Abstract
The current outbreak of coronavirus disease (COVID-19) has led to creating a public health emergency conditions since 2019. COVID-19, which is caused by SARS-CoV-2, is spread via human-to-human transmission by direct contact or droplets. Through conducting this study, we were looking for detecting SARS-CoV-2 in wastewater produced in Iran country (Ardabil, Nir, Khalkhal, and Kowsar) (wastewater collection network, wastewater treatment plant, and hospital wastewater). In this research, samples (n=76) were collected from influent and effluent of municipal and hospital wastewater treatment plants, and some samples were also collected from Ardabil municipal wastewater manholes. The sampling duration included the white (lower risk of COVID-19) and red (high risk of COVID-19) conditions. Samples were stored at −20 °C for further diagnostic tests. The specific primer and probe real-time reverse transcriptase-polymerase chain reaction (real-time PCR) targeting ORF1ab and N genes (nucleoprotein gene) were applied to detect viral genomes of the SARS-CoV-2 virus in the wastewater samples. Out of 76 samples, a total of 15 samples (19.73%) collected from wastewater in Ardabil province (Ardabil, Nir, Khalkhal, and Kowsar), were positive in terms of SARS-CoV-2. Wastewater epidemiology can facilitate detection of the incidence of pathogens through metropolises, measurement of population prevalence without direct testing, and provision of information to the public health system about the efficiency of intervening efforts.
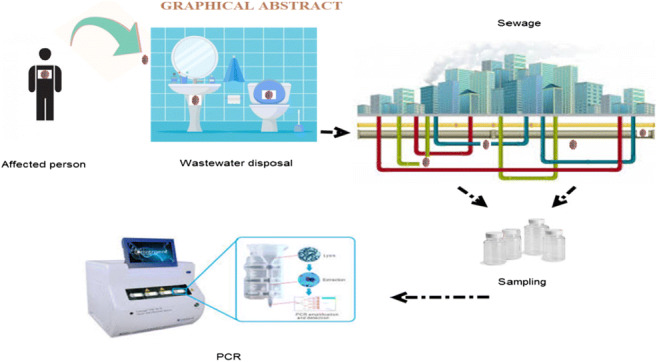
Graphical abstract
Keywords: SARS-CoV-2, Municipal wastewater, Coronavirus, Wastewater treatment plant, Collection network, Hospital wastewater
Introduction
Coronaviruses are single-stranded positive-sense RNA viruses, which are a member of the family Coronaviridae and the order of Nidovirales and are widely distributed in human and mammalian species (Kalan et al. 2020a; Nghiem et al. 2020; Sarailoo et al. 2021). Despite the mildness of most coronavirus infections, outbreaks of both beta-coronaviruses have been associated with acute respiratory syndrome coronavirus (SARS-CoV) (Drosten et al. 2003; Karami et al. 2021;Ksiazek et al. 2003; Kuiken et al. 2003) and Middle East acute respiratory syndrome coronavirus (MERS-CoV) (Dargahi et al. 2021b; de Groot et al. 2013; Zaki et al. 2012). These two diseases have led to the death of a large number of people (>10,000) in the last two decades (Zandian et al. 2021). Their death rates are reported to be 10% and 37%, respectively. In December 2019, a series of unknown cases of pneumonia, which had medical symptoms similar to viral pneumonia appeared in Wuhan, China. According to advanced sequencing analysis of lower respiratory tract samples, a new coronavirus known as the new coronavirus 2019 was detected (Dargahi et al. 2021a; Dargahi et al. 2021b; Munster et al. 2020). In the early stages of this pneumonia, symptoms of severe acute respiratory infection appeared, which in some patients developed acute respiratory distress syndrome (ARDS), severe respiratory failure, and other complications (Kalan et al. 2020b). As reports have indicated, clinical symptoms of patients with COVID-19 include cough, fever, sore throat, diarrhea, fatigue, and shortness of breath (Chen et al. 2020; Michael-Kordatou et al. 2020; Vosoughi et al. 2021).
