Abstract
Objective:
The TYR (Tyrosinase) and MC1R (Melanocortin 1 receptor) genes are recognized as important genes involved in plumage pigmentation in Korean native chickens. Specifically, most color patterns in chicken result from differential expression of the TYR gene. In this study, the co-segregation of the pigmentation and sequence of the TYR and MC1R genes was investigated through intercrosses between red (R1q1), red with black and black plumage color types of native Korean chickens.
Materials and Methods:
Using DNA, RNA, and tissue by plumage color of each Korean native chickens, the role of major genes in pigmentation of pheomelanin was evaluated. Reverse transcription polymerase chain reaction, sequencing, western blot, and immunohistochemical were performed to determine the effect of TYR and MC1R genes on plumage pigmentation in Korean native chickens.
Results:
The KCO line (Korean chicken Ogol: Black-line) with an EEC _ genotype exhibited black feathers, whereas red and red mixed with black chicken with EeC genotype exhibited white feathers. There were notable differences between the base sequences of MC1R and TYR in three Korean chicken breeds, with the highest variation in TYR. Perhaps this is the key characteristics of Korean chicken. Further, we analyzed the expression patterns of MC1R and TYR genes in each type of Korea native chicken and observed that TYR expression was high in feather follicle (R1q2) of KCO tissue. However, native red (Korean chicken red) and native red with black (Korean chicken red dark) chickens have increased TYR expression in the tissue. However, the expression of MC1R was much different from that of TYR.
Conclusion:
In this study, our results suggest that the differences in position and TYR expression levels exert more influence on plumage pigmentation in native Korean chicken breeds than changes in MC1R expression levels.
Keywords: TYR, MC1R, melanin, chicken, plumage pigmentation
Introduction
The number of chicken varieties raised in Korea has rapidly decreased since 1952 with a number of improvements. The livestock chickens originated from Red Jungle Fowl (Gallus gallus) around 5400 BC [1,2]. There are various hypotheses about domestication of chickens. Melanin pigmentation determining the feather color of a chicken depends on sex differentiation and geographic location, and a complex association of melanin formation mechanisms and many genetic variations of MC1R (melanocortin 1 receptor) genes has been previously reported [3,4]. In particular, variation in the E region of chromosome 1 containing the locus of MC1R gene (R1q5) is an important factor in determining chicken feather color [3,5]. Extension genotype locus (the E locus in birds) encoding MC1R is also common in some mammalian species [6-8]. Korean native chickens have been endangered due to continuous improvement since 1952 [9]. Therefore, genetic mechanism analysis of Korean native chickens and the influence of major genes is very important. In the process of domestication of Red Jungle Fowl (G. gallus), the ancestor of chickens, various feather colors were produced due to local influences and environmental changes. Studies of genes related to plumage colors may provide major gene markers for breed-line identification; however, available genetic data on Korean native chickens is limited. The native chickens include all the indigenous species generated through interbreeding between Korean native chickens and foreign-introduced chicken varieties, which settled in the area for a long period of time, as well as native species with pure blood [10,11].
Recent studies have identified genotypes of Korean chickens using next generation sequencing technology, verifying phenotypes for H-types with brown color and L-types with black lines, but lack research on exact gene mechanisms [12]. Up until now, the most common approach to identify and group Korean chickens is via assessment of MC1R expression to determine the levels of pheomelanin or eumelanin in relation to chicken color [13,14]. However, an effective method to identify the color pattern of Korean native chickens is not yet available. Therefore, this study analyzes the nucleotide sequence variations in DNA based on the results of genotypic changes of MC1R [15] and TYR (tyrosinase) [16] in three domestic chickens from Gyeonggi-do, Korea. Moreover, we analyzed the differences of gene expression patterns in chicken feathers to establish basic data on the differences in feather colors of Korean chickens.
Materials and Methods
Collection of animal samples
For this study, black (KCO: Korean chicken Ogol), native red with black (KCRD: Korean chicken red dark) and native red (KCR: Korean chicken red) chickens, which are pure-blood lines raised at the National Institute of Animal Science (KOR), were selected and used in the experiment (Animal Experimentation Permission Number: 2020-3) (Fig. 1). The phenotypic profiles of developmental changes for three different patterns were observed at hatch and at 16 weeks of age. 10 chickens were randomly selected from each blood-line groups according to Kang’s research method, and samples were collected by slaughter [10]. After plumages were removed from each sample to extract deoxyribonucleic acid (DNA), ribonucleic acid (RNA), proteins and tissues, tissues including the dermis and epidermis of the back were collected.
