Abstract
Intrinsic cardiorespiratory fitness (iCRF) indicates the CRF level in the sedentary state. However, even among sedentary individuals, a wide interindividual variability is observed in the iCRF levels, whose associated molecular characteristics are little understood. This study aimed to investigate whether serum and skeletal muscle metabolomics profiles are associated with iCRF, measured by maximal power output (MPO). Seventy sedentary young adults were submitted to venous blood sampling, a biopsy of the vastus lateralis muscle and iCRF assessment. Blood serum and muscle tissue samples were analyzed by proton nuclear magnetic resonance (1H NMR) spectroscopy. Metabolites related to iCRF were those supported by three levels of evidence: (1) correlation with iCRF, (2) significant difference between individuals with low and high iCRF, and (3) metabolite contribution to significant pathways associated with iCRF. From 43 serum and 70 skeletal muscle analyzed metabolites, iCRF was positively associated with levels of betaine, threonine, proline, ornithine, and glutamine in serum and lactate, fumarate, NADP+, and formate in skeletal muscle. Serum betaine and ornithine and skeletal muscle lactate metabolites explained 31.2 and 16.8%, respectively, of the iCRF variability in addition to body mass. The results suggest that iCRF in young adults is positively associated with serum and skeletal muscle metabolic levels, indicative of the amino acid and carbohydrate metabolism.
Keywords: metabolomics, intrinsic cardiorespiratory fitness, biomarkers
1. Introduction
Cardiorespiratory fitness (CRF) reflects the ability of the circulatory and respiratory systems to capture, transport, and supply oxygen to muscles during exercise training and can be directly assessed by measuring maximal oxygen uptake (V̇O2MAX) or represented indirectly by maximal power output (MPO), maximal running speed, and time to exhaustion obtained in incremental exercise tests.1−3 Low CRF levels are associated with a higher incidence of cardiovascular diseases and death from all causes.4,5
For a better understanding of CRF level effects on health, it is important to distinguish intrinsic CRF (iCRF) from acquired CRF. iCRF indicates the level of CRF in the sedentary state, while acquired CRF shows the level gained after exposure to regular exercise programs.6 However, there is no substantial relationship between iCRF levels and the magnitude of CRF gains acquired post-training.7
Even in sedentary individuals without a history of substantial involvement in regular exercise programs, large interindividual differences are observed in iCRF levels.6,8,9 In healthy and sedentary adults aged 17–41 years, the distribution of intrinsic CRF may vary from 14 to 58 mL·kg–1·min–1 when measured by V̇O2MAX.6 As seen in data from the HERITAGE Family Study, several factors may contribute to this variation, with heredity explaining 51% of the variation in V̇O2MAX adjusted for age, sex, body weight, lean body mass, and body fat.10
Although physiological and genetic aspects of iCRF have received some attention,6,8−11 little is known about molecular determinants and pathways related to variation in iCRF. Recently, studies have used comprehensive methods such as metabolomics to identify biomarkers associated with CRF.12−16
Metabolomics allows comprehensive identification and quantification of metabolites, which are small molecules resulting from cellular biochemical and physiological processes.17,18 Metabolites are the end products of interactions between genome, transcriptome, proteome, and cellular/tissue environments, which can provide accurate information about complex phenotypes such as CRF.19 Particularly, nuclear magnetic resonance (NMR) spectroscopy is one of the most widely employed metabolomics platforms for detecting and measuring metabolites related to physical activity, exercise, and health.20,21 The metabolomics based on NMR is known for its reproducibility, minimal sample preparation requirements, and nondestructive nature.22
Previous metabolomics-based studies have reported differences in the metabolic profile for different CRF levels,12,13,15,16 where young adults with elevated CRF presented lower baseline plasma levels of metabolites γ-tocopherol,13 isoleucine, 4-aminobutyrate, proline, 4-hydroxyproline, serine, phenylalanine, lysine,15 and choline,12 as well as higher baseline plasma levels of phosphatidylcholine,12 creatinine, and docosahexaenoic acid13 when compared to individuals with low CRF. In addition, significant associations of CRF with baseline serum (piperine, pyridoxine, glycerol, 3-methoxytyrosine, and 2-hydroxyisobutyrate)14 and urinary (cis-aconitate, tyrosine, guanidinoacetate, uracil, lactate, 3-aminoisobutyrate, trans-aconitate) metabolic levels have been demonstrated,16 whose combination of these metabolites were able to explain the CRF variance in 80% for an unadjusted model including only males14 and 17.6%16 for a model adjusted for age, sex, menopausal status, and lean body mass, respectively. On the other hand, a study with an animal model showed lower levels of glutamine, O-acetylcarnitine, citrate, and proline and higher levels of glycerol, alanine, and methionine in the skeletal muscle of rats with high CRF when compared to those in rats with low CRF.23 However, to the best of our knowledge, no previous study has investigated the relationship between the metabolic profile of skeletal muscle and interindividual variation of iCRF in humans.
In this sense, studying systemic integrative molecular bases and skeletal muscle through a metabolomics profile has the potential to clarify the mechanisms linked with heterogeneous levels of iCRF among individuals with similar phenotypic traits. The identification of serum molecules associated with iCRF has implications for the discovery of easily accessible future biomarkers, possibly enabling an assessment of individual health status without exhaustive assessments of high cost and complexity. While the identification of skeletal muscle metabolites contributes to elucidating the underlying physiology and biochemistry of iCRF. Therefore, this study aimed to investigate whether the baseline serum and skeletal muscle metabolic profile is associated with iCRF levels among sedentary young males.
2. Materials and Methods
2.1. Participants
Participated in this study seventy healthy young men from the TraInability and MEtabolomicS study (TIMES study). Additional information and details can be found in a previous publication.24 Briefly, participants were sedentary, defined as not engaged in regular exercises lasting 30 min·wk–1 or more, involving an energy expenditure of 6 metabolic equivalents (METS) or more in the previous 4 months.3,24,25 All participants provided a detailed medical history and received a medical examination that included an electrocardiogram at rest. All participants were free from smoking, hypertension (blood pressure > 140/90 mmHg), diabetes (fasting glucose > 7.0 mmol·L–1), obesity (defined as body mass index > 33 kg·m–2), dyslipidemia (based on medication use), heart diseases, metabolic disorders, significant chronic respiratory conditions, or musculoskeletal problems interfering with exercise.24 The sample size was sufficient to promote statistical power above 80% in analyses of association with values of r2 ≥ 0.2.26,27 The study was conducted following the Declaration of Helsinki. The protocol was approved by the Research Ethics Committee of the University (number, 2.717.688; CAAE, 52997216.8.0000.5404), and all participants gave their informed consent for inclusion before they participated in the study.
2.2. Experimental Design
First, blood samples and biopsy of the vastus lateralis muscle were collected after 12 h overnight fasting following a standardized meal as recommended for metabolomics studies.28 The standardized meal was balanced (60% carbohydrates, 25% lipids, and 15% proteins) and based on 30% of estimated daily energy.28−30 After 72 h, body composition was evaluated through plethysmography, followed by cardiorespiratory assessment and retest after 48 h.31
2.3. Blood and Muscle Tissue Collection
Samples of venous blood and muscle tissue were collected between 7 and 10 am. After that, the blood samples were centrifuged at 5000 rpm for 10 min and then serum aliquots were stored in a freezer at −80 °C. After blood collection, a biopsy of the vastus lateralis muscle of the dominant lower limb was performed according to previously described procedures.32 Prior to tissue extraction, the area was shaved and cleaned with an antiseptic. A small area of the selected region was anesthetized with 2% xylocaine injected subcutaneously. After anesthesia, a small incision (∼5 mm) was performed until muscle fascia using a surgical scalpel. The biopsy needle was then inserted into the muscle (∼3 cm) to obtain the muscle tissue sample. After removing the tissue, the incision was closed and covered with a bandage. The samples were cleaned (free from blood and excess connective tissue), separated in aliquots, immediately frozen in liquid nitrogen, and stored at −80 °C for further analyses.24
2.4. Body Composition Assessment
Participants were instructed to drink only water and not to eat or exercise for a period of 2 h before the assessment. All measurements were performed with participants without shoes and metallic accessories, wearing only swimsuits and shower caps. Body mass and height were measured using a digital scale and a stadiometer (BOD POD, Cosmed, Chicago), respectively. Then, body density was assessed by a whole-body plethysmograph that was calibrated under standard conditions of room temperature and humidity, according to the manufacturer’s recommendations (BOD POD; Body Composition System; Life Measurement Instruments; Concord, CA).33 Then, body density was converted into body fat percentage using the Siri equation.34
2.5. Cardiorespiratory Assessment
The cardiorespiratory assessment was performed using a cycle ergometer with electromagnetic braking (Corival 400, Quinton Instrument Co., Groningen, The Netherlands). After 5 min rest (sitting position), blood pressure was measured using the auscultatory method by a mercury column sphygmomanometer (Narcosul, Brazil). Then, the test started with a 3 min warm-up at 50 W, followed by increments of 25 W·min–1 until exhaustion.35 The pedaling rate was 70–80 rpm. The test was interrupted when the participant was unable to continue or keep a minimum rate of 70 rpm, despite verbal encouragement.36
Heart rate (HR) was continuously monitored during the whole test using a cardiofrequency monitor (S810, Polar, Keple, Finland), and the ratings of perceived exertion were recorded in the final 15 s of each exercise stage using Borg’s scale.37 The cycle ergometer was calibrated according to the manufacturer’s recommendations before and after each test.
