Abstract
Elevated serum prostate-specific antigen (PSA) levels in body fluids may indicate prostate cancer (PCa), but it is noted that the clinical performance is rather poor. Specificity and sensitivity values of 20 and 94% at a cutoff value of 4.1 ng/mL, respectively, result in overdiagnosis and unnecessary interventions. Previous exploratory studies have indicated that the glycosylation of PSA potentially leads to improved PCa diagnosis based on qualitative analyses. However, the applied methods are not suited for a quantitative evaluation or implementation in a medical laboratory. Therefore, in this proof-of-principle study, we have evaluated the use of hydrophilic interaction liquid chromatography (HILIC) in combination with targeted quantitative mass spectrometry for the sialic acid linkage-specific analysis of PSA glyco-proteoforms based on either trypsin or ArgC peptides. The efficiency of PSA proteolysis was optimized as well as the glycopeptide separation conditions (buffer type, strength, and pH). The HILIC-based analysis of PSA glyco-proteoforms presented here has the potential for the clinical validation of patient cohorts. The method shows the feasibility of the use of a HILIC stationary phase for the separation of isomeric glycopeptides to detect specific glyco-proteoforms. This is the first step toward the development and evaluation of PSA glyco-proteoforms for use in a clinical chemistry setting aiming for improved PCa diagnosis or screening.
Keywords: prostate-specific antigen, N-glycosylation, glycopeptide, quantitative bottom-up proteomics, prostate cancer, urine, HILIC, MRM−MS, clinical chemistry, glyco-proteoforms
Introduction
Prostate cancer (PCa) is one of the most prevalent cancers in men.1 Since the introduction of serum prostate-specific antigen (PSA) as a marker for PCa, the number of men diagnosed with PCa at an early stage has increased substantially.2 Nevertheless, this growth in (early) diagnoses has not reduced mortality rates. An elevated PSA level can indicate the presence of a tumor, but it may also be caused by prostate enlargement, benign prostate hyperplasia, or inflammation.3 Moreover, an increased PSA level is not prognostic with regard to disease severity: Both aggressive and indolent courses of PCa progression are observed, where the latter may not require any clinical intervention. The limited clinical performance of total PSA has led to a worldwide debate concerning the clinical need for alternative biomarkers for PCa screening.4−6
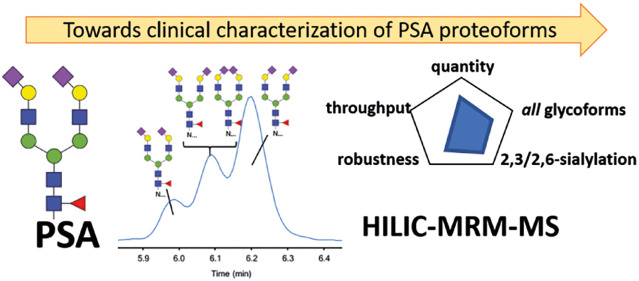
It is now widely acknowledged that there is a need for better tests with improved clinical performance specifications that can distinguish aggressive forms of PCa from clinically less significant forms of the disease. A promising candidate is glyco-proteoform analysis of PSA itself. Various studies have demonstrated aberrant glycosylation profiles in PCa patients, originating from the N-glycosylation site at Asn-45, which provides potential leads toward improving the specificity of the PSA test.7−18 The analysis of glycosyltransferase levels has furthermore demonstrated that sialic acids attached to glycoproteins are involved in cancer progression.19,20 So far, the best diagnostic potential for PCa was observed for the specific analysis of α2,3-linked sialic acids on urinary PSA using (lectin-based) immunoassays (IAs).8,11,14 These assays, however, summarize multiple glyco-proteoforms into one readout, whereas the quantification of a single species is preferred for an anticipated lab-developed test. The routine IA allows quantification with excellent sensitivity and high robustness, however, with no differentiation between various glyco-proteoforms (Figure 1). Because the diversity of PSA glycans is large, with at least 75 different structures reported from mass spectrometry (MS)-based glycopeptide analysis,21 the quantification of a specific glyco-proteoform is not trivial. For further validation and to demonstrate the clinical utility of PSA glyco-proteoforms, robust and quantitative analytical platforms are needed that include the separation of α2,3- and α2,6-linked sialic acid isomers.22 To this end, translation from biomarker discovery to a medical test has been subdivided into three tiers, of which tier 1 tests are suitable for clinical chemistry.23 Keeping these strict requirements in mind, the so-far preferred analytical strategy for MS-based protein quantitation in the medical laboratory is targeted MS by liquid chromatography (LC) coupled to multiple reaction monitoring (MRM) on a triple quadrupole instrument.24
Figure 1.
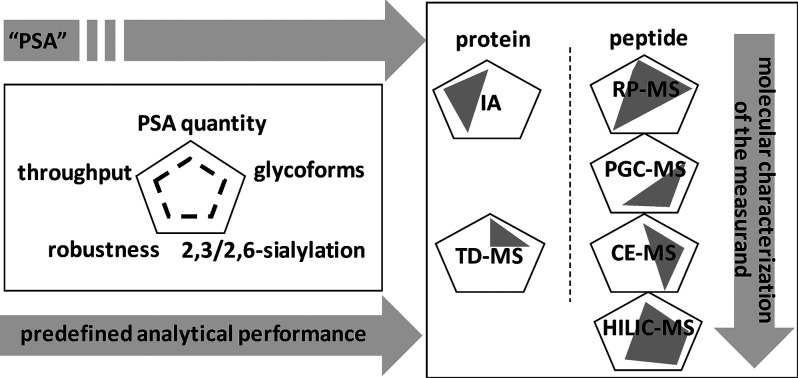
Overview of the capabilities of different strategies for the analysis of PSA and its sialylated glycopeptide linkage isomers. Six strategies typically used for PSA analysis are evaluated for their ability for absolute quantitation, throughput, robustness, glycoform profiling, and sialic acid linkage isomer-specific analysis. The gray figure indicates the capability per strategy. IA, immunoassay; TD-MS, top-down mass spectrometry; RP-MS, reversed-phase LC–MS; PGC-MS, porous graphitized carbon LC–MS; CE–MS, capillary electrophoresis coupled to MS; HILIC–MS, hydrophilic interaction liquid chromatography coupled to MS.