Figure 1. Classification of plumage color of Korean native chickens. A: KWC (Korean wild chicken), B: KCR (Korean native chicken Red), C: KCRD (Korean native chicken red with black), D: KCO (Ogol ; Korean native chicken black) [28].
Phenotype and genotype of plumage colors
Genetic lucus type were analyzed by the polymerase chain reaction (PCR) amplification and single nucleotide polymorphism of both TYR (GenBank: DQ118701/DQ118702) and MC1R DNA (GenBank: AY220303, AY220304, and AY220305). The segregation of plumage pigmentation and genetic polymorphisms in TYR and MC1R genes was randomly analyzed across three phenotypes. We designed a pair of primers for MC1R genotyping analysis by PCR-RFLP using BalI restriction sites; different pairs of primers are designed for TYR genotype analysis [16] (Table 1).
Table 1. Primer for sequencing and real-time RT-PCR analysis.
| No. | Primer name | Sequences (5' to 3') |
|---|---|---|
| 1 | MC1R-gF1 | GCCATCCTCAAGAACAGGAA |
| 2 | MC1R-gR1 | GCAGATGAGCATGTCGATGA |
| 3 | TYR-CC-F1 | CAAAACCATAAATAGCACTGGAAATAG |
| 4 | TYR-mL-F1 | CCTCTGGCTCTATTTGACTACACAGT |
| 5 | TYR-R1 | TTGAGATACTGGAGGTCTTTAGAAATG |
| 6 | MC1R-qF2 | GCCCTTCTTCTTCCACCTCAT |
| 7 | MC1R-qR2 | GCTCCGGAAGGCATAGATCA |
| 8 | TYR-qF2 | TGGTTTGCATAATGCCCTTCA |
| 9 | TYR-qR2 | AACCACCGCTCAAAAATGCT |
| 11 | β-actin F | GAGAAATTGTGCGTGACATCA |
| 12 | β-actin R | CCTGAACCTCTCATTGCCA |
F = forward; R = reverse, 1Primers for genotyping, 2Primers for real time RT-PCR.
MC1R and TYR gene sequencing
The MC1R and TYR genes were amplified by PCR from the DNA of each sample groups and then separated by electrophoresis on a 2% agarose gel, and the amplicons were purified using the QIAamp DNA Kit (QIAGEN, Valencia, CA). The purified target DNA samples was sequenced using ABI 3100 Sequencer (Applied Biosystems, Foster City, CA), and the nucleotide sequence was analyzed by the Sequencing Analysis Version 3.3 (Applied Biosystems, USA).
Complementary DNA (cDNA) synthesis and relative quantitative reverse transcription polymerase chain reaction (RT-PCR)
The total messenger RNA (mRNA) was extracted from the chicken tissues according to the TRIzol reagent (Invitrogen, Carlsbad, CA) method, and after purification using DNAase (Ambion, Austin, TX), cDNA was synthesized using Super-Script II (Invitrogen, Grand Island, NY). The RNA primers used for real time RT-PCR are shown in Table 1. RT-PCR amplification of mRNA genes were analyzed using the SYBR RT-PCR kit (TaKaRa, Shiga, Japan). Results were analyzed using cycle thresholds (Ct) using Rotor-Gene Real-Time Software 6.0 (BIOER, Tokyo Japan) to evaluate semi-log amplification plots. Finally, relative gene expression patterns were analyzed with β-actin mRNA expression levels as control groups (2-ΔΔCt method).
