Abstract
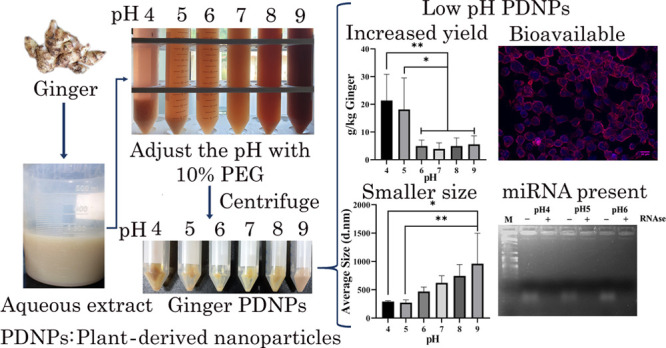
Plant-derived nanoparticles (PDNPs) are naturally occurring exosome-like nanovesicles derived from dietary plants containing key plant bioactives. Ginger-derived PDNPs have a therapeutic effect on alcohol-induced liver injury, inflammatory bowel disease, and colon cancer. PDNPs are conventionally purified by differential ultracentrifugation, a technique not amenable for scale up. We have recently developed a polyethylene glycol (PEG) 6000-based method for cost-effective purification of ginger PDNPs, with comparable efficiency to differential ultracentrifugation (Sci. Rep.2020, 10 (1), 4456.). Herein, we report a 4–5-fold higher ginger PDNP recovery when PEG precipitation was carried out in low pH conditions (pH 4 and 5). Low pH-derived ginger PDNPs were smaller in size without an overt change in zeta potential. The spontaneous intracellular entry and protection against oxidative stress in A431 cells were similar between ginger PDNPs purified under low, neutral, and alkaline pH. Low-pH purified ginger PDNPs had higher levels of total polyphenolic content compared to PDNPs purified under neutral and alkaline pH. Recently, ginger PDNP-derived microRNAs have been shown to exhibit cross-kingdom regulation by targeting human, gut microbiome, and viral transcripts. Using qRT-PCR, we also verified the presence of miRNAs that were predicted to target SARS-CoV-2 in ginger PDNPs purified under low pH. Thus, we have developed a method to purify ginger PDNPs in high yields by using low-pH conditions without affecting the major bioactive contents of PDNPs.
Introduction
Plant-derived nanoparticles (PDNP) are membrane-bound nanoscaled vesicles that are isolated from dietary fruits and vegetables such as grapefruit, lemon, ginger, broccoli, and orange. PDNPs are also known as exosome-like nanoparticles since they are morphologically similar to mammalian exosomes with a size range between 100 and 500 nm.1 These are composed of uni/multilamellar lipid bilayers with encapsulated proteins, small RNAs, and other phytochemicals as their key bioactive components. Plant bioactives, isolated in the form of PDNPs, show enhanced bioavailability in gastrointestinal (GI) tract. Moreover, PDNPs are naturally non-toxic, demonstrate more efficient intracellular uptake in difficult to transfect cell lines, and are biocompatible.2,3 Due to these features, PDNPs are also used as excellent nanocarriers for in vivo delivery of a range of biological cargo such as chemotherapeutic drugs, siRNAs, and phytochemicals.2,4,5
PDNPs isolated from ginger rhizomes have been shown to possess both therapeutic as well as nanocarrier potential. Oral administration of ginger PDNPs protected mice against alcohol-induced liver damage by activating nuclear factor erythroid 2-related factor 2 (Nrf2), resulting in the induction of genes of the antioxidant pathway and inhibition of reactive oxygen species (ROS) synthesis.6,7 Ginger PDNPs also contain high levels of 6-gingerol and 6-shagaol, the major bioactives of ginger rhizome. In mouse models, oral gavage of ginger PDNPs led to their selective uptake by intestinal epithelial cells (IEC) and macrophages, leading to amelioration of acute/chronic colitis and colitis-associated cancer by suppression of pro-inflammatory and induction of anti-inflammatory cytokines.7 Ginger PDNPs also exhibit potent inhibitory effect on the nucleotide-binding domain and leucine-rich repeat-containing family, pyrin domain-containing 3 (NLRP3) inflammasome.8
