ABSTRACT
IRON‐REGULATED TRANSPORTER 1 (IRT1) is a central iron transporter responsible for the uptake of iron from the rhizosphere to root epidermal cells. This study uses immunohistochemistry, histochemistry, and fluorometry to show that this gene’s promoter is also active in the aboveground parts, specifically in phloem companied cells. Promoter activity here was regulated by iron as it was in the roots. The promoter of the close IRT2 homolog was root-specific and only weakly active in the stem pits. RT-PCR showed the presence of a long splicing form exclusively in iron-deficient roots. The short splicing form was present in all organs regardless of the presence of iron. Immunohistology exhibited labeling on the periphery of the epidermal cells in matured root zone and intracellular patches in the meristematic cells. In the aboveground organs, the protein was seen in the whole volume of companion cells and in neighboring sieve elements as bodies. The fluorescent protein technique revealed the short IRT1 form to be present in the patches accumulated mainly around the nucleus and the long form as a continuous layer along the cells periphery. These results suggest that IRT1 has a role also in the aboveground organs.
KEYWORDS: IRON‐REGULATED TRANSPORTER 1, promoter activity, protein localization, gene expression, phloem companion cells, Arabidopsis
Introduction
Iron is a central component of electron chains and a co-factor of many enzymes in living organisms. In addition, plants require iron for photosynthesis and chlorophyll synthesis.1 Although this element is abundant in the soil, plants are unable to absorb all its forms efficiently. They have developed several strategies to uptake iron and maintain its balance in their organs. Facilitating iron absorption is based on iron reduction and chelation.2,3 Grasses secrete phytosiderophores (PS) which form complexes with Fe3+, and these are easily transported to roots. Non-graminaceous plants utilize reduction of Fe3+ to Fe2+, and divalent iron is transported through the plasma membrane of root epidermal cells by Fe2+ transporters. In the Arabidopsis , IRON‐REGULATED TRANSPORTER 1 (IRT1) is considered to be a central Fe transporter.4 However, this protein may also uptake other divalent metals such as Zn, Mn, Cd, Co or Ni.4–7
Studies on three independent mutant irt1 alleles demonstrated that the absence of IRT1 function in Arabidopsis causes severe defects resembling those induced by iron deficiency. They include chlorosis, disturbed normal tissue development and chloroplast differentiation, a drastic reduction in growth rate and fertility or significant alteration of photosensitivity and chlorophyll fluorescence parameters.4,6,8 These effects lead to plants’ premature death when they grow in the soil, but this can be overcome by excessive fertilization with iron.4,8 Interestingly IRT2, a highly homologous protein to IRT1, is not essential for plant survival, and the mutant shows no iron starving phenotype.8 IRT proteins are ZIP family members, and they possess several transmembrane domains at N- and C-termini.9 In a pioneering study Vert with colleagues 4, demonstrated through promoter analysis and in situ hybridization that IRT1 was expressed in the root’s external cell layers, specifically in response to iron starvation. They also showed by protoplast transient expression assay that protein was targeted to the plasma membrane.
Here we indicate that the IRT1 promoter in Arabidopsis is active in aboveground parts of plants as well. Besides, the promotor is regulated here by iron, although not to the same extent as in the roots. The staining is restricted to the vasculature, especially the phloem companion cells. These specific cells accumulate IRT1 protein as well. IRT2, a close homolog, is more root-specific. IRT1 more likely plays an important role also in non-root parts.
Material and methods
Plant material and growth conditions
Arabidopsis thaliana, ecotype Columbia (Col-0) plants or transgenic plants with Col-0 background were standardly cultivated in pots with soil substrate (50% peat moss, 30% perlite, 20% sand) in cultivation room under temperature 22 °C, humidity of 40% - 60% and 14 h light/10 h dark photoperiod. Illumination was provided by white-colored LED panels at an intensity of 150 µmol m-2 s−1. For the experiment, three weeks old pIRT1-GUS, pIRT2::GUS and wild plants were carefully removed from the soil substrate; roots were washed with tap water and planted in pots (diameter 6 cm) with perlite. They were watered one week with rainwater and then every 24 h with 15 mL solution containing 1/10 MS10 macro and microelements, or with the same solution with omitting or reducing iron sulfate to 0.27 mg/L. After three weeks, samples from all organs were harvested and used for cryotomy, fluorometry, western blot analysis and RT-PCR analysis. For the in vitro experiment, pIRT1::GUS and pIRT2::GUS seeds were surface sterilized with 1% sodium hypochlorite and germinated in Petri dishes on a solid growth medium (1/2 MS salts, minimal organics, 1% sucrose, solidified with 0.7% agar). Petri dishes were kept in the cultivation room under conditions as mentioned above. After three weeks, seedlings were moved to 1-L glass jars containing the same medium or medium with omitting iron. After six weeks, organ samples were harvested for fluorometry, western blotting and cryotomy. For confocal microscopy, seeds of pIRT1::IRT1::Dendra2 lines were germinated and grown in Petri dishes on the solid growth medium without iron in the vertical position. Wild seedlings were cultivated the same way and on the complete solid growth medium for western blot assay.
Vector construction and plant transformation
For the construction of the pIRT1::GUS and pIRT2::GUS transcription fusions, about 2000 bp long regulation sequences located upstream of the start codon of IRT genes were amplified by PCR from genomic DNA with the Q5® Hot Start High-Fidelity DNA Polymerase and inserted into pPCV820 vector between BglII and XmaI restriction sites. For preparation pIRT1::IRT1::Dendra2 fusions, genomic IRT1 sequences encoding short and long splicing IRT1 RNA forms, including around the 2000 promoter region, were amplified by PCR from genomic DNA with the same DNA polymerase. Dendra2 was PCR amplified from a Gateway Dendra2-At-N-entry clone (Evrogen). Fragments were inserted sequentially into the pAMPAT-MSC vector (GenBank: AY436765.1) between AscI and BamHI sites. The DNA sequence encoding Dendra2 was attached to the 3ʹ end of genomic DNA encoding IRT1.1, and IRT1.2 splicing forms such a way that stop codon was replaced by DNA stretch encoding Gly-Ala-Gly tripeptide. Sequences of primers are accessible in Table 1. All enzymes were purchased from New England BioLabs (Frankfurt, Germany). Sequence and reading frames were proved by sequencing. Plasmids were transformed into Arabidopsis thaliana (Col-0) through GV3101 (pMP90RK) Agrobacterium tumefaciens11 with the floral-dip method.12 Selection of homozygous plants with one insert was made with 7.5 mg/L phosphinothricin for the pAMPAT-MSC plasmid and with 15 mg/L hygromycin for the pPCV812 plasmid (both chemicals were from Duchefa, Haarlem, The Netherland). Resistant seedlings growing on the Fe depleted medium were screened under an epifluorescence microscope to select lines expressing IRT1::Dendra2 fusion protein or by histochemistry to detect lines with GUS activity.
