ABSTRACT
Infectious diseases are a leading cause of morbidity and mortality worldwide, and human pathogens have long been recognized as one of the main sources of evolutionary pressure, resulting in a high variable genetic background in immune-related genes. The study of the genetic contribution to infectious diseases has undergone tremendous advances over the last decades. Here, focusing on genetic predisposition to fungal diseases, we provide an overview of the available approaches for studying human genetic susceptibility to infections, reviewing current methodological and practical limitations. We describe how the classical methods available, such as family-based studies and candidate gene studies, have contributed to the discovery of crucial susceptibility factors for fungal infections. We will also discuss the contribution of novel unbiased approaches to the field, highlighting their success but also their limitations for the fungal immunology field. Finally, we show how a systems genomics approach can overcome those limitations and can lead to efficient prioritization and identification of genes and pathways with a critical role in susceptibility to fungal diseases. This knowledge will help to stratify at-risk patient groups and, subsequently, develop early appropriate prophylactic and treatment strategies.
KEYWORDS: host immune response, fungal infections, polymorphisms, genetic predisposition, immunology, functional genomics
HUMAN GENETIC SUSCEPTIBILITY TO INFECTIOUS DISEASES
Although there has been tremendous progress in medical research and health care, infectious diseases remain a leading cause of morbidity and mortality worldwide (1). Ever-increasing global connectivity together with human demographics and environmental changes has contributed to the emergence of new infectious diseases, such as the recent pandemic with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (2), and the reemergence of existing ones, such as Candida auris infection (3). Human infectious diseases are characterized by an extensive variation in clinical phenotypes among individuals infected by the same agent, indicating that genetic and nongenetic factors determine this variation. Many genetic epidemiological studies in the last half century, ranging from observational studies to more sophisticated twin or segregation studies, pointed to the importance of host heritable factors in susceptibility to infectious diseases. One of the first discovered single-gene traits influencing susceptibility to infection was the sickle hemoglobin variant as a major resistance factor for malaria (4). Stronger evidence came from several early twin studies reporting higher concordance rates in monozygotic than in dizygotic twins for genetic susceptibility to various infectious diseases (5–9). Also, follow-up studies of adopted children in the late 1980s showed they had a markedly increased risk of death from an infectious disease if one of the biological parents had died prematurely from an infectious cause rather than other causes, such as cancer or cardiovascular diseases (10).
Infectious pathogens, which elicit the host immune response, have long been recognized as the main source of evolutionary pressure (11, 12). Immune-related genes are the most abundant and diverse genes in the human genome (13), suggesting an evolutionary advantage of a varied immunological response to a wide range of infectious pathogens. The study of the genetic contribution to infectious diseases has undergone revolutionary advances over the last decades in line with the development of novel technologies in the field. Traditional linkage studies identified a few important candidate genes (14). With the advent of the genomic era, genome-wide association studies (GWASs) have identified numerous genetic loci in autoimmune diseases (15), but only with limited success in the field of genetics of infectious disease (16). High-throughput technologies and the generation of multi-omics data sets have enabled a powerful multilevel study of the genetics of complex diseases, including infectious disease, to offer a better understanding of the interplay between host, invading pathogen, and environment.
Here, we provide an overview of the available approaches for studying human genetic susceptibility in fungal infections, reviewing current methodological and practical limitations. We will also discuss the use of a systems genomics approach to understanding genetics and molecular pathways underlying the human host defense against fungal infections.
BURDEN OF FUNGAL DISEASES ON GLOBAL HEALTH
Human are constantly exposed to fungi: some are colonizing the human host, the so-called commensal fungi, and some are ubiquitous in the environment, the so-called environmental fungi. A fully functional host immune system has effective mechanisms for preventing severe fungal infections, but when the immune system fails, human-pathogenic fungi can cause potentially “opportunistic” life-threatening diseases (17). The burden of fungal diseases on global health is expanding in parallel with an increase in individuals with acquired immune deficiencies or those receiving immune suppressive therapies or myeloablative treatments (18). Human fungal infections cause more than 1.5 million deaths every year (19) and affect more than one billion individuals worldwide (20). The steady increase in the incidence of nosocomial invasive fungal infections has significantly contributed to health-related costs (21). Despite the increasing numbers and the recent outbreak of emerging C. auris infections (3), the impact of fungal diseases on human health still remains underestimated (22, 23). The majority of human fungal infections are caused by Candida, Aspergillus, and Cryptococcus spp. (19). These fungi are ubiquitous, but Cryptococcus and Aspergillus spp. are also environmental (24), whereas Candida spp. are commensal colonizers of mucocutaneous surfaces and the gastrointestinal tract (25).