Maximal heart rate (HRMAX) was obtained from the mean values in the final 10 s of the test. Maximal power output (MPO) was calculated as Wcompleted + [25·(t/60)], where Wcompleted is the last fully completed workload level and t is the number of seconds in the final workload.38 The highest MPO value recorded between tests and its respective time to exhaustion was considered for the analyses and defined as the measure of iCRF. MPO’s test–retest presented high reproducibility (coefficient of variation = 2.8% and intraclass correlation coefficient = 0.98).24
2.6. Blood Sample Preparation for Metabolomics Analysis
Prior to the metabolomics analysis by 1H NMR spectroscopy, 3 kDa filters (Amicon Ultra) were washed with 500 μL of Milli-Q water, followed by centrifugation at 14 000 rpm for 10 min at 4 °C. This process was repeated five times. After the fifth wash, spinning (filter reverse and rotation at 8000 rpm for 5 s) was performed to eliminate any residue of Milli-Q water. After that, 500 μL of blood serum was added to the filter and it was centrifuged at 14 000 rpm for 45 min at 4 °C. Then, 250 μL of filtered serum was transferred to a 5 mm NMR tube (Wilmad Standard Series 5 mm, Sigma-Aldrich) containing 60 μL of phosphate buffer [(monobasic sodium phosphate, NaH2PO4 H20, 137.99 g·mol–1; dibasic sodium phosphate, Na2HPO3, 141.96 g·mol–1), TMSP-d4 (3-(trimethylsilyl)-2,2′,3,3′-tetradeuteropropionic acid), at 50 mmol·L–1 in D2O (6.06 μL) (internal reference)] and 290 μL of D2O (99.9%; Cambridge Isotope Laboratories Inc.).21,24,39
2.7. Muscle Tissue Sample Preparation for Metabolomics Analysis
Muscle tissue samples were processed as described in a previous publication.24 Briefly, muscle tissue fragments (∼40 mg) were weighed and added to a cold methanol/chloroform solution (2:1 v/v, total 2.5 mL). Then, the tissues were homogenized on ice (for 30 s, three times, alternating with a 10 s pause) and sonicated (for 1 min, three times, alternating with a 10 s pause). After that, a cold solution of chloroform/Milli-Q water (1:1 v/v, total 2.5 mL) was added to the samples. The samples were briefly vortexed to form an emulsion and centrifuged (2000g, for 30 min, at 4 °C). The upper phase of the mixture (containing methanol, water, and polar metabolites) was collected and evaporated in a vacuum concentrator (miVac Duo Concentrator, Genevac, U.K.). The remaining solid phase was rehydrated in 0.6 mL of deuterium oxide containing phosphate buffer (0.1 M, pH 7.4) and 0.5 mM TMSP-d4 and then added to a 5 mm NMR tube (Wilmad Standard Series 5 mm, Sigma-Aldrich) for scanning and acquisition of the spectrum on the spectrometer.
2.8. Spectrum Acquisition and Metabolite Quantification
Each spectrum of 1H NMR was acquired using VnmrJ software (Varian NMR Systems) and a Varian Inova 1H NMR spectrometer (Agilent Technologies Inc., Santa Clara), operating at 600 MHz frequency and a constant temperature of 298 K (25 °C). A total of 256 free induction decays (FIDs) were collected over a spectral width of 8000 Hz, with an acquisition time of 4 s and relaxation delay intervals of 1.5 s.
After spectrum acquisition, baseline corrections, identification, and quantification of metabolites present in the samples were conducted using Suite 7.6 Chenomx software (Chenomx Inc., Edmonton, AB, Canada) by the TMSP signal (known concentration) as an internal reference to quantify other metabolites. All spectra were processed with 0.5 Hz line broadening (lb) to attenuate the noise in the spectral signals (Figures S1 and S2). Metabolites involved in the sample collection/preparation process (ethanol and acetone in serum, methanol in skeletal muscle) were excluded from the analysis, as well as those previously reported as presenting low reproducibility in serum (2-aminobutyrate, 2-hydroxybutyrate, 2-oxoglutarate, acetate, acetoacetate, fumarate, glucose, methylamine, and oxypurinol).24
2.9. Statistical analysis
For all variables, the normal distribution of data was verified. When appropriate (skewness values > 3.0), logarithmic transformations (log2) were applied to improve normal distribution.24
First, the variables of sample characterization were correlated to iCRF using Pearson’s correlation test (r). Then, partial Pearson correlations (order 1) were performed between baseline concentrations of serum and skeletal muscle metabolites with iCRF, controlling the effects of body mass. All metabolites presenting coefficient r ≥ |0.2| were retained for subsequent analyses—this threshold for the correlation coefficient was selected as it represents a potentially important effect expected for molecular predictors of cardiorespiratory fitness.16,24 Next, the participants were ranked and subdivided into two groups based on the first tertile (low iCRF) and third tertile (high iCRF) of the distribution of iCRF values. For the comparison of retained metabolites (r ≥ |0.2|) between low-iCRF and high-iCRF groups, a univariate general linear model (ANCOVA) was used, assuming group as a fixed factor and body mass as a covariate. These analyses were conducted using PASW Statistics software version 18.0 (SPSS, Chicago, IL). Given the large number of tests performed in this study, the level of significance was 1%, assuming that Bonferroni correction would be very conservative, leading to a high rate of false-negative results. To supplement this approach, the 95% confidence intervals of the effect size (ES: mean difference divided by pooled standard deviation from all subjects) of each metabolite concentration between low-iCRF and high-iCRF groups were calculated. Then, if the confidence intervals did not cross zero, the difference also was considered significant.40
From retained metabolites (r ≥ |0.2|), metabolic pathways were analyzed by over-representation and topology based on the “Homo sapiens” library using Hypergeometric Test for Over Representation Analysis and Relative-Betweenness Centrality for Test Pathway Topology Analysis.41 For the identification of significant metabolic pathways related to iCRF, a false discovery rate of 0.142 was used to control type I errors (Table S1).43 These analyses were conducted using MetaboAnalyst 4.0 (http://www.metaboanalyst.ca).
Afterward, to determine the metabolites associated with the levels of iCRF, only those metabolites supported by three levels of evidence were selected: (1) correlations with iCRF, (2) differences between the groups of participants with low iCRF and high iCRF, and (3) contribution to significant pathways associated with iCRF. This approach was used to minimize the occurrence of metabolites occasionally related to the studied phenotype.24 Finally, to determine the overall predictors of iCRF, stepwise multiple linear regression models were performed separately in serum and skeletal muscle, including body mass as a covariate and those metabolites retained by the three levels of evidence. Variables entered and remained in the model only if they were statistically significant (P < 0.05) after inclusion. To validate the models, the assumption of multicollinearity of measures between the independent variables was assessed by the variance inflation factor (VIF ∼ 1); the normality of residue distribution was determined by inspecting the frequency histograms, and the global influence of each case in the model was analyzed by inspecting the standardized residues and Cook’s distance.
3. Results
3.1. Participants
Table 1 shows the sample characterization data. No significant difference was observed between participants with low and high iCRF for variables age, height, body fat (%), fasting glucose, systolic and diastolic blood pressure, maximal heart rate, heart rate at rest, and ratings of perceived exertion (P > 0.01 for all variables). However, participants with low iCRF had lower body mass and body mass index (BMI) when compared to participants with high iCRF (P < 0.01). Besides, as expected, participants with low iCRF also presented lower maximal power output and time to exhaustion (P < 0.001 for both). Then, subsequent comparative analyses were performed using body mass as a covariate.
Table 1. Characteristics of Participantsa,b.
| variables | all participants (n = 70) | low iCRF (n = 24) | high iCRF (n = 24) |
|---|---|---|---|
| age (years) | 23.4 ± 3.1 | 22.9 ± 2.6 | 23.7 ± 3.0 |
| height (m) | 1.74±0.06 | 1.72 ± 0.06 | 1.74 ± 0.06 |
| body mass (kg) | 72.7 ± 10.8 | 65.5 ± 9.7 | 75.2 ± 8.8f |
| body fat (%) | 20.9 ± 7.2 | 20.0 ± 7.0 | 20.0 ± 7.4 |
| BMI (kg·m–2) | 24.0 ± 3.0 | 22.0 ± 2.7 | 24.8 ± 2.5f |
| fasting glucose (mmol·L–1)c | 4.1 ± 0.5d | 4.0 ± 0.5e | 4.2 ± 0.6 |
| systolic BP (mmHg) | 115.6 ± 12.0 | 111.0 ± 11.1 | 116.5 ± 11.3 |
| diastolic BP (mmHg) | 72.8 ± 9.8 | 69.8 ± 9.0 | 73.9 ± 8.5 |
| HR at rest (beats·min–1) | 70.8 ± 8.3 | 72.2 ± 8.1 | 70.6 ± 8.3 |
| HRMAX (beats·min–1) | 192.3 ± 8.6 | 191.7 ± 8.4 | 195.2 ± 7.3 |
| ratings of perceived exertion (points) | 19.3 ± 0.9 | 19.1 ± 0.9 | 19.4 ± 0.9 |
| time to exhaustion (s) | 577.0 ± 83.5 | 492.8 ± 42.8 | 672.6 ± 40.5f |
| maximal power output (W) | 239.2 ± 34.5 | 204.6 ± 17.7 | 278.7 ± 16.8f |
Data are mean ± standard deviation.
BMI, body mass index; BP, blood pressure; HR, heart rate; HRMAX, Maximal heart rate; iCRF, intrinsic cardiorespiratory fitness.
Fasting glucose levels were derived from 1H NMR analysis.
(n = 69).
(n = 23).
P < 0.01 when compared to low iCRF.