MS-based protein glycosylation studies generally apply a variety of separation strategies depending on whether the target analytes are released glycans or glycopeptides,25 with the latter being required for clinical chemistry purposes. With regard to PSA glycopeptide analysis, reversed-phase (RP) LC has been reported; however, this method does not distinguish different sialic acid linkages.10 The separation of sialic acid linkage isomers in PSA glycopeptides has been reported using capillary electrophoresis (CE), but this approach lacks robustness with regard to clinical chemistry requirements (Figure 1).21,26 Hitherto, a quantitative LC method for the separation of sialic acid linkage glycopeptide isomers of PSA has not been developed. With regard to glycopeptide analysis, porous graphitized carbon (PGC) and hydrophilic interaction liquid chromatography (HILIC) provide an alternative for RP LC, with both allowing for the separation of glycan linkage isomers (Figure 1).27−30However, with regard to simultaneous PSA peptide and glycopeptide analysis, HILIC is the preferred platform. HILIC material has successfully been applied for the solid-phase extraction of glycopeptides from complex mixtures,25 and recently, HILIC has been applied as a stationary phase for the analysis of isomeric N-glycopeptides from bovine fetuin and human IgG, including the separation of sialylated N-glycan isomers differing in α2,3 and α2,6 linkages.31 However, the analysis of proteotypic peptides and sialic acid linkage glycopeptide isomers using HILIC in a single run has not been described. Here we report the use of HILIC–MRM–MS for the combined absolute quantitation of PSA using “standard” proteotypic peptides and the relative quantitation of isomeric glycopeptides varying in sialic acid linkages.
Experimental Section
Chemicals, Reagents, and Enzymes
Ammonium acetate, ammonium formate (AF), ethylenediaminetetraacetic acid (EDTA), iodoacetamide (IAM), sodium deoxycholate (DOC), and tris(hydroxymethyl)aminomethane (TRIS) were obtained from Sigma-Aldrich (Zwijndrecht, The Netherlands). Ammonia solution 32%, calcium chloride, hydrochloric acid 37%, and phosphate-buffered saline (PBS) were purchased from Merck-Millipore (Burlington, MA), and tris(2-carboxyethyl)phosphine (TCEP) was purchased from Thermo Scientific (Rockford, IL). Ammonium bicarbonate (ABC) and formic acid (FA) were obtained from Honeywell (Morristown, NJ). Milli-Q water (MQ) was generated from a QGard2 system (at ≥18 MΩ) from Merck-Millipore (Burlington, MA). Sequencing-grade modified porcine trypsin and sequencing-grade ArgC were purchased from Promega (Madison, WI). Neuramidase A and S were obtained from New England Biolabs (Ipswich, MA). HPLC-grade solvents methanol (MeOH) and acetonitrile (ACN) were purchased from Biosolve (Valkenswaard, The Netherlands). In all experiments a standard PSA sample was used from Lee Biosolutions (St. Louis, MO) derived from a pool of human semen. Stable-isotope-labeled (SIL, heavy amino acid is indicated with an asterisk) and nonlabeled peptide standards were synthesized in-house, dissolved in 5% MeOH, and stored at −80 °C until further use.
Trypsin Digestion of PSA
The protocol for trypsin digestion of PSA was based on our in-house-developed method for serum apolipoproteins,32 with modifications. In brief, 200 μL of so-called reduction mix was prepared, containing 10 mM TCEP in 50 mM ABC (pH 8.0) and the two SIL-peptides FLRPGDDSSHDLMLLR* and LSEPAELTDAVK*. Then, 20 μg of human PSA was incubated in 60 μL of this reduction mix at 56 °C for 30 min to allow for disulfide bond reduction. Carbamidomethylation was performed by adding 20 μL of 10 mM IAM in 50 mM ABC with subsequent incubation at room temperature in the dark for 30 min. For digestion, 20 μg of trypsin was dissolved in 50 mM ABC to a volume of 2 mL. Proteolysis was performed at a 1:35 (w/w) trypsin–PSA ratio at 37 °C in a total volume of 200 μL. Digestion kinetics were followed by sampling after 30 min, 45 min, 60 min, 90 min, 3 h, 6 h, 9 h, 12 h, 18 h, and 24 h of incubation. Digestion was quenched using 200 μL of 0.6% (v/v) FA and 5% (v/v) MeOH in MQ. The digested sample was transferred to an LC–MS vial for analysis.
ArgC Digestion of PSA
The ArgC digestion method was based on the previously mentioned protocol for tryptic digestion, with optimization experiments performed in different buffers, namely, TRIS, ABC, sodium bicarbonate, and PBS.33 The use of EDTA and CaCl2 (recommended by the supplier) was tested (5, 20, and 50 mM) as well as the concentration TRIS (10, 50, and 100 mM). The digestion kinetics were optimized by taking a sample after 30 min, 45 min, 60 min, 90 min, 3 h, 6 h, 9 h, 12 h, 18 h, and 24 h of incubation. These optimizations resulted in a 100 mM TRIS-containing reduction mix (pH 7.8) that was used for S–S reduction and subsequent carbamidomethylation. For digestion, 10 μg of lyophilized ArgC was dissolved in 50 mM TRIS (pH 7.8) to a volume of 1 mL, and the digestion was performed at a 1:35 (w/w) ArgC–PSA ratio. The sample was incubated at 37 °C for 3 h, and the digestion was subsequently quenched using 200 μL of 0.6% (v/v) FA and 5% (v/v) MeOH in MQ. The digested sample was transferred to an LC–MS vial for analysis.
Exoglycosidase Digestion of PSA-Glycopeptides
Neuramidase A (α2,3/6/8/9-linked sialic acid cleavages) and neuramidase S (α2,3-linked sialic acid cleavages) were used to identify specific linkages in isomeric glycopeptides. In short, 3 μL of 10× GlycoBuffer 1 (50 mM CaCl2, 0.5 M sodium acetate, pH 5.5, New England Biolabs) was added to 20 μL of PSA tryptic digest to increase the pH to 4.0. Two microliters of neuramidase A or S was added to the PSA samples, and the mixture was incubated for 1 h at 37 °C. The samples were transferred to an LC–MS vial for LC–MRM–MS analysis.
Measurement of PSA Glycosylation from a Native Urine Matrix
PSA standard (Lee Biosolutions) was spiked into a PSA-deficient human urine pool at concentrations of 50, 250, and 1000 μg/L to prepare urine matrix samples. Biotinylated antihuman PSA antibodies were kindly provided by Roche (Penzberg, Germany) and were coupled to streptavidin-coated magnetic beads (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions at a concentration of 0.1 mg/mL for 10 min. 60 μL of beads was washed five times using PBS and subsequently incubated with 1 mL of urine matrix sample for 1 h. The beads were washed three times, followed by overnight on-bead trypsin digestion. Here 24 μL of 10 mM TCEP in 25 mM NH4HCO3 containing the two SIL peptides FLRPGDDSSHDLMLLR* and LSEPAELTDAVK* was added to the PSA-bound beads. Upon incubation at 56 °C for 30 min, carbamidomethylation was performed by adding 4 μL of 10 mM IAM in 25 mM NH4HCO3 with subsequent incubation at room temperature in the dark for 30 min. Proteolysis was performed using 0.1 μg of trypsin in 3 μL of 25 mM NH4HCO3 at 37 °C in a total volume of 30 μL. Digestion was quenched using 10 μL of 4.0% (v/v) FA and 10% (v/v) MeOH in MQ. The digested sample was transferred to an LC–MS vial for analysis.