Western blot analysis
To extract the total protein from each sample, the PRO-PREPTM kit (American Intron Biotechnology) was used. After, the total amount of each protein was quantified under the guidelines of Bradford Protein Analysis (Bio-Rad, CA, USA) and protein samples were kept at −80°C until they were used in the analysis. 30 μg of total protein extracted from each sample was separated on a 13% sodium dodecyl sulphate-polyacrylamide gel and transferred to a Immunoblot polyvinylidene fluoride membranes (Bio-Rad, USA). It was then detected using secondary antibody (anti-rabbit and/or anti-mouse secondary antibody; diluted 1:5,000; Abcam, MA) after inducing antigen antibody reactions using MC1R (diluted 1:1,000; TA308794, OriGene, USA, MD), TYR (diluted 1:1,000; AB6211, Abcam, UK, Cambridge) and β-actin (diluted 1:5,000; AB49900, Abcam) in the membrane where the protein was transferred. The membrane was then fluorescently reacted with ECL and analyzed after 1–5 min of exposure in diagnostic films.
Immunohistochemistry
The paraffin sections were de-paraffinized in a xylene (Polyclear solvent; Polysciences, Warrington, PA), and antigen unmasking step were performed with 10 mm sodium citrate (pH 6.0). It was then detected using secondary antibody after inducing antigen antibody reactions using MC1R and TYR in each tissue sections for 1 h at room temperature. After antigen-antibody reaction, protein expression was detected with ABC reagent (Vactor, USA) and diaminobenzidene according to the manufacturer’s instructions. Counter staining to clearly confirm protein expression was performed using Harris hematoxylin (Fisher, Pittsburgh, PA, USA). Afterwards, the section slides were sealed with Permount (Fisher) and analyzed under an optical microscope.
Statistical analysis
All the analysis results were repeated more than three times, and statistical significance was analyzed using SAS (Statistical Analysis System Institute, Version 9.4, Cary, NC). The data are represented by an average ± SD, and the significant difference between sample groups was determined at p < 0.05.
Results
Phenotypic and genotypic evaluation
The comparison results of Korea’s KCO, KCR, and KCRD breed-lines are shown in Table 2. The plumage color of the Korean native chickens was classified by phenotypes. Expectably, there was no apparent sexual dimorphism for the color of the Korean native chicken’s plumage. As a result of MC1R / TYR gene analysis, the pigmentation phenotype of the feathers of the group with the “EEC_” genotype in Korean chickens showed a black pattern, and the pigmentation phenotype of the feathers of the group with the “E_Cc” genotype was belonged to red and red with black.
Table 2. Plumage color and MC1RTYR genotype distributions in Korean native chickens.
| Penotypin | MC1R–TYR genotype | ||||
|---|---|---|---|---|---|
| EECC | EeCC | EECc | EeCc | Total | |
| KCO: Black | 7 | 3 | 10 | ||
| KCR: Red with brown | 7 | 3 | 10 | ||
| KCRD: Red with yellow and black | 6 | 1 | 3 | 10 | |
MC1R genetic distance of each group
The results of analyzing the base sequence of MC1R through DNA sequence analysis are shown in Figure 2. MC1R in AY220303 has 945 bp, KCO has 953 bp, KCR has 945 bp, and KCRD has 953 bp nucleotide sequences. A total of 11 nucleotide sequence variants exist between each group, and KCR and KCRD have 212 (C/T) and 274 (A/G) regions in which there is an existence of nucleotide variations at the same time. In addition, nucleotide added with KCO and KCRD are 314 (-/C) and 324 (-/C). However, unlike other groups, KCO has nucleotide variation in the intervals of 291 (-/T), 292 (T/G), 293 (G/C), 332 (C/G), and 337 (T/C), also nucleotide from 245 to 247 were removed. In addition, there was no significant difference between the base sequences, but KCO and KCRD had the most C-based sequence (36%) compared to other groups.
Figure 2. The identified SNP positions and haplotypes using MC1R gene control region in Korean native chicken.
Differences in TYR nucleotide sequences among each groups
Figure 3 shows the results of analysis of TYR CC sequences in Korean native chickens. There were many differences the TYR gene when Korean species sequence was compared to the DQ11870 sequence, but all three Korean species had similar sequences among themselves. KCO showed a lot of difference from the nucleotide sequences of the other two species, KCR and KCRD, and there were differences in 200 nucleotide sequences. DQ118701 and KCO had the highest proportion of T (U) (31%) in the ratio of each base, and this result was different from base sequences of other species. In other words, Korean native species showed a lot of changes in the base of TYR CC relative to foreign species.