3.2. Associations between Baseline Metabolic Levels and MPO
No significant correlation was observed between the characteristics of the participants (age, height, body fat, BMI, fasting glucose, systolic and diastolic blood pressure, maximal heart rate and heart rate at rest) and iCRF (P > 0.01 for all variables), except for body mass (r = 0.307, P = 0.0097). Then, subsequent correlational analyses were performed to control the effects of body mass.
For blood serum, of all 43 metabolites identified, 13 had baseline levels correlated with iCRF at r ≥ |0.2|: 2-hydroxy-isocaproate, asparagine, betaine, choline, dimethylamine, glutamine, glycine, histidine, ornithine, proline, succinate, threonine, and valine. For muscle tissue, of all 70 metabolites identified, 10 were correlated with MPO at r ≥ |0.2|: formate, glutamate, NADP+, O-acetylcarnitine, taurine, trimethylamine, 3-methylxanthine, acetate, fumarate, and lactate (Table 2).
Table 2. Partial Pearson’s Correlation Coefficients (r) and P Values for the Association between iCRF and Baseline Concentration Levels of Serum and Skeletal Muscle Metabolitesa.
| serumc | rb | P-value | skeletal musclec | rb | P-value |
|---|---|---|---|---|---|
| amino acids | alcohols and polyols | ||||
| alanine | 0.09 | 0.481 | ethylene glycol | –0.03 | 0.820 |
| asparagine | 0.31 | 0.011 | myo-inositol | 0.12 | 0.321 |
| glutamine | 0.31 | 0.010 | amino acids | ||
| glycine | 0.24 | 0.052 | alanineLT | 0.07 | 0.578 |
| histidine | 0.22 | 0.078 | anserineLT | –0.14 | 0.264 |
| isoleucine | 0.10 | 0.425 | β-alanineLT | 0.17 | 0.171 |
| lysine | 0.06 | 0.626 | glutamateLT | 0.22 | 0.079 |
| methionine | 0.13 | 0.273 | glutamineLT | 0.12 | 0.325 |
| phenylalanine | 0.04 | 0.756 | glycineLT | –0.01 | 0.947 |
| proline | 0.27 | 0.028 | histidineLT | 0.10 | 0.424 |
| threonine | 0.23 | 0.061 | isoleucineLT | 0.07 | 0.552 |
| tyrosine | 0.11 | 0.365 | leucineLT | 0.08 | 0.544 |
| valine | 0.20 | 0.098 | phenylalanineLT | –0.06 | 0.611 |
| carboxylic acids | prolineLT | 0.02 | 0.881 | ||
| betaine | 0.42 | <0.001 | threonineLT | –0.03 | 0.783 |
| creatinine | 0.11 | 0.362 | tyrosineLT | 0.19 | 0.123 |
| guanidinoacetate | 0.13 | 0.309 | valine | –0.01 | 0.937 |
| N,N-dimethylglycine | 0.14 | 0.242 | carboxylic acids | ||
| ornithine | 0.37 | 0.002 | acetate | 0.22 | 0.080 |
| succinate | 0.28 | 0.022 | betaine | 0.00 | 0.969 |
| creatine | –0.10 | 0.437 | citrate | 0.13 | 0.281 |
| creatinephosphate | 0.09 | 0.459 | creatineLT | 0.04 | 0.768 |
| formate | –0.09 | 0.482 | creatinephosphateLT | 0.05 | 0.711 |
| fatty acids | creatinine | –0.15 | 0.219 | ||
| 2-hydroxy-isocaproate | 0.21 | 0.091 | formateLT | 0.25 | 0.045 |
| 2-hydroxy-isovalerate | 0.03 | 0.821 | fumarate | 0.28 | 0.023 |
| methylsuccinate | –0.01 | 0.939 | glutathioneLT | –0.07 | 0.585 |
| O-acetylcarnitine | 0.02 | 0.890 | isobutyrateLT | 0.05 | 0.719 |
| hydroxy acids | isocitrate | –0.17 | 0.182 | ||
| 3-hydroxybutyrate | –0.13 | 0.290 | maleate | 0.03 | 0.831 |
| lactate | 0.08 | 0.498 | malonateLT | 0.11 | 0.359 |
| glycolate | 0.07 | 0.551 | N,N-dimethylglycine | 0.17 | 0.184 |
| imidazopyrimidines | N-acetylaspartate | 0.05 | 0.662 | ||
| hypoxanthine LT | 0.03 | 0.839 | N-acetylglutamine | 0.15 | 0.223 |
| xanthine | –0.03 | 0.823 | nicotinurate | –0.06 | 0.638 |
| organic carbonic acids | ornithineLT | 0.12 | 0.320 | ||
| N-methylhydantoin | 0.17 | 0.164 | succinateLT | 0.07 | 0.596 |
| urea | 0.15 | 0.236 | π-methylhistidineLT | 0.05 | 0.701 |
| organic oxygen compounds | τ-methylhistidineLT | 0.02 | 0.884 | ||
| glycerol | –0.06 | 0.616 | fatty acids | ||
| carnitine | 0.11 | 0.361 | 2-hydroxy-isocaproateLT | 0.07 | 0.561 |
| choline | 0.23 | 0.062 | 3-hydroxy-isovalerateLT | 0.02 | 0.892 |
| citrate | 0.12 | 0.343 | O-acetylcarnitineLT | 0.36 | 0.003 |
| dimethyl sulfone | 0.12 | 0.350 | hydroxy acids | ||
| trimethylamine | 0.14 | 0.256 | glycolate | 0.04 | 0.721 |
| propyleneglycol | 0.19 | 0.118 | lactate | 0.30 | 0.015 |
| unclustered | imidazopyrimidines | ||||
| dimethylamine | –0.22 | 0.075 | 3-methylxanthine | –0.30 | 0.015 |
| inosine | 0.03 | 0.822 | oxypurinolLT | –0.13 | 0.307 |
| pyruvate | 0.01 | 0.936 | theophylline | 0.03 | 0.827 |
| nucleosides and nucleotides | |||||
| ADP | –0.13 | 0.287 | |||
| AMPLT | 0.15 | 0.219 | |||
| ATPLT | 0.15 | 0.245 | |||
| NAD+ | 0.02 | 0.878 | |||
| NADP+LT | 0.21 | 0.094 | |||
| organic oxygen compounds | |||||
| 2-phosphoglycerate | 0.18 | 0.138 | |||
| glucoseLT | 0.08 | 0.527 | |||
| glycerol | 0.13 | 0.293 | |||
| organic nitrogen compounds | |||||
| carnitine | 0.18 | 0.147 | |||
| choline | 0.03 | 0.822 | |||
| dimethylamineLT | 0.19 | 0.131 | |||
| histamineLT | 0.15 | 0.243 | |||
| methylamineLT | 0.12 | 0.335 | |||
| N-nitrosodimethylamine | 0.09 | 0.484 | |||
| trimethylamineLT | 0.23 | 0.061 | |||
| trimethylamine-N-oxideLT | –0.04 | 0.745 | |||
| tartrate | –0.08 | 0.508 | |||
| unclustered | |||||
| 2-hydroxyphenylacetate | –0.07 | 0.586 | |||
| acetamide | 0.14 | 0.267 | |||
| carnosine | 0.07 | 0.582 | |||
| dimethyl sulfone | –0.09 | 0.487 | |||
| niacinamide | 0.16 | 0.206 | |||
| pyrimidineLT | –0.11 | 0.366 | |||
| pyruvate | –0.01 | 0.956 | |||
| taurineLT | 0.21 | 0.086 |
iCRF, intrinsic cardiorespiratory fitness. LTData log-transformed before analysis. Bold values are correlation coefficients (r) above |0.2|.
Correlation coefficient adjusted for body mass.
Chemical taxonomy of metabolites was based on classes and subclasses of the Human Metabolome Database.
3.3. Differences between Participants with Low and High CRF
For blood serum, participants with high iCRF presented higher levels in baseline concentrations of metabolites: betaine (P = 0.003), glutamine (P = 0.025), ornithine (P = 0.004), proline (P = 0.014), and threonine (P = 0.061) when compared to participants with low iCRF (Table 3). On the other hand, in muscle tissue, higher levels were observed in baseline concentrations of formate (P = 0.023), NADP+ (P = 0.074), O-acetylcarnitine (P = 0.041), fumarate (P = 0.047), and lactate (P = 0.038), as well as lower levels of 3-methylxanthine (P < 0.01), in participants with high iCRF (Table 3). Comparisons whose P values were greater than 0.01 had their significance confirmed by analyzing the confidence intervals of measurements of effect size that did not cross zero (Table 3).