Ginger PDNP-derived lipids have been exploited as a nano vector for systemic delivery of siRNAs to treat ulcerative colitis. Oral administration of ginger PDNPs carrying siRNA against CD98 was effectively delivered to colon tissues resulting in reduced expression of CD98.9 Similarly, ginger PDNP-derived lipids were used as nanocarriers for oral delivery of siRNA against divalent metal-ion transporter (Dmt1) to treat hereditary hemochromatosis.10 Ginger PDNP-derived nano vectors have also been employed for the targeted delivery of cargos to cancer cells in vivo, via co-encapsulating cancer-targeting ligands such as folic acid (FA), highlighting the potential of ginger PDNPs as a cost-effective nanocarrier.11,12
MicroRNAs (miRNAs) are small, endogenously derived 18–25 nt RNAs which regulate target mRNAs by translational repression and/or transcript degradation. Three independent studies have shown the presence of miRNAs in ginger PDNPs with potential therapeutic benefits. Xiao et al., (2018) detected the presence of 32 different miRNA species in ginger PDNPs, from which miR-1078 was predicted to target leptin (LEP), a key regulator of systemic inflammation.13 Ginger PDNP-derived miRNAs also target pathogenic bacterial mRNAs from Lactobacillus rhamnosus and Porphyromonas gingivalis in the gut and oral microbiome, thereby modulating oral and gut inflammation.14,15
The gold standard method for isolating PDNPs is through differential ultracentrifugation of plant extract, an expensive and non-scalable process. We have recently developed a cost-effective polyethylene glycol (PEG6000)-based precipitation method for the isolation of ginger PDNPs, which eliminates the need for ultracentrifugation.16 The overall yield and biochemical and biophysical characteristics of ginger PDNPs isolated in this method are similar to ultracentrifugation. Notably, we have also demonstrated that ginger PDNPs isolated by the PEG method also contain a small RNA population, including putative SARS-CoV-2-targeting miRNAs.16,17 Herein, we have improved this method further by lowering the pH during PEG precipitation. Under low pH conditions (pH 4 and 5), a higher yield of ginger PDNPs can be obtained compared to that under neutral/alkaline pH. The ginger PDNPs purified under low-pH are similar in biophysical and biochemical characteristics to those purified under native pH (7.0) and also retain key miRNAs shown to be associated with ginger PDNPs.
Results
Effect of pH on Ginger PDNP Precipitation by PEG6000 and Its Biophysical Characteristics
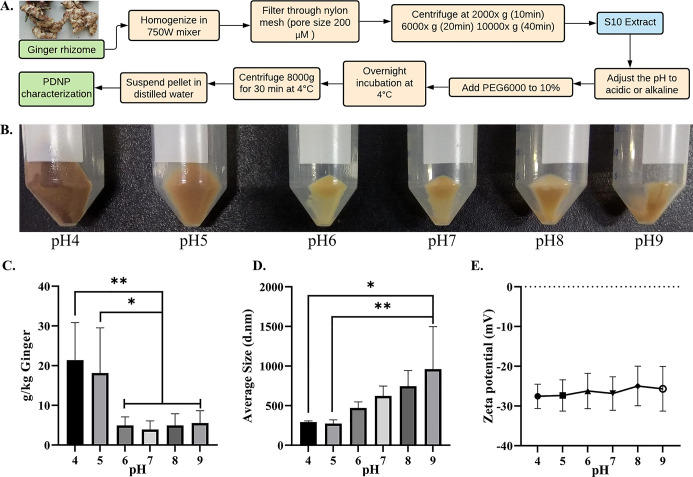
To investigate the effect of pH on ginger PDNP isolation by the PEG method, the pH of the S10 supernatant was adjusted to acidic (pH 4, 5, and 6), neutral (pH 7.0), and alkaline (pH 8 and 9) prior to PEG precipitation (Figure 1A). We noted a significant increase in ginger PDNP obtained in acidic pH (pH 4 and 5) compared to that under neutral and alkaline pH conditions (pH 6–9) (Figure 1B). In multiple batches, we observed a consistent increase in yield, up to 5-fold, in pH 4 and pH 5 (Figure 1C). A slight increase in PDNP yield was noted under alkaline pH conditions, although it was not significant. We further measured the size and zeta potential of ginger PDNPs purified under different pH. The ginger PDNP isolated under pH 4 and pH 5 showed a significant reduction in size when compared to the ginger PDNPs isolated under alkaline pH (Figures 1D and S1). The zeta potential of PDNPs did not show significant change across the pH ranges attempted (Figures 1E and S1).
Figure 1.
Isolation and characterization of ginger PDNPs under different pH. (A) Experimental flowchart depicting the purification of ginger PDNPs under different pH conditions. (B) Photomicrographs of ginger PDNP pellets obtained by PEG precipitation of S10 extract under different pH. Pictures taken by the first author of this article. (C) Total yield of ginger PDNPs isolated under different pH conditions PEG precipitation. (D) Average size of ginger PDNPs isolated under different pH. (E) Average zeta potential of ginger PDNPs isolated under different pH. Results presented are average of four independent experiments. *P < 0.05 and **P < 0.01.