Figure 3. The identified SNP positions and haplotypes using TYR gene control region in Korean native chicken.
Expression analysis of MC1R and TYR gene in each group
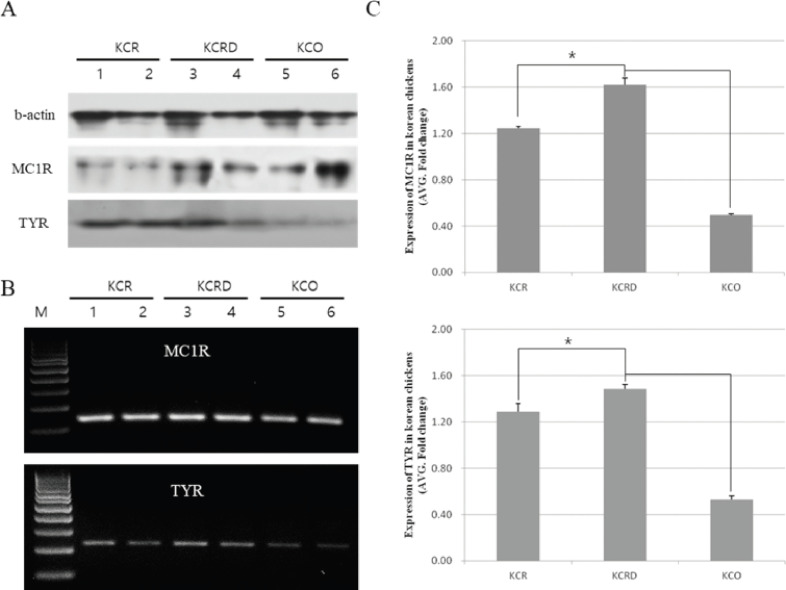
Gene expression analysis of MC1R and TYR in three Korean native chicken species is shown in Figures 4 and 5. Furthermore, we check the expression levels of MC1R and TYR at the mRNA level and observed that MC1R and TYR was highly expressed in KCRD, but was significantly lower in KCO. However, the expression patterns of proteins were different. MC1R expression was relatively high in KCO and relatively low in KCR. The expression patterns of TYR were relatively high in KCR and KCRD and relatively low in KCO (Fig. 4). Figure 3 shows the results of MC1R and TYR expression in the tissues of the outer feathers of each species. Expression of both genes was confirmed to be high in the hair follicles (Hf) and arrector pili muscle (Am) sections. In particular, the expression of MC1R was high in the melanocyte section present in the Hf of KCO. In the case of KCR and KCRD, the expression pattern was confirmed at the same site, but expression of KCRD was much higher than that of other groups. The expression of TYR was higher than that of MC1R. Unlike the expression of MC1R, the expression of TYR in Hf region was relatively high and the expression in KCRD was relatively lowly (Fig. 5).
Figure 4. Gene expression analysis of MC1R and TYR in the skin samples of Korean native chickens. A: Western blot, B: QPCR analysis, C: Real-time PCR analysis. *Significant difference (p < 0.05).
Figure 5. Localization of MC1R, TYR protein by skin samples in Korean native chickens. A figure magnification 40×, 100×, and 400×. A–C: MC1R expression, D–F: TYR expression, A, D: KCO, B, E: KCR, C,F: KCRD. The black arrow indicates protein expression position. Am: Arrector pili muscle, Hf: Hair follicles, At: Adipose tissue.
Discussion
Several studies showed that even though the genotypes of MC1R have the same phenotype, there are differences in protein combinations [16-18]. These results suggest that mutations or base substitutions of SNPs can affect various phenotypes of chicken feather color. In other words, all three Korean native chicken breeds have mutations in the MC1R gene, which may play an important role in changing chicken feather color [19-21]. In addition, similar variations were observed in species with red color, as Ellett et al. [19] findings indicate. Also, that the TYR was confirmed that the three species have a lot of differences compared to the existing nucleotide sequence, which can be said that the formation of TYR in the melanin metabolism process to control the increase or decrease of metabolism compared to MC1R [22]. That is, the expression of TYR seems to have a phenotypic effect on alterations of feather characteristics and some genes [22-24], suggesting that the expression of TYR may change depending on the mode of action of TYR, even MC1R expression is low. In particular, the mutation of the C locus located in intron 4 of TYR has several implications [25]. Moreover, it is possible to control the expression and variation of TYR, which seems to play a very important role in the determination of chicken feather color [16].