Table 3. Baseline Differences in Serum and Skeletal Muscle Metabolite Concentration Levels between Participants with Low iCRF and High iCRFa,b.
| serum (mM)c | low iCRF (n = 23) | high iCRF (n = 24) | ESf | 95% CI | skeletal muscle (mM·g–1)c | low iCRF (n = 23) | high iCRF (n = 22) | ESf | 95% CI | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2-hydroxy-isocaproate | 0.0608 ± 0.0164 | 0.0701 ± 0.0150 | –0.43 | –1.02 | 0.16 | formateLT | 0.7362 ± 0.4102 | 1.0209 ± 0.5012d | –0.78 | –1.40 | –0.16 |
| asparagine | 0.0362 ± 0.0109 | 0.0435 ± 0.0143 | –0.43 | –1.02 | 0.16 | glutamateLT | 0.7029 ± 0.2472 | 0.8668 ± 0.3464 | –0.29 | –0.89 | 0.31 |
| betaine | 0.0471 ± 0.0121 | 0.0555 ± 0.0133e | –1.02 | –1.64 | –0.40 | NADP+LT | 0.0143 ± 0.0079 | 0.0230 ± 0.0107d | –0.63 | –1.24 | –0.02 |
| choline | 0.0049 ± 0.0026 | 0.0062 ± 0.0020 | –0.54 | –1.14 | 0.05 | O-acetylcarnitineLT | 0.4828 ± 0.3175 | 0.6001 ± 0.2802d | –0.69 | –1.31 | –0.07 |
| dimethylamine | 0.0058 ± 0.0049 | 0.0039 ± 0.0031 | 0.40 | –0.19 | 0.99 | taurineLT | 4.5220 ± 1.8308 | 5.4847 ± 1.9589 | –0.50 | –1.10 | 0.11 |
| glutamine | 0.3228 ± 0.0919 | 0.3871 ± 0.0818d | –0.77 | –1.38 | –0.17 | trimethylamineLT | 0.0290 ± 0.0194 | 0.0373 ± 0.0256 | –0.32 | –0.92 | 0.28 |
| glycine | 0.2179 ± 0.0448 | 0.2427 ± 0.0619 | –0.40 | –0.99 | 0.19 | 3-methylxanthine | 0.1015 ± 0.0484 | 0.0704 ± 0.0320e | 1.16 | 0.51 | 1.80 |
| histidine | 0.0873 ± 0.0112 | 0.0965 ± 0.0183 | –0.47 | –1.06 | 0.12 | acetate | 0.3834 ± 0.2223 | 0.4900 ± 0.1677 | –0.61 | –1.22 | 0.00 |
| ornithine | 0.0250 ± 0.0127 | 0.0343 ± 0.0087e | –1.00 | –1.63 | –0.38 | fumarate | 0.0480 ± 0.0205 | 0.0569 ± 0.0193d | –0.67 | –1.28 | –0.05 |
| proline | 0.1033 ± 0.0536 | 0.1541 ± 0.0688d | –0.84 | –1.45 | –0.23 | lactate | 3.7562 ± 1.5430 | 4.7007 ± 1.7428d | –0.72 | –1.33 | –0.10 |
| succinate | 0.0077 ± 0.0030 | 0.0093 ± 0.0031 | –0.50 | –1.10 | 0.09 | ||||||
| threonine | 0.1141 ± 0.0252 | 0.1327 ± 0.0308d | –0.63 | –1.23 | –0.03 | ||||||
| valine | 0.2659 ± 0.0436 | 0.2961 ± 0.0580 | –0.42 | –1.01 | 0.18 | ||||||
Data are mean ± standard deviation.
iCRF, intrinsic cardiorespiratory fitness; ES, effect size calculated as the standardized difference between participants with low iCRF and high iCRF in standard deviation units; LTdata log-transformed before analysis, including ES when necessary. Participants with low and high iCRF were stratified by the first and third tertiles, respectively, from the distribution of the values of maximal power output.
Group of metabolites with correlation coefficients (r) ≥ |0.2|.
Significant differences when compared to low iCRF (determined by 95% CI of ES that did not cross zero) are represented in bold.
Significant difference in low iCRF (based on P < 0.01 and 95% CI for ES). Body mass was included as a covariate in the analysis of variance.
Effects values adjusted for body mass.
3.4. Metabolic Pathways
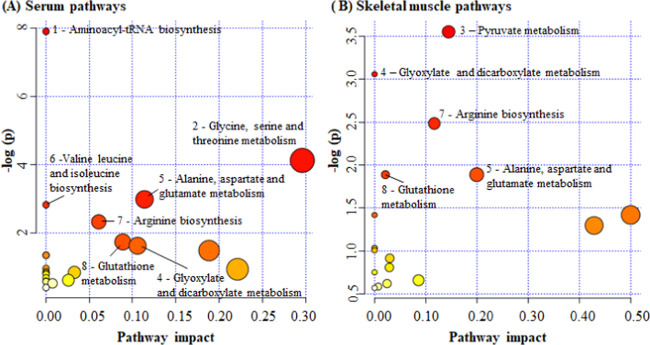
For pathway analysis, baseline metabolites that were correlated at r ≥ |0.2| with iCRF were used (serum, 13 metabolites; muscle tissue, 10 metabolites). From a total of 40 pathways indicated by the retained metabolic profile, 7 pathways for serum and 5 pathways for muscle tissue were significantly enriched and associated with iCRF, considering a false discovery rate of 0.1. From all observed significant pathways, four were related to iCRF in both serum and muscle tissue: glyoxylate and dicarboxylate metabolism; alanine, aspartate, and glutamate metabolism; arginine biosynthesis; and glutathione metabolism. On the other hand, aminoacyl-tRNA biosynthesis; glycine, serine, and threonine metabolism; and valine, leucine, and isoleucine biosynthesis were significant only in serum, while pyruvate metabolism was significant only in muscle tissue (Figure 1; Table S1).
Figure 1.
Summary of serum and skeletal muscle pathways related to iCRF. The numbers in the figures refer to pathways that were most enriched for both serum and muscle tissue. All numbered pathways had a false discovery rate of 0.1. The pathway impact on the horizontal axis represents the relative contribution of all matched metabolites concerning all metabolites in the given pathway. (1) Aminoacyl-tRNA biosynthesis (A, asparagine, histidine, glutamine, glycine, valine, threonine, and proline); (2) glycine, serine, and threonine metabolism (A, choline, betaine, glycine, and threonine); (3) pyruvate metabolism (B, lactate, fumarate, and acetate); (4) glyoxylate and dicarboxylate metabolism (A, glycine and glutamine; B, glutamate, acetate, formate); (5) alanine, aspartate, and glutamate metabolism (A, asparagine, glutamine, and succinate; B, glutamate and fumarate); (6) valine, leucine, and isoleucine biosynthesis (A, threonine and valine); (7) arginine biosynthesis (A, glutamine and ornithine; B, glutamate and fumarate); and (8) glutathione metabolism (A, glycine and ornithine; B, glutamine and NADP+).
3.5. Summary of Metabolites and Pathways Associated with MPO
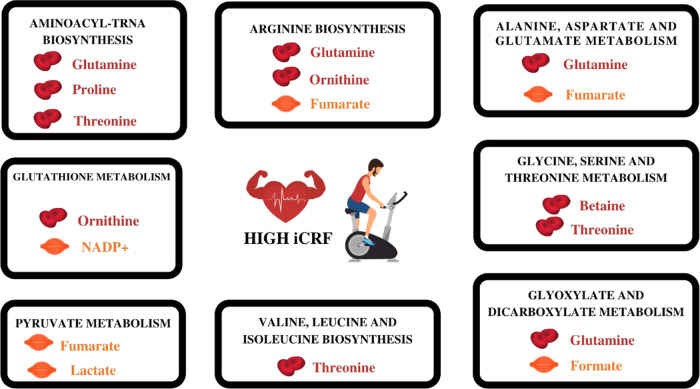
The baseline metabolites most associated with iCRF were identified from the three levels of evidence described above: (1) correlations with iCRF (r ≥ |0.2|), (2) differences between low and high iCRF, and (3) contributions in significant pathways related to iCRF.
The metabolites supported by all three levels of evidence were betaine (a), glutamine (b), ornithine (c), proline (d), and threonine (e) in serum; and formate (f), fumarate (g), lactate (h), and NADP+ (i) in muscle tissue. The most significant pathways and their metabolites associated with iCRF suggested by this metabolic profile were aminoacyl-tRNA biosynthesis (b, d, e); arginine biosynthesis (b, c, g); alanine, aspartate, and glutamate metabolism (b, g); glutathione metabolism (c, i); glycine, serine, and threonine metabolism (a, e); glyoxylate and dicarboxylate metabolism (b, f); pyruvate metabolism (g, h); and valine, leucine, and isoleucine biosynthesis (e) (Table 4; Figure 2).
Table 4. Levels of Evidence for the Identification of Metabolites in Serum and Skeletal Muscle Associated with iCRF.
| metabolites | association with iCRFa | significant pathways associated with iCRF | difference between low iCRF and high iCRF |
|---|---|---|---|
| Serum | |||
| 2-hydroxy-isocaproate | 0.21 | ||
| asparagine | 0.31 | aminoacyl-tRNA biosynthesis; alanine, aspartate, and glutamate metabolism | |
| betaine | 0.42 | glycine, serine, and threonine metabolism | low iCRF < high iCRF |
| choline | 0.23 | glycine, serine, and threonine metabolism | |
| dimethylamine | –0.22 | ||
| glutamine | 0.31 | aminoacyl-tRNA biosynthesis; alanine, aspartate, and glutamate metabolism; glyoxylate and dicarboxylate metabolism | low iCRF < high iCRF |
| glycine | 0.24 | aminoacyl-tRNA biosynthesis; glycine, serine, and threonine metabolism; glyoxylate and dicarboxylate metabolism; glutathione metabolism | |
| histidine | 0.22 | aminoacyl-tRNA biosynthesis | |
| ornithine | 0.37 | arginine biosynthesis; glutathione metabolism | low iCRF < high iCRF |
| proline | 0.27 | aminoacyl-tRNA biosynthesis | low iCRF < high iCRF |
| succinate | 0.28 | alanine, aspartate, and glutamate metabolism | |
| threonine | 0.23 | aminoacyl-tRNA biosynthesis; glycine, serine, and threonine metabolism | low iCRF < high iCRF |
| valine | 0.20 | aminoacyl-tRNA biosynthesis; valine, leucine, and isoleucine biosynthesis | |
| Skeletal Muscle | |||
| 3-methylxanthine | –0.30 | low iCRF > high iCRF | |
| acetate | 0.22 | pyruvate metabolism | |
| formate | 0.25 | glyoxylate and dicarboxylate metabolism | low iCRF < high iCRF |
| fumarate | 0.28 | alanine, aspartate, and glutamate metabolism; arginine biosynthesis; pyruvate metabolism | low iCRF < high iCRF |
| glutamate | 0.22 | alanine, aspartate, and glutamate metabolism; arginine biosynthesis; glutathione metabolism; glyoxylate and dicarboxylate metabolism | |
| lactate | 0.30 | pyruvate metabolism | low iCRF < high iCRF |
| NADP+ | 0.21 | glutathione metabolism | low iCRF < high iCRF |
| O-acetylcarnitine | 0.36 | low iCRF < high iCRF | |
| taurine | 0.21 | ||
| trimethylamine | 0.23 | ||
Metabolites selected with a correlation coefficient of r ≥ |0.2|.