FTIR Analysis of Ginger PDNPs Purified under Different pH
Overall FTIR absorption spectra were similar between ginger PDNPs isolated under different pH (Figure S2). The FTIR data set could not be used for the detection of lipids since the −CH2– functionality is also found in PEG6000, which remains in PDNP preparation despite the dialysis procedure.16 However, we did observe intensified peaks for pH 4 and 5 at 3300, 1531, and 1638 cm–1. The absorption peaks at 1638 and 1531 cm–1 correspond to amide I and amide II bands (C=O stretching), respectively, suggesting that PDNPs isolated under pH 4 and 5 may contain higher proportion of proteins.
Effect of pH on the Lipid Composition and Spontaneous Intracellular Uptake of Ginger PDNPs
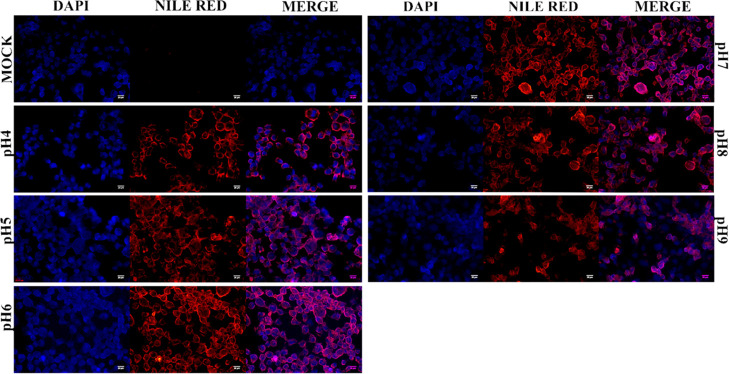
Total lipids isolated from ginger PDNPs were resolved through TLC. The overall lipid profile was similar between ginger PDNPs purified under different pH conditions, although lipid bands were more intense at pH 4, indicating the isolation of a higher amount of PDNPs (Figure S3). To assess the bioavailability of ginger PDNPs purified under different pH, A431 keratinocytes were incubated with an equal concentration of ginger PDNPs prelabeled with Nile Red, a lipophilic fluorescent dye. Unbound PDNPs were removed by washing the cells with PBS. Within 5 h of incubation, detectable fluorescence was observed in A431 cells incubated with ginger PDNPs but not in mock-treated cells (Figure 2). No significant difference in intensity or number of fluorescent cells were detected between PDNPs isolated under different pH, highlighting that PDNPs isolated under low pH are equally bioavailable.
Figure 2.
Intracellular uptake of ginger PDNPs purified under different pH. A431 cells were either mock-treated or incubated with Nile red-labeled ginger PDNPs for 5 h. Cells were fixed and counterstained with DAPI. Cytoplasmic fluorescence was detectable in cells treated with ginger PDNPs without significant difference in fluorescence intensity or number of fluorescing cells between PDNPs isolated under different pH conditions. Scale bar-20 μm.
Total Polyphenolic Content and in Vitro Antioxidant Activity of Ginger PDNPs Isolated under Different pH
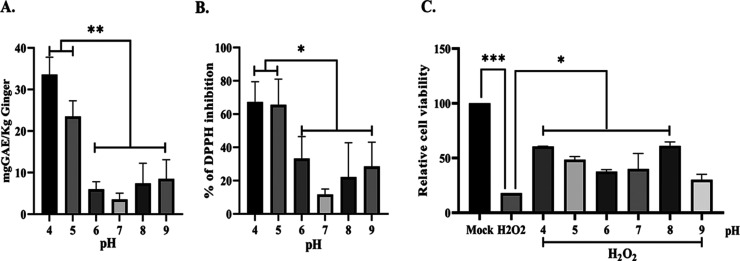
Total polyphenolic content (TPC) was measured to ascertain the presence of bioactives in ginger PDNPs derived under low pH conditions. In agreement with increased yield, ginger PDNPs purified under pH 4 and pH 5 displayed 3–4-fold higher levels of TPC compared to those purified under neutral and alkaline pH (Figure 3A). Furthermore, the in vitro antioxidant activity, assayed by measuring DPPH-free-radical scavenging activity, showed significantly higher inhibition of DPPH by low-pH-derived (pH 4 and 5) PDNPs compared to the rest (Figure 3B). To confirm this further, we tested the ability of ginger PDNPs derived under acidic, neutral, and alkaline pH to protect cells from H2O2-induced oxidative stress. Incubation of A431 keratinocytes with H2O2 led to a significant decrease in cell viability within 6 h post treatment (Figure 3C). Co-treatment of ginger PDNPs isolated under pH 4–8 led to the rescue of cell death induced by H2O2 treatment, although no significant difference in this rescue was observed between PDNPs isolated under different pH conditions (Figure 3C).