In our study, mutations in C locus were also identified, but other gene mutations were very high. Mutations in these genes were similar to the observation by Choi et al. [13], and the genetic variation of TYR and MC1R appears to be due to the environment and long-term settlement of Korean native chickens [25]. Based on these results, we found a very important difference in the analysis of the expression patterns of TYR and MC1R in this study, it can be confirmed that the TRY expression can affect the feather color even if the overall MC1R expression is low [22,26]. In our study, MC1R expression was high in the black system but low in the red line. The expression of TYR in the histological analysis was in contrast to the protein expression of MC1R and was high in the Hf of species with red color feathers. This suggests that feather color can be determined by the high expression of TYR in low stimulation of MC1R in Korean species in relation to MC1R and TYR [16,18]. However, the expression of MC1R and TYR genes in the black-line showed a lot of difference from previous studies. These results showed that MC1R gene was highly expressed, but TYR gene expression was relatively low, unlike previous studies on gene expression in black-line. This result is different from the fact that the high secretion of TYR increases the color of the dark hair, but unlike the red-colored species, the black-colored species has very high expression in Hf, Saha et al. [24] shows a similar pattern. In other words, our findings revealed genetic variation of MC1R and TYR and confirmed that there could be a very different pattern of protein expression in Korean chickens contrary to previous studies.
This study focused on the mutation of the existing MC1R and the deposition of feather color according to the region, but the results of other studies tended to focus on the large difference in mutation according to the environmental effect, which is a breeding program [24,27]. In addition, in terms of geographic ontogeny, the results of the association between MC1R and TYR as shown by Yang et al. [28] research suggest that even chickens of the same lineage may have different phenotypes. However, the results of this study suggest that TYR genetic variation may be formed according to MC1R mutations, and dynamic variation of TYR genetic sequence seem to play an important role in the regulation of plumage color in Korean native chicken. These results shows that variation in MC1R and TYR genes expressed within target tissues where feathers are formed can control the color determination of plumage color and the very complex properties of melanogenesis in Korean native chicken.
Conclusion
In this study, we found that due to major gene mutations and melanogenesis mechanisms, differences in feather color in native chickens bred in each region may differ according to metabolic processes caused by unique mutations in the MC1R and TYR genes. In addition, this result is considered to be very important as a basic study to confirm that there are many differences in the color variation of native chickens in different countries and regions.
List of Abbreviations
TYR: Tyrosinase; MC1R: Melanocortin 1 receptor; KCO: Korean chicken Ogol; KCRD: Korean chicken Red dark; KCR: Korean chicken Red; Am: Arrector pili muscle; At: Adipose tissue; Hf: Hair follicles; EEC, ECc, CC : E and C locus Genotype of TYR gene in chickens plumage color; KOR: Korea; RFLP: Restriction fragment length polymorphism; ECL: Enhanced chemiluminescence; SNPs: Single nucleotide polymorphism.
Acknowledgment
This study was conducted materially in the Laboratory of the Hankyong National University in Korea.
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
ISN have participated in developing the protocol, the sample, and in drafting the manuscript. ISN and MGO participated in the identification of the development of the database. ISN, MSN and WSK contributed to the translation of the manuscript. WSK supervised the analysis.
