ABSTRACT
Streptococcus oralis is a commensal viridans group streptococcus of the human oral cavity and a frequent cause of endovascular infection. Here, we report the complete whole-genome sequence of S. oralis strain SF100, which was originally isolated from the blood of a patient with infective endocarditis. This strain contains the lysogenic bacteriophage SM1, which enhances the virulence of SF100 in animal models of endocardial infection.
ANNOUNCEMENT
Streptococcus oralis is one of the most frequent causes of infective endocarditis (1, 2). Strain SF100 was originally isolated in 1980 from the blood of a patient with this disease, using standard clinical methods. It was originally identified as Streptococcus mitis, based on biochemical testing and 16S RNA gene sequencing (3). However, we now report that strain SF100 more closely resembles members of the species S. oralis, based on whole-genome sequencing.
SF100 contains the lysogenic bacteriophage SM1, which encodes several important virulence factors, including PblA, PblB, and lysinSM1 (4–6). Disruption of these genes results in a significant reduction in the binding of SF100 to human platelets and reduced virulence in animal models of endocarditis (5, 7). Of note, these adhesins appear to be widely prevalent among S. oralis strains of the human oral microbiome (8–10).
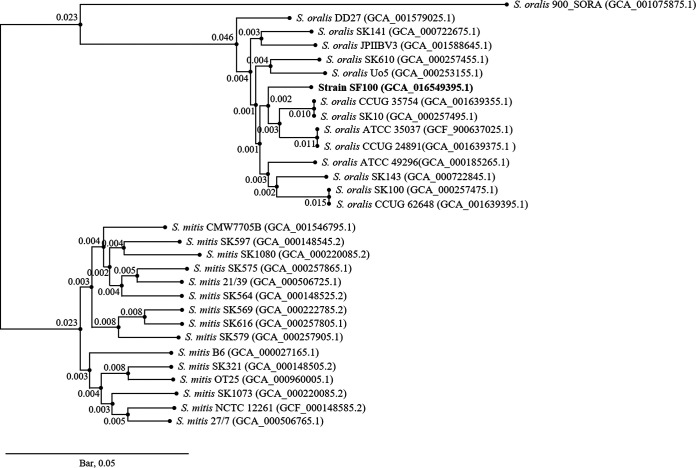
For whole-genome sequencing, strain SF100 was grown in Todd-Hewitt broth at 37°C to an optical density (600 nm) of 0.8 to 1.0. Genomic DNA was isolated using the G-spin genomic DNA extraction kit (Intron, Seoul, South Korea). Approximately 100 ng of isolated genomic DNA was sequenced using the RS II platform (Pacific Biosciences [PacBio], Menlo Park, CA, USA) at Macrogen Co., Ltd. (Seoul, South Korea). The DNA library was prepared using a single-molecule real-time (SMRT) Cell 8Pac v3 and the P6 DNA polymerase binding kit (PacBio). Reads were filtered and assembled using Canu v1.7 (correctedErrorRate = 0.015) (11). The sequencing yielded 140,974 subreads (1.11 Gbp). The mean subread length and N50 value were 7,905 bp and 12,037 bp, respectively. After assembly, strain SF100 contained one contig of 1,969,104 bp, with a G+C content of 41.5 mol%. To assess the completeness of genome assembly, Benchmarking Universal Single-Copy Orthologs (BUSCO) v3.0 analysis of the bacteria_odb9 data set (148 orthologs) was performed (12). This showed 96.6% complete orthologs and 3.4% missing orthologs. The assembled genome contained 1,909 genes annotated by the Prokaryotic Genome Annotation Pipeline (PGAP) v5.1 of the National Center for Biotechnology Information (13, 14). Protein sequences of each single-copy ortholog group were aligned using MAFFT v7.435 with the auto option (15). A phylogenetic tree of 1,010 single-copy ortholog genes was built by the maximum likelihood method using RAxML v8.2.12 with the -m PROTGAMMAAUTO -p 1000 options (16). In the phylogenetic tree (Fig. 1), strain SF100 clustered with the S. oralis group. The results of the phylogenetic tree indicate that SF100 is a member of S. oralis. Default parameters were used for all software unless otherwise specified.
FIG 1.