Table 1.
The sequences of DNA oligomers used in this study
| Oligomer | Sequence |
|---|---|
| For pAtIRT1::GUS construct: | |
| pAtIRT1-BglII-F | 5′-CACAGATCTTCAAATCATACACACTTCATCCTG-3′ |
| pAtIRT1-XmaI-R | 5′-TTTCCCGGGTTTTTTTTTTTTTCTTTTTTCTTTTGGATTG-3′ |
| pAtIRT2-BglII-F | 5′-GACAGATCTAACTTCTAAACTCTTCCTCATCTC-3′ |
| pAtIRT2-XmaI-R | 5′-TATCCCGGGGTAGTATTGAGATTGTTTTATAATATATGATG-3′ |
| For pAtIRT1::gAtIRT1::Dendra2 constructs: | |
| gIRT1.2-PstI-R | 5′-ATTAGCTGCAGAGCCCATTTGGCGATAATCGACATTC-3′ |
| gIRT1.1-PstI-R | 5′-GTAATCTGCAGTTTAAAATACAATGATATTCGGTATGTATA-3′ |
| Dendra2-PstI-F | 5′-GATACTGCAGGGAGCTGGTATGAACACTCCTGGAATCAATCTC-3′ |
| Dendra2-BamHI-R | 5′-CAAGGATCCTCACCAAACCTGTGATGGGAGAG-3′ |
| pAtIRT1-AscI-F | 5′-CGGGAGGCGCGCCAAATCATACACACTTCATCCTGACAA-3′ |
| For RT-PCR: | |
| IRT1.1–279/2-313-F | 5′-TCGTAATCCTCATTGCAAGCATGA-3′ |
| IRT1.1–415/2-449-F | 5′-TATGCACGTTTTACCTGATTCTTTC-3′ |
| IRT1.1–551/2-585-F | 5′-ACGAGCCTATACACCAGCAAGA-3′ |
| IRT1.2/1368-R | 5′-TGGGGCCTATCACATTTACATATCTACC-3′ |
| IRT1.2/1149-R | 5′-CGAGAAGAGCCGCGATTAAACA-3′ |
| IRT1.1/723-R | 5′-CGGTATGTATATATGTGCGTACGAACC-3′ |
| IRT1.2/753-R | 5′-CTATGATCCCAAGTTCCAAGACC-3′ |
| IRT1.1/746-R | 5′-GTCATTTAAAATACAATGATATTCGGTATGTA-3′ |
| AT4G05320.1–908-F | 5′-GGAAAACAATTGGAGGATGGTC-3′ |
| AT4G05320.1–1238-R | 5′-ACGAGATTTAGAAACCACCACGA-3′ |
| AT2G28390-1905-F | 5′-AAAGGATTGGGACCCCACAA-3′ |
| AT2G28390-2250-R | 5′-TCTCTCAAGGGTTTCTGGGTACA-3′ |
Cryosectioning
Different parts of plants were fixed with freshly prepared 1% formaldehyde from paraformaldehyde powder in MTBS buffer13 [50 mM PIPES, 5 mM MgSO4, 5 mM EGTA, pH 7.1,] for histochemistry or with 4% paraformaldehyde in the same buffer for 3 hrs for immunohistochemistry. After washing three times with MTSB, samples were infiltrated with 30% sucrose and then immersed into FSC 22 Frozen Section Media (Leica Biosystems, Buffalo Grove, Illinois, USA) using plastic tissue mold and frozen in liquid nitrogen. Frozen blocks with samples were mounted on holders, and 10 µM thick sections were cut with Reichert-Jung Cryo-cut II cryostat at −20 °C. Section ribs were moved into cooled 2 mL Eppendorf microcentrifuge tubes and then used for immunolocalization or GUS histochemistry.
GUS histochemistry and fluorometry
For whole-mount histochemistry, samples were incubated in GUS staining solution containing 50 mM phosphate buffer, 10 mM EDTA, 2 mM K3Fe(CN)6, 2 mM K4Fe(CN)6, 10% methanol, 0.1% Triton X-100 and 1 mM 5-Bromo-4-chloro-3-indolyl-ß-D-glucuronic acid (X-GlcA, Duchefa, Haarlem, Netherlands) at 37 °C in the dark. After appearing blue staining, the samples were treated with ethanol to remove chlorophyll, infiltrated with glycerol and examined.
In histochemistry using cryosections, microcentrifuge tubes were removed from the cryostat, and 1 mL of MTSB was added to the sections. When the sections settled to the bottom of the tube, the buffer was carefully pipetted off and replaced with a new one. The process was repeated twice. Then the sections were treated with GUS histochemical solution, the composition of which is given above. After that, the solution was removed, and sections were counterstained with 0.1% basic fuchsine for 10 min. After washing with water, sections were infiltrated with glycerol, transferred on a microscopic slide and covered with a coverslip.
For GUS fluorometry assay, samples of different organs were homogenized in protein extraction buffer (50 mM phosphate buffer, 10 mM EDTA, 0.1% Triton X-100, 0.1% SDS) in 1.5 mL Eppendorf microcentrifuge tubes with LLG metal micro pestle fitted in a hand drill. Extracts were then centrifuged at 16,000 x g and 4 °C, and protein concentration in the supernatant was determined with a DC protein assay kit (Bio-Rad, Hercules, California, USA). Afterward, samples were diluted with extraction buffer to equal protein concentration. The reaction was carried out in 50 µL extraction buffer containing 1 µg total proteins and 2 mM 4-Methylumbelliferyl-ß-D-glucuronide trihydrate (4-MUG, Duchefa, Haarlem, Netherlands) in black 96 well-plate (BRAND, Wertheim, Germany). The plate was incubated in the dark at 37 °C, and after 1 h the reaction was stopped by adding 200 μL 0.2 M Na2CO3. Fluorescence was measured with a microplate reader Fluoroskan Ascent® FL (Thermo Fisher Scientific, Waltham, USA), employing 355 nm excitation and 485 nm emission filters.
Immunohistochemistry
Sections prepared and washed in Eppendorf microcentrifuge tubes as was described above were treated for 1 h with anti-GUS (Sigma-Aldrich, G5420, 1:300) or anti-IRT1 (Agrisera, AS11 1780, 1:300) antibodies in MTSB buffer supplemented with 1% (w/v) bovine serum albumin (fraction V, fatty acid-free, Roche). After washing three times with MTSB, sections were treated for 1 h with Alexa Fluor® 488 (Abcam, ab150077, 1:150) secondary antibody in buffer mentioned above, again washed three times, infiltrated with glycerol supplemented with p-phenylenediamine,13 transferred on a microscopic slide and covered with a coverslip.