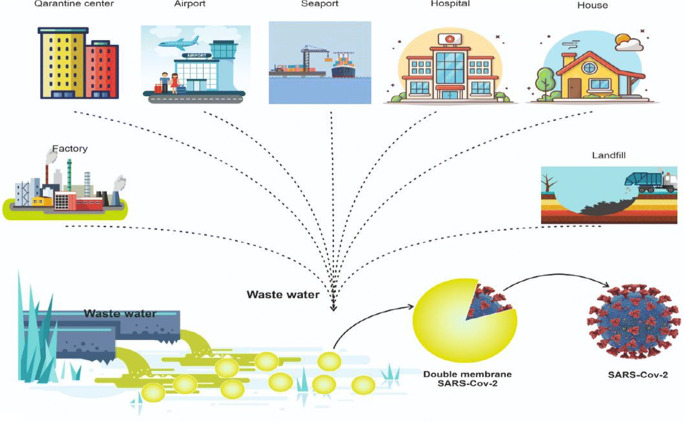
The occurrence of current epidemics around the world has been associated with growing knowledge about the risk of exposure to emerging pathogens during the collection and treatment of wastewaters. The entrance of emerging pathogens into wastewater systems may occur through the following ways including pathogen shedding in human waste, release of contaminated wastewater, illegal activity, animal farming, and hospital effluents, or surface water runoff from a wide area following a biological incident. Some emerging pathogens (e.g., Ebola [EBOV] virus and SARS-CoV-2) are serious health threats, so that serious health consequences may be raised due to their discharge to the sewage system (Lahrich et al. 2020; Mallapaty 2020). Therefore, the assessment of the possibility of exposure to virus and its transmission through sanitation systems is of great importance. Previous studies have been indicative of the detection of SARS-CoV-2 and its RNA in sewage excreted from people’s saliva, sputum, and feces (Waggoner et al. 2020). The inhalation by person-to-person transmission and aerosol/droplet, as well as fomite and hand contamination have been mentioned as foremost pathways for transmission of virus (Manoj et al. 2020). Nonetheless, current data suggest that a better understanding of the role of wastewater, which is considered as a possible source of epidemiological data and as a risk factor for public health, is an important subject. Detecting the SARS-CoV-2 in feces has led to conducting multiple research projects throughout the world to encourage the examination of wastewater for evaluating its circulation in populations (Lodder and de Roda Husman 2020; Mallapaty 2020).
Scientific evidence of SARS-CoV-2 ribonucleic acid (RNA) has also been perceived not only in the feces of patients with symptoms but also in asymptomatic patients with COVID-19, and its transfer to wastewater has been confirmed (Cai et al. 2020; Gao et al. 2020; Holshue et al. 2020; Wu et al. 2020). These studies have shown the possibility of the presence of virus in municipal sewage of communities infected by virus. Wastewater-based epidemiology (WBE) has been introduced as an auspicious way for realizing the prevalence of viruses for a population at wastewater treatment plants (WWTPs) because the wastewater contains the viruses that enter the wastewater treatment plant and are excreted in the environment from the individual with and without symptoms (Sinclair et al. 2008; Xagoraraki and O’Brien 2020). According to previous reports provided by the studies conducted on intestinal viruses (e.g., norovirus, hepatitis A virus, and poliovirus), the early warning of disease outbreaks and awareness of the effectiveness of public health interventions is possible using mentioned approach, i.e., WEB (Asghar et al. 2014; Hellmér et al. 2014). In Iran, this disease was first announced in February 2019. During the conducting this study and its completion (June 6, 2021), Iran with over 3,167,741 confirmed COVID-19 infections and more than 83,845 deaths was ranked 13th in the world in terms of coronavirus (https://www.worldometers.info/coronavirus/). Due to the impossibility of simultaneous testing of all members of the community, dozens of research groups around the world are examining wastewater analysis methods as a way for estimating the total number of individuals infected with the new coronavirus. Scientists believe that these methods can be of great value, especially if COVID-19 returns to communities. In addition, the virus in the sewage can be transmitted to humans in various ways and cause disease again in exposed individuals. The studies showed that coronavirus was able to survive in faces for 3 days and in raw chlorine-free water and hospital wastewater for 2 days at 20 °C. If wastewater is not treated, the virus can enter surface and groundwater and thereby cause environmental pollution and disease in humans (Gundy et al. 2009; Lodder and de Roda Husman 2020; Medema et al. 2020; Naddeo and Liu 2020).
Although several studies in different countries of the world (such as the Netherlands, the USA, and Australia) have reported molecular detection of SARS-CoV-2 in wastewater treatment plant samples (Lodder and de Roda Husman 2020; Medema et al. 2020; Waggoner et al. 2020; Wu et al. 2020), no reports for detecting SARS-CoV-2 in Iranian wastewater have been provided. As no study has so far been conducted on the identification of SARS-CoV-2 in wastewater collection network, wastewater treatment plants, and hospital wastewater, the aim of conducting this study was the identification of SARS-CoV-2 in wastewater produced in Ardabil, Nir, Khalkhal, and Kowsar (wastewater collection network, wastewater treatment plant, and hospitals wastewater).
