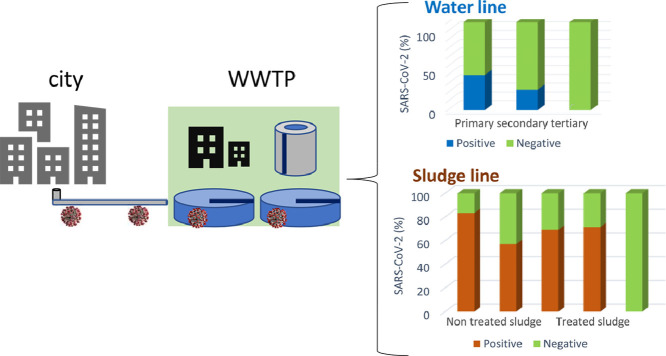
Abstract
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is shed in the feces of infected people. As a consequence, genomic RNA of the virus can be detected in wastewater. Although the presence of viral RNA does not inform on the infectivity of the virus, this presence of genetic material raised the question of the effectiveness of treatment processes in reducing the virus in wastewater and sludge. In this work, treatment lines of 16 wastewater treatment plants were monitored to evaluate the removal of SARS-CoV-2 RNA in raw, processed waters and sludge, from March to May 2020. Viral RNA copies were enumerated using reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) in 5 different laboratories. These laboratories participated in proficiency testing scheme and their results demonstrated the reliability and comparability of the results obtained for each one. SARS-CoV-2 RNA was found in 50.5% of the 101 influent wastewater samples characterized. Positive results were detected more frequently in those regions with a COVID-19 incidence higher than 100 cases per 100,000 inhabitants. Wastewater treatment plants (WWTPs) significantly reduced the occurrence of virus RNA along the water treatment lines. Secondary treatment effluents showed an occurrence of SARS-CoV-2 RNA in 23.3% of the samples and no positive results were found after MBR and chlorination. Non-treated sludge (from primary and secondary treatments) presented a higher occurrence of SARS-CoV-2 RNA than the corresponding water samples, demonstrating the affinity of virus particles for solids. Furthermore, SARS-CoV-2 RNA was detected in treated sludge after thickening and anaerobic digestion, whereas viral RNA was completely eliminated from sludge only when thermal hydrolysis was applied. Finally, co-analysis of SARS-CoV-2 and F-specific RNA bacteriophages was done in the same water and sludge samples in order to investigate the potential use of these bacteriophages as indicators of SARS-CoV-2 fate and reduction along the wastewater treatment.
Keywords: SARS-CoV-2, COVID-19, Wastewater, WWTP, Sludge
Graphical abstract
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), is responsible for the COVID-19 disease, a pandemic declared by the World Health Organization (WHO) in March 2020. SARS-CoV-2 belongs to the Coronaviridae family. Coronaviruses are enveloped, RNA single-stranded and positively charged viruses (at neutral pH). This family includes other human pathogenic viruses, such as SARS-CoV and MERS-CoV.
COVID-19 induces respiratory symptoms leading to a critical disease such as pneumonia in 14-17% of the cases (Jones et al., 2020). Besides, other symptoms include fever, cough, dyspnea and gastrointestinal symptoms (nausea, vomit, diarrhea). Accordingly, SARS-CoV-2 RNA has been detected in the respiratory tract, blood and feces of infected people (Jones et al., 2020). SARS-CoV-2 shedding by infected people has been observed in asymptomatic individuals (Park et al., 2020), patients during clinical symptomatology and after patient's recovery for a mean of 12.5 days and up to 33 days after respiratory samples reported negative results (van Doorn et al., 2020). However, the viral infectivity in these samples is still not well understood. For instance, Wölfel et al., (2020), suggested that no infective viral particles can be found in patients feces while they were present in the respiratory tract. In addition, Zang et al. (2020) demonstrated that SARS-CoV-2 released into the intestinal lumen were inactivated by simulated human colonic fluid, and infectious virus was not recovered from the stool specimens of COVID-19 patients. On the contrary, (Wang et al., 2020) and (Xiao et al., 2020) recovered infectious virus from stool samples tested positive for SARS-CoV-2 RNA.
The shedding of SARS-CoV-2 RNA in COVID-19 patients feces leads to the occurrence of viral genetic material in wastewater. The presence of SARS-CoV-2 RNA in untreated wastewater has been observed in different studies worldwide (Medema et al., 2020; Wurtzer et al., 2020; La Rosa et al., 2020; Nemudryi et al., 2020; Chavarria-Miró et al., 2021; Ahmed et al., 2020a)). The measurement of SARS-CoV-2 RNA in wastewater may provide helpful information regarding the COVID-19 epidemic trends in particular areas (Lodder and De Roda-Husman, 2020), through the approach named wastewater-based epidemiology, whose application has been suggested for surveillance of previous virus outbreak such as poliovirus (Bancroft, 1957; Wiley, 1962). A good correlation between SARS-CoV-2 RNA occurrence in wastewater and the reported COVID-19 incidence has been shown in different areas (Medema et al., 2020; Weidhaas et al. 2021; Peccia et al. 2020; Zahedi et al. 2020). Furthermore, SARS-CoV-2 RNA in wastewater has been detected in areas with low COVID-19 incidence (Randazzo et al., 2020) or in wastewater samples collected before the first cases were reported (Chavarria-Miró et al., 2021; Medema et al., 2020), highlighting the potential SARS-CoV-2 sewer surveillance as an early warning system (Peccia et al., 2020).
Although preliminary studies performed by Westhaus et al., (2021) suggest that wastewater is unlikely to be a major route of transmission for SARS-CoV-2 to humans, fecal transmission of SARS-CoV-2 has been suggested by two epidemiological studies (Kang et al., 2020; Yuan et al., 2020). However, the body of evidence is still low and to the best of the authors’ knowledge, there is to date no reported excess risk of SARS-CoV-2 infection in sewage workers. Improving the knowledge on the fate of SARS-CoV-2 genetic material in WWTPs would be useful to better characterize the risk related with discharges to the environment or wastewater reuse (Lesimple et al., 2020). The effectiveness of wastewater treatment technologies in reducing viruses has been previously documented (Oakley and Mihelcic, 2019), but information on the reduction of specific virus types such as SARS-CoV-2 is still scarce. Randazzo et al., (2020) determined the occurrence of SARS-CoV-2 RNA after secondary treatment, but it was not detected after tertiary treatment. Similarly, other authors highlighted the efficiency of disinfection treatment such as chlorination in the inactivation of SARS-CoV-2 (Collivignarelli et al., 2020).
Considerably less information is available concerning SARS-CoV-2 RNA presence in sludge. Few studies reported the occurrence of SARS-CoV-2 RNA in primary sludge (Balboa et al., 2021; Peccia et al., 2020). Besides, SARS-CoV-2 RNA has been also detected in both, primary and activated sludge in different WWTPs from Turkey (Alpaslan Kocamemi et al., 2020).