Microscopy
Specimens treated by histochemistry, immunohistochemistry and whole roots expressing IRT proteins tagged with Dendra2 were estimated with an Olympus FV1000 confocal laser scanning microscope (Olympus, Tokyo, Japan). For IRT localization, seedlings of pAtIRT1::AtIRT1::Dendra2 transgenic lines were mounted on microscopic slides covered with a medium as described previously.14 Roots of these seedlings and cryo-section samples prepared by immunohistochemistry were imaged using UPlanFL 20x/0.50, UPLanSApo 40x/0.90, or UPlanSApo 60x/1.35 Oil objectives. The green signal was excited with a 488 nm line of an argon laser, and the signal was collected with a 505–525 nm band-pass filter and a 488/543/633 nm dichroic mirror. Bright light images of samples treated by histochemistry were captured with Levenhuk M1400 PLUS Microscope Digital Camera and LevenhukLite software.
cDNA synthesis and semi-quantitative RT-PCR
Total RNA was extracted using the RNeasy Plant MiniKit (Qiagen) from 100 mg of roots and upper ground parts of seedlings growing three weeks on medium with or without Fe or upperground organs of the perlite-derived plants growing with or without Fe as described above. After rigorous elimination of the possible genome DNA contamination with TURBO DNA-free™ Kit (Thermo Fisher Scientific), the cDNA was prepared in a total volume of 20 µL from 100 ng total RNA with FIREScript RT cDNA Synthesis Kit (Solis BioDyne, Estonia) according to the manufacturer’s instructions and using a mixture of attached oligo(dT) and random primers. After eliminating the enzyme by heating, the original reaction mixture was diluted with 60 µL water, and 2 µL of cDNA mix was used for PCR reaction in a volume of 15 µL. PCR was accomplished with Q5® High-Fidelity DNA Polymerase (New England Biolabs, UK, Ltd) on an Eppendorf MastercyclerPro instrument (Eppendorf). The amplification program was: 98 °C for 45 s, then 35 or 50 cycles at 98 °C for 10 s, 65 °C for 20 s, 72 °C for 30 s, and then one cycle at 72 °C for 5 min. The primers are listed in Table 1. POLYUBIQUITIN10 (At4G05320) and MONENSIN SENSITIVITY1 (At2G28390) were used as reference genes.15 The experiment was independently repeated twice.
Western blot analysis
Total protein was extracted from 100 mg of plant organs with 100 µL double concentrated RIPA buffer (100 mM Tris-HCl pH 7.4, 300 mM NaCl, 2 mM EDTA, 2 mM EGTA, 2% Triton X-100, 1% sodium deoxycholic acid, 0.2% SDS, 2 mM PMSF and plant protease inhibitor) or Pierce ™ Plant Total Protein Extraction Kit (Thermo Scientific). Homogenization of tissue was done in 1.5 mL Eppendorf microcentrifuge tubes with LLG metal micro pestle fitted in a hand drill. After centrifugation (16,000x g, 20 min, 4 °C), the supernatant was used for Western blotting. For membrane protein extraction, we used Minute™ Plant Plasma Membrane Protein Isolation Kit (Invent, Biotechnologies INC) following the manufacturer’s instructions. The pellet was resuspended in 1x RIPA buffer. Proteins were separated with SDS-PAGE using (10% -polyacrylamide) in Tris-Gly buffer (25 mM Tris, 192 mM Gly, and 0.1% SDS, pH 8.3) using Mini-PROTEAN® Tetra electrophoresis system (Bio-Rad). After electrophoresis, the proteins were transferred onto Immun-Blot® PVDF Membrane (Bio-Rad) using blotting buffer (25 mM Tris, 192 mM Gly, and 20% ethanol, pH 8.3). Then the membrane was blocked for 1 h with 5% skim milk (Serva), in TBST buffer (20 mM Tris-HCl, pH 7.4, 180 mM NaCl, and 0.1% Tween 20), before incubation in the primary antibody in TBST containing 5% skim milk. Subsequently, the membrane was washed three times in TBST for 10 min and incubated in secondary antibody in TBST containing 5% skim milk. After three washes in TBST for 10 min and two washes in TBS, for 5 min, the membrane was treated with Clarity Western ECL Substrate (Bio-Rad). Signal was recorded on photographic paper, which was developed and fixed in the normal way. In the western blotting studies, we used polyclonal anti-GUS (Sigma-Aldrich, G5420, 1:2,000), Polyclonal anti-IRT1 (Agrisera, AS11 1780, 1:3,000), Monoclonal anti-Actin (Sigma-Aldrich, A0480-25UL, 1:10,000) primary and goat anti-rabbit IgG:HRP (BioRad, 403005, 1:10,000) and goat anti-mouse IgG:HRP[BioRad, 103005, 1:10,000) secondary antibodies.
Results
In a pioneering study by Vert et al. 4, the Arabidopsis IRT1 promoter was shown to be active in the root epidermis. We wanted to analyze the promoter’s strength in the roots in detail and examine whether the promoter activity is root-specific. We prepared transcriptional fusion constructs with about 2 kbp long promoter region of IRT1 with DNA sequences for the E. coli GUS reporter. For comparison, we also prepared a construct for IRT2, a close homolog to IRT1. We isolated several homozygous lines with a single insert and performed a classical histochemical analysis of GUS activity in advanced seedlings of transgenic plants growing in vitro on ½ MS medium. In strongly expressing transgenic lines, roots in the zone of maturation with visible root hairs were stained within one hour. However, surprisingly after extended treatment, a blue coloration was also seen in veins of cotyledons and developing leaves. This finding inspired us to carry out a more detailed study. We analyzed the plants grown in the soil, and the results were analogous. Promoter activity pattern was similar in all transgenic lines; poorly expressing plants only required more extended treatment. The IRT1 promoter was active except for roots in the vascular bundles of leaves and stems (Figure 1a) and was also very active inside pistils and at the junction of anthers and filaments (Figure 1g). When we analyzed promotor activity at the tissue level on hand sections, the signal was detected in the phloem part of the vascular bundles of all aboveground organs. For comparison, we analyzed the promoter of close IRT2 homolog. This promotor was active only in the roots, and a weak reaction was seen in parenchymatous pith cells (Figure 1m). Based on histochemical estimation, we selected for quantitative analysis three transgenic lines with high, medium, and low GUS expression. Fluorometric measurement of GUS enzyme activity showed a different strength of promoter among organs. The roots showed the strongest signal (Figure 2a). Considerable differences in promotor activity were between three lines, but the proportion of signal intensities between organs in all lines was maintained. As expected IRT2 promotor was very active in roots, and a weak signal was registered in stem extract (Figure 2a). The pioneering study by Vert et al. 4 also showed that the IRT1 expression was regulated by iron.