References
- [1].West B, Zhou BX. Did chickens go north? New evidence for domestication. J Archaeol Sci. 1989;15(5):205–18. https://doi.org/10.1079/WPS19890012. [Google Scholar]
- [2].Crawford RD. Origin, history, and distribution of commercial poultry. In: Hunton P, editor. Poultry Production. Amsterdam, The Netherlands: Elsevier; 1999. pp. 1–21. 5. [Google Scholar]
- [3].Dunn PO, Whittingham LA, Pitcher TE. Mating systems, sperm competition and the evolution of sexual dimorphism in birds. Evolution. 2001;55(1):161–75. doi: 10.1111/j.0014-3820.2001.tb01281.x. https://doi.org/10.1111/j.0014-3820.2001.tb01281.x. [DOI] [PubMed] [Google Scholar]
- [4].Zhang GW, Liao Y, Zhang WX, Wu Y, Liu A. A new dominant haplotype of MC1R gene in Chinese black plumage chicken. Anim Genet. 2017;48(5):624. doi: 10.1111/age.12576. https://doi.org/10.1111/age.12576. [DOI] [PubMed] [Google Scholar]
- [5].Yang CW, Ran JS, Yu CL, Qiu MH, Zhang ZR, Du HR, et al. Polymorphism in MC1R, TYR and ASIP genes in different colored feather chickens. 3 Biotech. 2019;9(5):203. doi: 10.1007/s13205-019-1710-z. https://doi.org/10.1007/s13205-019-1710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Klungland H, Vage DI, Gomez-Raya L, Adalsteinsson S, Lien S. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm Genome. 1995;6(9):636–9. doi: 10.1007/BF00352371. https://doi.org/10.1007/BF00352371. [DOI] [PubMed] [Google Scholar]
- [7].Kijas JMH, Wales R, Törnsten A, Chardon P, Moller M, Andersson L. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics. 1998;150(3):1177–85. doi: 10.1093/genetics/150.3.1177. https://doi.org/10.1093/genetics/150.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Väge DI, Klungland H, Lu D, Cone RD. Molecular and pharmacological characterization of dominant black coat colour in sheep. Mamm Genome. 1999;10(1):39–43. doi: 10.1007/s003359900939. https://doi.org/10.1007/s003359900939. [DOI] [PubMed] [Google Scholar]
- [9].Sang BD, Kong HS, Kim HK, Choi CH, Kim SD, Cho YM, et al. Estimation of genetic parameters for economic traits in Korean native chickens. Anim Biosci. 2006;19(3):319–23. https://doi.org/10.5713/ajas.2006.319. [Google Scholar]
- [10].Kang BS. Restoration of native chicken and industrialization of Korean chicken. Korea Poult Assoc. 2010;42(12):141–3. [Google Scholar]
- [11].Korea National Institute of Animal Science, Poultry Stabilization Committee; 2008. Breeding of domestic chickens and establishing certification standards. [Google Scholar]
- [12].Eck SH, Benet-Pages A, Flisikowski K, Meitinger T, Fries R, Strom TM. Whole genome sequencing of a single bos taurus animal for single nucleotide polymorphism discovery. Genome Biol. 2009;10(8):R82. doi: 10.1186/gb-2009-10-8-r82. https://doi.org/10.1186/gb-2009-10-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Choi JA, Lee JH, Jang HJ, Lee KT, Kim TH, Lee HJ, et al. Genetic variations of chicken TYR gene and associations with feather color of Korean native chicken (KNC) Korean J Poult Sci. 2014;41(1):7–14. https://doi.org/10.5536/KJPS.2014.41.1.7. [Google Scholar]
- [14].Dávila SG, Gil MG, Resino-Talaván P, Campo JL. Association between polymorphism in the melanocortin 1 receptor gene and E locus plumage color phenotype. Poult Sci. 2014;93(5):1089–96. doi: 10.3382/ps.2013-03611. https://doi.org/10.3382/ps.2013-03611. [DOI] [PubMed] [Google Scholar]
- [15].Xu JG, Xie MG, Zou SY, Liu XF, Li XH, Xie JF, et al. Interactions of allele E of the MC1R gene with FM and mutations in the MLPH gene cause the five-gray phenotype in the Anyi tile-like gray chicken. Genet Mol Res. 2016;15(2):1–8. doi: 10.4238/gmr.15027633. https://doi.org/10.4238/gmr.15027633. [DOI] [PubMed] [Google Scholar]