Given the widespread detection of SARS-CoV-2 RNA in raw wastewaters, there is the need for comprehensive studies to understand the role of wastewater treatment in the elimination of SARS-CoV-2 RNA and the protection of water bodies receiving the treated effluents. The present work reports the occurrence and quantification of SARS-CoV-2 RNA along the treatment lines of 16 WWTPs in Spain and France receiving wastewaters from populations presenting different COVID-19 incidence rates. Data are provided on the efficiency of different water treatment technologies in eliminating SARS-CoV-2 RNA, also the fate of SARS-CoV-2 RNA in sludge lines is discussed.
2. Material and methods
2.1. Sample collection
Sixteen WWTPs (8 wastewater treatment facilities in France and 8 in Spain) were sampled. Considering all WWTPs jointly, their water treatment capacity is higher than 10 million population equivalents. Samples were taken from mid-March until the end of May 2020, at different steps of the water treatment process. The characteristics of the monitored WWTPs are specified in the supporting information; Table S1 specifies the water and sludge treatment technology for each monitored WWTP as well as the samples collected at each site. Overall, considering all WWTPs investigated in the study, 164 water samples were analyzed including 101 non-treated and 63 treated wastewater samples, originating from primary, secondary and tertiary effluents. In addition, 107 sludge samples were analyzed, including 56 non-treated sludge and 51 treated sludge samples (thickened sludge, digested sludge and digested sludge plus thermal hydrolysis).
For water analysis, 0.5 L was collected from the WWTP influents and primary and secondary treatment effluents, whereas 20 L were obtained from tertiary treatment effluents. Influent water was collected with automatic samplers as 24 h integrated samples, and the rest of the water samples were collected using grab sampling techniques. Regarding sludge, 0.4 L was sampled from primary, secondary and mixed sludge, whereas 100 g were collected for solid sludge analysis. Both, water and sludge samples were transported under refrigerated conditions and stored at 5 ± 3°C until sample processing. Samples were processed within 24-48 hours after collection.
2.2. Sample processing
Sample concentration and RNA extraction were performed in five different laboratories depending on the sample's origin. The samples obtained from the WWTPs located in France were analyzed at the LCPME (CNRS and University of Lorraine, Nancy, France) and the CIRSEE (Suez, Le Pecq, France) laboratories while those from Spanish plants were analyzed in three different laboratories, i) University of Barcelona (Spain) for samples from the Barcelona area; ii) University of Santiago de Compostela (Spain), for samples from Ourense and iii) Labaqua (Alicante, Spain) for samples from Alicante, Sabadell and Murcia regions.
Slightly different protocols were followed for water and liquid sludge concentration, depending on the laboratory. Wastewater and liquid sludge samples from the French WWTPs were processed according to the paper by Bertrand et al. (2021), Spanish samples (except Ourense) were processed according to Hjelmsø et al., (2017). Finally, Ourense samples were processed as reported in Balboa et al., (2021). Details on the different processing protocols for wastewater, liquid and solid sludge are reported as supporting information.
RNA extraction from concentrated wastewater and sludge samples was performed in each laboratory according to the corresponding equipment manufacturer's instructions. Briefly, RNA extraction was done using NucliSENS® (BioMérieux) kit for those water samples obtained from French WWTPs. The RNA extraction from the Barcelona area WWTPs samples was done using a Maxwell ® RSC PureFood GMO and Authentication kit (Promega). For the rest of Spanish water samples RNA was extracted using QIAmp Viral RNA mini kit by (QIAGEN), except for Ourense WWTP samples that were extracted using STARMag 96 × 4 Universal Cartridge Kit (Seegene, Seoul, South Korea).
2.3. RT-qPCR analysis
SARS-CoV-2 detection and quantification was done through RT-qPCR analysis at the five different laboratories depending on samples origin (as mentioned in section 2.2).
For the French WWTPs, viral RNA samples were processed with the RNA UltraSense™ One-Step Quantitative RT-PCR System kit (invitrogen) and the SuperScript III One-Step RT-PCR System (Invitrogen) on a Stratagene MX 3005P real time PCR system (Agilent). The used SARS-CoV-2-related genes primer set included, “E” set developed by Corman et al. (2020) targeting the envelope protein E and “RdRp-IP4” set targeting part of the ORF1ab developed by Pasteur Institute (Paris, France; WHO, 2020). Both genes were quantified according to the method described in Bertrand et al. (2021). Samples from the WWTPs at the Barcelona area were analyzed using the IP4, E and N1 targets with the RNA UltraSense™ One-Step Quantitative RT-PCR System kit (Invitrogen). Samples obtained from Murcia, Sabadell and Alicante WWTPs were processed by an Applied Biosystems 7500 PCR system using the reaction mix Taqman Virus 1-step Master Mix (ThermoFisher; 4444436) combined with TaqMan 2019-nCoV Kit (ThermoFisher; A47532) for the analysis of three target genes, ORF1ab (RNApol), S gene, N gene. Finally, samples obtained from Ourense WWTP were characterized for viral RNA determination by a one-step multiplex RT-qPCR Allplex system™ 2019-nCoV (Seegene, Seoul, South Korea); the target genes were RdRP, N gene and E gene. The specificity of the target genes used in each laboratory has been previously reported in bibliography (references are described at table 1 for each target gene). All target genes had a high specificity for SARS-CoV-2, except for E gene that is specific for SARS-Coronavirus (Barra et al., 2020). Therefore, to minimize the risk of false positives results, a minimum of two out of all target genes used in each laboratory should give positive results to consider the sample as positive. For quantification purposes of SARS-CoV-2 genomic copies, ten-fold serial dilutions of a quantified SARS-CoV-2 RNA reference material (Twist synthetic SARS-CoV-2 RNA control 2 (MN908947.3; Ref.: 102024; Twist biosience) was used to prepare the standard curves to compare the Cycle threshold (Ct) values obtained in the analyzed samples. The highest concentration detected from the different targeted genes was reported for each positive sample.
Table 1.
Analytical performance parameters from the five laboratories involved in the study. Recoveries, detection and quantification limits, standard curves R2 and efficiency are reported as mean values per laboratory.