Figure 1.
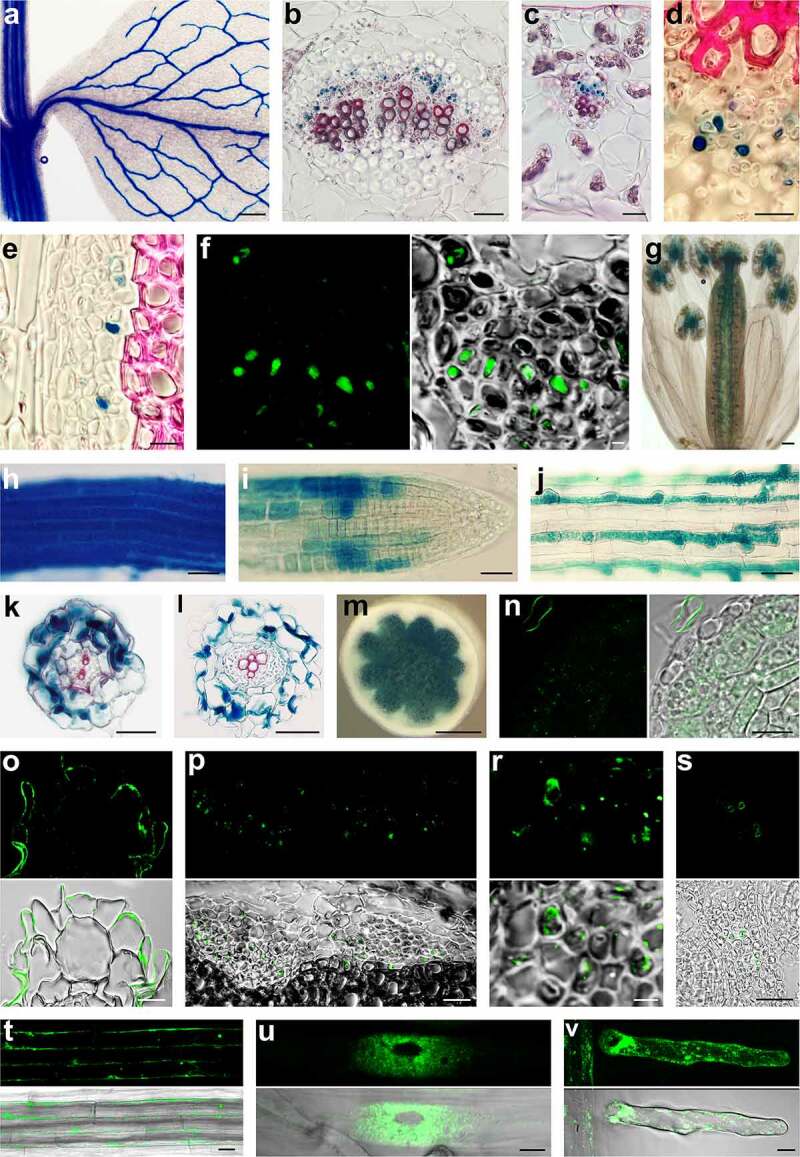
Gus histochemistry and IRT1 protein localization. (a): Young shoot segment with developing leaf of pIRT1::GUS line. The GUS histochemistry reaction resulted in blue color is visible in the vasculature of leaf and stem. (b) Cross-section through leaf major vein and (c) minor vein of the pITR1::GUS plant shows that IRT1 promoter is active in the phloem part of the vasculature. (d) Detail view of phloem in pITR1::GUS plant leaf vein and stem vascular bundle (e) showing GUS activity in the companion cells and feeble reaction in sieve elements. (f) Immunohistochemical localization of GUS protein in phloem companion cells of pITR1::GUS plant using the antibody against GUS. (g) IRT1 promotor is active in pistil and junctions between filaments and anthers. Maturation (h) and meristem (i) zone of the root of pIRT1::GUS seedlings growing without iron. (j) Signal at the beginning of mature root zone of pIRT1::GUS seedlings when growing on iron supplemented medium is mainly detected in trichoblasts. Cryo-sections through maturation (k) and elongation (l) zone of pIRT1::GUS roots demonstrate the activity of IRT1 promotor in epidermis and cortex but not in the central cylinder. (m) Hand section of the stem of pITR2::GUS line showing GUS activity in the pith. (n) Immunolocalisation of IRT1 protein on cryosections in meristematic zone of roots of wild seedlings growing on iron-free medium. Labeling is seen in intracellular patches but not at the cell periphery. Note labeling at the periphery of root hair of another root accidentally appearing in the vicinity during embedding. (o) IRT1 labeling with IRT1 antibody on cryosection through the maturation root zone detect signal mainly at the periphery of root epidermal cells. (p) Histoimmunology assay on cryosections shows cells labeled with IRT1 antibody scattered in the phloem part of the vascular bundle, and detailed view (r) shows the signal in the phloem companion cells. (s) In pistil, IRT1 was located at the periphery of several cells in the septum transmitting tract. Parts of ovules are on the left and right sides. (t) Long version of IRT protein with attached Dendra2 at its C-terminus is localized at the periphery of epidermal cells in matured root zone and in intracytoplasmic bodies. (u) Short IRT version of IRT1 with Dendra2 attached to its C-terminus is localized to numerous patches surrounding the nucleus (u, image prepared with Image J program represent Z projection of 3 slices) and patches at the periphery as seen in root hair (v, image represent Z projection of 5 slices)
Scale bars: 500 µm (a and m); 100 µm (g); 50 µm (b,c,h,i,j,k,l and p); 25 µm (d,e,n,o,s,t and u); 10 µm (f and r).
Figure 2.