Figure 3.
Ginger PDNPs purified under acidic pH have higher TPC and protects cells upon oxidative stress. (A) Total polyphenolic content was extracted from ginger PDNPs and measured by Folin-Ciocalteau method. Ginger PDNPs isolated under acidic conditions (pH 4 and 5) possessed significantly higher TPC compared to neutral or alkaline conditions. (B) In vitro antioxidant activity measured by DPPH assay. Low pH-derived ginger PDNPs showed higher antioxidant activity. (C) Relative cell viability of A431 cells treated with 500 μM H2O2 alone or co-treated with ginger PDNPs. *P < 0.05, **P < 0.01, and ***P < 0.001.
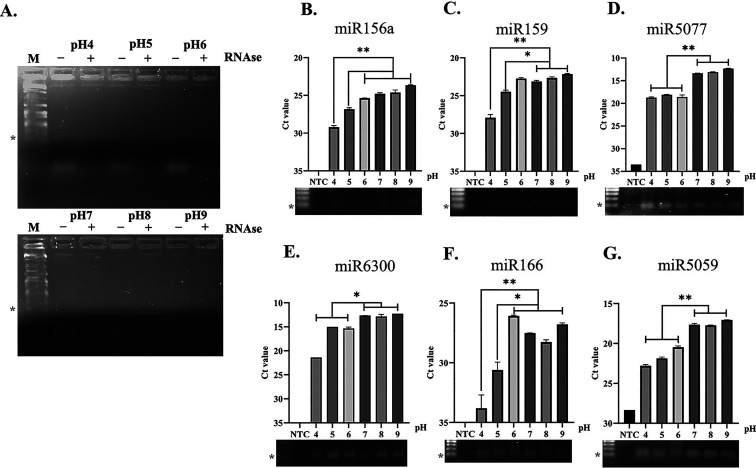
Effect of pH on Ginger PDNP-Derived Small RNA Population and SARS-CoV-2-Targeting miRNAs
We have recently shown the presence of intact small RNA populations in PEG-derived ginger PDNPs, which also include some key miRNAs that could potentially target SARS-CoV-2.17 To verify the presence of a small RNA population and miRNAs in ginger PDNPs purified under low pH conditions, we isolated total RNA from PDNPs and resolved it through agarose gel electrophoresis. The small RNA population isolated from different pH-derived PDNPs were of identical size and were susceptible to RNAse A treatment16,17 (Figure 4A). Furthermore, we also quantified the relative abundance of six miRNAs (miR-156a, miR-159, miR-5077, miR-6300, miR-166, and miR-5059) in ginger PDNPs isolated under different pH. All six miRNAs showed detectable amplification in all PDNP samples, while the control reaction lacking the template cDNA did not show any amplification (Figure 4B–G, lower panels). However, ginger PDNPs isolated under acidic conditions (<pH 6) showed reduced levels of all six miRNAs (Figure 4B–G) with the lowest levels of miRNAs observed at pH 4. This effect was prominent for miR-166 probably due to its naturally lower expression level in PDNPs (Figure 4F). Nevertheless, PDNPs isolated at pH 5 had higher expression levels of miRNAs compared to pH 4 with three out of six miRNAs (miR-5077, miR-6300, and miR-5077) still showing relative Ct values between 15 and 25 (Figure 4D–G).
Figure 4.