| Laboratory | Target genes (reference) | Standard curvesSlope | Inhibition assessment | Recovery (%) | Reference material | Detection limit complete process |
|---|---|---|---|---|---|---|
| Labaqua | ORF1ab (Lue et al., 2020) S gene (Lu et al., 2020) N gene (Lue et al., 2020) |
-3.22 -3.13 -3.49 |
Human RNasaP | 11-55% (n=158) | Accuplex SARS-CoV-2 Reference material kit | 7,2 × 102 gc/L |
| University of Barcelona | RdRp IP4, (Institut Pasteur, Paris) E (Charité, Berlin) N1 gene (CDC, Atlanta) |
-3.40 -3.44 -3.51 |
Dilution analysis* | 2.53% ± 0.17% (n=70) | SARS-CoV-2 synthetic RNA (MN908947.3, Twist Bioscience) | 1 × 102 gc/L for IP4, E and N1 genes. |
| LCPME (University of Lorraine) | RdRp IP4 (Pasteur Institute, Paris, France) E gene (Corman et al., 2020) |
-3.61 -3.28 |
FRNAPH GII | 55.8-64.0 % (n=4) | RdRP-IP4: RNA extracted from tested positive patients Gene E: eurofins genomics |
2 × 102 -2 × 103 for RdRp IP4 genes. 2 × 102 gc/L for E genes. |
| CIRSEE | RdRp IP4 (Pasteur Institute, Paris, France) E gene (Corman et al., 2020) |
-3.21 -3.47 |
FRNAPH GII | 12-116% (n=7) | RdRp -IP4 and E amplicons in genscript plasmid from in vitro transcribed RNA derived from strain BetaCov_Wuhan_WIV04_2019 (NC_004718.3) (Pasteur institute donation). (Pezzi et al., 2020) | 2 × 102 -2 × 103 for RdRp IP4 genes. 2 × 102 gc/L for E genes. |
| Univ. Santiago de Compostela | E, RdRp and N (not provided by kit) |
-3.24 | 33.3 % ± 15.6%. (n=50) | RT-qPCR Allplex system™ 2019-nCoV Reference material kit, | 1 × 103 gc/L |
Testing two replicates of the direct sample and two replicates of the one tenth dilution
To account for role of potential inhibitors blocking amplification, Amplification Controls, internal (IAC) or external (EAC) were included in each sample. Briefly, in the case of Labaqua samples a fragment of the human RNase P was amplified simultaneously with each specific RNA target (TaqManTM 2019nCoV Assay Kit v1). A mean value for RNase P was obtained by analyzing at least 30 negative samples. Then an acceptation interval was stablished as the media value of +/-3 times the standard deviation. Samples with RNase P outside this interval were considered inhibited (ISO 22174 2005). Regarding the University of Santiago de Compostela, internal control provided by the RT-qPCR kit (Allplex 2019-nCoV Assay test) was used in raw and diluted samples. In the University of Barcelona laboratory, TGV virus was used, as well as logarithmic dilutions (1/10). For French samples the estimation of RT-qPCR inhibition in the wastewater samples was based on the concentrations of F-specific RNA phages of genogroup II (FRNAPH GGII) detected by using the primers set “VTB4-F-GII” published by (Wolf et al., 2010). For raw wastewater, logarithmic dilutions (1/10 and 1/100) were performed in PCR grade water following viral RNA extraction. The RT-qPCR assay was then carried out on both undiluted and diluted RNA extracts. The percentage of inhibition was estimated for undiluted and 1/10 samples by taking the concentration obtained from the 1/100 samples as a reference, due to the high dilution of potential inhibitors. For sludges, values of concentrations of FRNAPH GGII phages were used as quality control to take in account role of potential inhibitors of the sample matrix.
In addition to SARS-CoV-2 RNA, F-specific bacteriophages RNA was characterized in some samples. Briefly, for the FRNAPH GGII genome, the VTB4-Fph GII set published by Wolf et al. (2010) was used with the SuperScript III One-Step RT-PCR System with 1 µM of each primer, 0.275 µM of probe in 20 µl final reaction volume with 2µl of RNA and 0,75 µl of Superscript III RT/Platinum Taq mix. On the Stratagene system, the RT was conducted at 55°C during 20 min followed by an initial denaturation at 94°C during 3 min and 45 cycles including denaturation step at 94°C during 15s, hybridization step at 58°C during 30s. Final cooling is done at 40°C during 30s.
2.4. Proficiency testing scheme
To evaluate the performance of the laboratories involved in the present work and to assess the reliability of the different methods used, a Proficiency Testing (PT) scheme was performed. The test was coordinated by ielab, a company accredited by Entidad Nacional de Acreditación (ENAC) for organizing PT schemes based on ISO/IEC17043 standard and the (CGA-ENAC-PPI 2020) guide. In order to perform the test, the supplied material included two homogeneous and stable samples in liquid format (1 mL vial each) containing natural SARS-CoV-2 and one plastic bottle containing the matrix (500 mL of secondary wastewater effluent). Regarding the analysis of the provided sample vials, one of them (vial A) had to be analyzed directly, to evaluate RNA extraction and RT-qPCR steps, while the other vial (B) had to be used to inoculate the provided matrix. In this case, the evaluation comprised the concentration of the inoculated sample matrix, RNA extraction and RT-qPCR steps.
All laboratories analyzed the two materials in triplicate providing qualitative and quantitative results. The PT scheme data pre-treatment was performed according to Murtula et al., 2012, including, (i) eliminate laboratories reporting false positive or negative results; (ii) make a logarithmic conversion (log10) of the results, which allows obtaining a Gaussian distribution (normal), and (iii) detect the outlier laboratories. The performance of the laboratories was assessed through a Z-score approach, as required by the (ISO 13528 2015) standard. The z-scores were calculated from the laboratory results as follows:
where x is the average of the logarithmic values of the results of a laboratory; X is the assigned value; and σpt is the Standard Deviation for Proficiency Assessment (SDPA). To establish the assigned value the participant's PT results were used (Murtula et al., 2012). SDPA defines the acceptable range variation of the laboratories on each assay. In this PT scheme, the SDPA value has been fixed to 0.75 Log by ielab Steering Committee to adequate it as much as possible to the fitness for purpose. This value was determined considering the standard deviation that is found in the concentration step and in the RT-qPCR/RT-PCR step.
The Z-score indicates how far away the score of each participant is from the mean in units of standard deviations and in which direction (positive or negative). The interpretation of the z-score is the following: | Z | ≤ 2 = satisfactory results; 2 <| Z | ≤ 3 = questionable results; and | Z |≥ 3 = unsatisfactory results (ISO 13528:2015).
2.5. Data treatment
Log removal values were calculated as follows:
Influent concentration refers to the SARS-CoV-2 genetic copies / L in the influent of each WWTP. Only those samples showing positive results for SARS-CoV-2 RNA in the influent were considered for the log removal calculation. Effluent concentration refers to SARS-CoV-2 genetic copies / L in the effluent of each corresponding treatment step. For solid sludge samples, the RNA concentration was expressed in genetic copies / Kg. Log removal was calculated for each day of analysis and the values are presented as mean ± standard deviation for each treatment analyzed. When virus RNA was not detected at the effluent of a given treatment, a value of 2 logs was used for log removal calculation corresponding to the average limit of quantification of all methods.
3. Results and discussion
3.1. Quantitative detection of SARS-CoV-2 in wastewater samples
The performance of the different RT-qPCR methodologies applied in the study is reported in Table 1. All methodologies provided good performance in terms of the slope of the standards, recoveries and detection limits.
3.2. Proficiency testing for the quantification of virus RNA in wastewater
The PT scheme included a total of 36 laboratories using a variety of different SARS-CoV-2 RNA detection and quantification RT-qPCR methods. The 5 laboratories participating in the present work, correctly reported qualitative results (positive/negative) values. Besides, from the 36 laboratories participating in the PT scheme, 19 reported quantitative results. In order to evaluate the performance of the participants regarding the quantified results reported, the Z-score was estimated. The obtained absolute Z-score value for all the laboratories involved in the present work was lower than 2 for vial A, which is intended to evaluate the RNA extraction and RT-qPCR steps. The same result (a Z-score lower than 2) was obtained also for all the laboratories participating in the present work when analyzing and quantifying vial B, which evaluates the concentration process, RNA extraction and RT-qPCR analysis. Therefore, the reliability of the results obtained for all the laboratories and the suitability of their analytical methods for SARS-CoV-2 RNA quantification in wastewater was confirmed as established in the (ISO 13528 2015) standard.