- [16].Liu WB, Chen SR, Zheng JX, Qu LJ, Xu GY, Yang N. Developmental phenotypic-genotypic associations of tyrosinase and melanocortin 1 receptor genes with changing profiles in chickens plumage pigmentation. Poult Sci. 2010;89(6):1110–4. doi: 10.3382/ps.2010-00628. https://doi.org/10.3382/ps.2010-00628. [DOI] [PubMed] [Google Scholar]
- [17].Oh JD, Lee KW, Seo OS, Cho BW, Jeon GJ, Lee HG, et al. Estimation of genetic characteristics and cumulative power of discrimination in Korean native chicken and Korean native commercial chicken. J Life Sci. 2010;20(7):1086–92. https://doi.org/10.5352/JLS.2010.20.7.1086. [Google Scholar]
- [18].Kim SH. Identification of genetic association among different colors of Korean native chicken breeds through the RAPD-PCR method. J Anim Health Prod. 2021;9(1):33–9. https://doi.org/10.17582/journal.jahp/2021/9.1.33.39. [Google Scholar]
- [19].Ellett AE, Okimoto R. Melanocortin 1-receptor (MC1R-R) gene polymorphisms associated with chickens E locus alleles. Univers Student J Inquiry. 2000;1:37–41. [Google Scholar]
- [20].Boichard M, Coville JL, Coquerelle G. Polymorphism of the MC1R gene and feather color in the chicken: between breeds diversity and within family analysis. Proc. 27th International Conference an Animal Genetics (ISAG); 2000. p. 342. [Google Scholar]
- [21].Ling MK, Lagerström MC, Fredriksson R, Okimoto R, Mundy NI, Takeuchi S, et al. Association of feather colour with constitutively active melanocortin 1 receptors in chicken. Eur J Biochem. 2003;270(7):1441–9. doi: 10.1046/j.1432-1033.2003.03506.x. https://doi.org/10.1046/j.1432-1033.2003.03506.x. [DOI] [PubMed] [Google Scholar]
- [22].Saha B, Singh SK, Sarkar C, Mallick S, Bera R, Bhadra R. Transcriptional activation of tyrosinase gene by human placental sphingolipid. Glycoconj J. 2006;23(3–4):259–68. doi: 10.1007/s10719-006-7931-5. https://doi.org/10.1007/s10719-006-7931-5. [DOI] [PubMed] [Google Scholar]
- [23].Murisier F, Guichard S, Beermann F. The tyrosinase enhancer is activated by Sox10 and Mitf in mouse melanocytes. Pigment Cell Res. 2007;20(3):173–84. doi: 10.1111/j.1600-0749.2007.00368.x. https://doi.org/10.1111/j.1600-0749.2007.00368.x. [DOI] [PubMed] [Google Scholar]
- [24].Saha B, Singh SK, Mallick S, Bera R, Datta PK, Mandal M, et al. Sphingolipid-mediated restoration of Mitf expression and repigmentation in vivo in a mouse model of hair graying. Pigment Cell Melanoma Res. 2009;22(2):205–18. doi: 10.1111/j.1755-148X.2009.00548.x. https://doi.org/10.1111/j.1755-148X.2009.00548.x. [DOI] [PubMed] [Google Scholar]
- [25].Chang CM, Coville JL, Coquerelle G, Gourichon D, Oulmouden A, Tixier-Boichard M. Complete association between a retroviral insertion in the tyrosinase gene and the recessive white mutation in chickens. BMC Genomics. 2006;7:19–46. doi: 10.1186/1471-2164-7-19. https://doi.org/10.1186/1471-2164-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Le Pape EL, Wakamatsu K, Ito S, Wolber R, Heraing VJ. Regulation of eumelanin/pheomelanin synthesis and visible pigmentation in melanocytes by ligands of the melanocortin 1 receptor. Pigment Cell Melanoma Res. 2008;21(4):477–86. doi: 10.1111/j.1755-148X.2008.00479.x. https://doi.org/10.1111/j.1755-148X.2008.00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hoque MR, Jin S, Heo KN, Kang BS, Jo C, Lee JH. Investigation of MC1R SNPs and their relationships with plumage colors in Korean native chicken. Asian-Australas J Anim Sci. 2013;26(5):625–962. doi: 10.5713/ajas.2012.12581. https://doi.org/10.5713/ajas.2012.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang CW, Ran JS, Yu CL, Qiu MH, Zhang ZR, Du HR, et al. Polymorphism in MC1R, TYR and ASIP genes in different colored feather chickens. 3 Biotech. 2019;9(5):203. doi: 10.1007/s13205-019-1710-z. https://doi.org/10.1007/s13205-019-1710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]