HILIC–MRM–MS Analysis
An Agilent 1290 ultra-high-performance LC system equipped with a 20 μL sampling loop was used in combination with an Agilent 6495 triple quadrupole mass spectrometer (QQQ-MS, Agilent Technologies, Santa Clara, CA) operating in positive ionization mode. Three different HILIC columns were evaluated, namely, an Acquity UPLC glycan BEH amide column (130 Å 2.1 × 100 mm, 1.7 μm particle size from Waters), a TSKgel amide 80 column (100 Å, 5 μm, 4.6 × 250 mm from Tosoh Bioscience), and an InfinityLab Poroshell 120 HILIC narrow-bore LC column (2.1 × 150 mm, 1.9 μm from Agilent). The Acquity UPLC glycan BEH amide column was selected for further analysis at a column oven temperature of 40 °C. Eluent A consisted of 10 mM AF in MQ, whereas eluent B consisted of 10 mM AF in 10% MQ and 90% ACN. The pH was set at 4.2 for both the tryptic and ArgC digest. Upon the injection of 2 μL of sample, peptides were separated using the following gradient: plateau of 1 min at 80% eluent B, followed by a linear gradient to 20% eluent B at 10 min, followed by a plateau of 2 min at 80% eluent B, all at a flow of 0.3 mL/min. For peptide identification, the system was first operated in full-scan mode with m/z values ranging from 300 to 1500 to generate a full MS1 scan of the digested peptides. Then, the QQQ-MS was operated in product-ion scan mode to obtain reference fragmentation spectra of the PSA peptides. Peptides were measured in dynamic MRM mode with a 1 min window. The cycle time was set at 500 ms. Doubly or triply charged precursor ions were selected for peptides, and one quantifier and two qualifier ion transitions were monitored in unit resolution. For glycopeptides, triply and quadruply charged precursor ions were selected, together with one quantifying and one qualifying transition. For each transition, the collision energy was optimized (detailed in the Supporting Information). To ensure that the LC–MS instrumentation is performing accurately during the sample analysis, a system suitability testing (SST) procedure was designed and run in association with all digestion optimization experiments. For this purpose, a system suitability sample consisting of three nonlabeled and three labeled synthetic peptides, each at 0.15 μmol/L in 5% (vol/vol) MeOH and 0.6% (vol/vol) FA in MQ, was prepared. Five microliters of this sample was then analyzed five times prior to a test run as well as five times afterward. A blank sample followed every five samples to assess the carryover. Criteria for accurate performance were defined.
MS Parameter Optimization
The initial parameter settings on the QQQ-MS were: gas flow, 15 L/min; gas temperature, 250 °C; sheath gas temperature, 250 °C; nozzle voltage, 650 V; high-pressure ion funnel RF voltage, 150 V; fragmentor voltage, 380 V; cell accelerator voltage, 5 V; capillary voltage, 3500 V; and nebulizer pressure, 30 psi. During the optimization of the MS parameters, the following conditions were tested: gas flow, 10, 13, and 15 L/min; gas temperature, 100, 150, 200, and 250 °C; sheath gas temperature, 100, 150, 200, and 250 °C; nozzle voltage, 300, 500, 650, 800, and 1000 V; high-pressure ion funnel RF voltage, 80, 100, 125, and 150 V; fragmentor voltage, 250, 300, 350, and 380 V; cell accelerator voltage, 4, 5, and 6 V; capillary voltage, 3000, 3500, and 4000 V; and nebulizer pressure, 25, 30, and 35 psi. The optimized MS conditions were: gas flow, 15 L/min; gas temperature, 250 °C; sheath gas temperature, 250 °C; nozzle voltage, 1000 V; high-pressure ion funnel RF voltage, 125 V; fragmentor voltage, 350 V; cell accelerator voltage, 4 V; capillary voltage, 4000 V; and nebulizer pressure, 25 psi.
Data Analysis
LC–MS/MS data were processed using Mass Hunter workstation software, version B.06.00 (Agilent Technologies). Signal intensities were obtained from the peak areas, and all transitions (both quantifying and qualifying) were individually evaluated. Initial data quality control was performed by assessing the ion ratios between quantifying and qualifying transitions, which were required to be within 15% accuracy. The SST was passed for all analyses performed in this study.
Results and Discussion
Both trypsin and ArgC were considered for the proteolysis of PSA because of the common usage and specific peptide length of the glycopeptide, respectively. Methods were developed for the simultaneous analysis of peptides and glycopeptides obtained with both proteases. Trypsin is the most widely used and best characterized protease for quantitative purposes,34,35 but in the case of PSA, the resulting glycopeptides have a rather short peptide backbone of only two amino acids that limits peptide chromatographic retention. Larger tryptic PSA glycopeptides have previously been reported that contain a missed cleavage (e.g., ref (10)); however, we did not observe such glycopeptides (with backbone NKSVILLGR) in our study.
MRM Transition Development and MS Source Optimization
Transitions were developed for PSA peptides and glycopeptides, both from the digestion with trypsin and with ArgC. For both proteases, two peptides were selected for protein quantitation, namely, FLRPGDDSHDLMLLR and LSEPAELTDAVK for trypsin and FLRPGDDSHDLMLLR and KWIKDTIVANP for ArgC. Fragmentation spectra were generated to identify proteotypic peptides and develop transitions. For peptides, three major fragments were selected, whereas for glycopeptides, two of the oxonium ions m/z 274 (sialic acid–H2O), m/z 366 (HexNAcHex), and m/z 204 (HexNAc) were selected, depending on the glycopeptide identity. Collision energies were optimized for each of the transitions; see Tables S1 and S2 for the final lists of transitions. The MS source and ion-transfer parameters required optimization for glycopeptide analysis because triple quadrupole instruments are generally tuned for small molecules and peptides, whereas glycopeptides are larger structures that exhibit an inherently lower signal intensity.36 Specifically, the optimal source parameters for our system were gas flow, 15 L/min; gas temperature, 250 °C; nozzle voltage, 1000 V; nebulizer pressure, 25 psi; and capillary voltage, 4000 V. The optimal fragmentor voltage was 350 V, and the best cell accelerator voltage was 4 V. Using these parameters, the signal intensity of the glycopeptides roughly doubled, with a slight increase in the signal intensity of the peptides.