Material and method
Approval statement
This study was confirmed to by the ethical code IR.ARUMS.REC.1399.249 in agreement to the ethical concept and the general custom and standards for accompanying medical research in Iran.
Specimen collection, storage, and transfer
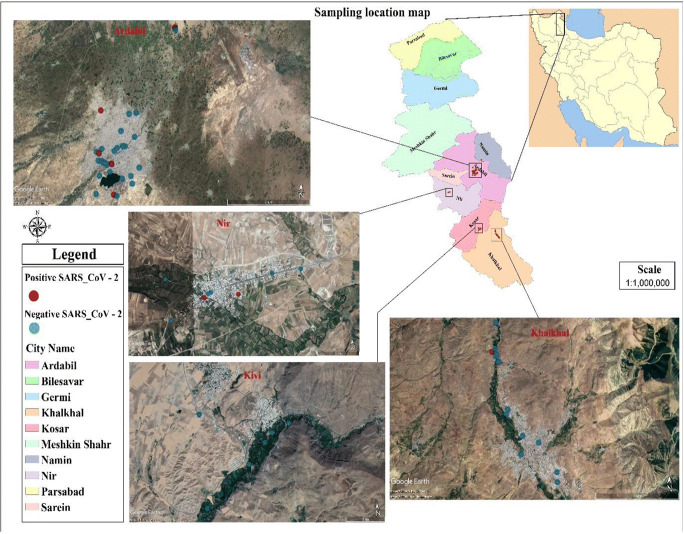
In this project, wastewater samples were collected from influent and effluent of municipal and hospital wastewater treatment plants, and some samples were also collected from Ardabil metropolitan wastewater manholes. Ardabil province is one of the northwestern Iran with a human population of about 625000 people. Some parts of the province are affected by the coronavirus and are often considered as red zones alert. Ardabil province has 10 cities, which are listed in Fig 1.
Fig. 1.
Position of different sampling points in this study
The sampling period included the white (lower risk of COVID-19) and red (high risk of COVID-19) situations. The location of sampled points in the present study are presented in Fig 1. To collect samples, staff used standard personal protective equipment (PPE) (such as long pants, security boots, hardhat, goggles, and protection gloves) in agreement with the health procedure for sampling from sewage (Waggoner et al. 2020). The sample size for each infected manhole was 250 ml that was dispensed and preserved in special closed glass bottles. It should be noted that all sampling steps were accomplished according to the standard technique (Federation and Association 2005). The sample size was selected based on the number of Ardabil towns, hospitals and wastewater treatment plants. We also followed the sampling procedures of similar studies to determine the number of samples needed. In the present study, 76 samples were taken from the main manholes and important points of all Ardabil cities. Samples were collected using the composite sampling method (by sampling from a specific point at different times 10 am to 2 pm and combining them together). The samples were transferred to the laboratory on ice and stored at the −20° C in order to deactivate the bacterial activity and subsequently the degradation for further analyses. A 24-h sample of raw wastewater was taken from the wastewater and pasteurized at 60 °C for 90 min to inactivate the virus according to the hygienic protocol for sewage sampling.
RNA extraction method
The sample was then filtered through 0.45-μm pore size (Millipore Sigma) to remove bacterial cells, and supernatant of pre-centrifugation is applied to the filter device. Pre-centrifugation that is commonly used in concentration, to remove larger particles and fragments, could affect the recovery. To concentrate the samples isolated from the wastewater, 100–200 mL of the sample were centrifuged (Universal 320R) at 4750 g for 30 min, and then the supernatant was carefully removed without disturbing the stagnation of the material. Supernatant was then centrifuged at 3500 g for 15 min along with the centrifugal filter with a cutoff of 10 kDa. The containers including the concentrated sample were placed upside down on top of the sample filter cup and centrifuged at 1000 rpm for 2 min. We took 250 μL of the concentrated sample from the collection cup with a pipette and extracted the sample directly using the RNA extraction kit using the (high pure viral nucleic acid kit). Bacteriophage ø6 is used as external control in order to determine the effectiveness of RNA extraction. Appropriate primers and probes were designed for N and RDRP genes to perform real-time PCR. The following steps were performed for virus identification: At the first stage, the appropriate concentrations of reagents, temperature cycle, and sufficient number of replication cycles were observed for the initial screening stage. The specific primer and probe real-time reverse transcriptase-polymerase chain reaction (real-time PCR) targeting ORF1ab and N genes (nucleoprotein gene) were applied to detect viral genomes of the SARS-CoV-2 virus in the wastewater samples. Applied Biosystems™ Real-Time PCR System 7500 with software v2.0.5 was used to run the real-time PCR. Appropriate concentrations of the synthesis reaction for real-time PCR MasterMix are as follows: H2O (RNAse free) 1.1 μL, 2x reaction mix 12.5 μl, MgSO4 (50 mM) 0.4 μL, BSA (1 mg/mL) 1 μL, primer RdRP_SARSr-F, and 2019-nCoV_N F (10-μM stock solution) 1.5 μl, primer RdRP_SARSr-R and 2019-nCoV_NR (10-μM stock solution) 2 μL, probe RdRP_SARSr-P and 2019-nCoV_N P (10-μM stock solution) 0.5 μ, SSIII/Taq EnzymeMix 1 μL, total reaction mix 20 μL, template RNA, add 5 μL, and total volume 25 μL. Thermal cycler: 55°C 10′ 94°C 3′ 94°C 15″ 58°C 30″, 45× (Table 1). (Corman et al. 2020; Waggoner et al. 2020).
