Dear Editor
The coronavirus disease 2019 (COVID-19)1 originated in Wuhan, China, and has subsequently spread to 192 countries and territories. As of June 30, 2021, it has caused approximately 180 million infections and 3.9 million deaths worldwide. The impact of this emerging infectious disease on global health is unprecedented in recent history, and the social and economic challenges it poses to humanity are daunting.
While the pandemic is in progress, being able to timely forecast when COVID-19 outbreaks will peak would be highly valuable for the planning of resource allocation and procuring supplies for disease treatment by health authorities. Forecasting would also help the general public better understand the current situation and plan for the future. Infectious disease experts often rely on mathematical models of various types to study the mechanisms by which diseases spread and to predict the future course of an outbreak.2, 3, 4 These models depend heavily on modeling assumptions and require large amounts of information regarding transmission dynamics. Moreover, they are often too complicated to be used for routine monitoring purposes.
Here, we demonstrate that by simply monitoring the growth of the epidemic curve, one can make a timely forecast of the peaks of multiwave COVID-19 outbreaks.
An epidemic curve is the plot of the number of new cases against the date reported. At a particular date, t, we calculate the growth rate of the epidemic curve at t-4 as the natural logarithm of the ratio of the cumulative number of new cases from t-3 to t and that from t-8 to t-5. The growth rate (× 100%) at a particular time can be interpreted as the symmetric percentage change5 in the number of new cases immediately before and after that time point. The resulting growth rate curve lags behind the epidemic curve by 4 days and may appear rugged. To smooth the curve and nowcast the growth rate at t, we applied a weighted linear regression of the daily growth rate estimates of the most recent 5 weeks (t-38 to t-4) against time (the weights used in the regression are proportional to the inverse variances of the daily growth rate estimates).
We analyzed COVID-19 data from ten countries (United States, Spain, Italy, United Kingdom, Germany, Netherlands, Korea, Russian, Mexico, and Japan), and monitored the peaks of the epidemic curves for these countries from April 1, 2020, to June 30, 2021.
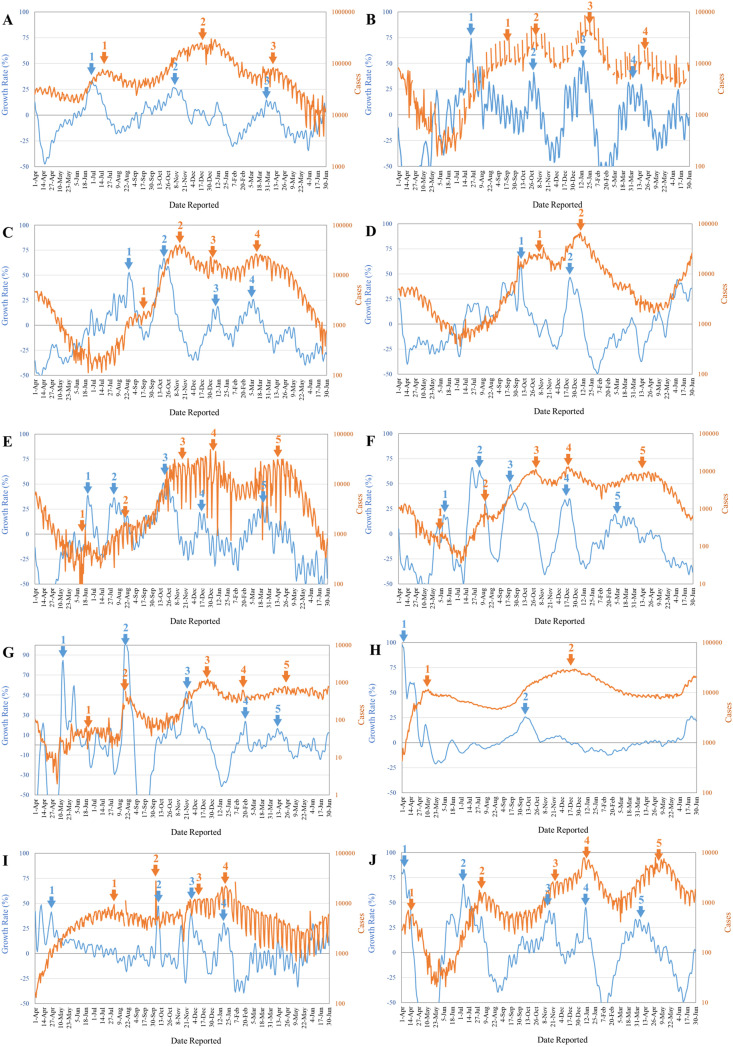
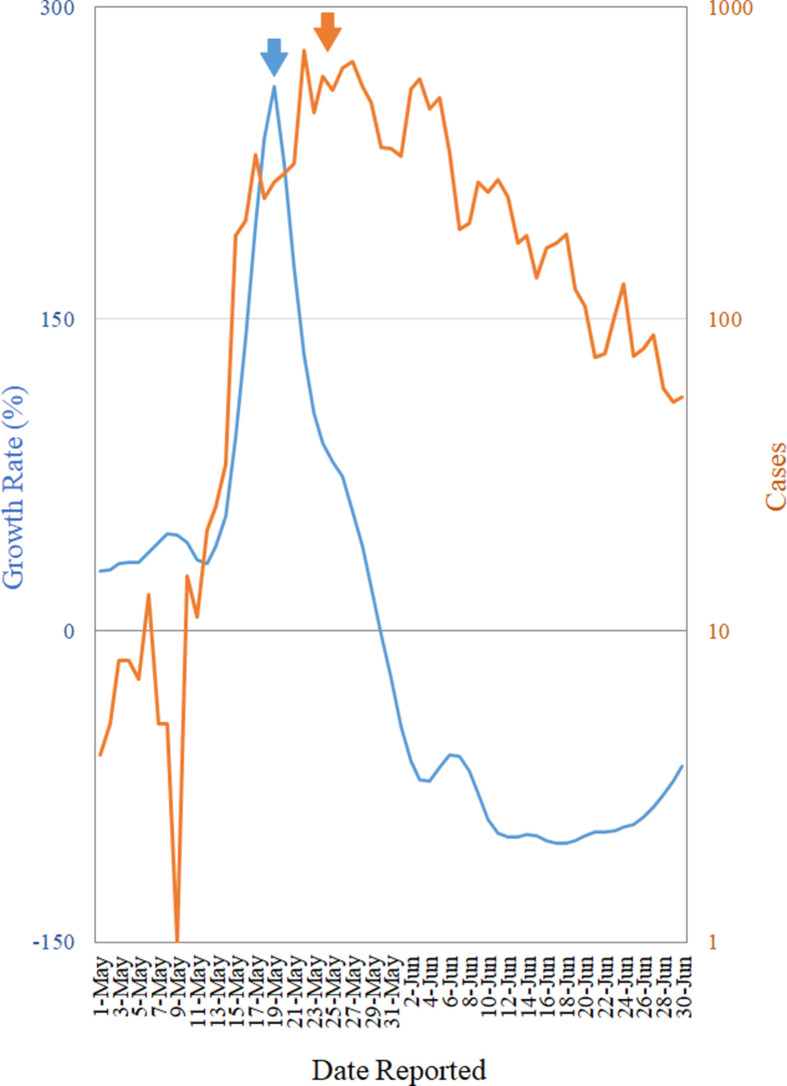
The growth rate curves provide early warnings for the majority of the COVID-19 peaks in these countries several weeks ahead (Fig. 1 ; comparing the blue and the orange arrows). Some early warnings do not correspond to peaks but leveling (the first arrows in panels B and G, and the third arrows in panels E, I, and F) or inflections (the first arrows in panel C, and the second arrows in panels E and F) in the epidemic curves. Nonetheless, while the epidemic curves are on the sharp decline and then slightly rebound, the warnings come a bit late (the first arrows in panel F, the second arrows in panel I, the third arrows in panel C, and the fourth arrows in panel G). The growth rate curve also provides a week-ahead warning for the peak of the recent COVID-19 outbreak in Taiwan (eFig. 1).