Figure 2.
Summary of serum and skeletal muscle metabolites matching the three levels of evidence and their pathways associated with iCRF. All of these metabolites presented higher concentration levels in individuals with high iCRF compared to low iCRF. Created in Canva.com.
3.6. Variance in iCRF Explained by Serum and Skeletal Muscle Metabolites
When all metabolites supported by all three levels of evidence were included in multiple linear regression models, the variation in the iCRF levels was explained by 31.2% in the serum metabolites model (P < 0.001) and 16.8% in the muscle tissue model (P = 0.003) in addition to body mass (Table 5).
Table 5. Results of Multiple Linear Regression Models for the iCRF Levelsa.
| model | B | β | CI 95% for B | P-value | partial R2 | model R2 | adjusted R2 |
|---|---|---|---|---|---|---|---|
| Serum | |||||||
| (constant) | 81.35 | 18.4–144.3 | 0.012 | 0.312 | 0.280 | ||
| body mass (kg) | 1.29 | 0.41 | 0.6–2.0 | 0.000 | 0.093 | ||
| betaine | 877.83 | 0.33 | 282.6–1473.1 | 0.004 | 0.162 | ||
| ornithine | 720.41 | 0.25 | 99.9–1341.0 | 0.024 | 0.057 | ||
| Muscle Tissue | |||||||
| (constant) | 133.81 | 73.0–194.6 | 0.000 | 0.168 | 0.142 | ||
| body mass (kg) | 1.13 | 0.36 | 0.4–1.9 | 0.003 | 0.087 | ||
| lactate | 5.43 | 0.29 | 1.1–9.8 | 0.015 | 0.081 | ||
B, slope; β, standardized slope; iCRF, intrinsic cardiorespiratory fitness.
4. Discussion
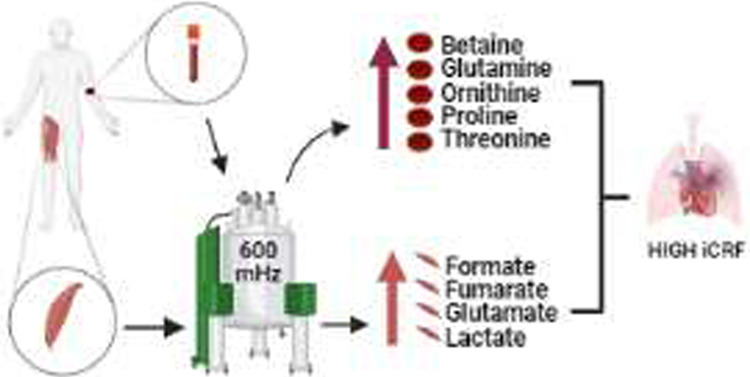
This study investigated whether baseline serum and skeletal muscle metabolic levels are associated with iCRF. The results were based on the identification of metabolites supported by three levels of evidence: (1) association with iCRF, (2) differences between participants with high iCRF and low iCRF, and (3) metabolite contribution to significant pathways associated with iCRF. The main findings of this study include (i) the presence of wide heterogeneity in the levels of iCRF among sedentary young male individuals, ranging from 170 to 315 W; (ii) identification of metabolites whose baseline levels in serum (betaine, threonine, proline, ornithine, and glutamine) and skeletal muscle (lactate, fumarate, NADP+, and formate) were positively associated with iCRF; (iii) higher concentration levels of these metabolites in individuals with high iCRF compared to low iCRF, regardless of their body mass; (iv) identification of metabolic pathways associated with iCRF, being the most impacted pathways (impact >0) related to amino acids (alanine, aspartate and glutamate metabolism; arginine biosynthesis; glycine, serine, and threonine metabolism; and glutathione metabolism) and carbohydrates (pyruvate metabolism and glyoxylate and dicarboxylate metabolism); and finally (v) identification of serum (betaine and ornithine) and skeletal muscle (lactate) metabolites that was able to explain 31.2 and 16.8% of the variability inherent in iCRF, respectively, in addition to body mass.
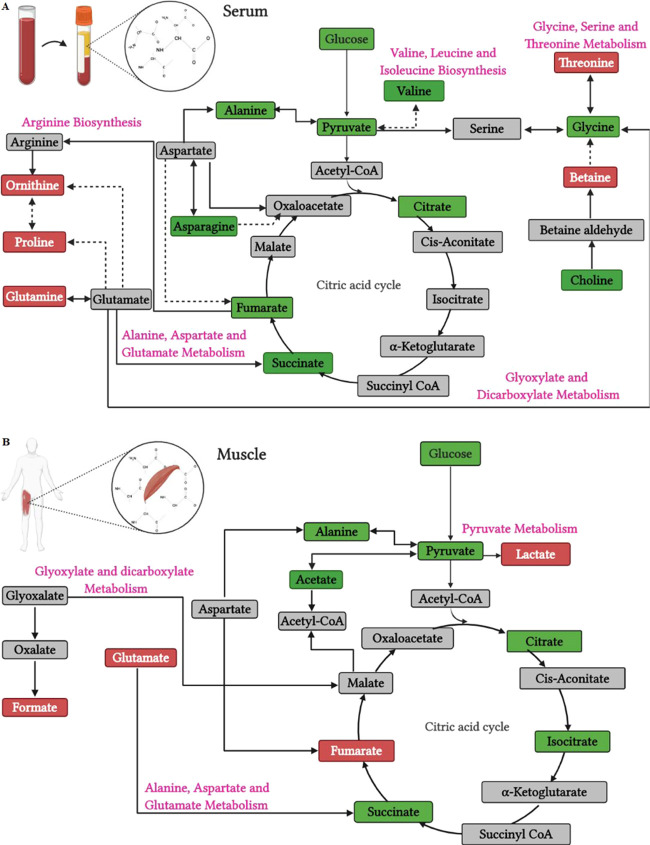
Of note, a baseline serum and skeletal muscle metabolic regulation was observed, depending on the iCRF levels. A summary of the origin and pathways of the identified metabolites is provided in Figure 3. Regarding blood serum, individuals with high iCRF showed higher levels of baseline concentration of metabolites betaine, glutamine, ornithine, proline, and threonine when compared to individuals with low iCRF. In particular, betaine is derived from glycine and its circulating fasting levels can be attributed to endogenous synthesis in the liver and kidneys.44 Betaine acts as an organic osmolyte to protect cells under stress or as an intermediate in the production of methionine, a precursor of creatine whose function is related to fast energy supply in skeletal muscle.45,46 Prior studies demonstrated that higher baseline levels of circulating betaine are associated with better performance in aerobic exercises.47 Threonine and proline are blood glucose precursor amino acids that can be converted into pyruvate (threonine) or intermediates in the citric acid cycle (both), possibly reflecting a more accelerated mechanism of maintenance of the glycemic level in individuals with high iCRF.48 In contrast, Morris et al. observed higher circulating levels of threonine and proline in women with low CRF when compared to high CRF.15 Although these discrepancies have not been fully explained, these women with low CRF, when compared to those with high CRF, had greater evidence of insulin resistance,15 which has been associated with an increase in circulating proline49 and threonine,50,51 and did not have the physical activity reported level, limiting direct comparisons with the results based on iCRF in our study. Regarding ornithine, it is synthesized in the cytoplasm from arginine and plays a critical role in the urea cycle and mitochondrial metabolic processes, producing indirectly proline among other products.52 Perhaps, a higher level of serum ornithine in individuals with high iCRF reflects an increased rate of the urea cycle, required to inhibit an increase in circulating ammonia, transferred between tissues by glutamine, due to the degradation of amino acids53 like threonine and proline, which presented increased levels. There is evidence that dietary supplements with ornithine have a fatigue-reducing effect while practicing aerobic exercises as it improves energy supply and ammonia excretion.53 Finally, glutamine is a precursor to the synthesis of glutamate, which plays an important role in energy supply through the citric acid cycle,54 responsible for the transfer of nitrogen between organs, detoxification of ammonia, and maintenance of the acid–base balance in the kidneys.55−58 Increased circulating levels of glutamine also have been observed in parallel with increases in cardiorespiratory fitness induced by endurance training.59
Figure 3.
Pathways and origin of the identified metabolites built based on Kyoto Encyclopedia of Genes and Genomes (KEGG) and Small Molecule Pathway Database (SMPD) libraries. Serum (A) and skeletal muscle (B) metabolites are color-coded as follows: red boxes, metabolites within the three levels of evidence (higher concentration levels in individuals with high iCRF compared to those in individuals with low iCRF); green boxes, identified metabolites without difference between individuals with high and low iCRF; gray boxes, undetected metabolites. Solid and dashed black lines indicate direct and indirect conversion, respectively. Created in BioRender.com.