Table 1.
| Organisms | Target gene | Sequence (5′–3′) | Cycling parameters |
|---|---|---|---|
| SARS-CoV-2 |
Probe and primer ORF1a/b |
FACAGGTGGAACCTCATCAGGAGATGC-BBQ F-GTGARATGGTCATGTGTGGCGG R-CARATGTTAAASACACTATTAGCATA |
55°C 10′ 94°C 3′ 94°C 15″ 58°C 30″ 45x. |
|
Primer and probe N gene |
F-AAATTTTGGGGACCAGGAAC R-TGGCAGCTGTGTAGGTCAA PFAM-ATGTCGCGCATTGGCATGGA-BHQ |
55°C 10′ 94°C 3′ 94°C 15″ 58°C 30″ 45x |
Positive and negative control
Positive Control2019-nCoV-qPCR was used to monitor whether the qRT-PCR process worked properly in each detection run. Negative Control2019-nCoVqPCR was used to monitor whether there was any contamination for the RT-PCR course in each detection run. An internal control in the extraction step was used by Wuhan coronavirus N-gene kit (TIB Molbiol, Berlin, Germany) to authorize the extraction and PCR amplification procedure.
Running real-time PCR and data analysis
The micro tube containing extracted genome along with other reaction components to detect the SARS-CoV-2 virus was placed in the real-time thermal cycler, and then the real-time PCR results were interpreted based on data analysis criteria by the operator.
Results and discussion
Table 2 shows the specifications of all samples of SARS-CoV-2 collected in the wastewater treatment plants. SARS-CoV-2 samples collected in the wastewater treatment plants in two different white and red conditions were examined. According to the results presented in Table 2, when the area was a white zone for corona virus, the results of all the samples examined from different parts of the wastewater treatment plant were negative, and in the case of the red alert condition, the results of the samples were also positive. The results taken from hospitals wastewater are summarized in Table 2. A total of 15 contaminated samples of SARS-CoV-2 were collected from hospitals wastewater in Ardabil province (Ardabil, Nir, Khalkhal, and Kowsar) (Table 3). Samples taken from hospital wastewater in white condition in terms of coronavirus were examined. According to the presented results, two of the studied samples (Imam Khomeini Corona wards and Ardabil City Health Center) in the hospital wastewater were positive for the presence of coronavirus.
Table 2.
Identification of COVID-19 virus in wastewater treatment plants
| Wastewater treatment plant | Coron virus status | ||||
|---|---|---|---|---|---|
| White | Red | ||||
| Results | Results | Ct-ORF1ab gene | Ct-N gene | ||
| Ardabil and Nir | Inlet wastewater | Negative | Positive | 35.12 | 32.21 |
| Aeration basin | Negative | Positive | 36.31 | 32.68 | |
| Secondary sedimentation basin 1 (before chlorination) | Negative | Positive | 35.95 | 33.5 | |
| Secondary sedimentation basin 2 (before chlorination) | Negative | Positive | 35.72 | 32.3 | |
| Outlet wastewater (after chlorination) | Negative | Positive | 37.8 | 34.01 | |
| Khalkhal and Kowsar | Inlet wastewater | Negative | Positive | 36.31 | 33.17 |
| Secondary sedimentation basin | Negative | Positive | 35.46 | 31.61 | |
| Sludge drained into the bed | Negative | Positive | 36.73 | 32.48 | |
| Outlet wastewater (after chlorination) | Negative | Positive | 34.66 | 30.57 | |
Table 3.