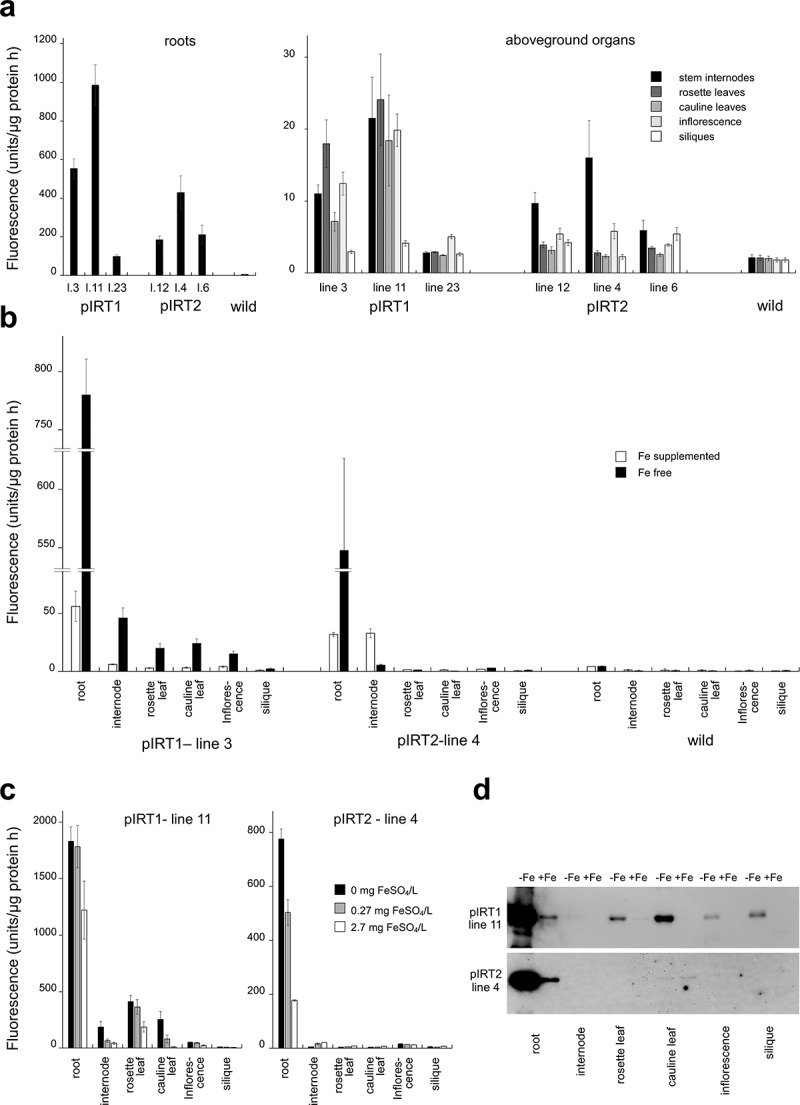
IRT1 and IRT2 promotor activity evaluated by GUS fluorometry. (a) Activity of IRT promoters in different organs of three lines growing in soil substrate. Wild represent extracts from nontransgenic plants. (b) Iron deficiency in the medium increases the activity of IRT1 promotors in all organs and activity of IRT2 promotor in roots but not in shoots. (c) Activity of IRT1 promotor in all organs and activity of IRT2 in roots are iron concentration-dependent. Their activities decrease when iron becomes available. (d) GUS protein analysis by western blotting confirming regulation of promotors by iron
We wanted to know if this also applies to the aboveground parts. We used two plant cultivation methods, in in vitro conditions and pots with perlite with controlled watering. The results were similar. When plants were cultured in vitro, the activity of the IRT1 promoter was strongly affected in the roots by the availability of iron (Figure 2b). If iron was available, the promoter shows approximately ten times lower activity than in plants growing on an iron-free medium. When we analyzed the aboveground parts, we also noticed iron’s effect, but this was smaller than in the roots.
Experiments with plants growing in pots with perlite revealed that the activity of IRT1 promotors was dependent on iron concentration (Figure 2c). In the case of roots, we also noticed a qualitative difference. In roots growing on the medium with iron, the promoter was active mainly in the trichoblasts of the mature root zone (Figure 1j). However, when plants grew without iron, the promoter was active in all cells, not only in the maturation root zone (Figure 1h) but also in the root’s tip (Figure 1i), but not in old root parts. The IRT2 promotor’s activity was also regulated by iron in the same manner as the IRT1 promotor. Interestingly, in the shoot extract, the activity of the IRT2 promoter was higher when iron was available than it was absent (Figure 2c). The same was also seen in the case of cultivation in vitro (Figure 2b). To rule out that the signal is not specific for GUS reporter, we performed a western blot analysis with plants growing in vitro using an antibody against E. coli GUS protein. The protein was detected in all organs of the pIRT1::GUS transgenic plant grown on the medium without iron and a considerable amount in the roots (Figure 2d). Only week signals if even were registered in extracts when plants were grown on iron supplemented medium. In the pIRT2::GUS line, the GUS protein was also abundant in roots, and a low level was present in shoot extract (Figure 2d).
Then we wish to analyze the promotor’s activities at tissue and sub-tissue levels. For detailed histochemical and histoimmunological analysis, we used cryosections. After overnight treatment with GUS substrate, blue staining was observed in root epidermal cells and the cortex, however not in the central cylinder (Figure 1k,l). When we perform a reaction on cryosection of aboveground organs, blue coloring was seen in cells scattered in the phloem of all organs (Figure 1b,c). Detail observation revealed robust staining in phloem companion cells and weak staining in sieve elements (Figure 1d,e). This finding was confirmed alternatively by immunohistochemistry using an antibody against GUS (Figure 1f).
To confirm the results at the transcription level, we employed semi-quantitative RT-PCR. According to the Tair database, IRT1 encodes two splicing forms; therefore, we analyzed the abundance of both forms in the roots and rosette leaves of the seedlings and in aboveground organs of the perlite grown plants. In the seedlings after 35 cycles, the transcript corresponding to the long IRT1 RNA splicing form was detected only in the roots of the seedlings cultured on the Fe depleted medium (Figure 3a). This result was obtained with three independent combinations of primers. In no case, we detected a signal corresponding to genomic DNA. Since the correct combination of primers can distinguish both splicing forms, we tried to analyze the short form. We confirmed the short transcript in the roots and the rosette leaves with two pairs of primers (Figure 3c). Abundance was similar regardless of whether the seedlings grow in the presence or absence of iron. References transcripts are shown in Figure 3e. When we analyzed aboveground organs of plants growing on the perlite substrate, we detected a signal with the primers for the short IRT1 splicing form after 35 multiplication cycles regardless of the presence or absence of iron (Figure 3d). References transcripts are shown in Figure 3f. After 50 cycles, the bands corresponding to the long IRT1 RNA splicing form were identified in aboveground organs; however, no bands confirming the presence of genomic DNA contamination were detected (Figure 3b).
Figure 3.