Validation of bioactive miRNAs in ginger PDNPs purified with different pH. (A) Total RNA isolated from ginger PDNPs were resolved through agarose gel electrophoresis (AGE) and visualized by ethidium bromide staining. Pretreatment of samples led to the disappearance of the band corresponding to a small RNA population (<100 bp), confirming the RNA nature of the sample. M-100bp DNA ladder. (B–G) Bar graphs showing the raw Ct values of each miRNA in ginger PDNPs isolated under different pH conditions. RT-PCR without template cDNA was used as a control (NTC). The RT-PCR amplicons obtained for each miRNA were further confirmed by resolving through 1.6% AGE. Asterisks next to the marker lane indicate 100 bp. *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
In this investigation, we have demonstrated that acidification during PEG precipitation significantly increases the yield of ginger PDNPs. Ginger PDNPs purified under acidic conditions, specifically at pH 4 and 5, had smaller-sized vesicles, greater phenolic and lipid content, and comparable antioxidant activity in vitro. The increase in size for PDNPs purified under alkaline conditions is an undesirable feature for therapeutic use.5 The change in PDNP size under acidic conditions is in line with previous studies wherein changes in the size of PDNPs were noted for both ginger and grape PDNPs when subjected to stomach-like acidic solution.18 From earlier reports, it is apparent that acidic condition is the suitable environment for the existence and isolation of exosomes. Two independent reports demonstrate that acidic pH could increase the stability of exosomes in vitro, resulting in a higher yield of exosome isolation.19,20 Since PDNPs are closely related in structure and function to mammalian exosomes, lowering the pH is likely to enhance PDNP precipitation in a similar manner. PEG-based isolation of nanovesicles is known to conserve the integrity of nanovesicles through its ability to entrap them in a mesh-like net formation.16,21 Hence, PEG precipitation can increase the PDNP yield much higher in lower pH without losing the integrity and key bioactives of PDNP.22,23
We also noted that the relative levels of bioactive miRNA content were lesser in PDNPs purified under both pH 4 and 5 compared to other conditions. This could be due to the ability of PEG to precipitate more proteins at acidic pH which may impact the quality of total RNA.24 In support, FTIR results showed increased protein content in low-pH-derived ginger PDNPs. Taking into account the 5-fold higher yield of PDNPs with pH 5, this decreased level of key miRNAs can be counterbalanced by administering 5 times more PDNPs to achieve a particular therapeutic benefit.20
Taken together, we have observed that lowering the pH, especially to pH 5, increases the PDNP yield with higher bioactive content along with key miRNAs compared to pH 4. Hence, isolation of PDNPs under low pH conditions may aid in the production of PDNPs in scalable quantities to be used for both therapeutic and nanocarrier purposes.
Materials and Methods
Cell Culture
Epidermoid carcinoma cell line, A-431, was cultured in Dulbecco’s modified Eagle’s medium (Sigma Aldrich) supplemented with 10% fetal bovine serum (Sigma Aldrich) and antibiotics (Penicillin and Streptomycin). The cells were maintained in a 5% CO2 environment at 37 °C.
Isolation of Ginger PDNPs by PEG Method under Different pH Conditions
Fresh ginger (Zingiber officinale) variety Nadia brought from the Devraja market (Mysore) was washed thoroughly and homogenized using a mixer grinder at medium speed for 3 min. The excess fiber was filtered through a nylon mesh (pore size 200 μm). The filtrate was subjected to low- (2000×g for 10 min, 6000×g for 20 min) and medium-speed (10,000×g for 40 min) centrifugation. The supernatant obtained after 10,000×g step (S10) was equally divided, and the pH was adjusted to pH 4, pH 5, pH 6, pH 7, pH 8, and pH 9 using either 11 N HCl or 1 N NaOH. The pH-adjusted S10 supernatant was mixed with PEG6000 (Sigma Aldrich) to reach a final concentration of 10% (weight by volume) and incubated overnight at 4 °C. Extracts were centrifuged at 8000×g for 30 min to retrieve ginger PDNPs as described earlier.16 The tube is inverted on a piece of tissue paper to remove excess supernatant, and the pellet was suspended in sterile water to reach a final concentration of 0.5 mg/μL. The sample was dialyzed overnight against milli-Q water using a dialysis membrane (Himedia) with a pore size of 10 kDa.
Particle Size and Surface Charge of Ginger PDNPs
Nanoparticle size and surface potential were measured using a Malvern zeta sizer nano ZS (Malvern Instruments, Malvern, UK). The sample was diluted 100–1000-fold in milli-Q water and triplicate measurements were taken for each sample at room temperature. Particle size and zeta potential of at least three independent batches were measured, and its mean ± standard deviation was calculated.
Fourier-Transform Infrared Spectroscopy Analysis
Attenuated total reflection Fourier-transform infrared (FTIR) spectroscopy was performed with ginger PDNPs purified under different pH as per earlier methods.16 Measurements were taken at ambient conditions, and the spectral range was collected between 4000 and 400 cm–1. Background subtraction, baseline correction, and spectrum smoothening were performed as described earlier.25
Lipid Extraction and Characterization
Total lipid extraction and separation of lipids by thin-layer chromatography (TLC) was carried out as described earlier.16 Briefly, lipids were extracted from PDNPs by mixing with equal volumes of chloroform and methanol. After centrifugation, the organic phase containing the lipids was resolved through silica gel 60 F254 TLC plates (Merck). The mobile phase was a mixture of chloroform/methanol/acetic acid (95:4.5:0.5 volume by volume ratio). Plates were dried, and lipids were detected by staining with 10% copper sulfate and 8% phosphoric acid, followed by charring.
