qiu Y., He J.S. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.