HILIC Separation Optimization
The HILIC retention mechanism largely relies on the partitioning of analytes to the water-rich layer that surrounds the hydrophilic stationary phase.37 The major characteristics that influence HILIC separation are the stationary phase,38,39 the type40 and concentration of salt,41 and the organic solvent.42,43 We aimed to optimize each of these parameters for the separation of peptides and glycopeptides from PSA (Table 1, Figure 2). Amide-based HILIC stationary phases44 or sulfobetaine-based zwitterionic (ZIC) HILIC stationary phases28 are most commonly used for glycan separation, typically using solvent systems containing ACN in AF solution. Three columns using these conditions were evaluated: a neutral glycan BEH amide column (Waters), a neutral TSKgel amide 80 column (Tosoh Bioscience), and a zwitterionic Poroshell 120 HILIC column (Agilent). The most suitable results for both tryptic and ArgC glycopeptides were obtained using the Waters BEH amide column, and further optimization was performed using this column. Subsequently, two types of buffer were tested: ACN/water with 10 mM ammonium acetate (at pH 4.0) and ACN/water with 10 mM AF (at pH 4.0); the best separation was obtained using AF, which was used for further optimization. Next, the solvent pH was optimized using a 10 mM AF buffer at different pH values ranging from 3.6 to 5.2. The performance with regard to glycopeptide separation was not altered within this pH range; however, the retention time decreased slightly with increasing pH. The retention time of peptides, on the contrary, was more affected, with the retention of some peptides increasing and that of some peptides decreasing, irrespective of the pKi of the peptide. The optimal pH for peptide and glycopeptide isomer separation was 4.2 for the ArgC digests, whereas pH 4.4 performed slightly better for tryptic glycopeptides. Furthermore, the buffer concentration is known to affect the HILIC retention, albeit not as much as the pH. Therefore, various AF concentrations were evaluated (between 5 and 50 mM). Interestingly, limited effects of the buffer concentration on (glyco)peptide retention were observed. However, there was an inverse relation between the concentration of AF and the signal intensity in the MS, which is in line with the reported ion suppression by AF in MS.45 A 10 mM concentration of AF was shown to provide stable retention times and peak shape and was therefore chosen for further method optimization. Lastly, the starting ACN concentration was optimized. Whereas higher concentrations of ACN typically result in better retention of hydrophilic compounds, we also aimed to separate less hydrophilic peptides, which could precipitate at high acetonitrile concentrations. 90% ACN provided optimal glycopeptides and peptide separation; a further increase in ACN content resulted in the increased retention of glycopeptides but not most peptides, thus decreasing the separation efficiency. Overall, the optimal conditions for the separation of the tryptic PSA digest were solvent A, 10 mM AF, pH 4.4 in water and solvent B, 90% ACN in 10 mM AF, pH 4.4, whereas the optimal conditions for the ArgC digest were solvent A, 10 mM AF, pH 4.2 in water and solvent B, 90% ACN in 10 mM AF, pH 4.2, as outlined in Table 1. A typical chromatogram of the trypsin digest is also shown in Figure 2.
Table 1. Optimization of HILIC LC–MS Separation of PSA Peptides and Glycopeptidesa.
| protease | level | % AcN (50–95%) | pH (3.6–5.2) | [AF] (5–50 mM) |
|---|---|---|---|---|
| trypsin | peptides | increased retention with increased % AcN, minimum 90% | higher pH gives better peptide separation | no effects on retention, lower signals intensity with higher AF concentration |
| glycopeptides | increased retention with increased % AcN, minimum 80% | higher pH gives less retention, but slightly better signal intensity | no effects on retention, lower signals intensity with higher AF concentration | |
| optimal | 90% can | 4.4 | 10 mM | |
| ArgC | peptides | increased retention with increased % AcN, minimum 90% | lower pH gives better peptide separation | variable effects on retention, lower signal intensity with higher AF concentration |
| glycopeptides | increased retention with increased % AcN, minimum 80% | higher pH gives less retention, but slightly better signal intensity | no effects on retention, lower signals intensity with higher AF concentration | |
| optimal | 90% can | 4.2 | 10 mM |
Effects of changes in solvent composition are indicated together with the eventual optimal solvent conditions.
Figure 2.
Optimization of the HILIC LC–MS separation of PSA glycopeptides. HILIC–MRM–MS chromatograms obtained for peptide and glycopeptide separation of the tryptic PSA digest (left) and the similar analysis of tryptic PSA digest after immunocapture of PSA from the urinary sample (right). MRM ion intensities are depicted on the y axis in arbitrary units.
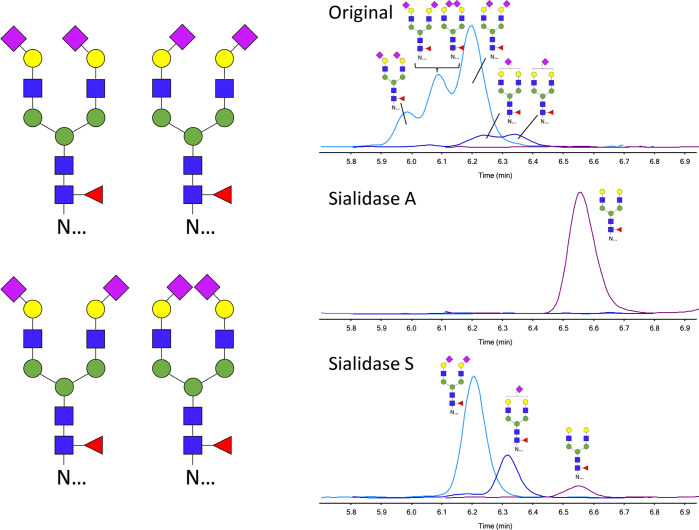
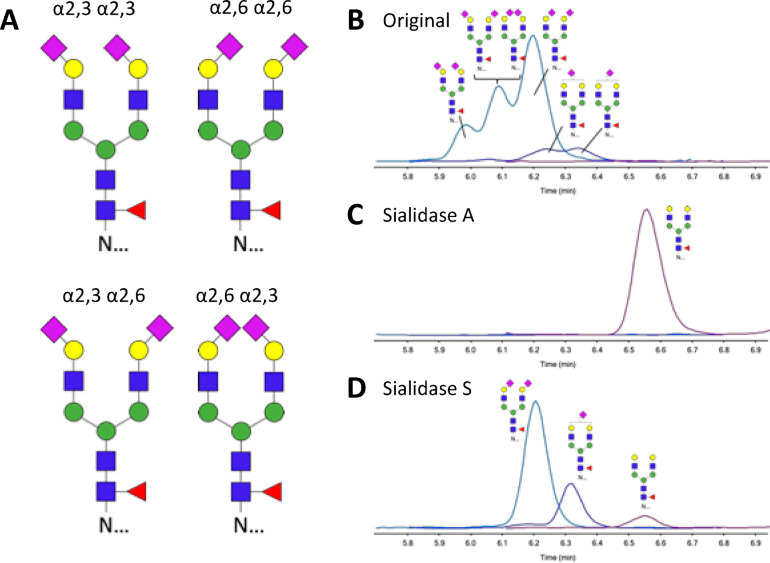
Identification of Sialic Acid Linkages
To identify the origin of the three signals retained by HILIC–LC–MS, exosialidases were used. The results of the neuraminidase treatments for ArgC glycopeptides are shown in Figure 3, and the same retention order was observed for tryptic glycopeptides. Sialidase A has a broad specificity and cleaves both α2,3- and α2,6-linked terminal sialic acids. Treatment of PSA glycopeptides results in the complete removal of both α2,3- and α2,6-linked sialic acids; see Figure 3C. Sialidase S specifically catalyzes the hydrolysis of α2,3-linked sialic acids but leaves α2,6-linked sialic acids unaffected. Figure 3D shows PSA glycopeptides with α2,6-linked sialic acids upon treatment with Sialidase S, indicating that glycans with α2,3-linked sialic acids elute slightly earlier with HILIC than α2,6-linked sialic acids.