Maximum likelihood phylogenetic tree of single-copy ortholog genes in strains of S. oralis, S. mitis, and SF100. The GenBank assembly accession number for each strain is shown in parentheses. The numbers indicate the length. The phylogenetic tree showed that the S. oralis group and the S. mitis group were clearly distinguished, and strain SF100 was closely related to the S. oralis group. The scale bar indicates 0.05 substitution per nucleotide position.
Data availability.
The whole-genome sequence of S. oralis SF100 has been deposited in DDBJ/EMBL/GenBank under the accession number CP066172. The BioProject, BioSample, and SRA accession numbers are PRJNA685325, SAMN17082796, and SRR13854333, respectively.
ACKNOWLEDGMENTS
This work was funded by National Institutes of Health grants R01 AI41513 and R01 AI106987 (to P.M.S.), National Research Foundation of Korea grants NRF-2018K2A206023828 and NRF-2020M2D8A3094054 (to H.S.S.), and a Nuclear R&D Program of the Ministry of Science, ICT, and Future Planning grant (to H.S.S.).
Contributor Information
Paul M. Sullam, Email: paul.sullam@ucsf.edu.
David Rasko, University of Maryland School of Medicine.
REFERENCES
- 1.Baddour LM. 1998. Infective endocarditis caused by β-hemolytic streptococci. Clin Infect Dis 26:66–71. doi: 10.1086/516266. [DOI] [PubMed] [Google Scholar]
- 2.Moreillon P, Que YA, Bayer AS. 2002. Pathogenesis of streptococcal and staphylococcal endocarditis. Infect Dis Clin North Am 16:297–318. doi: 10.1016/s0891-5520(01)00009-5. [DOI] [PubMed] [Google Scholar]
- 3.Bensing BA, Rubens CE, Sullam PM. 2001. Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect Immun 69:1373–1380. doi: 10.1128/IAI.69.3.1373-1380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seo HS, Xiong YQ, Mitchell J, Seepersaud R, Bayer AS, Sullam PM. 2010. Bacteriophage lysin mediates the binding of Streptococcus mitis to human platelets through interaction with fibrinogen. PLoS Pathog 6:e1001047. doi: 10.1371/journal.ppat.1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell J, Siboo IR, Takamatsu D, Chambers HF, Sullam PM. 2007. Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol Microbiol 64:844–857. doi: 10.1111/j.1365-2958.2007.05703.x. [DOI] [PubMed] [Google Scholar]
- 6.Bensing BA, Siboo IR, Sullam PM. 2001. Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage. Infect Immun 69:6186–6192. doi: 10.1128/IAI.69.10.6186-6192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo HS, Sullam PM. 2011. Characterization of the fibrinogen binding domain of bacteriophage lysin from Streptococcus mitis. Infect Immun 79:3518–3526. doi: 10.1128/IAI.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, Angly FE, McDole T, Mariella RP, Jr, Rohwer F, Haynes M. 2010. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci U S A 108(Suppl 1):4547–4553. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Hebshi NN, Baraniya D, Chen T, Hill J, Puri S, Tellez M, Hasan NA, Colwell RR, Ismail A. 2019. Metagenome sequencing-based strain-level and functional characterization of supragingival microbiome associated with dental caries in children. J Oral Microbiol 11:1557986. doi: 10.1080/20002297.2018.1557986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Kamel A, Baraniya D, Al-Hajj WA, Halboub E, Abdulrab S, Chen T, Al-Hebshi NN. 2019. Subgingival microbiome of experimental gingivitis: shifts associated with the use of chlorhexidine and N-acetyl cysteine mouthwashes. J Oral Microbiol 11:1608141. doi: 10.1080/20002297.2019.1608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 13.Kitts PA, Church DM, Thibaud-Nissen F, Choi J, Hem V, Sapojnikov V, Smith RG, Tatusova T, Xiang C, Zherikov A, DiCuccio M, Murphy TD, Pruitt KD, Kimchi A. 2016. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res 44:D73–D80. doi: 10.1093/nar/gkv1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome sequence of S. oralis SF100 has been deposited in DDBJ/EMBL/GenBank under the accession number CP066172. The BioProject, BioSample, and SRA accession numbers are PRJNA685325, SAMN17082796, and SRR13854333, respectively.