TPC Estimation of Ginger PDNPs
TPC was extracted from PDNPs by methanol extraction as described earlier.16 PDNPs were vortexed with 100 μL of absolute methanol and incubated at room temperature for 10 min followed by centrifugation at 10,000×g for 5 min. The supernatant was mixed with 400 μL of Folin-Ciocalteu reagent (HiMedia laboratories) and 800 μL of 7.5% sodium carbonate. After incubation at room temperature for 30 min, TPCs were measured using a colorimeter at 765 nm. Gallic acid was used to draw the standard curve, and values are represented as gallic acid equivalents in mg.
1,1-Diphenyl-2-picrylhydrazyl (DPPH) Assay for Antioxidant Activity
100 μL of methanolic extract purified from ginger PDNPs was mixed with 900 μL of DPPH (0.2 mM) and incubated at room temperature for 30 min. The absorbance was measured at 517 nm using an ELISA plate reader. DPPH mixed with an equal volume of methanol served as a control. The antioxidant activity of the sample is calculated using the following formula
Intracellular Uptake of Ginger PDNPs
To track the intracellular uptake, ginger PDNPs were labeled with a lipophilic dye, Nile Red (Sigma Aldrich). Nile red was added to the S10 extract with a final concentration of 1 μM prior to pH adjustment so that the unbound dye gets removed during the centrifugation step. For intracellular uptake experiment, A431 cells (50,000 cells) were seeded into a 24-well TC plate containing glass coverslips. After 24 h, cells were treated with an equal concentration of ginger PDNPs (500 μg in 500 μL media) and incubated for 5 h. The cells were washed twice with PBS and fixed with 4% paraformaldehyde for 20 min, counterstained with DAPI (Sigma Aldrich), and mounted onto a glass slide with FluorSave fluorescent mounting media (Sigma Aldrich). Images were acquired in an Olympus IX73 fluorescence inverted microscope under 20× magnification.
In Vitro Antioxidant Activity
The in vitro antioxidant activity of Ginger PDNPs was evaluated using the hydrogen peroxide (H2O2)-induced oxidative stress on A431 cells as described earlier.26 Briefly, A431 cells were seeded onto 96-well plates at a density of 15,000 cells per well. After 24 h, the cells were washed with PBS and treated with either H2O2 alone (500 μM) or co-treated with ginger PDNPs (150 μg/200 μL media). After incubation for 6 h, cells were washed and relative cell viability was measured through MTT (3-(4,5-dimethylthiazol-2-yl)-w,5-diphenyltetrazolium bromide) reagent as per standard protocols.
Total RNA Extraction and miRNA Quantification
To isolate total RNA from ginger PDNPs, 100 μL of PDNPs were mixed with 500 μL of TRI reagent (Sigma) and 200 μL of chloroform and vortexed vigorously. After centrifugation at 10,000×g for 10 min, the aqueous layer was collected and precipitated with an equal volume of chilled isopropanol. The obtained total RNA pellet was washed twice with 70% ethanol and suspended in 30 μL of nuclease-free water. 1 μg of total RNA was treated with or without RNase A and resolved through 1.5% agarose gel to confirm the integrity of the small RNA population. Quantification of mature miRNAs was carried out as per Kalarikkal and Sundaram (2021). Briefly, 100 ng of total RNA was polyadenylated using poly A polymerase. The polyadenylated miRNAs were reverse transcribed using oligo (dT) primers containing adapter sequence.17 The cDNA samples were diluted to 10-fold, and mature miRNAs were quantified using miRNA-specific forward primer and reverse primer containing an adapter sequence as described earlier.17
Statistical Methods
The data described here are the average results of three or more independent experiments with minimum triplicate measurements performed in each assay. Data are plotted using GraphPad Prism software. Statistical testing between samples was conducted with the ANOVA algorithm in GraphPad, with Turkey’s multiple testing correction.
Acknowledgments
The authors would like to acknowledge the Director CSIR-CFTRI for the facilities and support. Academy of Scientific and Innovative Research (AcSIR) is acknowledged for the fellowship received by A.P.S. This work was supported by the Start-up Research Grant, Department of Science and Technology, Science and Engineering Research Board, New Delhi, India (SRG/2019/000584).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c02162.
Representative size distribution and zeta potential profiles of ginger PDNPs isolated under different pH conditions; FTIR absorbance spectrum of ginger PDNPs isolated under different pH conditions; and thin-layer chromatographic image showing the distribution of total lipids extracted from ginger PDNPs isolated under different pH conditions (PDF)
Author Contributions
G.M.S., A.P.S. conceptualized the study, participated in research design and methods, validated the hypothesis, and wrote the original draft. S.P.K. performed the RNA isolation and qRT-PCR and B.P. performed FTIR experiments and data analysis. G.M.S. was responsible for project administration and funding acquisition.