3.3. SARS-CoV-2 RNA in untreated wastewater and relationship with COVID-19 cases
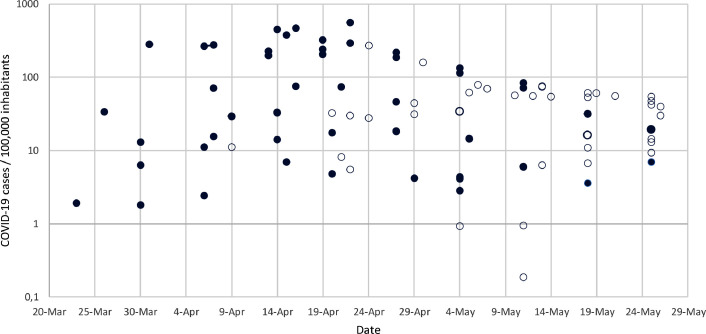
The occurrence and concentration of SARS-CoV-2 RNA in wastewater are expected to be related with the COVID-19 incidence rate in the corresponding area. It is hypothesized that the amount of SARS-CoV-2 RNA in wastewater is depending on the viral load shed in the corresponding area due to both, symptomatic and asymptomatic cases. Figure 1 shows all the influent wastewater samples collected from the different WWTPs in which SARS-CoV-2 RNA was either detected (filled dots) or not detected (empty dots) and the Y-axis indicates COVID-19 prevalence rate in the corresponding area at the time of sampling. Besides, Figure S1 shows the evolution of SARS-CoV-2 RNA quantification at the influent of each WWTP over time with the changes in the reported COVID-19 incidence values.
Fig. 1.
Influent wastewater samples from the different targeted WWTP. Y-axis represents the COVID-19 prevalence rate in the corresponding area at the time of sampling. Filled dots represent SARS-CoV-2 RNA positive samples whereas empty dots represent no SARS-CoV-2 RNA occurrence.
From the 101 wastewater influent samples collected, half of them (50.5%) showed positive results for SARS-CoV-2 RNA occurrence. Most of the wastewater influent samples (89.5%) collected from WWTPs located in areas with a COVID-19 incidence higher than 100 cases per 100,000 inhabitants presented positive results of SARS-CoV-2 RNA. The number of positive samples from regions with a COVID-19 incidence higher than 100 cases per 100,000 inhabitants was significantly higher than the ones reporting lower COVID-19 incidence, between 1 to 100 cases per 100,000 inhabitants (p-value < 0.05, Chi-squared test). Samples collected in these areas (COVID-19 incidence rate between 1 to 100 cases per 100,000 inhabitants) showed high variability, being about half of the samples collected positive for SARS-CoV-2 RNA. Finally, no positive samples were obtained when the incidence was lower than 1 case per 100,000 inhabitants, however only three samples were characterized under this COVID-19 prevalence.
The evolution of SARS-CoV-2 RNA concentration at the influent is represented for each WWTP with the corresponding incidence value at supporting information, Figure S1. Most of the plants showed a decreasing number of SARS-CoV-2 RNA copies along the sampling time. This decreasing trend could be related in most cases with a decrease in the reported COVID-19 cases in the corresponding regions along the sampling period. However, this trend did not apply to all target sites. It is important to note that the testing capacity to report the COVID-19 incidence, was more limited at the beginning of the pandemic (when samples were taken in France and Spain for the present study) than in the later months. For instance, the reported COVID-19 cases in French regions are explained at the department level, which in some cases, the disease trend may not be representative of the local variation of the WWTP sanitation area. Besides, some factors could provoke variability in the reported SARS-CoV-2 RNA concentration in wastewater. For instance, in a thorough assessment of uncertainty in SARS-CoV-2 prevalence estimation by wastewater-based epidemiology, Li et al (2021) pinpointed precipitation and the decay in sewers as major obstacles to prevent higher accuracy. Correlation between higher COVID-19 incidence and positive samples in wastewater has been observed in previous studies, although high variability in detection has been reported. Hart and Halden (2020) performed a detailed computational analysis, showing that the detection limits would range from 1 in 114 individuals to 1 in 2,000,000 individuals (spanning 5 orders of magnitude). Despite the fact that wastewater based epidemiology is out of the scope of this study, the obtained results are in line with previous works that indicated an association between health data and viral RNA quantification in wastewater (Medema et al., 2020; Randazzo et al., 2020; (Weidhaas, 2021)).
To further evaluate the evolution of SARS-CoV-2 RNA in wastewater treatment, only those WWTPs showing positive results in the influent were considered in the following sections.
3.4. SARS-CoV-2 RNA in WWTP water lines
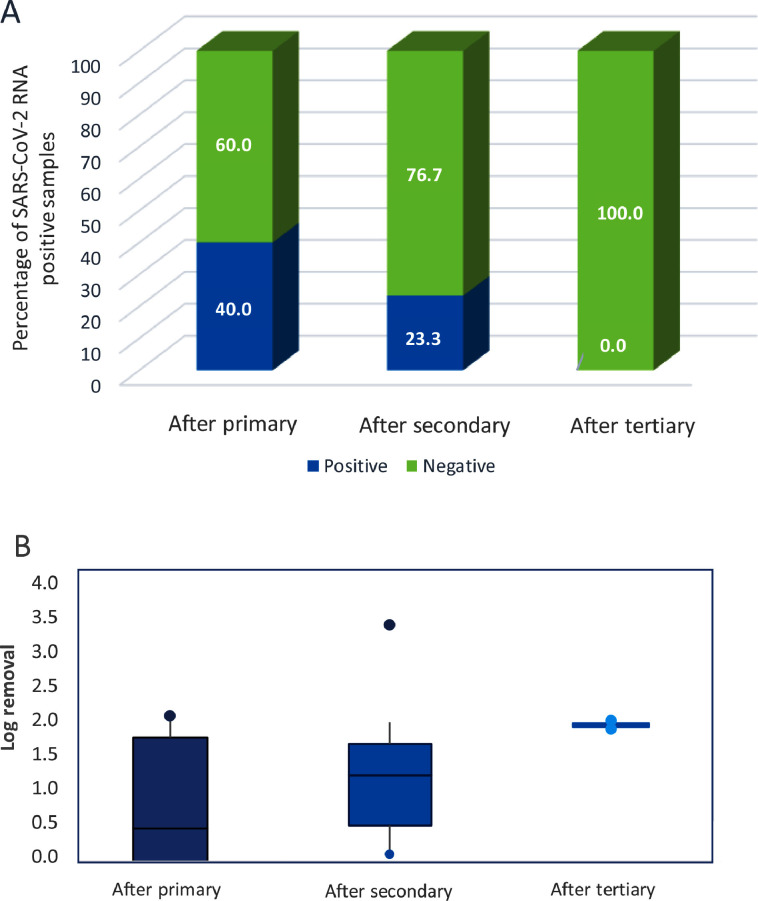
In order to evaluate the removal of SARS-CoV-2 RNA in WWTPs water lines, samples were taken at the effluent of different WWTPs secondary treatment (which considers the removal of primary plus secondary treatment) as it is the most common configuration for WWTPs worldwide. Besides, to understand the role of each treatment step, few samples were obtained after primary treatment only, whereas some of them were obtained after tertiary treatment (which considers primary, secondary and tertiary treatment). Figure 2 shows the occurrence of SARS-CoV-2 RNA at the different wastewater treatment steps, as well as the log removal.