On the other hand, in skeletal muscle, higher levels of baseline concentration of metabolites, lactate, fumarate, NADP+, and formate, were observed in participants with high iCRF when compared to those in participants with low iCRF. Lactate is the end product of anaerobic glucose breakdown and has an important physiological role, serving at least three purposes: a source of energy for mitochondrial respiration, the main gluconeogenic precursor, and a signaling molecule.60 There is evidence that glycolysis proceeds to lactate under fully aerobic conditions in healthy humans.60,61 In this case, considering that serum lactate levels were not different between participants with high and low iCRF, we suggest the greatest availability of skeletal muscle lactate in high-iCRF participants to be useful in providing a source of energy for mitochondria by converting it to pyruvate and then to oxaloacetate, an intermediate of the citric acid cycle.60,61 This reasoning is supported by the significant involvement of the skeletal muscle lactate in pyruvate metabolism, which was a pathway positively associated with iCRF, evidencing a more active carbohydrate metabolism for resting and fasting conditions in the high-iCRF group. Regarding fumarate, it is one of the intermediates in the citric acid cycle. An increased concentration of intramuscular fumarate indicates an augmented rate of the citric acid cycle flux at rest in high-iCRF individuals. This result corroborates with previous studies that demonstrated increased citric acid cycle flux, evidenced by higher levels of fumarate, after exercise to exhaustion62 and in individuals with higher aerobic performance.63 NADP+ (nicotinamide adenine dinucleotide phosphate) is a coenzyme that acts as a cofactor and substrate for many enzymes promoting the maintenance of cellular redox homeostasis. Deficient NADP+ levels cause a disturbance in the redox status of the cell and metabolic homeostasis, leading to oxidative stress, energy stress (impairment of ATP turnover conditions), and eventually pathological states.64 In this study, higher levels of NADP+ were associated with higher iCRF, suggesting cellular redox homeostasis as a relevant factor for iCRF. Finally, formate is an indirect product of glyoxylate (Figure 3), glycine, choline, serine, or methanol.65 In our study, formate was related to glyoxylate and dicarboxylate metabolism and positively associated with iCRF. In the glyoxylate and dicarboxylate metabolism, formate is an indirect product of glyoxylate that can be converted into malate, an intermediate of the citric acid cycle (Figure 3). Although the relationship of formate with iCRF or physical exercise is little known,66 previous studies with animal models corroborate the results obtained here, showing greater activation of glyoxalate and dicarboxylate metabolism in rats with high iCRF compared to those with low iCRF,23 in addition to a positive association with increased fatigue resistance in rats submitted to exhaustive aerobic exercise.67
In summary, these results suggest that, at rest, individuals with high iCRF probably have a more active mechanism for the supply of cellular energy via carbohydrate and amino acid metabolism and are supported by a better excretion system of byproducts, for example, through the urea cycle. It is supported by the skeletal muscle metabolic pathways related to carbohydrate (pyruvate metabolism and glyoxylate and dicarboxylate metabolism) and amino acid (alanine, aspartate, and glutamate metabolism and arginine biosynthesis) metabolism positively associated with iCRF; increased levels of serum glucose precursor amino acids (threonine and proline); increased serum metabolites related to transport of byproducts of amino acid metabolism (glutamine); and increased serum urea cycle intermediate (ornithine).
Some strengths
and limitations must be considered in the present
study. This is an observational study based on a specific cohort of
participants (young healthy sedentary men) analyzed. Therefore, causal
relationships and extrapolation of the findings from this study to
other populations should be avoided. The low and high iCRF terminologies
must be taken with caution when comparing studies, as they reflect
the context and the distribution of iCRF values in the population
studied herein. On the other hand, we should point out our results
are based on a considerable cohort size (70 participants), which was
characterized by highly standardized clinical and physiological examinations,
diet control 12 h prior to blood and muscle tissue collection, and
a strictly scheduled experimental setting. Our exploratory analysis
was based on only one metabolomics technique; therefore, a relatively
small number of metabolites were detected. Although the current number
of metabolites is comparable in quantity to most of the studies previously
reported on this topic,12,13,15,16 other complementary measures
such as mass spectrometry can help to identify metabolites undetected
in this study also linked to iCRF. Most of the previous studies have
shown associations between baseline metabolism and ACR measured by 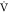 O2MAX14,16 but not necessarily referencing
MPO or iCRF, which makes it difficult
to compare with our results. However, MPO as a surrogate of CRF is
known to have a high correlation (r = 0.94) with
O2MAX14,16 but not necessarily referencing
MPO or iCRF, which makes it difficult
to compare with our results. However, MPO as a surrogate of CRF is
known to have a high correlation (r = 0.94) with 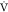 O2MAX using a similar maximal
incremental test in sedentary men.68 In
addition, MPO is a better predictor of endurance performance compared
to
O2MAX using a similar maximal
incremental test in sedentary men.68 In
addition, MPO is a better predictor of endurance performance compared
to 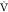 O2MAX69 since the power output provides
a direct and immediate measure of
work rate, as opposed to the subject’s perceptual or cardiovascular
response to exercise intensity.70 Finally,
MPO presents a lower typical measurement error compared to
O2MAX69 since the power output provides
a direct and immediate measure of
work rate, as opposed to the subject’s perceptual or cardiovascular
response to exercise intensity.70 Finally,
MPO presents a lower typical measurement error compared to 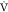 O2MAX71,72 which is suitable for tracking
real small differences between groups
with distinct iCRF but similar physical characteristics, as in the
present study.
O2MAX71,72 which is suitable for tracking
real small differences between groups
with distinct iCRF but similar physical characteristics, as in the
present study.
5. Conclusions
This study showed that baseline levels of iCRF were positively associated with serum and skeletal muscle metabolites, with the most relevant metabolic pathways related to the metabolism of amino acids (alanine, aspartate, and glutamate metabolism; arginine biosynthesis; glycine, serine, and threonine metabolism; and glutathione metabolism) and carbohydrates (pyruvate metabolism; and glyoxylate and dicarboxylate metabolism) in young healthy sedentary men. Regardless of body mass, individuals with high iCRF presented higher baseline serum levels of betaine, threonine, proline, ornithine, and glutamine and higher levels in the skeletal muscle of lactate, fumarate, NADP+, and formate when compared to individuals with low iCRF. Furthermore, serum betaine and ornithine and skeletal muscle lactate metabolites were able to explain 31.2 and 16.8% respectively, of the variability inherent in iCRF in addition to the body weight.
Acknowledgments
The authors thank the NMR Facility at the Brazilian Biosciences National Laboratory (LNBio) for the use of the NMR spectrometer (600 MHz, Varian Inova) and the Integrated Laboratory of Education and Research (LABFEF) at the University of Campinas for the Body Composition Tracking System (BOD POD, Cosmed, USA). The authors also thank Espaço da Escrita – Pró-Reitoria de Pesquisa—UNICAMP—for the language services provided.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00905.
Representation of the serum 1D 1H NMR spectrum region where the main metabolites were quantified for a single participant (Figure S1), representation of a skeletal muscle 1D 1H NMR spectrum region where the main metabolites were quantified for a single participant (Figure S2), and summary of serum and skeletal muscle pathways related to iCRF (Table S1) (PDF)
Author Contributions
Conceptualization, A.C. and M.P.T.C.M.; methodology, A.C., M.P.T.C.M., and C.R.C.; software, A.C. and R.G.D.; validation, A.C. and M.P.T.C.M.; formal analysis, A.C., L.M.S. and R.G.D.; investigation, A.C., R.G.D., M.L.V.F., A.A.L.L., and L.M.S.; resources, M.P.T.C.M.; data curation, A.C.; writing—original draft preparation, A.C.; writing—review and editing, R.G.D., C.R.C., C.F.B. and M.P.T.C.M.; supervision, M.P.T.C.M.; project administration, A.C. and M.P.T.C.M.; funding acquisition, M.P.T.C.M. All authors have read and agreed to the published version of the manuscript.
This research was funded by São Paulo Research Foundation (grant number 2016/057417). A.C. was funded by the National Council for Scientific and Technological Development (grant numbers 149201/2015-0, 140302/2018-2) and the Brazilian Federal Agency for Support and Evaluation of Graduate Education (grant number 88881.135219/2016-01).
The authors declare no competing financial interest.