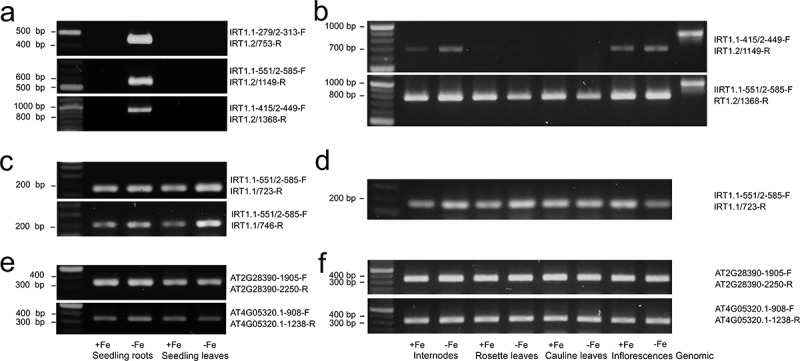
Semi-quantitative RT-PCR analysis of IRT1. (a) Long IRT1 splicing form was detected with three independent primers combinations in iron-deficient seedling roots but not leaves after 35 cycles. (b) After 50 cycles long splicing form was also detected in aboveground organs. We failed to distinguish genomic contamination in cDNA samples. Bands obtained from genomic DNA under the same PCR conditions are presented for fragment size comparison. (c) Short IRT1 splicing form was detected in seedling leaves and roots regardless of iron status and in all aboveground organs of mature plants (d). (e and f) represent reference transcripts after 35 cycles
In the next step, we analyzed the presence of IRT1 protein in different parts of plants by western blot assay. We utilized a commercially available anti-IRT antibody that has been employed in the previous study.16 We detected a single band in different aboveground parts of the plants in the presence and absence of iron (Figure 4a). We then focused on analyzing the amount of protein in the seedlings growing in vitro. The protein was present in rosette leaves regardless seedlings were grown in the presence or absence of iron; however, in the roots, IRT1 was abundant in seedlings grown on the iron-depleted medium (Figure 4b). We also noticed that the product’s molecular size isolated from leaves is lower than that detected in root lysate, which is around 35 kD and corresponds to the predicted size of the long IRT1 form. Then, we analyzed which root and rosette leaf fractions contain IRT1. We found IRT1 protein to be very abundant in root plasma membrane and organelle membrane fractions when seedlings were grown on an iron-free medium (Figure 4c). In leaves, IRT1 protein was present mainly in organelle fraction regardless of the iron status (Figure 4c).
Figure 4.
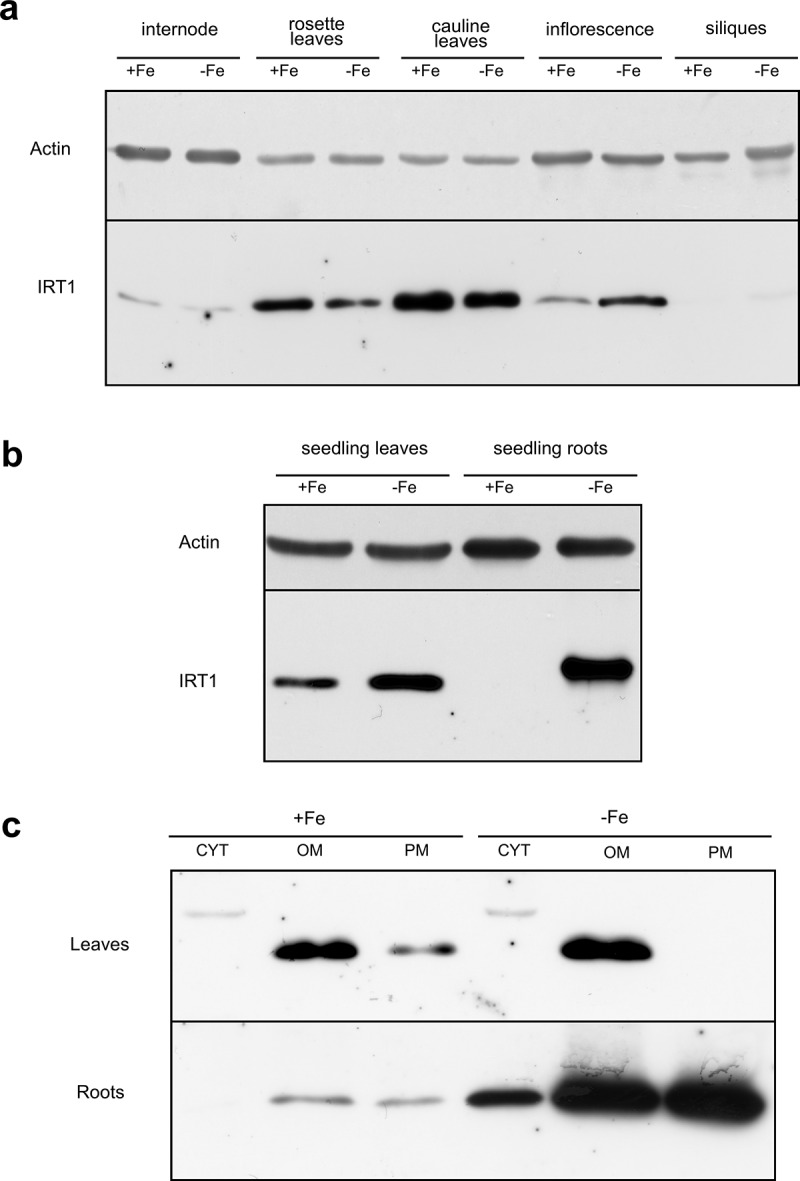
Western blotting IRT1 detection. (a) IRT1 protein abundance in aboveground parts of plants grown on perlite and watered with iron-containing (13.5 mg/L) or iron-depleted 1/10 MS solution. (b) Abundance of IRT1 protein in leaf rosette leaves (l) and roots (r) of seedlings grown in vitro on 1/2 MS or the same medium but with omitting iron sulfate. (c) IRT1 in different fractions of leaves and roots of seedlings grown in vitro on 1/2 MS or the same medium but with omitting iron sulfate. CYT = cytosol fraction, OM = organelle membrane fraction, PM = plasma membrane fraction
Further, we wished to confirm the presence of IRT1 protein in situ. Using histoimmunology using a specific antibody against IRT1, we documented the protein’s presence in the peripheral layer of the epidermal cells of the root mature zone (Figure 1o). However, when we analyzed the cells in the meristematic zone, we did not observe a signal at the periphery, but the weak labeling was present in the intracytoplasmic patches (Figure 1n). When we analyzed other organs, we registered the fluorescence in phloem tissue (Figure 1p), especially in cells with cell wall ingrowths (Figure 1r). The signal was present in the whole intercellular space. However, we also noticed a signal in neighboring sieve elements in the form of few bodies (Figure 1r). In pistils, IRT1 was located at the periphery of several cells in the septum transmitting tract (Figure 1s).