Fig. 2.
The graph shows integrated results of all WWTPs monitored. Primary (n=5), secondary (n=30), tertiary (n=2). A) Percentage of positive and negative samples for SARS-CoV-2 RNA occurrence in each water treatment step and B) SARS-CoV-2 RNA log removal in each treatment step comparing influent and effluent concentration in each treatment.
The common configuration of WWTPs, primary plus secondary treatment, eliminated SARS-CoV-2 RNA from the 76,7% of the positive samples occurring at the influent (only 7 out of the 30 secondary treatment samples analyzed showed positive results) with a virus RNA removal up to 1.50 ± 0.80 logs (Figure 2, table S2). From this removal, a significant part might correspond to the primary treatment which allowed to eliminate SARS-CoV-2 RNA from the 60% of the positive samples analyzed (only 2 out of the 5 samples analyzed showed presence of SARS-CoV-2 RNA) with a mean log removal of 0.48 ± 1.17 log of magnitude (figure 2, table S2). Finally, no occurrence of virus RNA was determined after tertiary treatments (2 samples analyzed) achieving more than 1.97 ± 0.08 logs of reduction.
SARS-CoV-2 RNA occurrence was significantly reduced after the water treatment steps. However, our study showed that about 23% of the effluent samples from secondary treatments were tested positive for SARS-CoV-2 RNA. Previous studies also reported positive samples of SARS-CoV-2 RNA after secondary treatments, including two WWTPs from Israel (Abu Ali et al., 2020), two WWTPs in Spain (Randazzo et al., 2020) and one WWTP in Japan (Haramoto et al., 2020). On the contrary, other studies reported no occurrence of SARS-CoV-2 RNA after secondary treatment, despite positive samples measured in the inflow (Kumar et al., 2020; Sherchan et al., 2020). These results suggest that secondary treatments are not able to eliminate SARS-CoV-2 RNA in all cases. However, virus detection after treatment can be strongly influenced by the influent viral concentration, as well as by the nature of the secondary treatment applied. Abu Ali et al., (2020) detected SARS-CoV-2 RNA at the effluent of secondary treatment when the virus influent concentration was close to 6 log copies / L. A similar influent concentration was reported in one WWTP in Spain when positive samples for SARS-CoV-2 RNA in the effluent were detected. However, an additional positive effluent sample was reported in Spain when the influent concentration was lower than 2 log copies / L (Randazzo et al., 2020). Therefore, the presence of RNA in effluent from secondary treatment has been reported in the literature at various levels of RNA concentration at the influent (between 2 and 6 log copies / L). It is likely that the secondary treatment plays an important role in the final SARS-CoV-2 RNA elimination rate. However, the scarcity of reported data on secondary treatment inlet/outlet measurements hinders a more accurate analysis of the fate of SARS-CoV-2 RNA in this section, which would be needed to cope with the potential variability due to the different analytical methods applied or the variation of the wastewater characteristics from the different studies.
In order to evaluate potential differences in the elimination of SARS-CoV-2 RNA within different secondary treatments, the three secondary treatment technologies covered in the study, namely activated sludge, activated sludge plus nutrient removal and membrane bioreactor (MBR) were compared. For each of them, the detection occurrence and the log removal of SARS-CoV-2 RNA are shown in Figure 3 .
Fig. 3.
The graph shows integrated results of all WWTPs monitored. Activated sludge (n=11), activated sludge plus nutrient removal (n=11), MBR (n=8). A) Percentage of positive and negative samples for SARS-CoV-2 RNA occurrence in each treatment process and B) SARS-CoV-2 RNA log removal in each treatment process comparing influent and effluent concentrations after each treatment.
SARS-CoV-2 RNA was not detected in 63.6% of the samples after activated sludge followed by clarification (4 out of the 11 activated sludge effluent samples showed positive results). Detection in the effluent was less frequent in activated sludge plus nutrient removal followed by clarification where SARS-CoV-2 RNA was not detected in 81% of the samples analyzed after this treatment (2 out of the 11 activated sludge plus nutrient removal effluent samples showed positive results). Finally, there were no SARS-CoV-2 RNA positive samples in the effluent from the MBR treatment (8 samples analyzed) (Figure 3A). The same trend was observed when the log removals were calculated for each treatment; samples obtained after activated sludge and clarification, showed a mean value of 1.03 ± 0.59, followed by activated sludge plus nutrient removal and clarification, with 1.37 ± 0.72 and the highest log removal was obtained after MBR treatment being more than 1.97 ± 0.93 log (Figure 3, Table S2). Our results suggested MBR as the most effective secondary treatment in the elimination of SARS-CoV-2 RNA from the water line. However, a high variability was observed in the measures (presented as standard deviation) which may difficult to determine significant differences concerning the efficiency in the elimination of SARS-CoV-2 RNA within the different treatments.
Scarce information is available regarding the efficiency of water treatment technologies in the elimination of SARS-CoV-2. For instance, positive samples of SARS-CoV-2 RNA were observed after conventional activated sludge treatment in three WWTPs from Germany (Westhaus et al., 2021). Primary settling followed by activated sludge treatment provided an elimination rate close to 2 logs for SARS-CoV-2 (Abu Ali et al., 2020), which is slightly higher than the one reported in the present work. Previous studies showed a higher efficiency of MBR treatment compared to activated sludge process in the elimination of adenovirus (Amoah et al. 2020). Removal for different types of viruses including adenovirus, norovirus and F+ coliphages during MBR treatment has been reported as being between 2 and 7 logs (Amoah et al. 2020). These results of other viruses’ elimination rate agree with the ones found for SARS-CoV-2 removal in the present work; MBR treatment has been shown to be more effective than conventional sludge with a log reduction for SARS-CoV-2 in a similar range than the ones reported for other type of viruses. Secondary treatments including activated sludge and MBR can be effective in reducing SARS-CoV-2 and other type of viruses from the water line, due to the adverse conditions that viruses are exposed to in these treatments (Lesimple et al., 2020) and because of virus sorption to organic particles and further elimination by settling (Bogler et al., 2020). Further virus removal can be achieved by MBR through virus particle retention by membrane and cake layer (Bhatt et al. 2020).