Figure 3.
Separation of isomeric glycopeptides by HILIC LC–MS and confirmation of sialic acid linkage type. (A) Structural representation of PSA glycopeptides with glycan composition H5N4F1S2 and identification of sialic acid linkage isomers by sialidase treatment. HILIC LC–MS chromatograms of (B) original, (C) sialidase-A-treated, and (D) sialidase-A-treated argC glycopeptides from PSA. Ion intensities from MRM are depicted on the y axis in arbitrary units.
The results indicate that HILIC provides sufficient separating power to distinguish 2,3- and 2,6-linked sialic acids at the glycopeptide level and may be suitable for application in a HILIC–MRM–MS-based test in the medical laboratory. Whereas it was previously reported that isomer separations may be achieved using CE–MS, this approach lacks robustness with regard to clinical chemistry requirements (Figure 1). The HILIC-based method allows the absolute quantitation of PSA through proteotypic peptides as well as glycopeptide separation in the same run, whereas PGC cannot deliver both aspects within the same analysis (Figure 1). Peptide-based quantitation can be achieved with RP-MRM-MS with similar performance as routine immunoassays but does not provide information on glycopeptide isomers (Figure 1). To demonstrate the feasibility of HILIC–MRM–MS-based PSA glycopeptide measurements from clinical material, a proof-of-principle PSA immunocapture experiment was performed from a urinary matrix (Figure 2).
System Suitability Testing
An SST was developed to ensure the accurate performance of the LC–MRM–MS system during experiments using a mixture of both nonlabeled and labeled synthetic peptides. All peptides consistently performed within the predefined acceptance criteria, with absolute abundances deviating <10%, relative abundances deviating <15%, and carryover <1% for all peptides.
Optimization of Digestion
To ensure the accurate quantitation of PSA using isotope dilution mass spectrometry, in which the peptide signal intensity is quantified relative to an SIL peptide, consistent digestion results are necessary.22 Whereas the protease trypsin has already been widely characterized46 and has been consistently used for quantitative purposes,32,35,47,48 the protease ArgC has not. Therefore, the digestion conditions were optimized, starting from the manufacturer’s recommendations of a 50 mM TRIS buffer containing 50 mM CaCl2 and 2 mM EDTA at a protein–ArgC ratio of 1:35 (w/w) (Supplementary Figure S1). Increasing the protein–ArgC ratio did not increase the peptide recovery (Supplementary Figure S1, conditions 1–4), but the omission of EDTA and CaCl2 substantially increased the digestion efficiency (condition 7). The buffer type and concentration were then also evaluated. Increasing the TRIS concentration to 100 mM (condition 11) did not affect the peptide generation but increased the glycopeptide generation. Interestingly, the exchange of the TRIS buffer for 50 mM ABC (condition 12) performed equally well for most peptides but was not successful in the generation of glycopeptides. Therefore, the optimal digestion conditions selected were 100 mM TRIS without EDTA or CaCl2.
Evaluation of Digestion Efficiency
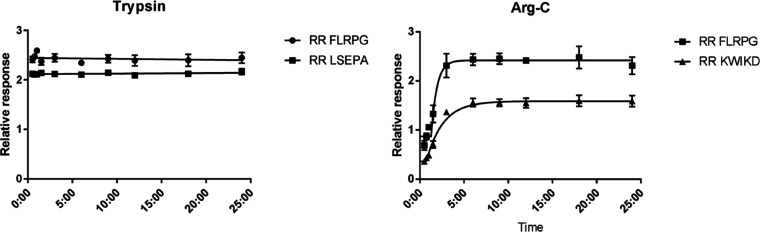
For quantitative clinical chemistry purposes, optimized digestion conditions do not necessarily result in a reproducible protein digestion. In this context, it is emphasized that the large majority of MS-based proteomics studies have focused on optimizing the number of protein identities rather than quantities. A prerequisite for true quantitative results is that the digestion reaches a plateau, which is preferable to even completeness.34,49 To this end, digestion time courses were made using the optimized digestion conditions for both trypsin and ArgC digestion. The results for peptides are shown in Figure 4 and are representative for the glycopeptides. Plateaus are reached within 30 min for trypsin and within 3 h for ArgC without major signal loss for the SIL peptides, indicating that stable digestion can be reached and most likely allowing for repeatable quantitative results.
Figure 4.
PSA digestion curves. After the optimization of digestion conditions with regard to buffer and additives, digestion curves were generated to evaluate the progress of digestion and the stability of the digest. An optimum was reached within 1 h for trypsin (left) and within 6 h for ArgC (right).
So far, further evaluation and proof of equimolarity are required to make a decision between the use of trypsin and ArgC for the development of a medical test, as both proteases have positives and negatives. The longer peptide backbone generated using ArgC (NKSVILLGR vs NK with trypsin) will likely provide better analytical specificity without a loss of separation power on the HILIC stationary phase. Trypsin, on the contrary, is preferred for clinical chemistry applications because it is well-characterized, widely used, and widely available. Therefore, both proteases should be evaluated during further test development.