The diagnosis of fungal infections can be problematic due to clinical challenges in fungal isolation and identification (26, 27). Therapeutic challenges are raised by the fact that no vaccines are yet available, current antifungal therapeutic options remain limited, and, on top of that, multidrug-resistant fungal species are arising (28). As a result, the mortality rate of patients with invasive fungal infection remains unacceptably high, reaching 40% to 50% (29). Risk factors to develop fungal infections have been well described (30–33), and certain high-risk groups of patients can be further classified according to specific risk scores, which include a large panel of clinical and laboratory parameters linked to disease susceptibility or clinical phenotype evolution (34–37). However, not all patients at risk develop fungal disease, and a large variability in clinical evolution has been reported among patients with the same predisposing factors. This observation suggests that human genetic variation plays roles in the susceptibility to fungal infections and in the severity of the outcome. Indeed, several monogenetic disorders resulting in severe primary immunodeficiencies, as well as mutations and common polymorphisms in immune genes, have been associated with an increased susceptibility to mucosal and/or invasive fungal infections, which have been reviewed elsewhere (38). Despite significant advances over the last few years in identifying genetic variations leading to immune imbalances, which lead to increased susceptibility to fungal infections, there are still many challenges to fully understand the genetic architecture of fungal infections. To overcome these challenges, a systems genomics approach has been followed to identify risk loci and molecular pathways underlying host immune defense and disease pathogenesis. By integrating multiple molecular data sets that reveal interindividual variability, it is possible to prioritize and identify genes and pathways with a critical role in susceptibility to fungal diseases. Ultimately, this knowledge will help to stratify at-risk patient groups and, subsequently, develop early appropriate prophylactic and treatment strategies against opportunistic fungal infections.
OVERVIEW OF HOST IMMUNE RESPONSE TO FUNGAL PATHOGENS
Opportunistic fungal infections are characterized by the interaction between the host, the invading fungus, and the environment, which is sustained by a complex and dynamic equilibrium of several interconnected factors. The microbiological and environmental factors taking part in this delicate interaction, such as the role of the commensal microbiome, the dynamic fungal morphological adaptations, and genomic mutations, are well reviewed elsewhere (39). Despite the differences in the pathogeneses of infection between environmental and commensal fungal species, there are several common host immune defense mechanisms. To infect the human host, the fungal pathogen must be able to overcome three levels of host defense; the first is a physical barrier that consists of the skin and mucosa. The second barrier, presented by the innate immune system, is largely dependent on the recognition of evolutionarily conserved fungal cell wall components (pathogen-associated molecular patterns [PAMPs]). These PAMPs are recognized by various pattern recognition receptors (PRRs) circulating, such as pentraxin-3 (PTX3) or mannose binding lectin (MBL), or present on the surface of innate immune cells, such as macrophages, monocytes, NK cells, and neutrophils. In particular, the mannan cell wall component is mainly recognized by the macrophage mannose receptor (MMR), the C-type lectin-like receptor dectin-2, and Toll-like receptor 4 (TLR4). TLR2 binds to phospholipomannan, and the dectin-1 receptor recognizes β-glucan. Coordinated engagement of PRRs and following intracellular signaling pathways mediated by several kinases and adaptor molecules, such as spleen tyrosine kinase (Syk) and caspase recruitment domain-containing