In addition to the secondary treatment evaluation, our study provided insights on the role of primary treatment only in the removal of SARS-CoV-2 RNA, as well as the addition of a tertiary process in the water line. The obtained results on the primary treatment SARS-CoV-2 RNA removal of 0.48 ± 1.17 log are in the range of previously reported elimination for WWTPs primary treatment (Abu Ali et al. 2021) that observed an elimination close to 1 log. Whereas no detection of SARS-CoV-2 RNA was determined after the addition of chlorination, applied as tertiary treatment. There is a general agreement that tertiary treatments are effective in the elimination of SARS-CoV-2 from the water lines (Bogler et al., 2020; Lesimple et al., 2020; Randazzo et al., 2020). Specifically, chlorination has been previously determined as an effective treatment for the elimination of SARS-CoV-2 (Abu Ali et al., 2020). Nevertheless, further characterization of SARS-CoV-2 RNA occurrence after primary treatment only and the addition of a tertiary treatment should be performed to confirm the insight provided in the present work.
3.5. SARS-CoV-2 RNA in WWTP sludge lines
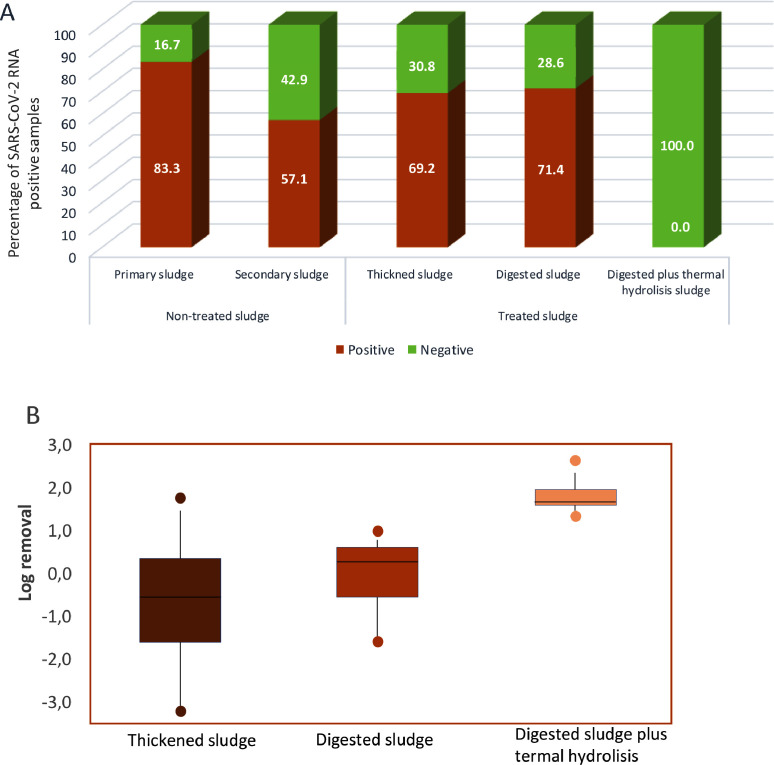
Sludge samples were obtained from those WWTPs showing SARS-CoV-2 RNA positive results in the water influent samples. Firstly, non-treated sludge was obtained from primary and secondary treatments and secondly, treated sludge was collected after sludge thickening, anaerobic digestion and anaerobic digestion plus thermal hydrolysis (Figure 4 ).
Fig. 4.
The graph shows integrated results of all WWTPs monitored. Primary sludge (n=6), secondary sludge (n=14), thickened sludge (n=13), digested sludge (n=7), digested sludge plus thermal hydrolysis (n=5) A) Percentage of positive and negative samples for SARS-CoV-2 RNA in sludge samples and B) Boxplot showing first and third quartiles and the median value of SARS-CoV-2 RNA log removal in each sludge matrix comparing influent and effluent samples after each treatment.
Non-treated sludge samples presented a high percentage of SARS-CoV-2 RNA occurrence, being 83% of the samples positive. However, a small percentage of positive samples at the influent were negative for SARS-CoV-2 RNA occurrence in primary sludge, that might be related to heavy rainfall and low solids content in the influent, as no inhibition was detected in those samples. In the case of secondary sludge, the 57% of the samples were positive (Figure 4A). These percentages of occurrence are consistent with the elimination observed in water treatments (with a detection in only 40% of the effluents after primary treatment and 23% after secondary treatment). This can be explained by the fact that virus particles can adsorb to solids and colloidal material because of the lipid bilayer surrounding the SARS-CoV-2 protein capsid (Balboa et al., 2021). In accordance, previous studies have already reported an important occurrence of SARS-CoV-2 RNA in primary and secondary sludge (Alpaslan-Kocamemi et al., 2020; Balboa et al., 2021; Peccia et al., 2020), and in some cases with concentrations higher than in water samples, which highlights SARS-CoV-2 affinity for solids.
Similarly to non-treated sludge, thickened and digested sludge also presented a high percentage of SARS-CoV-2 RNA positive samples, being 69% and 71%, respectively. In both, thickened and digested sludge, the reported viral log removal was negative, being mean values of -0.47 ± 1.20 and -0.17 ± 0.89, respectively (table S2). A negative log removal indicates higher viral RNA concentration after the treatment when compared to the entrance. The elimination of water at these steps certainly contributes to the increase in viral RNA concentration in these matrices. Previous studies reported an important occurrence of SARS-CoV-2 in thickened sludge and pinpointed it as a hotspot of virus RNA in the plant (Balboa et al., 2021). Besides, digested sludge showed an occurrence of virus RNA in a similar proportion than thickened sludge. To the best of the authors’ knowledge, no previous information regarding SARS-CoV-2 RNA in digested sludge is available. Previous studies with other human coronaviruses showed the occurrence of these types of viruses in sludge samples after anaerobic digestion (Bibby et al., 2011; Bibby and Peccia, 2013). These results pinpoint that human coronavirus genome can be found after digestive phases, in line with the results of the present work with SARS-CoV-2.
The only group of sludge samples with no detection of SARS-CoV-2 RNA was after anaerobic digestion followed by thermal hydrolysis. The high temperatures applied during this treatment (150–160°C saturated steam) allowed the complete elimination of viral RNA with a log removal higher than 1.69 ± 0.27. Therefore, sludge thermal hydrolysis enabled a complete inactivation of SARS-CoV-2 and guarantees sludge safeness if it is applied for agricultural purposes.