The authors declare no competing financial interest.
Supplementary Material
References
- Yu L.; Deng Z.; Liu L.; Zhang W.; Wang C. Plant-Derived Nanovesicles: A Novel Form of Nanomedicine. Front. Bioeng. Biotechnol. 2020, 8, 1–6. 10.3389/fbioe.2020.584391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Ren Y.; Mu J.; Egilmez N. K.; Zhuang X.; Deng Z.; Zhang L.; Yan J.; Miller D.; Zhang H.-G. Grapefruit-Derived Nanovectors Use an Activated Leukocyte Trafficking Pathway to Deliver Therapeutic Agents to Inflammatory Tumor Sites. Cancer Res 2015, 75, 2403. 10.1158/0008-5472.CAN-14-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Zhuang X.; Mu J.; Deng Z.-B.; Jiang H.; Zhang L.; Xiang X.; Wang B.; Yan J.; Miller D.; Zhang H.-G. Delivery of Therapeutic Agents by Nanoparticles Made of Grapefruit-Derived Lipids. Nat. Commun. 2013, 4, 11347. 10.1038/ncomms2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Zhuang X.; Deng Z.-B.; Jiang H.; Mu J.; Wang Q.; Xiang X.; Guo H.; Zhang L.; Dryden G.; Yan J.; Miller D.; Zhang H.-G. Targeted Drug Delivery to Intestinal Macrophages by Bioactive Nanovesicles Released from Grapefruit. Mol. Ther. 2014, 22, 522–534. 10.1038/mt.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.; Zhang M.; Merlin D. Advances in Plant-Derived Edible Nanoparticle-Based Lipid Nano-Drug Delivery Systems as Therapeutic Nanomedicines. J. Mater. Chem. B 2018, 6, 1312–1321. 10.1039/C7TB03207B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X.; Deng Z.-B.; Mu J.; Zhang L.; Yan J.; Miller D.; Feng W.; McClain C. J.; Zhang H.-G. Ginger-Derived Nanoparticles Protect against Alcohol-Induced Liver Damage. J. Extracell. vesicles 2015, 4, 28713. 10.3402/jev.v4.28713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Viennois E.; Prasad M.; Zhang Y.; Wang L.; Zhang Z.; Han M. K.; Xiao B.; Xu C.; Srinivasan S.; Merlin D. Edible Ginger-Derived Nanoparticles: A Novel Therapeutic Approach for the Prevention and Treatment of Inflammatory Bowel Disease and Colitis-Associated Cancer. Biomaterials 2016, 101, 321–340. 10.1016/j.biomaterials.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Zhou Y.; Yu J. Exosome-like Nanoparticles from Ginger Rhizomes Inhibited NLRP3 Inflammasome Activation. Mol. Pharm. 2019, 16, 2690–2699. 10.1021/acs.molpharmaceut.9b00246. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Wang X.; Han M. K.; Collins J. F.; Merlin D. Oral Administration of Ginger-Derived Nanolipids Loaded with SiRNA as a Novel Approach for Efficient SiRNA Drug Delivery to Treat Ulcerative Colitis. Nanomedicine 2017, 12, 1927–1943. 10.2217/nnm-2017-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Zhang M.; Flores S. R. L.; Woloshun R. R.; Yang C.; Yin L.; Xiang P.; Xu X.; Garrick M. D.; Vidyasagar S.; Merlin D.; Collins J. F. Oral Gavage of Ginger Nanoparticle-Derived Lipid Vectors Carrying Dmt1 SiRNA Blunts Iron Loading in Murine Hereditary Hemochromatosis. Mol. Ther. 2019, 27, 493–506. 10.1016/j.ymthe.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Yang C.; Yan X.; Sung J.; Garg P.; Merlin D. Highly Biocompatible Functionalized Layer-by-Layer Ginger Lipid Nano Vectors Targeting P-Selectin for Delivery of Doxorubicin to Treat Colon Cancer. Adv. Ther. 2019, 2, 1900129. 10.1002/adtp.201900129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Xiao B.; Wang H.; Han M. K.; Zhang Z.; Viennois E.; Xu C.; Merlin D. Edible Ginger-Derived Nano-Lipids Loaded with Doxorubicin as a Novel Drug-Delivery Approach for Colon Cancer Therapy. Mol. Ther. 2016, 24, 1783–1796. 10.1038/mt.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J.; Feng S.; Wang X.; Long K.; Luo Y.; Wang Y.; Ma J.; Tang Q.; Jin L.; Li X.; Li M. Identification of Exosome-like Nanoparticle-Derived MicroRNAs from 11 Edible Fruits and Vegetables. PeerJ 2018, 6, e5186 10.7717/peerj.