Identification of COVID-19 virus in hospitals wastewater
| City | Hospitals | Results | Ct-ORF1ab gene | Ct-N gene |
|---|---|---|---|---|
| Ardabil and Nir | Imam Khomeini (Corona wards( | Positive | 38.45 | 33.68 |
| Imam Khomeini (Public wards( | Negative | - | - | |
| Imam Reza | Negative | - | - | |
| Ghaem | Negative | - | - | |
| Alavi | Negative | - | - | |
| Bouali | Negative | - | - | |
| Fatemi | Negative | - | - | |
| Isar | Negative | - | - | |
| Sabalan | Negative | - | - | |
| Ardabil City Health Center | Positive | 38.19 | 34.4 | |
| Nir City Health Center | Negative | - | - | |
| Khalkhal and Kowsar | Imam Khomeini Khalkhal | Negative | - | - |
| Imam Khomeini Kowsar (infectious wards) | Negative | - | - | |
| Imam Khomeini Kowsar (public wards) | Negative | - | - |
The results obtained from the identification of the SARS-CoV-2 virus in wastewater collection network are summarized in Table 4. A total of 4 contamination samples of SARS-CoV-2 were collected from wastewater collection network in Ardabil province (Ardabil, Nir, Khalkhal, and Kowsar) (Table 4). In fact, the samples collected from the sewage collection network were examined in white condition of coronavirus threat. According to the presented results, all the studied samples of Ardabil province wastewater collection network (Ardabil, Nir, Khalkhal, and Kowsar cities) (except 4 cases) were affirmed to be negative in terms of the presence of coronavirus.
Table 4.
Identification of COVID-19 virus in wastewater collection network
| City | Sample code | Results | Ct- ORF1ab gene | Ct- N gene | City | Sample code | Results | Ct-ORF1ab gene | Ct-N gene |
|---|---|---|---|---|---|---|---|---|---|
| Ardabil | 1 | Negative | - | - | Nir | 3 | Negative | - | - |
| 2 | Negative | - | - | 4 | Negative | - | - | ||
| 3 | Negative | - | - | 5 | Negative | - | - | ||
| 4 | Positive | 38.51 | 36.71 | 6 | Negative | - | - | ||
| 5 | Negative | - | - | Khalkhal | 7 | Negative | - | - | |
| 6 | Negative | - | - | 8 | Positive | 37.69 | 33.4 | ||
| 7 | Negative | - | - | 1 | Negative | - | - | ||
| 8 | Negative | - | - | 2 | Negative | - | - | ||
| 9 | Negative | - | - | 3 | Negative | - | - | ||
| 10 | Negative | - | - | 4 | Negative | - | - | ||
| 11 | Positive | 39.47 | 36.57 | 5 | Negative | - | - | ||
| 12 | Negative | - | - | 6 | Negative | - | - | ||
| 13 | Negative | - | - | 7 | Negative | - | - | ||
| 14 | Negative | - | - | 8 | Negative | - | - | ||
| 15 | Negative | - | - | 9 | Negative | - | - | ||
| 16 | Negative | - | - | 10 | Negative | - | - | ||
| 17 | Negative | - | - | Kowsar | 1 | Negative | - | - | |
| 18 | Negative | - | - | 2 | Negative | - | - | ||
| 19 | Negative | - | - | 3 | Negative | - | - | ||
| 20 | Negative | - | - | 4 | Negative | - | - | ||
| Nir | 1 | Negative | - | - | 5 | Negative | - | - | |
| 2 | Positive | 37.85 | 36.26 | 6 | Negative | - | - |
When the prevalence of coronavirus in the community is low, if the virus sample becomes positive in the community sewage, surveying the sewage contamination with the virus can be a means to monitor the spread of the virus in the community (Medema et al. 2020b). Despite the fact that RT-PCR detects SARS-CoV-2 in a large proportion of fecal samples, there are only two researches that have recorded infectious virus culture from stool (Wang et al. 2020). As a result, sewage does not appear to be a major transmission route for SARS-CoV-2. These findings have significant implications to determine how WBE may help identify diseases across communities and predict community incidence without the need for separate tests, as well as educate public health officials on the effectiveness of treatments. According to the records available, SARS-CoV-2 seems to have a poor level of environmental stability. These viruses can be inactivated or removed using traditional wastewater treatment methods. The viral RNA, on the other hand, can still be present in the treated wastewater (Amoah et al. 2020).