3.6. SARS-CoV-2 and F-specific RNA bacteriophages load along WWTP processes
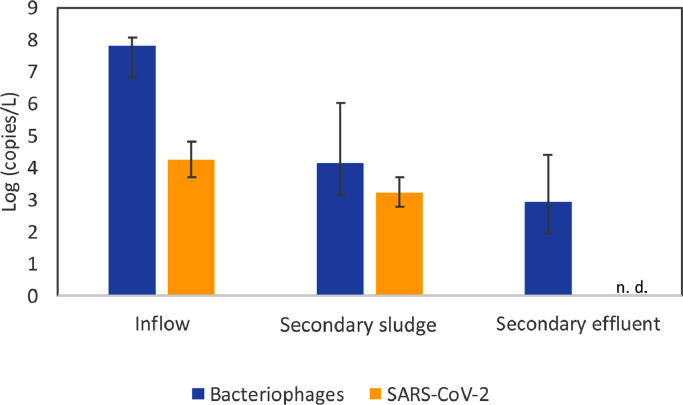
Bacteriophages are commonly used as viral indicators to determine the efficiency of water treatments and guaranty safe water use and reuse (Withey et al., 2005). Figure 5 shows the concentration of genome of both, SARS-CoV-2 and F-specific RNA bacteriophages in the WWTP14. This WWTP has a secondary treatment based on activated sludge. Water was sampled in the influent of the plant and in the effluent (after activated sludge and clarification). Secondary sludge was collected from the activated sludge process (Figure 5). Paired samples were taken from 4 different days in the WWTP target processes.
Fig. 5.
SARS-CoV-2 RNA and F-specific RNA bacteriophages concentration Log (copies/L) along the different steps of a selected WWTP. n. d. = not detected. Bars represent mean values (n=4) with the corresponding standard deviation.
The concentration of bacteriophages in the influent of the WWTP was higher than the concentration of SARS-CoV-2, with mean values of 7.80 ± 0.26 log copies/L and 4.26 ± 0.56 log copies/L, respectively. Bacteriophages presented also a higher concentrations than SARS-CoV-2 in activated sludge, although the difference in concentration between both viruses was reduced at this step, bacteriophages and SARS-CoV-2 presenting respectively a mean concentration of 4.19 ± 1.9 log copies/L and 3.23 ± 0.46 log copies/L in this matrix (Figure 5, Table S3). At the effluent of the plant (after activated sludge and clarification), SARS-CoV-2 RNA was not detected in any of the samples characterized, whereas bacteriophages were still detected in all of them with a mean concentration of 2.94 ± 1.52 log copies/L. The wastewater treatment line was able to eliminate SARS-CoV-2 down to below detection limits, but not bacteriophages. However, bacteriophages presented an initial concentration about three orders of magnitude higher than SARS-CoV-2. F-specific RNA bacteriophages may be valid indicators of viral elimination in water treatment due to its high initial amounts and measurable concentrations after the treatment process. Nevertheless, further studies would be needed to determine the sensitivity of both virus along the wastewater treatment process; considering if possible, higher concentrations of SARS-CoV-2 in the influent, enabling a detection of the virus also in the effluent, in order to compare more precisely the reduction of both viruses in the same conditions.
4. Conclusions
WWTPs significantly reduced the occurrence and concentration of SARS-CoV-2 RNA in the water lines. However, a complete removal was not observed in all cases in this study after secondary treatment, and viral RNA could be still detected after activated sludge and activated sludge plus nutrient removal followed by clarification. On the contrary, SARS-CoV-2 RNA was not detected after MBR and chlorination applied as a tertiary treatment.
SARS-CoV-2 RNA was detected in large proportions of all types of sludge, including sludge from primary and secondary treatments and treated sludge after thickening and anaerobic digestion. These results highlight that these treatments are not effective in eliminating SARS-CoV-2 RNA from sludge, while, only after applying thermal hydrolysis SARS-CoV-2 RNA was not detected.
The presence of virus RNA in the analyzed samples (wastewater and sludge) does not imply infectivity nor integrity. Further studies are required to estimate the infectious state of the virus in the effluents from secondary treatments and after sludge thickening and digestion to verify whether additional treatments are needed to protect the receiving environments.
Finally, F-specific RNA bacteriophages presented high concentrations at the influent which makes these viruses suitable viral indicators to determine its reduction along treatment processes. However, further research should be done in order to determine the potential of bacteriophages as indicators of SARS-CoV-2 elimination in water and sludge treatments.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This study was financed by SUEZ company. The authors would like to thank the Área Metropolitana de Barcelona (AMB), the city halls of Sabadell, Alicante and Ourense. Also, thanks to Entidad de Saneamiento y Depuración de Aguas Residuales de la Región de Murcia (ESAMUR) and to the water authorities from the regions covering the present work. Authors also acknowledge operators and local authorities (Métropole de Lyon, Eurométropole de Strasbourg, Dijon métropole and Toulouse métropole) for the sampling effort made in the French WWTPS and the commitment of Suez Eau France Technical Direction (Hubert Dupont and Thierry Lebrun). The study was also supported by the French Scientific Group of interest named Obépine. Sabela Balboa, Miguel Mauricio-Iglesias and Juan M. Lema belong to the CRETUS Strategic Partnership (ED431E 2018/01) and to the Galician Competitive Research Group (ED431C2017/029). All these programs are co-funded by ERDF (EU).
Authors contribution
M. A., S. G., J. F. L., O. S., A. M. Y., M. P., B. G., G. S., A. P. M., J. M. L., C. M., and X. L. Designed and validated the experimental strategy and methods. S. C., S. Be., E. S. S., G. S., A. P. M., B. G., S. Ba., M. M., A. B., R. M. P., I. B., C. G. Coordinated sampling and performed extraction and analysis through q-PCR for SARS-COV-2 determination. M. F. and A. M. Y. contributed in interlaboratory assay. I. B., C. G., S. Be., S. C., performed bacteriophages analysis. A. S. C., S. Be., A. M. Y., wrote the manuscript. All authors revised and agreed with the manuscript content.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2021.117435.