Based on the assumption that the IRT1 gene encodes two RNA splicing forms, and proteins may have divergent functions and localizations, we prepared transcription-translation fusion pIRT1::IRT1::Dendra2 constructs. When we ligated the DNA sequence encoding Dendra2 to the genomic DNA encoding the short IRT1 form, the fusion protein in epidermal root cells was present as cytoplasmic patches enriched in the cytoplasm surrounding the nucleus (Figure 1u). Spots were also scattered at the cell periphery (Figure 1v). When DNA encoding the long IRT1 form was used, the fluorescence signal was seen as a layer along the cells’ periphery, and few patches were observed inside the cells (Figure 1t).
Discussion
IRT1 is a central transporter of iron and some other divalent metals in plant roots, as it has been demonstrated by genetic, molecular, cell biological, biochemical, and bioinformatics approaches in numerous studies. Evidence is such as the gene is expressed in the roots, its expression is regulated by iron deficiency, the absence of gene function causes an iron-deficient phenotype, the protein is localized in the root epidermal cell plasma membrane, and the gene can complement mutations in analog genes in yeast.4–8,17–20 The IRT1 gene expression must be strictly regulated to ensure the required iron balance in the plant. Transcription seems to be the most critical point where this regulation occurs.4,5,19 We confirmed this in the present study when the promoter was much more active in the roots in the absence of iron than when iron was available (Figure 2). Consequently in roots, the long transcript of IRT1 (Figure 3a) and protein (Figure 4b,c) were detected almost exclusively in seedlings grown on an iron-depleted medium.
However, there are indications that iron may regulate the abundance of IRT1 protein post-translationally, and protein is accumulated only in the roots when the iron is limiting regardless of the level of transcription.19 Such regulation at the post-translational level by iron was not confirmed later on,17 but non-iron metals surprisingly have such ability.20 Anyway, IRT1, like other proteins, is subject to continual turnover and undergoes relocalization in cells that may be induced by various factors, e.g., non-iron metal status.20 Much attention has been paid to protein internalization in this context, and post-translational modifications of the IRT1 protein and other factors that impact this process have been characterized.16–18,20,21
Previous studies by Barberon et al.17,18 also showed that, surprisingly, the protein was not detected on the membrane as expected owing to IRT1 transports iron and other metals across the plasma membrane into the cells. These studies were performed with ectopically overexpressed IRT1 under the 35S promoter and using the whole-mount immunological approach. The antibody has raised against two peptides located at the protein N-terminus and the centrally positioned intracytoplasmic loop. The protein was present only in the patches even if plants were grown in the absence of iron, and these were identified as trans-Golgi network/early endosomes [TGN/EE17]. Later studies with this tool also demonstrated localization of IRT1 in the cells’ outer lateral periphery when the plants were exposed to a deficiency of divalent metals other than iron but, surprisingly, not in iron-deficient conditions.18 This finding was confirmed with fluorescent protein technology.20 Membrane localization has also been noted when internalization of IRT1 was prevented by different approaches.17,18,20 We demonstrated by histoimmunology using commercial IRT1 antibody and cryosectioning clear localization of IRT1 at periphery of epidermal cells in the maturation root zone of wild seedlings grown under iron-deficient conditions (Figure 1o). We also saw the signal in the intracellular patches in meristem epidermal and cortex cells but not at their periphery (Figure 1n). However, our study also provides a new possible explanation for the appearance of IRT1 in intracellular patches based on the presence of two IRT1 isoforms.
We wish to note that the Tair database indicates that the Arabidopsis AT4G19690 gene encodes two IRT1 RNA splicing forms. Transmembrane domain (TMD) prediction programs can identify two regions with TMDs, one at the N-terminus and the other at the C-terminus of the long IRT1 protein variant. These regions are connected via an intracytoplasmic loop that contains residues with an affinity for metals.5,20 The short splicing form is predicted to encode a truncated protein lacking the C-terminal TMD region. According to the comprehensive RNA-seq analysis of Arabidopsis root tissues performed by Li with co-workers 22, both IRT1 transcripts are present in the root’s maturation zone, mainly in the trichoblasts, to a lesser extent, the atrichoblasts of the root epidermis. Further, this study has revealed that the long splicing form was a major transcript of which abundance prevails significantly over the short minor transcript. However, it is unknown whether and how the splicing process is regulated, for example, by iron status or by other factors and data for aboveground parts are missing. Based on the above evidence, we created transcription-translation constructs with genomic DNA sequences for long and short splice forms and the Dendra 2 fluorescent protein DNA sequence.23 Dendra2 tag has been used and characterized in our previous studies.14,24,25 As shown in Figure 2t–v, localization patterns of both populations are entirely different in iron-deficient conditions. The short form is present in patches mainly around the nucleus and also on the cell periphery but never in a form reminiscent of its plasma membrane localization as a transmembrane protein. In contrast, the long-form was located along the cell periphery as a continuous layer although clusters were also present in the intracellular space. We want to note that the antibody used in the previous study17,18 grew against peptides present in both IRT1 isoforms; therefore, it cannot distinguish complete and truncated IRT1 versions. Perhaps patches in ectopically expressed lines represent a short IRT1 isoform that cannot be targeted to the plasma membrane, as documented in the present study. However, further studies are needed to define the function of the truncated form of the protein.
An interesting finding of this study is that the IRT1 gene is also expressed in plants’ aboveground parts. We proved this by studying the IRT1 promoter and analyzing transcript and protein abundance. Although the overall activity of the IRT1 promoter was much lower in aboveground organs than in roots, it was evident in the case of strongly and moderately expressing pIRT1::GUS lines. Moreover, the activity of the IRT1 promoter was here similarly regulated by iron status as in the roots, as seen in Figure 2. Detailed analysis using GUS histochemistry and histoimmunology showed that this activity is seen only in phloem companion cells. This high cell specificity explains the relative weakness of the IRT1 promoter when compared aboveground organs with roots. In roots, we recorded the promoter activity in the epidermis and the cortex in the relative long root hair zone. Companion cells were positive in all pIRT1::GUS lines but not in the IRT2::GUS line, so the signal is not a consequence of a putative position effect and influence of some endogenous promoter present in the vicinity of the pIRT1::GUS insertion site.26 We can also rule out some endogenous GUS activity as we detected the GUS protein with a specific antibody against the GUS protein by western blotting and immunohistochemistry.