Since viral envelopes are susceptible to disconnection by detergents including bile salts, maximum enveloped viruses do not recruit infection in the gastrointestinal tract. While enteric coronaviruses are noteworthy exceptions, it is uncertain if these enveloped viruses will survive the harsh conditions of viral pathogenesis in the gastrointestinal tract. CoV’s RNA-producing machinery interacts with altered host membranes, such as double-membrane vesicles (DMVs), which are produced from membranes of coronavirus replication factories in the secretory pathway, which could contribute to coronavirus resistance in harsh environments. Convoluted membranes and DMVs, which are hallmarks of beta-CoV infections, as well as DMSs, which have not previously been identified for beta-coronaviruses, are among the interconnected modified membrane structures induced (Fig. 2). According to Figure 2, SARS-CoV-2 decant complete stools making a potential fecal-oral contamination of transmission to environment network. The finding of enteric envelope viruses in the environmental samples is exceptionally interesting. Most enveloped viruses do not initiate infection in the alimentary tract, because viral envelopes are susceptible to dissociation by detergents such as bile salts. Enteric coronaviruses are notable exceptions, but it is not known why these enveloped viruses can withstand the harsh conditions in the alimentary tract. The RNA synthesizing machinery of coronaviruses related with modified host membranes, such as double-membrane vesicles (DMVs) that are derived from membranes of the secretory pathway which may play a critical role in coronavirus resistance in harsh environmental conditions such as a wastewater. Bacteriophages share unique biological properties with the gastroenteric viruses and aid as replacements. Temperature is one of the major factors that manage the virion persistence in the environment. Several types of coronavirus wastewaters such as SARS-CoV-1, human coronavirus (HCoV 229E) and animal coronavirus as transmissible gastroenteritis (TGEV), feline infectious peritonitis virus (FIPV), and murine hepatitis virus A59 (MHV) to study their persistence at inconstant temperature conditions (4–25 °C). In vitro studies on SARS-CoV-1 in stool and urine sample, hospital and domestic wastewater network, and tap water exhibited virus persistence at moderately lower temperature condition (4 °C). SARS-CoV-1 keep at for 2 days at 20 °C, while for 14 days at 4 °C. On the conflicting, urine samples showed long time persistence (17 days) at 20 °C. Salt compounds existing in urine facilitated the virus to preserve osmotic pressure that is desirable for their persistence (Kumar et al. 2021).
Fig. 2.
Molecular mechanism persistent SARS-CoV-2 in wastewater treatment plant
Replication factories for coronavirus SARS-CoV-2 appear to be deactivated in water earlier than non-enveloped human enteric viruses that have been linked to the waterborne transmission. Temperature has been shown to be a key factor in viral survival in studies. While SARS-CoV-2 survives in medical and domestic wastewater for 2 days at 20 °C and 14 days at 4 °C, the future of SARS-CoV-2 in wastewater still seems to be unknown (Alygizakis et al. 2020). Recovery rates for enveloped viruses including SARS-CoV-2 are also acceptable. According to the findings of this research, all the different parts of Ardabil and Khalkhal wastewater treatment plants were red in corona red conditions. The main outputs of both cities were positive, which shows that following coronavirus through sewage is one of the best and most economical ways to detect coronavirus. Because through sewage, people with coronavirus can be identified with and without symptoms in different areas. Necessary precautions should be taken before different outbreaks in different areas. The lack of ideal external control criteria that could have the same characteristics as the SARS-CoV-2 was a significant shortcoming of the current protocol as defined.
As a limitation of this study, the lack of equipment such as ultracentrifuge and DNA sequencer can be further evaluated. More study is needed to adapt to enveloped viruses, as well as the sampling and concentration methods commonly carried out in aquatic media.
Conclusion
Our findings show that testing SARS-CoV-2 in wastewater is possible. These results can help decision-makers to make appropriate decisions on the main prevention measures such as social distancing and quarantine efforts based on wastewater catchment-level estimations of prevalence. The results showed that out of the total samples, 15 samples (19.73%) were positive for SARS-CoV-2. WBE is a suitable way to recognize the prevalence of viruses among the population based on wastewater treatment plants, because sewages contain the viruses from individuals with and without symptoms that enter the wastewater treatment plant.
Acknowledgements
This study was financially supported by Ardabil University of Medical Sciences (IR.ARUMS.REC.1399.249). Corresponding authors would like to express their gratitude to Iran National Science Foundation (INSF) for supporting this research under grant number (99004286).