Appendix. Supplementary materials
References
- Abu Ali H., Yaniv K., Bar-Zeev E., Chaudhury S., Shaga M., Ronen Z., Kushmaro A., Nir O. Tracking SARS-CoV-2 RNA through the wastewater 1 treatment process. medRxiv. 2020 doi: 10.1021/acsestwater.0c00216. 2020.10.14.20212837. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpaslan-Kocamemi B., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. medRxiv. 2020 doi: 10.1101/2020.05.12.20099358. 2020.05.12.20099358. [DOI] [Google Scholar]
- Amoah D.I., Kumari S., Bux F. Coronaviruses in wastewater processes: Source, fate and potential risks. Environment International. 2020;143 doi: 10.1016/j.envint.2020.105962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-CoV-2 in WWTPS points out the sludge line as a suitable spot for monitoring. Sci. Tot. Environm. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft P.M., Engelhard, W.E. Evans, C.A. Poliomyelitis in Huskerville (Lincoln) Nebraska; studies indicating a relationship between clinically severe infections and proximate fecal pollution of water. J Am Med Assoc. 1957;164:836–847. doi: 10.1001/jama.1957.02980080006002. [DOI] [PubMed] [Google Scholar]
- Barra GB, Santa Rita TH, Mesquita PG, Jácomo RH, Nery LFA. Analytical Sensitivity and Specificity of Two RT-qPCR Protocols for SARS-CoV-2 Detection Performed in an Automated Workflow. Genes (Basel) 2020;12(10):1183. doi: 10.3390/genes11101183. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand I., Challant J., Jeulin H., Hartard C., Mathieu L., Lopez S., Interest S., Obépine G., Schvoerer E., Courtois S., Gantzer C. Epidemiological surveillance of SARS-CoV-2 by genome quantification in wastewater applied to a city in the northeast of France: comparison of ultrafiltration- and protein precipitation-based methods. Int. J. Hyg. Environ. Health. 2021;113692 doi: 10.1016/j.ijheh.2021.113692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A., Arora P., Prajapati S.k. Occurrence, fates and potential treatment approaches for removal of viruses from wastewater: A review with emphasis on SARS-CoV-2. Journal of Environmental Chemical Engineering. 2020;8 doi: 10.1016/j.jece.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Viau E., Peccia J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett Appl Microbiol. 2011;52:386–392. doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler A., Packman A., Furman A., Gross A., Kushmaro A., Ronen A., Dagot C., Hill C., Vaizel-Ohayon D., Morgenroth E., Bertuzzo E., Wells G., Kiperwas H.R., Horn H., Negev I., Zucker I., Bar-Or I., Moran-Gilad J., Balcazar J.L., Bibby K., Elimelech M., Weisbrod N., Nir O., Sued O., Gillor O., Alvarez P.J., Crameri S., Arnon S., Walker S., Yaron S., Nguyen T.H., Berchenko Y., Hu Y., Ronen Z., Bar-Zeev E. Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nat. Sustain. 2020 doi: 10.1038/s41893-020-00605-2. [DOI] [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Martínez-Velázquez A., Vázquez-Portero M., Guix S., Paraira M., Galofré B., Sáanchez G., Pintó R., Bosch A. Time -evolution of SARS -CoV-2 in wastewater during the first 2 pandemic wave of COVID -19 in the metropolitan area of Barcelona. Applied Environmental Microbiology. 2021 doi: 10.1128/AEM.02750-20. Online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collivignarelli M.C., Collivignarelli C., Carnevale Miino M., Abbà A., Pedrazzani R., Bertanza G. SARS-CoV-2 in sewer systems and connected facilities. Process Saf. Environ. Prot. 2020;143:196–203. doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CGA-ENAC-PPI. Entidad Nacional de Acreditación. 2020. Organismos de Control de Instalaciones: requisitos de competencia técnica.
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O., Halden R. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Science of the Total Environment. 2020:730. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmsø M.H., Hellmér M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., Elsässer D., Aarestrup F.M., Löfström C., Bofill-Mas S., Abril J.F., Girones R., Schultz A.C. Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One. 2017;12:1–17. doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISO 13528 (2015) Statistical methods for use in proficiency testing by interlaboratory comparisons.
- ISO 22174: 2005. Microbiology of food and animal feeding stuffs - Polymerase chain reaction (PCR) for the detection of food-borne pathogens - General requirements and definitions.
- Jones D., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Wei J., Yuan J., Guo J., Zhang Y., Hang J., Qu Y., Qian H., Zhuang Y., Chen X., Peng X., Shi T., Wang J., Wu J., Song T., He J., Li Y., Zhong N. Probable Evidence of Fecal Aerosol Transmission of SARS-CoV-2 in a High-Rise Building. Ann. Intern. Med. 2020;1–8 doi: 10.7326/m20-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesimple A., Jasim S.Y., Johnson D.J., Hilal N. The role of wastewater treatment plants as tools for SARS-CoV-2 early detection and removal. J. Water Process Eng. 2020;38 doi: 10.1016/j.jwpe.2020.101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang S., Shi J., Lugy S.P., Jiang G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chemical Engineering Journal. 2021;415:1. doi: 10.1016/j.cej.2021.129039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., De Roda-Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhang F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;2020:19–21. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Murtula R., Soria E., Yáñez M.A., Catalán V. Proficiency testing schemes for the assessment of Legionella PCR methodologies. Accred Qual Assur. 2012 doi: 10.1007/s00769-012-0903-5. [DOI] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Vanderwood K., Wilkinson R., Wiedenheft B. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. Cell Reports Med. 100098. 2020 doi: 10.2139/ssrn.3664367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, S. and Mihelcic, J.R. 2019. Pathogen Reduction and Survival in Complete Treatment Works. In: J.B. Rose and B. Jiménez-Cisneros, (eds) Global Water Pathogen Project. http://www.waterpathogens.org (J.R. Mihelcic and M.E. Verbyla) (eds) Part 4 Management Of Risk from Excreta and Wastewater) http://www.waterpathogens.org/book/pathogen-reduction-and-survival-in-complete-treatment-works Michigan State University, E. Lansing, MI, UNESCO. https://doi.org/10.14321/waterpathogens.49.
- Park S.-K., Lee C.-W., Park D.-L., Woo H.-Y., Cheong H.S., Shin C.S., Ahn K., Kown M.-J., Joo E.-J. Detection of SARS-CoV-2 in Fecal Samples From Patients With Asymptomatic and Mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2020 doi: 10.1016/j.cgh.2020.06.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana. USA. Sci. Total Environ. 2020;15 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn A.S., Meijer B., Frampton C.M.A., Barclay M.L., de Boer N.K.H. Systematic review with meta-analysis: SARS-CoV-2 stool testing and the potential for faecal-oral transmission. Aliment. Pharmacol. Ther. 2020;52:1276–1288. doi: 10.1111/apt.16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xy Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA - J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas Jennifer, Aanderud, Zachary T. Roper, D. Keith. VanDerslice, James. Brown Gaddis, Erica. Ostermiller, Jeff. Hoffman, Ken. Jamal, Rubayat. Heck, Phillip. Zhang, Yue. Torgersen, Kevin. Vander Laan, Jacob. LaCross, Nathan Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Science of The Total Environment. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – Suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley John S., Chin, Tom D. Y. Gravelle, Clifton R. Robinson, Sherry Enterovirus in Sewage during a Poliomyelitis Epidemic. Water Pollution Control Federation. 1962;34:168–178. [Google Scholar]
- Withey S., Cartmell E., Avery L.M., Stephenson T. Bacteriophages–potential for application in wastewater treatment processes. Sci. Total Environ. 2005;339:1–18. doi: 10.1016/j.scitotenv.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Wolf S., Hewitt J., Greening G.E. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl. Environ. Microbiol. 2010;76:1388–1394. doi: 10.1128/AEM.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. EuroSurveill. 2020;25(50) doi: 10.2807/1560-7917.ES.2020.25.50.2000776. pii=200077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., LI H., Zhao J., Huang J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020;26(8):1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Chen Z., Gong C., Liu H., LI B., Li K., Chen X., Xu C., Jing Q., Liu G., Qin P., Liu Y., Zhong Y., Huang L., Zhu B.-P., Yang Z. Coronavirus Disease 2019 Outbreak Likely Caused by Sewage Exposure in a Low-income Community: Guangzhou, China, April 2020. Lancet. 2020 doi: 10.1093/cid/ciaa1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi A., Monis P., Deere D., Ryan U. Wastewater-based epidemiology – survillance of waterborne pathogens with a focus on SARS-CoV-2, Cryptosporidium and Giardia. Parasitology Research. 2020 doi: 10.1007/s00436-020-07023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc3582. eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.