protein 9 (CARD9), results in the activation of innate immune effector mechanisms. Those mechanisms include phagocytosis, generation of reactive oxygen species (ROS) by NADPH oxidase and reactive nitrogen species (RNS) by myeloperoxidase (MPO) that promote the killing of the fungus and, finally, to production of pro- and anti-inflammatory cytokines. Proinflammatory cytokines, such as interleukin-1 beta (IL-1β) and tumor necrosis factor alpha (TNF-α), have important roles in the host defense against fungal infections. IL-1β is transcribed as an inactive form (pro-IL-1β) and further processed into its active mature form via the NLRP3 inflammasome, a multiprotein complex, which is also crucial for antifungal host defense (40). TNF-α enhances antifungal activities by promoting phagocytosis and neutrophil recruitment (41, 42). In turn, the release of cytokines, combined with antigen presentation activity of myeloid cells, is crucial for activation of the adaptive T-cell immunity, in particular, Th1 and Th17 subsets (43), representing a third longer-term barrier against fungal infection (44). Gamma interferon (IFN-γ) produced by Th1 lymphocytes had been shown to have a central role in the resistance against systemic fungal infections (43); Th17 responses have been proven to be crucial for human anti-Candida mucosal host defense and granulocyte recruitment, but they can contribute to detrimental immunopathology during fungal infections (45, 46). In an immunocompetent host, the majority of the invading microorganisms are detected and destroyed within minutes or hours by the innate immune defense mechanisms. An overview of host immune responses against fungal infection is presented in Fig. 1. Invasive fungal infections are mainly found in patients with a weakened immune system, either due to reduced cellular immune effector mechanisms or defects in epithelial barriers.
FIG 1.
Overview of the mechanism of immune response toward a fungal infection. PMN, polymorphonuclear leukocyte; APC, antigen-presenting cell.
APPROACHES TO STUDY THE GENETICS OF FUNGAL INFECTIONS
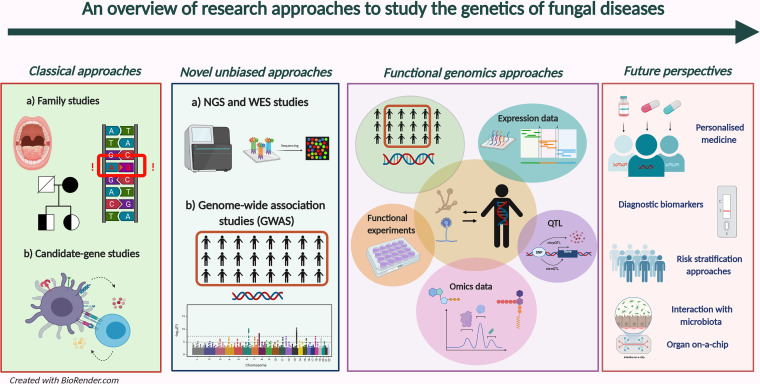
Although the above-mentioned factors are important, they do not explain all infections, and only a minority of patients at risk will actually develop disease, suggesting the critical role of genetics in determining disease susceptibility. Indeed, several approaches, from classical family-based and candidate-gene approaches, to novel ones, such as genome-wide association studies (GWASs) and integrative approaches, have attempted to decipher the genetic factors to mucosal and/or invasive fungal infections. An overview of these approaches is presented in Fig. 2.
FIG 2.
Classical and novel research approaches to study the genetics of fungal infections.
“Classical” approaches.
(i) Mendelian susceptibility to fungal infections: a family-based approach. Classical approaches, such as family-based approaches to study genetic factors, have captured rare mutations that confer a mendelian (monogenic) form of predisposition to fungal infections. Much of our understanding about genetic susceptibility to specific fungal pathogens has been achieved through family-based studies of certain rare primary immunodeficiencies presenting as clinical manifestation signs of a mucosal or invasive fungal infection (47, 48). A prototypical example is chronic granulomatous disease (CGD), a rare inherited disorder (frequency, ∼1/200,000) caused by mutations in genes encoding four of five protein subunits of the phagocyte NADPH oxidase, namely, the X-linked CYBB gene (gp91phox) and the autosomal recessive CYBA (p22phox), NCF1 (p47phox), and NCF2 (p67phox) genes (49). Patients with CGD fail to produce ROS and suffer from recurrent life-threatening bacterial and fungal infections, especially invasive aspergillosis (IA) (50), accounting for one-third of all deaths in CGD patients (51). Notably, patients with mutations in the NCF4 (p40phox) gene do not develop IA, as they are still able to produce ROS (52).