Conclusions
In this study, a HILIC-based LC–MRM–MS method for the quantitation of PSA was established, including the measurement of specific glycopeptide linkage isomers. The results indicate that HILIC provides sufficient separating power to distinguish 2,3- and 2,6-linked sialic acids at the glycopeptide level and may be suitable for application in a HILIC–MRM–MS-based test in the medical laboratory. With suitable external calibrators, this HILIC-based method allows the absolute quantitation of PSA through proteotypic peptides as well as glycopeptide separation in the same run. Peptide-based quantitation has been achieved with RP–MRM–MS with similar performance compared with routine IAs; however, it does not provide information on glycopeptide isomers, which is essential for precision oncology and refined PCa diagnosis. It is emphasized that the stable digestion of PSA is an essential prerequisite for an accurate test, and absolute quantitation may be achieved using either trypsin or ArgC. The inherently low abundance of PSA in serum, plasma, and urine will pose a challenge for translating these results into an improved PCa test, specifically due to the 100-fold signal intensity difference between the peptides and the glycopeptides (Figure 1). Therefore, the sensitivity of the HILIC–MRM–MS method warrants additional developments.
Notwithstanding the benefits of proteomics approaches, it is widely acknowledged that most (if not all) of the biomarker candidates do not find their way into clinical diagnostics applications. The careful evaluation of promising biomarkers with appropriate techniques, study designs, and samples is essential for successful translation.50,51 The HILIC-based LC–MS method that was developed in this study for the separation of PSA glycopeptide linkage isomers, specifically the attachment of 2,3- and 2,6-linked sialic acids, demonstrates the feasibility of quantifying individual glycopeptide isomers. Moreover, the focus on specific glyco-proteoforms using a targeted MS approach as outlined here provides the granularity that is necessary for the development of molecular tests for the detection and monitoring of PCa in this era of precision medicine.
Acknowledgments
We thank Roche (Penzberg, Germany) for supplying biotinylated anti-PSA antibody and Mr. Fred Romijn for performing proof-of-principle PSA immunocapture experiments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00050.
Table S1. Transitions developed for tryptic peptides and glycopeptides from PSA. Table S2. Transitions developed for ArgC peptides and glycopeptides from PSA. Figure S1. Optimization of digestion conditions. Figure S2. Repeatability of the LC–MRM–MS analysis of PSA (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Siegel R. L.; Miller K. D.; Jemal A. Cancer statistics, 2019. Ca-Cancer J. Clin. 2019, 69 (1), 7–34. 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Etzioni R.; Tsodikov A.; Mariotto A.; Szabo A.; Falcon S.; Wegelin J.; DiTommaso D.; Karnofski K.; Gulati R.; Penson D. F.; Feuer E. Quantifying the role of PSA screening in the US prostate cancer mortality decline. Cancer causes & control: CCC 2008, 19 (2), 175–81. 10.1007/s10552-007-9083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson I. M.; Ankerst D. P.; Chi C.; Lucia M. S.; Goodman P. J.; Crowley J. J.; Parnes H. L.; Coltman C. A. Jr. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/mL or lower. JAMA 2005, 294 (1), 66–70. 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- Vickers A. J. Redesigning Prostate Cancer Screening Strategies to Reduce Overdiagnosis. Clin. Chem. 2019, 65 (1), 39–41. 10.1373/clinchem.2018.287094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PSA position. Nature 2011, 478 ( (7369), ), 286. 10.1038/478286a. [DOI] [PubMed] [Google Scholar]
- Tikkinen K. A. O.; Dahm P.; Lytvyn L.; Heen A. F.; Vernooij R. W. M.; Siemieniuk R. A. C.; Wheeler R.; Vaughan B.; Fobuzi A. C.; Blanker M. H.; Junod N.; Sommer J.; Stirnemann J.; Yoshimura M.; Auer R.; MacDonald H.; Guyatt G.; Vandvik P. O.; Agoritsas T. Prostate cancer screening with prostate-specific antigen (PSA) test: a clinical practice guideline. BMJ. 2018, 362, k3581. 10.1136/bmj.k3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burgt Y. E. M.; Cobbaert C. M. Proteoform Analysis to Fulfill Unmet Clinical Needs and Reach Global Standardization of Protein Measurands in Clinical Chemistry Proteomics. Clin Lab Med. 2018, 38 (3), 487–497. 10.1016/j.cll.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Yoneyama T.; Ohyama C.; Hatakeyama S.; Narita S.; Habuchi T.; Koie T.; Mori K.; Hidari K. I.; Yamaguchi M.; Suzuki T.; Tobisawa Y. Measurement of aberrant glycosylation of prostate specific antigen can improve specificity in early detection of prostate cancer. Biochem. Biophys. Res. Commun. 2014, 448 (4), 390–6. 10.1016/j.bbrc.2014.04.107. [DOI] [PubMed] [Google Scholar]
- Peracaula R.; Tabares G.; Royle L.; Harvey D. J.; Dwek R. A.; Rudd P. M.; De Llorens R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology 2003, 13 (6), 457–470. 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- Haga Y.; Uemura M.; Baba S.; Inamura K.; Takeuchi K.; Nonomura N.; Ueda K. Identification of Multisialylated LacdiNAc Structures as Highly Prostate Cancer Specific Glycan Signatures on PSA. Anal. Chem. 2019, 91 (3), 2247–2254. 10.1021/acs.analchem.8b04829. [DOI] [PubMed] [Google Scholar]