Author contribution
Chiman Karami: conceptualization, methodology, validation, formal analysis, investigation, resources, supervision, funding acquisition. Abdollah Dargahi and Mehdi Vosoughi: methodology, validation, formal analysis, writing—original draft. Farhad Jeddi: methodology. Hamed Zandian: validation, formal analysis, supervision. Ali Normohammadi and Vahid Asghariazar: methodology. Ahamad Mokhtari: writing—original draft. Morteza Alighadri: formal analysis, investigation. Anoshirvan Sedigh: formal analysis, investigation.
Funding
Ardabil University of Medical Sciences (IR.ARUMS.REC.1399.249) and Iran National Science Foundation (INSF) (grant number: 99004286).
Availability of data and material
The dataset and analyzed during the current study are available from the corresponding authors on realistic demand.
Code availability
Not applicable
Declarations
Ethics approval
No ethical approval was required as all the data analyzed were publicly available.
Consent to participate
Not applicable
Consent for publication
Not applicable
Conflict of interest
The authors declare no competing interests.
Footnotes
Highlights
• Double-membrane vesicles may play a role in SARS-CoV-2 resistance in harsh conditions.
• 19.7 % of wastewater samples were positive for coronavirus.
• When the area was a white condition for coronavirus, the results of all the samples examined from different parts of the wastewater treatment plant were negative.
• In the case of the red condition, the results were positive.
• Two of the studied samples in the hospital wastewater were positive for the presence of coronavirus.
• All the samples of Ardabil province wastewater collection network except 4 cases were affirmed to be negative in terms of the presence of coronavirus.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Abdollah Dargahi, Email: a.dargahi29@yahoo.com.
Mehdi Vosoughi, Email: mvn_20@yahoo.com.
References
- Alygizakis N, Markou AN, Rousis NI, Galani A, Avgeris M, Adamopoulos PG, Scorilas A, Lianidou ES, Paraskevis D, Tsiodras S. Analytical methodologies for the detection of SARS-CoV-2 in wastewater: protocols and future perspectives. TrAC Trends Anal Chem. 2020;134:116125. doi: 10.1016/j.trac.2020.116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoah ID, Kumari S, Bux F. Coronaviruses in wastewater processes: source, fate and potential risks. Environ Int. 2020;143:105962. doi: 10.1016/j.envint.2020.105962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar H, Diop OM, Weldegebriel G, Malik F, Shetty S, El Bassioni L, Akande AO, Al Maamoun E, Zaidi S, Adeniji A. Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. J J Infect Diseases. 2014;210:S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Xu J, Lin D, Xu L, Qu Z, Zhang Y, Zhang H, Jia R, Wang X, Ge Y. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. J Clin Infect Diseases. 2020;71(6):1547–1551. doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V, Bleicker T, Brünink S, Drosten C, Zambon M (2020) Diagnostic detection of 2019-nCoV by real-time RT-PCR. World Health Organization 17
- Dargahi A, Jeddi F, Ghobadi H, Vosoughi M, Karami C, Sarailoo M, Hadisi A, Mokhtari SA, Haghighi SB, Sadeghi H. Evaluation of masks’ internal and external surfaces used by health care workers and patients in coronavirus-2 (SARS-CoV-2) wards. Environ Res. 2021;196:110948. doi: 10.1016/j.envres.2021.110948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargahi A, Jeddi F, Vosoughi M, Karami C, Hadisi A, Mokhtarie SA, Alighadri M, Haghighi SB, Sadeghi H. Investigation of SARS CoV-2 virus in environmental surface. Environ Res. 2021;195:110765. doi: 10.1016/j.envres.2021.110765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish ZA. Commentary: Middle east respiratory syndrome coronavirus (mers-cov): announcement of the coronavirus study group. J J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Günther S, Preiser W, Van Der Werf S, Brodt H-R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. J New England J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Federation, W.E.; Association, A . Standard methods for the examination of water and wastewater. Washington: American Public Health Association (APHA); 2005. [Google Scholar]
- Gao QY, Chen YX, Fang JY. 2019 novel coronavirus infection and gastrointestinal tract. J J Digestive Dseases. 2020;21:125–126. doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy PM, Gerba CP, Pepper IL. Survival of coronaviruses in water and wastewater. Food Environm Virol. 2009;1:10–14. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Hellmér M, Paxéus N, Magnius L, Enache L, Arnholm B, Johansson A, Bergström T, Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. J Appl Environ Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A. First case of 2019 novel coronavirus in the United States. J New England J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalan ME, Ghobadi H, Taleb ZB, Adham D, Cobb CO, Ward KD, Behaleh R, Fazlzadeh M (2020a) COVID-19 and beliefs about tobacco use: an online cross-sectional study in Iran. Environ Sci Pollut Res 7:1–9 [DOI] [PMC free article] [PubMed]