Supplementary Material
References
- Hill A. V.; Lupton H. Muscular exercise, lactic acid, and the supply and utilization of oxygen. QJM: Int. J. Med. 1923, os-16, 135–171. 10.1093/qjmed/os-16.62.135. [DOI] [Google Scholar]
- Hawkins M. N.; Raven P. B.; Snell P. G.; Stray-Gundersen J.; Levine B. D. Maximal oxygen uptake as a parametric measure of cardiorespiratory capacity. Med. Sci. Sports Exercise 2007, 39, 103–107. [DOI] [PubMed] [Google Scholar]
- Riebe D.; Ehrman Jk.; Liguori G.; Magal M.. ACSM’s Guidelines for Exercise Testing and Prescription.; 10th ed.; Wolters Kluwer: Philadelphia, 2018. [Google Scholar]
- Kodama S.; Saito K.; Tanaka S.; Maki M.; Yachi Y.; Asumi M.; Sugawara A.; Totsuka K.; Shimano H.; Ohashi Y.; Yamada N.; Sone H. Cardiorespiratory fitness as a quantitative predictor of all-cause mortality and cardiovascular events in healthy men and women: a meta-analysis. JAMA 2009, 301, 2024–2035. 10.1001/jama.2009.681. [DOI] [PubMed] [Google Scholar]
- Fardman A.; Banschick G. D.; Rabia R.; Percik R.; Segev S.; Klempfner R.; Grossman E.; Maor E. Cardiorespiratory Fitness Is an Independent Predictor of Cardiovascular Morbidity and Mortality and Improves Accuracy of Prediction Models. Can. J. Cardiol. 2021, 37, 241–250. 10.1016/j.cjca.2020.05.017. [DOI] [PubMed] [Google Scholar]
- Ghosh S.; Hota M.; Chai X.; Kiranya J.; Ghosh P.; He Z.; Ruiz-Ramie J. J.; Sarzynski M. A.; Bouchard C. Exploring the underlying biology of intrinsic cardiorespiratory fitness through integrative analysis of genomic variants and muscle gene expression profiling. J. Appl. Physiol. 2019, 126, 1292–1314. 1985 10.1152/japplphysiol.00035.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J. S.; Jaskólski A.; Jaskólska A.; Krasnoff J.; Gagnon J.; Leon A. S.; Rao D. C.; Wilmore J. H.; Bouchard C. Age, sex, race, initial fitness, and response to training: the HERITAGE Family Study. J. Appl. Physiol. 2001, 90, 1770–1776. 1985 10.1152/jappl.2001.90.5.1770. [DOI] [PubMed] [Google Scholar]
- Bouchard C.; Blair S. N.; Katzmarzyk P. T. Less Sitting, More Physical Activity, or Higher Fitness?. Mayo Clin. Proc. 2015, 90, 1533–40. 10.1016/j.mayocp.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Del Coso J.; Gu Z.; Gerile W.; Yang R.; Díaz-Peña R.; Valenzuela P. L.; Lucia A.; He Z. Interindividual Variation in Cardiorespiratory Fitness: A Candidate Gene Study in Han Chinese People. Genes 2020, 11, 555 10.3390/genes11050555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C.; Daw E. W.; Rice T.; Pérusse L.; Gagnon J.; Province M. A.; Leon A. S.; Rao D. C.; Skinner J. S.; Wilmore J. H. Familial resemblance for VO2max in the sedentary state: the HERITAGE family study. Med. Sci. Sports Exercise 1998, 30, 252–8. 10.1097/00005768-199802000-00013. [DOI] [PubMed] [Google Scholar]
- Lundby C.; Montero D.; Joyner M. Biology of VO2 max: looking under the physiology lamp. Acta Physiol. 2017, 220, 218–228. 10.1111/apha.12827. [DOI] [PubMed] [Google Scholar]
- Bye A.; Vettukattil R.; Aspenes S. T.; Giskeødegård G. F.; Gribbestad I. S.; Wisløff U.; Bathen T. F. Serum levels of choline-containing compounds are associated with aerobic fitness level: the HUNT-study. PLoS One 2012, 7, e42330 10.1371/journal.pone.0042330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorell E.; Svensson M. B.; Moritz T.; Antti H. Physical fitness level is reflected by alterations in the human plasma metabolome. Mol. BioSyst. 2012, 8, 1187–96. 10.1039/c2mb05428k. [DOI] [PubMed] [Google Scholar]
- Lustgarten M. S.; Price L. L.; Logvinenko T.; Hatzis C.; Padukone N.; Reo N. V.; Phillips E. M.; Kirn D.; Mills J.; Fielding R. A. Identification of serum analytes and metabolites associated with aerobic capacity. Eur. J. Appl. Physiol. 2013, 113, 1311–1320. 10.1007/s00421-012-2555-x. [DOI] [PubMed] [Google Scholar]
- Morris C.; Grada C. O.; Ryan M.; Roche H. M.; De Vito G.; Gibney M. J.; Gibney E. R.; Brennan L. The relationship between aerobic fitness level and metabolic profiles in healthy adults. Mol. Nutr. Food Res. 2013, 57, 1246–54. 10.1002/mnfr.201200629. [DOI] [PubMed] [Google Scholar]
- Kistner S.; Rist M. J.; Döring M.; Dörr C.; Neumann R.; Härtel S.; Bub A. An NMR-Based Approach to Identify Urinary Metabolites Associated with Acute Physical Exercise and Cardiorespiratory Fitness in Healthy Humans-Results of the KarMeN Study. Metabolites 2020, 10, 212. 10.3390/metabo10050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr M. Metabolomics: ready for the prime time?. Circ.: Cardiovasc. Genet. 2008, 1, 58–65. 10.1161/CIRCGENETICS.108.808329. [DOI] [PubMed] [Google Scholar]
- Patti G. J.; Yanes O.; Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–9. 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser G. A.; Larrouy-Maumus G.; de Carvalho L. P. Metabolomic strategies for the identification of new enzyme functions and metabolic pathways. EMBO Rep. 2014, 15, 657–669. 10.15252/embr.201338283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R. S.; Kelly M. P.; Kelly P. Metabolomics, physical activity, exercise and health: A review of the current evidence. Biochim. Biophys. Acta, Mol. Basis Dis. 2020, 1866, 165936 10.1016/j.bbadis.2020.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duft R. G.; Castro A.; Bonfante I. L. P.; Brunelli D. T.; Chacon-Mikahil M. P. T.; Cavaglieri C. R. Metabolomics approach in the investigation of metabolic changes in obese men after 24 weeks of combined training. J. Proteome Res. 2017, 16, 2151–2159. 10.1021/acs.jproteome.6b00967. [DOI] [PubMed] [Google Scholar]
- Wishart D. S. NMR metabolomics: A look ahead. J. Magn. Reson. 2019, 306, 155–161. 10.1016/j.jmr.2019.07.013. [DOI] [PubMed] [Google Scholar]
- Falegan O. S.; Vogel H. J.; Hittel D. S.; Koch L. G.; Britton S. L.; Hepple R. T.; Shearer J. High Aerobic Capacity Mitigates Changes in the Plasma Metabolomic Profile Associated with Aging. J. Proteome Res. 2017, 16, 798–805. 10.1021/acs.jproteome.6b00796. [DOI] [PubMed] [Google Scholar]
- Castro A.; Duft R. G.; Ferreira M. L. V.; Andrade A. L. L.; Gáspari A. F.; Silva L. M.; Oliveira-Nunes S. G.; Cavaglieri C. R.; Ghosh S.; Bouchard C.; Chacon-Mikahil M. P. T. Association of skeletal muscle and serum metabolites with maximum power output gains in response to continuous endurance or high-intensity interval training programs: The TIMES study - A randomized controlled trial. PLoS One 2019, 14, e0212115 10.1371/journal.pone.0212115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C.; Leon A. S.; Rao D. C.; Skinner J. S.; Wilmore J. H.; Gagnon J. The HERITAGE family study. Aims, design, and measurement protocol. Med. Sci. Sports Exercise 1995, 27, 721–729. 10.1249/00005768-199505000-00015. [DOI] [PubMed] [Google Scholar]
- Beck T. W. The importance of a priori sample size estimation in strength and conditioning research. J. Strength Cond. Res. 2013, 27, 2323–37. 10.1519/JSC.0b013e318278eea0. [DOI] [PubMed] [Google Scholar]
- Faul F.; Erdfelder E.; Lang A. G.; Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]
- Castro A.; Duft R. G.; Zeri A. C. M.; Cavaglieri C. R.; Chacon-Mikahil M. P. T. Commentary: Metabolomics-Based Studies Assessing Exercise-Induced Alterations of the Human Metabolome: A Systematic Review. Front. Physiol. 2020, 11, 353 10.3389/fphys.2020.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peake J. M.; Tan S. J.; Markworth J. F.; Broadbent J. A.; Skinner T. L.; Cameron-Smith D. Metabolic and hormonal responses to isoenergetic high-intensity interval exercise and continuous moderate-intensity exercise. Am. J. Physiol.: Endocrinol. Metab. 2014, 307, E539–52. 10.1152/ajpendo.00276.2014. [DOI] [PubMed] [Google Scholar]
- Shrestha A.; Müllner E.; Poutanen K.; Mykkänen H.; Moazzami A. A. Metabolic changes in serum metabolome in response to a meal. Eur. J. Nutr. 2017, 56, 671–681. 10.1007/s00394-015-1111-y. [DOI] [PubMed] [Google Scholar]
- Dideriksen K.; Mikkelsen U. R. Reproducibility of incremental maximal cycle ergometer tests in healthy recreationally active subjects. Clin. Physiol. Funct. Imaging 2017, 37, 173–182. 10.1111/cpf.12283. [DOI] [PubMed] [Google Scholar]
- Shanely R. A.; Zwetsloot K. A.; Triplett N. T.; Meaney M. P.; Farris G. E.; Nieman D. C. Human skeletal muscle biopsy procedures using the modified Bergström technique. J. Vis. Exp. 2014, 91, 51812 10.3791/51812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrory M. A.; Gomez T. D.; Bernauer E. M.; Molé P. A. Evaluation of a new air displacement plethysmograph for measuring human body composition. Med. Sci. Sports Exercise 1995, 27, 1686–1691. 10.1249/00005768-199512000-00016. [DOI] [PubMed] [Google Scholar]
- Siri W. E. Body composition from fluid spaces and density: analysis of methods. 1961. Nutrition 1993, 9, 480–492. [PubMed] [Google Scholar]