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram K.; Miller D. P.; Kumar A.; Teng Y.; Sayed M.; Mu J.; Lei C.; Sriwastva M. K.; Zhang L.; Yan J.; Merchant M. L.; He L.; Fang Y.; Zhang S.; Zhang X.; Park J. W.; Lamont R. J.; Zhang H.-G. Plant-Derived Exosomal Nanoparticles Inhibit Pathogenicity of Porphyromonas Gingivalis. iScience 2019, 21, 308–327. 10.1016/j.isci.2019.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y.; Ren Y.; Sayed M.; Hu X.; Lei C.; Kumar A.; Hutchins E.; Mu J.; Deng Z.; Luo C.; Sundaram K.; Sriwastva M. K.; Zhang L.; Hsieh M.; Reiman R.; Haribabu B.; Yan J.; Jala V. R.; Miller D. M.; Van Keuren-Jensen K.; Merchant M. L.; McClain C. J.; Park J. W.; Egilmez N. K.; Zhang H.-G. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 2018, 24, 637–652. 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalarikkal S. P.; Prasad D.; Kasiappan R.; Chaudhari S. R.; Sundaram G. M. A Cost-Effective Polyethylene Glycol-Based Method for the Isolation of Functional Edible Nanoparticles from Ginger Rhizomes. Sci. Rep. 2020, 10, 4456. 10.1038/s41598-020-61358-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalarikkal S. P.; Sundaram G. M. Edible Plant-Derived Exosomal MicroRNAs: Exploiting a Cross-Kingdom Regulatory Mechanism for Targeting SARS-CoV-2. Toxicol. Appl. Pharmacol. 2021, 414, 115425. 10.1016/j.taap.2021.115425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J.; Zhuang X.; Wang Q.; Jiang H.; Deng Z. B.; Wang B.; Zhang L.; Kakar S.; Jun Y.; Miller D.; Zhang H. G. Interspecies Communication between Plant and Mouse Gut Host Cells through Edible Plant Derived Exosome-like Nanoparticles. Mol. Nutr. Food Res. 2014, 58, 1561–1573. 10.1002/mnfr.201300729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban J.-J.; Lee M.; Im W.; Kim M. Low PH Increases the Yield of Exosome Isolation. Biochem. Biophys. Res. Commun. 2015, 461, 76–79. 10.1016/j.bbrc.2015.03.172. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Chen K.; Li H.; Wu H. Effect of PH on the Isolation of Urinary Exosome. Int. Urol. Nephrol. 2017, 49, 165–169. 10.1007/s11255-016-1408-7. [DOI] [PubMed] [Google Scholar]
- Deregibus M. C.; Figliolini F.; D’antico S.; Manzini P. M.; Pasquino C.; De Lena M.; Tetta C.; Brizzi M. F.; Camussi G. Charge-Based Precipitation of Extracellular Vesicles. Int. J. Mol. Med. 2016, 38, 1359–1366. 10.3892/ijmm.2016.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.; Zhou J.; Creyer M. N.; Yim W.; Chen Z.; Messersmith P. B.; Jokerst J. V. Phenolic-Enabled Nanotechnology: Versatile Particle Engineering for Biomedicine. Chem. Soc. Rev. 2021, 50, 4432–4483. 10.1039/D0CS00908C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q.-Q.; Xu X.-Y.; Cao S.-Y.; Gan R.-Y.; Corke H.; Beta T.; Li H.-B. Bioactive Compounds and Bioactivities of Ginger (Zingiber Officinale Roscoe). Foods 2019, 8, 185. 10.3390/foods8060185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R. L.; Morr C. V.; Reineccius G. A. Whey Protein Fractionation with Polyethylene Glycol. J. Dairy Sci. 1974, 57, 793–796. 10.3168/jds.S0022-0302(74)84966-0. [DOI] [Google Scholar]
- Mihály J.; Deák R.; Szigyártó I. C.; Bóta A.; Beke-Somfai T.; Varga Z. Characterization of extracellular vesicles by IR spectroscopy: Fast and simple classification based on amide and C H stretching vibrations. Biochim. Biophys. Acta, Biomembr. 2017, 1859, 459–466. 10.1016/j.bbamem.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Kumar R.; Balaji S.; Sripriya R.; Nithya N.; Uma T. S.; Sehgal P. K. In Vitro Evaluation of Antioxidants of Fruit Extract of Momordica charantia L. on Fibroblasts and Keratinocytes. J. Agric. Food Chem. 2010, 58, 1518–1522. 10.1021/jf9025986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.