- Kalan ME, Ghobadi H, Taleb ZB, Ward KD, Adham D, Matin S, Fazlzadeh M, Narimani S (2020b) Descriptive characteristics of hospitalized adult smokers and never-smokers with COVID-19. Tob Induc Dis 18:46. 10.18332/tid/122759 [DOI] [PMC free article] [PubMed]
- Karami C, Normohammadi A, Dargahi A, Vosoughi M, Zandian H, Jeddi F, Mokhtari SA, Moradi-Asl E. Investigation of SARS-CoV-2 virus on nozzle surfaces of fuel supply stations in North West of Iran. Sci Total Environ. 2021;780:146641. doi: 10.1016/j.scitotenv.2021.146641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W. A novel coronavirus associated with severe acute respiratory syndrome. J New England J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, Van Amerongen G, Van Riel D, Laman JD, De Jong T, Van Doornum G, Lim W. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Alamin M, Kuroda K, Dhangar K, Hata A, Yamaguchi H, Honda R. Potential discharge, attenuation and exposure risk of SARS-CoV-2 in natural water bodies receiving treated wastewater. npj Clean Water. 2021;4:1–11. [Google Scholar]
- Lahrich S, Laghrib F, Farahi A, Bakasse M, Saqrane S, El Mhammedi M. Review on the contamination of wastewater by COVID-19 virus: impact and treatment. Sci Total Environ. 2020;751:142325. doi: 10.1016/j.scitotenv.2020.142325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W, de Roda Husman AM. SARS-CoV-2 in wastewater: potential health risk, but also data source. J Lancet Gastroenterol Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Manoj M, Kumar MS, Valsaraj K, Sivan C, Vijayan SK. Potential link between compromised air quality and transmission of the novel corona virus (SARS-CoV-2) in affected areas. Environ Res. 2020;190:110001. doi: 10.1016/j.envres.2020.110001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A (2020) Presence of SARS-Coronavirus-2 in sewage. J MedRxiv [DOI] [PubMed]
- Michael-Kordatou I, Karaolia P, Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J J Environ Chem Eng. 2020;8:104306. doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A novel coronavirus emerging in China—key questions for impact assessment. J New England J Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- Naddeo V, Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ Sci: Water Research & Technology. 2020;6:1213–1216. [Google Scholar]
- Nghiem LD, Morgan B, Donner E, Short MD. The COVID-19 pandemic: considerations for the waste and wastewater services sector. J Case Stud Chem Environ Eng. 2020;1:100006. doi: 10.1016/j.cscee.2020.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarailoo M, Matin S, Vosoughi M, Dargahi A, Gholizadeh H, Rajabi Damavandi M, Abbasi-Ghahramanloo A, Kamran A. Investigating the relationship between occupation and SARS-CoV2. Work. 2021;68:27–32. doi: 10.3233/WOR-205066. [DOI] [PubMed] [Google Scholar]
- Sinclair RG, Choi CY, Riley MR, Gerba CP. Pathogen surveillance through monitoring of sewer systems. J Adv Appl Microbiol. 2008;65:249. doi: 10.1016/S0065-2164(08)00609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosoughi M, Karami C, Dargahi A, Jeddi F, Jalali KM, Hadisi A, Haghighi SB, Dogahe HP, Noorimotlagh Z, Mirzaee SA (2021) Investigation of SARS-CoV-2 in hospital indoor air of COVID-19 patients’ ward with impinger method. Environ Sci Pollut Res 1–9. 10.1007/s11356-021-14260-3 [DOI] [PMC free article] [PubMed]
- Waggoner JJ, Stittleburg V, Pond R, Saklawi Y, Sahoo MK, Babiker A, Hussaini L, Kraft CS, Pinsky BA, Anderson EJ. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:1633–1635. doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Xiao A, Zhang J, Gu X, Lee WL, Kauffman K, Hanage W, Matus M, Ghaeli N, Endo N (2020) SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. J medRxiv [DOI] [PMC free article] [PubMed]
- Xagoraraki I, O’Brien E (2020) Wastewater-based epidemiology for early detection of viral outbreaks. Women in Water Quality: Springer
- Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. J New England J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zandian H, Sarailoo M, Dargahi S, Gholizadeh H, Vosoughi M, Dargahi AJW. Evaluation of knowledge and health behavior of university of medical sciences students about the prevention of COVID-19. Work. 2021;68:543–549. doi: 10.3233/WOR-203395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset and analyzed during the current study are available from the corresponding authors on realistic demand.
Not applicable