Another example is myeloperoxidase (MPO) deficiency, which is the most common inherited phagocytic disorder (frequency, ∼1/2,000) (53). The vast majority of MPO-deficient patients are asymptomatic; however, a complete enzymatic deficiency predisposes them to invasive candidiasis (54). More recently, CARD9 deficiency has emerged as an important and fungus-specific susceptibility factor for both mucosal and invasive fungal infections (55), without predisposing individuals to other infectious or noninfectious sequelae. More than 15 missense and nonsense mutations in the CARD9 gene (56) result in Th17 deficiency and altered dectin-1 signaling as well as defective neutrophil recruitment to certain anatomical sites, including the central nervous system (CNS) (57). A few inborn monogenic disorders that predispose individuals to invasive fungal infections (IFIs) (but not fungal specific) were previously described: specific mutations in the transcription factor GATA2 cause the so-called “MonoMAC syndrome” characterized by monocytopenia, B-cell and natural killer (NK)-cell lymphopenias, myelodysplasia, and increased susceptibility not only to mycoses but also to papillomaviruses and nontuberculous mycobacteria (NTM) of low virulence potential (58). Genetic mutations in genes involved in the IL-12/IFN-γ signaling pathway, extensively reviewed in reference 29, have been shown to predispose individuals not only to NTM but also to fungal infections by intracellular fungi (59). Such fungal infections include especially those whose eradication relies on an effective interaction between phagocytes and Th1 lymphocytes (e.g., Histoplasma capsulatum, Paracoccidioides brasiliensis, and Cryptococcus neoformans) (59, 60).
An intact host mucocutaneous barrier depends on functional IL-17 signaling. Chronic mucocutaneous candidiasis (CMC) is another primary immunodeficiency characterized by recurrent or persistent skin, mucosal, or nail infections by Candida spp., mainly C. albicans. CMC refers to a heterogeneous group of disorders, all caused by impaired Th17 responses and subsequent defective mucosal and skin antifungal host defense mechanisms. CMC can be caused by direct mutation in IL-17R signaling resulting in mucosal but not systemic candidiasis, such as IL-17F and IL-17RC, which are specific for CMC, as well as IL-17RA and the adaptor ACT1 (TRAF3IP2), also predisposing individuals to bacterial infections (61–64). Other genetic mutations in several genes variously involved in Th17 differentiation can be causal for CMC and are generally associated with other syndromic manifestations. Such examples include the loss-of-function STAT3 mutation which causes hyper-IgE syndrome (65), biallelic mutations of the Th17 differentiation master regulator RORC (66), autosomal dominant STAT1 mutations, which lead to defective Th17 responses by indirectly impairing STAT3 activity (67, 68), and CARD9 mutations (56). Other CMC-associated monogenic diseases include (but are not limited to) autosomal recessive DOCK8 deficiency (69), X-linked severe combined immunodeficiency disorder (SCID), 22q11.2 deletion (athymic DiGeorge syndrome), and diseases from mutations of many other genes, nicely reviewed elsewhere (70). Interestingly, autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED) syndrome, caused by AIRE mutations and characterized by the presence of neutralizing autoantibodies against IL-17F and IL-22, presents CMC as the sole infectious consequence (71). To sum up, primary immunodeficiencies offer unique opportunities for a better understanding of the genetic and immunological components of fungal infections, which will help to develop novel immune-based therapeutic approaches against these infections.