- Buchfuhrer M. J.; Hansen J. E.; Robinson T. E.; Sue D. Y.; Wasserman K.; Whipp B. J. Optimizing the exercise protocol for cardiopulmonary assessment. J. Appl. Physiol. 1983, 55, 1558–1564. 10.1152/jappl.1983.55.5.1558. [DOI] [PubMed] [Google Scholar]
- Thompson P. D.; Arena R.; Riebe D.; Pescatello L. S. ACSM’s new preparticipation health screening recommendations from ACSM’s guidelines for exercise testing and prescription. Curr. Sports Med. Rep. 2013, 12, 215–217. 10.1249/JSR.0b013e31829a68cf. [DOI] [PubMed] [Google Scholar]
- Borg G.; Linderho H. Perceived exertion and pulse rate during graded exercise in various age groups. Acta Med. Scand. 1967, 181, 194–206. 10.1111/j.0954-6820.1967.tb12626.x. [DOI] [Google Scholar]
- Kuipers H.; Verstappen F. T.; Keizer H. A.; Geurten P.; van Kranenburg G. Variability of aerobic performance in the laboratory and its physiologic correlates. Int. J. Sports Med. 1985, 6, 197–201. 10.1055/s-2008-1025839. [DOI] [PubMed] [Google Scholar]
- Duft R. G.; Castro A.; Bonfante I. L. P.; Lopes W. A.; da Silva L. R.; Chacon-Mikahil M. P. T.; Leite N.; Cavaglieri C. R. Altered metabolomic profiling of overweight and obese adolescents after combined training is associated with reduced insulin resistance. Sci. Rep. 2020, 10, 16880 10.1038/s41598-020-73943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S.; Cuthill I. C. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol. Rev. 2007, 82, 591–605. 10.1111/j.1469-185X.2007.00027.x. [DOI] [PubMed] [Google Scholar]
- Xia J.; Wishart D. S. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–4. 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- Benjamini Y.; Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995, 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- van den Oord E. J.; Sullivan P. F. False discoveries and models for gene discovery. Trends Genet. 2003, 19, 537–42. 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Yancey P. H.; Clark M. E.; Hand S. C.; Bowlus R. D.; Somero G. N. Living with water stress: evolution of osmolyte systems. Science 1982, 217, 1214–22. 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- Craig S. A. Betaine in human nutrition. Am. J. Clin. Nutr. 2004, 80, 539–549. 10.1093/ajcn/80.3.539. [DOI] [PubMed] [Google Scholar]
- Stead L. M.; Brosnan J. T.; Brosnan M. E.; Vance D. E.; Jacobs R. L. Is it time to reevaluate methyl balance in humans?. Am. J. Clin. Nutr. 2006, 83, 5–10. 10.1093/ajcn/83.1.5. [DOI] [PubMed] [Google Scholar]
- Cholewa J. M.; Guimarães-Ferreira L.; Zanchi N. E. Effects of betaine on performance and body composition: a review of recent findings and potential mechanisms. Amino Acids 2014, 46, 1785–93. 10.1007/s00726-014-1748-5. [DOI] [PubMed] [Google Scholar]
- Nelson D.; Cox M.. Lehninger Principles of Biochemistry.; 6th ed.; Worth Publishers: New York, 2013. [Google Scholar]
- Seibert R.; Abbasi F.; Hantash F. M.; Caulfield M. P.; Reaven G.; Kim S. H. Relationship between insulin resistance and amino acids in women and men. Physiol. Rep. 2015, 3, e12392 10.14814/phy2.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte C. C.; Farsijani S.; Marliss E. B.; Gougeon R.; Morais J. A.; Pereira S.; Bassil M.; Winter A.; Murphy J.; Combs T. P.; Chevalier S. Plasma Amino Acids vs Conventional Predictors of Insulin Resistance Measured by the Hyperinsulinemic Clamp. J. Endocr. Soc. 2017, 1, 861–873. 10.1210/js.2016-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth C.; Kirchberg F. F.; Lass N.; Harder U.; Peissner W.; Koletzko B.; Reinehr T. Tyrosine Is Associated with Insulin Resistance in Longitudinal Metabolomic Profiling of Obese Children. J. Diabetes Res. 2016, 2016, 2108909 10.1155/2016/2108909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs H. A.; Henseleit K. Über die Harnsäuresynthese im Vogelorganismus. Biol. Chem. 1932, 210, 33–66. [Google Scholar]
- Sugino T.; Shirai T.; Kajimoto Y.; Kajimoto O. L-ornithine supplementation attenuates physical fatigue in healthy volunteers by modulating lipid and amino acid metabolism. Nutr. Res. 2008, 28, 738–743. 10.1016/j.nutres.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Rutten E. P.; Engelen M. P.; Schols A. M.; Deutz N. E. Skeletal muscle glutamate metabolism in health and disease: state of the art. Curr. Opin. Clin. Nutr. Metab. Care 2005, 8, 41–51. 10.1097/00075197-200501000-00007. [DOI] [PubMed] [Google Scholar]
- Pérez-Sala D.; Parrilla R.; Ayuso M. S. Key role of L-alanine in the control of hepatic protein synthesis. Biochem. J. 1987, 241, 491–8. 10.1042/bj2410491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood D. A.; Terjung R. L. Amino acid metabolism during exercise and following endurance training. Sports Med. 1990, 9, 23–35. 10.2165/00007256-199009010-00003. [DOI] [PubMed] [Google Scholar]
- Newsholme P.; Procopio J.; Lima M. M.; Pithon-Curi T. C.; Curi R. Glutamine and glutamate--their central role in cell metabolism and function. Cell Biochem. Funct. 2003, 21, 1–9. 10.1002/cbf.1003. [DOI] [PubMed] [Google Scholar]
- Yoo H. C.; Yu Y. C.; Sung Y.; Han J. M. Glutamine reliance in cell metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. 10.1038/s12276-020-00504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargotich S.; Keast D.; Goodman C.; Bhagat C. I.; Joske D. J.; Dawson B.; Morton A. R. Monitoring 6 weeks of progressive endurance training with plasma glutamine. Int. J. Sports Med. 2007, 28, 211–6. 10.1055/s-2006-924218. [DOI] [PubMed] [Google Scholar]
- Brooks G. A. Lactate as a fulcrum of metabolism. Redox Biol. 2020, 35, 101454 10.1016/j.redox.2020.101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks G. A. The Science and Translation of Lactate Shuttle Theory. Cell Metab. 2018, 27, 757–785. 10.1016/j.cmet.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Gibala M. J.; MacLean D. A.; Graham T. E.; Saltin B. Tricarboxylic acid cycle intermediate pool size and estimated cycle flux in human muscle during exercise. Am. J. Physiol.: Endocrinol. Metab. 1998, 275, E235–E242. 10.1152/ajpendo.1998.275.2.E235. [DOI] [PubMed] [Google Scholar]
- Lewis G. D.; Farrell L.; Wood M. J.; Martinovic M.; Arany Z.; Rowe G. C.; Souza A.; Cheng S.; McCabe E. L.; Yang E.; Shi X.; Deo R.; Roth F. P.; Asnani A.; Rhee E. P.; Systrom D. M.; Semigran M. J.; Vasan R. S.; Carr S. A.; Wang T. J.; Sabatine M. S.; Clish C. B.; Gerszten R. E. Metabolic signatures of exercise in human plasma. Sci. Transl. Med. 2010, 2, 33ra37 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W.; Wang R. S.; Handy D. E.; Loscalzo J. NAD(H) and NADP(H) Redox Couples and Cellular Energy Metabolism. Antioxid. Redox Signaling 2018, 28, 251–272. 10.1089/ars.2017.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzke M.; Meiser J.; Vazquez A. Formate metabolism in health and disease. Mol. Metab. 2020, 33, 23–37. 10.1016/j.molmet.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H. J.; Lee J. D.; Jeon H. S.; Kim A. R.; Kim S.; Lee H. S.; Kim K. B. Metabolic Profiling of Eccentric Exercise-Induced Muscle Damage in Human Urine. Toxicol. Res. 2018, 34, 199–210. 10.5487/TR.2018.34.3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X.; Xiao B.; Shui S.; Yang J.; Huang R.; Dong J. Metabolomics analysis of serum reveals the effect of Danggui Buxue Tang on fatigued mice induced by exhausting physical exercise. J. Pharm. Biomed. Anal. 2018, 151, 301–309. 10.1016/j.jpba.2018.01.028. [DOI] [PubMed] [Google Scholar]
- Storer T. W.; Davis J. A.; Caiozzo V. J. Accurate prediction of VO2max in cycle ergometry. Med. Sci. Sports Exercise 1990, 22, 704–12. 10.1249/00005768-199010000-00024. [DOI] [PubMed] [Google Scholar]
- Hawley J. A.; Noakes T. D. Peak power output predicts maximal oxygen uptake and performance time in trained cyclists. Eur. J. Appl. Physiol. Occup. Physiol. 1992, 65, 79–83. 10.1007/BF01466278. [DOI] [PubMed] [Google Scholar]
- Jobson S. A.; Passfield L.; Atkinson G.; Barton G.; Scarf P. The analysis and utilization of cycling training data. Sports Med. 2009, 39, 833–844. 10.2165/11317840-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Montero D.; Lundby C. Refuting the myth of non-response to exercise training: ’non-responders’ do respond to higher dose of training. J. Physiol. 2017, 595, 3377–3387. 10.1113/JP273480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J. S.; Wilmore K. M.; Jaskolska A.; Jaskolski A.; Daw E. W.; Rice T.; Gagnon J.; Leon A. S.; Wilmore J. H.; Rao D. C.; Bouchard C. Reproducibility of maximal exercise test data in the HERITAGE family study. Med. Sci. Sports Exercise 1999, 31, 1623–1628. 10.1097/00005768-199911000-00020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.