Figure 1.
Epidemic curves (Orange) and growth rate curves (Blue) for ten countries (A: United States; B: Spain; C: Italy; D: United Kingdom; E: Germany; F: Netherlands; G: Korea; H: Russian; I: Mexico; J: Japan).
The proposed method is straightforward. We encourage countries in COVID-19 outbreaks to begin monitoring the epidemic curves using our method to prepare early for the upcoming peaks.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Data on daily confirmed cases and deaths from COVID-19 by country were extracted from the online database provided by the National Center for High-performance Computing. Please refer to the link: https://covid-19.nchc.org.tw/.
Funding
This paper is supported by grants from the Ministry of Science and Technology, Taiwan (MOST 108-3017-F-002-001, MOST 108-2314-B-002-127-MY3), and the Innovation and Policy Center for Population Health and Sustainable Environment (Population Health Research Center, PHRC) from Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan (NTU-109L900308).
Authors' contributions
WCL conceptualized the hypothesis for this paper, analyzed the data, and wrote the manuscript. SYS prepared and analyzed the data, prepared the figure, and wrote the manuscript. All authors read and approved the final version of the manuscript.
Declaration of competing interest
The authors declare no potential conflicts of interest.
Acknowledgments
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmii.2021.07.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
figs1.
References
- 1.Fauci A.S., Lane H.C., Redfield R.R. Covid-19 - navigating the uncharted. N Engl J Med. 2020;382(13):1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauch C.T., Lloyd-Smith J.O., Coffee M.P., Galvani A.P. Dynamically modeling SARS and other newly emerging respiratory illnesses: past, present, and future. Epidemiology. 2005;16(6):791–801. doi: 10.1097/01.ede.0000181633.80269.4c. [DOI] [PubMed] [Google Scholar]
- 3.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang B., Wang X., Li Q., Bragazzi N.L., Tang S., Xiao Y., et al. Estimation of the transmission risk of the 2019-nCoV and its implication for public health interventions. J Clin Med. 2020;9(2):462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole T.J. Sympercents: symmetric percentage differences on the 100 log(e) scale simplify the presentation of log transformed data. Stat Med. 2000;19(22):3109–3125. doi: 10.1002/1097-0258(20001130)19:22<3109::aid-sim558>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data on daily confirmed cases and deaths from COVID-19 by country were extracted from the online database provided by the National Center for High-performance Computing. Please refer to the link: https://covid-19.nchc.org.tw/.