- Ferrer-Batalle M.; Llop E.; Ramirez M.; Aleixandre R. N.; Saez M.; Comet J.; de Llorens R.; Peracaula R. Comparative Study of Blood-Based Biomarkers, alpha2,3-Sialic Acid PSA and PHI, for High-Risk Prostate Cancer Detection. Int. J. Mol. Sci. 2017, 18 (4), 845. 10.3390/ijms18040845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C. J.; Tzai T. S.; Chen C. H.; Yang W. H.; Chen C. H. Analysis of Urinary Prostate-Specific Antigen Glycoforms in Samples of Prostate Cancer and Benign Prostate Hyperplasia. Dis. Markers 2016, 2016, 1. 10.1155/2016/8915809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Dong Z.; Sun C.; Wen F.; Wang H.; Guo H.; Gao X.; Xu C.; Yang C.; Sun Y. Alterations in expressed prostate secretion-urine PSA N-glycosylation discriminate prostate cancer from benign prostate hyperplasia. Oncotarget 2017, 8 (44), 76987–76999. 10.18632/oncotarget.20299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T.; Yoneyama T.; Tobisawa Y.; Hatakeyama S.; Kurosawa T.; Nakamura K.; Narita S.; Mitsuzuka K.; Duivenvoorden W.; Pinthus J. H.; Hashimoto Y.; Koie T.; Habuchi T.; Arai Y.; Ohyama C. An Automated Micro-Total Immunoassay System for Measuring Cancer-Associated alpha2,3-linked Sialyl N-Glycan-Carrying Prostate-Specific Antigen May Improve the Accuracy of Prostate Cancer Diagnosis. Int. J. Mol. Sci. 2017, 18 (2), 470. 10.3390/ijms18020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llop E.; Ferrer-Batalle M.; Barrabes S.; Guerrero P. E.; Ramirez M.; Saldova R.; Rudd P. M.; Aleixandre R. N.; Comet J.; de Llorens R.; Peracaula R. Improvement of Prostate Cancer Diagnosis by Detecting PSA Glycosylation-Specific Changes. Theranostics 2016, 6 (8), 1190–204. 10.7150/thno.15226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldova R.; Fan Y.; Fitzpatrick J. M.; Watson R. W.; Rudd P. M. Core fucosylation and alpha2–3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology 2011, 21 (2), 195–205. 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- Vermassen T.; Van Praet C.; Lumen N.; Decaestecker K.; Vanderschaeghe D.; Callewaert N.; Villeirs G.; Hoebeke P.; Van Belle S.; Rottey S.; Delanghe J. Urinary prostate protein glycosylation profiling as a diagnostic biomarker for prostate cancer. Prostate 2015, 75 (3), 314–22. 10.1002/pros.22918. [DOI] [PubMed] [Google Scholar]
- Drake R. R.; Jones E. E.; Powers T. W.; Nyalwidhe J. O. Altered glycosylation in prostate cancer. Adv. Cancer Res. 2015, 126, 345–82. 10.1016/bs.acr.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Kawamura S.; Sato I.; Wada T.; Yamaguchi K.; Li Y.; Li D.; Zhao X.; Ueno S.; Aoki H.; Tochigi T.; Kuwahara M.; Kitamura T.; Takahashi K.; Moriya S.; Miyagi T. Plasma membrane-associated sialidase (NEU3) regulates progression of prostate cancer to androgen-independent growth through modulation of androgen receptor signaling. Cell Death Differ. 2012, 19 (1), 170–9. 10.1038/cdd.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Wuhrer M.; Holst S. Serum sialylation changes in cancer. Glycoconjugate J. 2018, 35 (2), 139–160. 10.1007/s10719-018-9820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammeijer G. S. M.; Jansen B. C.; Kohler I.; Heemskerk A. A. M.; Mayboroda O. A.; Hensbergen P. J.; Schappler J.; Wuhrer M. Sialic acid linkage differentiation of glycopeptides using capillary electrophoresis - electrospray ionization - mass spectrometry. Sci. Rep. 2017, 7 (1), 3733. 10.1038/s41598-017-03838-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit N.; Van den Broek I.; Romijn F. P.; Haex M.; Deelder A. M.; Van der Burgt Y. E.; Van der Laarse A.; Cobbaert C. M. Quality requirements for quantitative clinical chemistry proteomics. Transl. Proteomics 2014, 2, 1–13. 10.1016/j.trprot.2013.10.00. [DOI] [Google Scholar]
- Carr S. A.; Abbatiello S. E.; Ackermann B. L.; Borchers C.; Domon B.; Deutsch E. W.; Grant R. P.; Hoofnagle A. N.; Huttenhain R.; Koomen J. M.; Liebler D. C.; Liu T.; MacLean B.; Mani D.; Mansfield E.; Neubert H.; Paulovich A. G.; Reiter L.; Vitek O.; Aebersold R.; Anderson L.; Bethem R.; Blonder J.; Boja E.; Botelho J.; Boyne M.; Bradshaw R. A.; Burlingame A. L.; Chan D.; Keshishian H.; Kuhn E.; Kinsinger C.; Lee J. S. H.; Lee S. W.; Moritz R.; Oses-Prieto J.; Rifai N.; Ritchie J.; Rodriguez H.; Srinivas P. R.; Townsend R. R.; Van Eyk J.; Whiteley G.; Wiita A.; Weintraub S. Targeted Peptide Measurements in Biology and Medicine: Best Practices for Mass Spectrometry- based Assay Development Using a Fit- for- Purpose Approach. Mol. Cell. Proteomics 2014, 13 (3), 907–917. 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak L. R.; Romijn F. P. H. T. M.; Smit N. P. M.; van der Laarse A.; Pieterse M. M.; de Maat M. P. M.; Haas F. J. L. M.; Kluft C.; Amiral J.; Meijer P.; Cobbaert C. M. Detecting molecular forms of antithrombin by LC-MRM-MS: defining the measurands. Clin. Chem. Lab. Med. 2018, 56 (10), 1704–1714. 10.1515/cclm-2017-1111. [DOI] [PubMed] [Google Scholar]
- Ruhaak L. R.; Xu G.; Li Q.; Goonatilleke E.; Lebrilla C. B. Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses. Chem. Rev. 2018, 118 (17), 7886–7930. 10.1021/acs.chemrev.7b00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammeijer G. S. M.; Nouta J.; de la Rosette J.; de Reijke T. M.; Wuhrer M. An In-Depth Glycosylation Assay for Urinary Prostate-Specific Antigen. Anal. Chem. 2018, 90 (7), 4414–4421. 10.1021/acs.analchem.7b04281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagesan K.; Everest-Dass A.; Kolarich D. Isomeric Separation and Characterisation of Glycoconjugates. Adv. Exp. Med. Biol. 2018, 1104, 77–99. 10.1007/978-981-13-2158-0_5. [DOI] [PubMed] [Google Scholar]
- Zauner G.; Deelder A. M.; Wuhrer M. Recent advances in hydrophilic interaction liquid chromatography (HILIC) for structural glycomics. Electrophoresis 2011, 32 (24), 3456–66. 10.1002/elps.201100247. [DOI] [PubMed] [Google Scholar]
- Palmisano G.; Larsen M. R.; Packer N. H.; Thaysen-Andersen M. Structural analysis of glycoprotein sialylation – part II: LC-MS based detection. RSC Adv. 2013, 3 (45), 22706–22726. 10.1039/c3ra42969e. [DOI] [Google Scholar]