(ii) Nonmonogenic susceptibility to fungal infections: a candidate gene approach. Another classical approach widely adopted in several genetic studies of complex diseases, including fungal diseases, is the candidate gene approach. The selection of the candidate genes usually relies on in vitro murine or patient’s experimental data by hypothesis-driven biological plausibility. The majority of candidate gene studies include a case-control design. To avoid any form of confounding and population heterogeneity, case and controls need to be accurately matched, and the sample size should be adequate to ensure reproducibility and statistical power (72). The vast majority of candidate gene studies for susceptibility to fungal infections have focused on immune-related genes involved in innate recognition of microbes, acquired immunity, intracellular signaling pathways, or different cytokines. Immune-related genes are a special case in the genome because, depending on the geographic region, the selective pressure on them has been different; that is the reason why most of those genes are highly polymorphic and, subsequently, highly prone to population stratification biases (73). Several single nucleotide polymorphisms (SNPs) in immune-related genes have been described that increase or decrease the risk of fungal diseases in patients with an acquired immunocompromised status (74–76). Two of the most studied pathological conditions characterized by an immunocompromised status are systemic candidiasis in the intensive care unit (ICU) and invasive aspergillosis (IA) in allogeneic hematopoietic stem cell transplant (HSCT) recipients, and most studies that identified SNPs associated with fungal infections have been conducted with these kinds of patients.
Since other excellent recent reviews already described in more detail SNPs influencing susceptibility to fungal infections (74–76), here and in Table 1 we report only some representative associations which have been described in the last 14 years supported by strong functional evidences. One of the most studied immune gene families that encode receptors on innate immune cells that recognize fungal antigens is that of the Toll-like receptors (TLRs). Three SNPs in the TLR1 gene were significantly associated with candidemia susceptibility (77), while SNPs in the TLR4 gene were associated with both IA (78, 79) and systemic candidiasis (80). A stop codon in DECTIN1 (Tyr238X) has been associated with increased risk for IA after HSCT (81) but not for invasive candidiasis after HSCT (82). The same stop codon polymorphism was further associated with CMC (83) and oral and gastrointestinal colonization by Candida species in HSCT patients (82). Two frequent polymorphisms (281A/G and 734A/C) in the PTX3 gene have been associated with increased risk of developing IA in both HSCT donors (84) and solid organ transplant recipients (85). These SNPs have also been functionally validated using in vitro studies with patients’ primary neutrophils, showing impaired Aspergillus phagocytosis and killing (84). SNPs in the NOD2 gene regulate susceptibility to IA after HSCT, and NOD2 deficiency affords resistance to IA (86). In addition, genetic variation for the monocyte/macrophage-targeted chemokine receptor CX3CR1 and the neutrophil-targeted chemokine receptor CXCR1 has been shown to be crucial for fungal infections, particularly those caused by Candida spp.: carrying the allele M280 in the CX3CR1 gene in homozygosity was associated with an increased risk for disseminated candidiasis but not mucosal or recurrent vulvovaginal candidiasis (RVVC) (87) in two different patient cohorts (88) and leads to impaired human monocyte trafficking and survival (89). The mutant CXCR1-T276 allele was associated with increased susceptibility to disseminated candidiasis and impaired neutrophil degranulation and fungal killing capacity (90). Last, but not least, genetic variation in pro- and anti-inflammatory cytokines has also been shown to be associated with susceptibility to fungal diseases. An important example is represented by IL-1 family genes: polymorphisms or certain haplotypes in IL-1β, IL-1α, and IL-1Ra were associated with an increased risk of developing IA in solid organ recipients (91) and in leukemic patients (92) as well as decreased Aspergillus fumigatus-induced cytokine production (91). Candidate gene studies have historically paved the way for personalized medicine and prophylactic antifungal treatment in high-risk patients. However, these studies present limitations, such as, to name a few, population stratification issues, lack of replication among different studies and across populations, poor functional evidence, and noncorrection for multiple testing as well as small sample size, which result in limited statistical power (73, 75).
TABLE 1.