We tried to analyze the presence of both transcripts by RT-PCR. We identified the long splicing IRT RNA form in roots after c 35 cycles, and the amount of transcript was clearly regulated by iron deficiency (Figure 2a). After much more cycles, the long IRT1 splicing form was detected also in aboveground organs. The short transcript has been identified after 35 cycles in leaves and roots of seedlings and organs of matured plants regardless of the iron status. However, since it is not possible to design primers for RT -PCR distinguishing short form and genomic sequence, it cannot be ruled out that the signal also represents a genomic sequence. However, we would like to emphasize that the RNA samples were thoroughly treated with DNAse, and the presence of genomic contamination was not distinguished with primer combination even after 50 cycles. Finally, we detected the signal in aboveground organs by western blotting (Figure 4a) and identified labeling in the phloem, specifically in companion cells attached to sieve elements (Figure 1p,r) by immunohistochemistry. Interestingly, the band in western blot analysis corresponded to a lower protein mass than that detected in the root lysate (Figure 2b). It can be speculated that in companion cells of aboveground organs, IRT1 is present as a truncated form encoded by the short IRT1 splicing form.
Although the expression of IRT1 is unexpected in the aboveground parts of plants, since IRT1 is considered to act mainly as an iron transporter in the root, a similar expression has been observed in rice.27 Rice belongs to plants that use chelating iron uptake strategies, but its genome also contains OsIRT1 and OsIRT2, homologs of AtIRT1.Ishimaru with co-workers27, analyzed the expression of OsIRT1 by promotor evaluation and transcript abundance and found that the promoter was active in root phloem accompanying cells in iron sufficient conditions. However, under iron deficiency, it was highly active in the root epidermis, the cortex’s inner layer’s cells, and the stele in the entire phloem area. Interestingly we did not notice the activity of IRT1 promoter in root companion cells in Arabidopsis (Figure 1p,r). Besides, in rice GUS histochemical reaction and transcript were detectable in phloem region of leaf and shoots, which agrees with our finding. The authors concluded that Fe2+ transporters participate in Fe distribution and partitioning in rice plants. Our study brings evidence that this statement may be applied in general and not just to rice. In phloem companion cells, the IRT1 protein may be involved in the translocation of Fe between organs when it is deficient, and the protein may be part of a long-distance Fe signaling network. Interestingly few other transporters such as SULTR1,3 sulfate transporter or several sucrose transporters are specifically expressed in companion cells[28, for review].
Funding Statement
This work was supported by the Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic and Slovak Academy of Sciences (VEGA) under Grant number 2/0115/17
Disclosure statement
No potential conflicts of interest were disclosed.
References
- 1.Schmidt W, Thomine S, Buckhout TJ.. Iron nutrition and interactions in plants. Front Plant Sci. 2020;10:1. doi: 10.3389/fpls.2019.01670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brumbarova T, Bauer P, Ivanov R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2015;20(2):124–11. doi: 10.1016/j.tplants.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Morrissey J, Guerinot ML. Iron uptake and transport in plants: the good, the bad, and the ionome. Chem Rev. 2009;109(10):4553–4567. doi: 10.1021/cr900112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14(6):1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA. 1996;93(11):5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henriques R, Jásik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C. Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol. 2002;50(4):587–597. doi: 10.1023/A:1019942200164. [DOI] [PubMed] [Google Scholar]
- 7.Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol. 1999;40(1):37–44. doi: 10.1023/A:1026438615520. [DOI] [PubMed] [Google Scholar]
- 8.Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D. The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J. 2002;31(5):589–599. doi: 10.1046/j.1365-313X.2002.01381.x. [DOI] [PubMed] [Google Scholar]
- 9.Guerinot ML. The ZIP family of metal transporters. Biochimica Et Biophysica Acta (Bba)-biomembranes. 2000;1465(1–2):190–198. doi: 10.1016/S0005-2736(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 10.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 11.Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet MGG. 1986;204(3):383–396. doi: 10.1007/BF00331014. [DOI] [Google Scholar]
- 12.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 13.Vitha S, Baluška F, Jasik J, Volkmann D, Barlow PW. Steedman’s wax for F-actin visualization. In: Staiger CJ., Baluška F, Volkmann D, Barlow PW, editors. Actin: a dynamic framework for multiple plant cell functions.: Kluwer Academic Publishers; 2000. p. 619–636. [Google Scholar]
- 14.Jásik J, Boggetti B, Baluška F, Volkmann D, Gensch T, Rutten T, Altmann T, Schmelzer E. PIN2 turnover in Arabidopsis root epidermal cells explored by the photoconvertible protein Dendra2. PLoS One. 2013;8(4):e61403. doi: 10.1371/journal.pone.0061403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139(1):5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov R, Brumbarova T, Blum A, Jantke AM, Fink-Straube C, Bauer P. SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. Plant Cell. 2014;26(3):1294–1307. doi: 10.1105/tpc.113.116244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G. Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA. 2011;108(32):E450–E458. doi: 10.1073/pnas.1100659108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barberon M, Dubeaux G, Kolb C, Isono E, Zelazny E, Vert G. Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc Natl Acad Sci USA. 2014;111(22):8293–8298. doi: 10.1073/pnas.1402262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connolly EL, Fett JP, Guerinot ML. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell. 2002;14(6):1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubeaux G, Neveu J, Zelazny E, Vert G. Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol Cell. 2018;69(6):953–964. doi: 10.1016/j.molcel.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Shin LJ, Lo JC, Chen GH, Callis J, Fu H, Yeh KC. IRT1 degradation factor1, a ring E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell. 2013;25(8):3039–3051. doi: 10.1105/tpc.113.115212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, Yamada M, Han X, Ohler U, Benfey PN. High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev Cell. 2016;39(4):508–522. doi: 10.1016/j.devcel.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24(4):461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 24.Jásik J, Schmelzer E. Internalized and newly synthesized Arabidopsis PIN-FORMED2 pass through brefeldin A compartments: a new insight into intracellular dynamics of the protein by using the photoconvertible fluorescence protein Dendra2 as a tag. Mol Plant. 2014;7(10):1578–1581. doi: 10.1093/mp/ssu052. [DOI] [PubMed] [Google Scholar]
- 25.Jásik J, Bokor B, Stuchlík S, Mičieta K, Turňa J, Schmelzer E. Effects of auxins on PIN-FORMED2 (PIN2) dynamics are not mediated by inhibiting PIN2 endocytosis. Plant Physiol. 2016;172(2):1019–1031. doi: 10.1104/pp.16.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peach C, Velten J. Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol Biol. 1991;17(1):49–60. doi: 10.1007/BF00036805. [DOI] [PubMed] [Google Scholar]
- 27.Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, et al. Rice plants take up iron as an Fe3+‐phytosiderophore and as Fe2+. Plant J. 2006;45(3):335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- 28.Otero S, Helariutta Y. Companion cells: a diamond in the rough. J Exp Bot. 2017;68(1):71–78. doi: 10.1093/jxb/erw392. [DOI] [PubMed] [Google Scholar]