- Kozlik P.; Goldman R.; Sanda M. Hydrophilic interaction liquid chromatography in the separation of glycopeptides and their isomers. Anal. Bioanal. Chem. 2018, 410 (20), 5001–5008. 10.1007/s00216-018-1150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Nie Y.; Boyes B.; Orlando R. Resolving Isomeric Glycopeptide Glycoforms with Hydrophilic Interaction Chromatography (HILIC). Journal of biomolecular techniques: JBT 2016, 27 (3), 98–104. 10.7171/jbt.16-2703-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek I.; Romijn F. P.; Nouta J.; van der Laarse A.; Drijfhout J. W.; Smit N. P.; van der Burgt Y. E.; Cobbaert C. M. Automated Multiplex LC-MS/MS Assay for Quantifying Serum Apolipoproteins A-I, B, C-I, C-II, C-III, and E with Qualitative Apolipoprotein E Phenotyping. Clin. Chem. 2016, 62 (1), 188–97. 10.1373/clinchem.2015.246702. [DOI] [PubMed] [Google Scholar]
- Giansanti P.; Tsiatsiani L.; Low T. Y.; Heck A. J. Six alternative proteases for mass spectrometry-based proteomics beyond trypsin. Nat. Protoc. 2016, 11 (5), 993–1006. 10.1038/nprot.2016.057. [DOI] [PubMed] [Google Scholar]
- van den Broek I.; Smit N. P.; Romijn F. P.; van der Laarse A.; Deelder A. M.; van der Burgt Y. E.; Cobbaert C. M. Evaluation of interspecimen trypsin digestion efficiency prior to multiple reaction monitoring-based absolute protein quantification with native protein calibrators. J. Proteome Res. 2013, 12 (12), 5760–74. 10.1021/pr400763d. [DOI] [PubMed] [Google Scholar]
- Fu Q.; Kowalski M. P.; Mastali M.; Parker S. J.; Sobhani K.; van den Broek I.; Hunter C. L.; Van Eyk J. E. Highly Reproducible Automated Proteomics Sample Preparation Workflow for Quantitative Mass Spectrometry. J. Proteome Res. 2018, 17 (1), 420–428. 10.1021/acs.jproteome.7b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Q.; Ruhaak L. R.; Totten S. M.; Smilowitz J. T.; German J. B.; Lebrilla C. B. Label-free absolute quantitation of oligosaccharides using multiple reaction monitoring. Anal. Chem. 2014, 86 (5), 2640–7. 10.1021/ac404006z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert A. J. Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. J. Chromatogr. 1990, 499, 177–96. 10.1016/S0021-9673(00)96972-3. [DOI] [PubMed] [Google Scholar]
- Jandera P.; Janas P. Recent advances in stationary phases and understanding of retention in hydrophilic interaction chromatography. A review. Anal. Chim. Acta 2017, 967, 12–32. 10.1016/j.aca.2017.01.060. [DOI] [PubMed] [Google Scholar]
- Ikegami T.; Tomomatsu K.; Takubo H.; Horie K.; Tanaka N. Separation efficiencies in hydrophilic interaction chromatography. J.Chromatogr.A 2008, 1184 (1–2), 474–503. 10.1016/j.chroma.2008.01.075. [DOI] [PubMed] [Google Scholar]
- McCalley D. V. Understanding and manipulating the separation in hydrophilic interaction liquid chromatography. Journal of chromatography. A 2017, 1523, 49–71. 10.1016/j.chroma.2017.06.026. [DOI] [PubMed] [Google Scholar]
- Alpert A. J. Effect of salts on retention in hydrophilic interaction chromatography. Journal of chromatography. A 2018, 1538, 45–53. 10.1016/j.chroma.2018.01.038. [DOI] [PubMed] [Google Scholar]
- Alagesan K.; Khilji S. K.; Kolarich D. It is all about the solvent: on the importance of the mobile phase for ZIC-HILIC glycopeptide enrichment. Anal. Bioanal. Chem. 2017, 409 (2), 529–538. 10.1007/s00216-016-0051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Heaton J. C.; McCalley D. V. Practical investigation of the factors that affect the selectivity in hydrophilic interaction chromatography. Journal of chromatography. A 2013, 1276, 33–46. 10.1016/j.chroma.2012.12.037. [DOI] [PubMed] [Google Scholar]
- Ruhaak L. R.; Huhn C.; Waterreus W. J.; de Boer A. R.; Neususs C.; Hokke C. H.; Deelder A. M.; Wuhrer M. Hydrophilic interaction chromatography-based high-throughput sample preparation method for N-glycan analysis from total human plasma glycoproteins. Anal. Chem. 2008, 80 (15), 6119–26. 10.1021/ac800630x. [DOI] [PubMed] [Google Scholar]
- Johnson D.; Boyes B.; Orlando R. The use of ammonium formate as a mobile-phase modifier for LC-MS/MS analysis of tryptic digests. Journal of biomolecular techniques: JBT 2013, 24 (4), 187–97. 10.7171/jbt.13-2404-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek I.; Romijn F. P.; Smit N. P.; van der Laarse A.; Drijfhout J. W.; van der Burgt Y. E.; Cobbaert C. M. Quantifying protein measurands by peptide measurements: where do errors arise?. J. Proteome Res. 2015, 14 (2), 928–42. 10.1021/pr5011179. [DOI] [PubMed] [Google Scholar]
- Hoofnagle A. N.; Becker J. O.; Wener M. H.; Heinecke J. W. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin. Chem. 2008, 54 (11), 1796–804. 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bros P.; Delatour V.; Vialaret J.; Lalere B.; Barthelemy N.; Gabelle A.; Lehmann S.; Hirtz C. Quantitative detection of amyloid-beta peptides by mass spectrometry: state of the art and clinical applications. Clin. Chem. Lab. Med. 2015, 53 (10), 1483–93. 10.1515/cclm-2014-1048. [DOI] [PubMed] [Google Scholar]
- Shuford C. M.; Walters J. J.; Holland P. M.; Sreenivasan U.; Askari N.; Ray K.; Grant R. P. Absolute Protein Quantification by Mass Spectrometry: Not as Simple as Advertised. Anal. Chem. 2017, 89 (14), 7406–7415. 10.1021/acs.analchem.7b00858. [DOI] [PubMed] [Google Scholar]
- Monaghan P. J.; Lord S. J.; St John A.; Sandberg S.; Cobbaert C. M.; Lennartz L.; Verhagen-Kamerbeek W. D.; Ebert C.; Bossuyt P. M.; Horvath A. R. Test Evaluation Working Group of the European Federation of Clinical, C.; Laboratory, M., Biomarker development targeting unmet clinical needs. Clin. Chim. Acta 2016, 460, 211–9. 10.1016/j.cca.2016.06.037. [DOI] [PubMed] [Google Scholar]
- Horvath A. R.; Lord S. J.; StJohn A.; Sandberg S.; Cobbaert C. M.; Lorenz S.; Monaghan P. J.; Verhagen-Kamerbeek W. D.; Ebert C.; Bossuyt P. M. Test Evaluation Working Group of the European Federation of Clinical Chemistry Laboratory, M., From biomarkers to medical tests: the changing landscape of test evaluation. Clin. Chim. Acta 2014, 427, 49–57. 10.1016/j.cca.2013.09.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.