Selected allelic polymorphisms influencing susceptibility to fungal infections discovered in the last 14 years using a classical approach and supported by functional validations
| Gene(s) | Polymorphism(s) | Chromosome location | Reported associations | Functional evidence | Reference |
|---|---|---|---|---|---|
| TLR1 | rs4833095, rs5743618, rs5743611 | 4p14 | Increased candidemia susceptibility | Impaired cytokine release by primary monocytes | 77 |
| TLR4 | rs4986790, rs4986791 | 9q33.1 | Increased susceptibility to IA | Delayed immune cell reconstitution after HSCT (78) | 78 |
| Validation study in a separate cohort | 79 | ||||
| Increased candidemia susceptibility | Increased C. albicans-induced IL-10 in PBMCs | 80 | |||
| CLEC7A (Dectin-1) | rs16910526 | 12p13.2 | Increased susceptibility to IA | Diminished A. fumigatus-induced IFN-γ and IL-10 in PBMCs | 81 |
| Higher oral and gastrointestinal Candida colonization, no increased risk of candidemia | Diminished C. albicans-induced IL-1β in PBMCs and reduced amplification of TLR2 signaling | 82 | |||
| Mucocutaneous fungal infections | Lower β-glucan-induced IL-6 in monocytes and lower Candida binding | 83 | |||
| PTX3 | rs2305619, rs3816527 | 3q25.32 | Increased susceptibility to IA after HSCT | Lower phagocytosis efficiency and A. fumigatus killing in neutrophils | 84 |
| NOD2 | rs2066842 | 16q12.1 | Reduced susceptibility to IA after HSCT | Lower A. fumigatus-induced cytokine production in PBMCs | 86 |
| CX3CR1 | rs3732378 | 3p22.2 | Increased candidemia susceptibility | C. albicans-induced renal failure in Cx3cr1–/– mice | 88 |
| Impaired AKT and ERK signaling and decreased blood monocyte counts | 89 | ||||
| CXCR1 | rs2234671 | 2q35 | Increased candidemia susceptibility | Impaired C. albicans killing and neutrophil degranulation | 90 |
| CLEC1A (MelLec) | rs2306894 | 12p13.2 | Increased susceptibility to IA after HSCT | Lower A. fumigatus-induced IL-1 β and IL-8 production in macrophages | 14 |
| IL1B | rs16944 | 2q14.1 | Increased invasive mold infection | Reduced Aspergillus-induced IL-1β, TNF-α, and IL-22 production in PBMCs | 91 |
| IL1RN | rs419598 | 2q14.1 | Increased invasive mold infection | Reduced Aspergillus-induced IL-1β and TNF-α production in PBMCs | 91 |
| IFNG | rs2069705 | 12q15 | Decreased susceptibility to IA after HSCT | Improved Aspergillus killing and higher IFN phytohemagglutinin-induced IFN-γ production in PBMCs | 144 |
“Novel” approaches.
For decades, studies of genetic susceptibility to infectious diseases have been looking at inherited monogenic defects causing spontaneous infections and have been screening for single polymorphisms in candidate genes. However, such studies were performed in relatively small patient cohorts and were usually based on hypothesis-driven in vitro or previous knowledge in the field.
(i) Moving to unbiased genome-wide approaches to study genetics of fungal infections. The advent of the genomic era with advances, such as the mapping of human genetic variation compiled by the international HapMap project (93) and the 1000 Genomes project (94), and the development of several high-throughput sequencing (HTS) platforms for (deep) sequencing and imputation tools have all contributed to a better understanding of genetics in various human complex diseases in diverse populations. Such advances have also been applied to fungal infections. For example, next-generation sequencing (NGS) and whole-exome sequencing (WES), which sequences all of the protein-coding regions of genes in a genome, have become among the most widely used, unbiased, “hypothesis-generating” novel methods for studying the rare monogenic defects underlying susceptibility to fungal infections. For example, van der Veerdonk et al. identified the STAT1 gene as a cause of chronic mucocutaneous candidiasis by using an NGS approach (68), and this was validated by Liu et al. who identified heterozygous germ line mutations in the STAT1 gene in 47 patients with autosomal dominant chronic mucocutaneous candidiasis by using WES (95). WES in a case of a leukemic patient presenting an unusual invasive mucormycosis has revealed several putative polymorphisms in immune-related genes (e.g., PTX3, TLR6, NOD2, DDX58 (RIG-I), and CCR5) potentially influencing mucormycosis infection (96). Moreover, exome sequencing has been implemented as a discovery tool for genetic diagnosis of primary immunodeficiencies (PIDs) manifested as fungal infections described in a Dutch hospital (97). Collectively, these studies show that WES is a promising and affordable approach for discovering novel disease-causing genes and allelic polymorphisms influencing disease susceptibility targeting a small number of individuals or even single patients. In addition, sequencing of just the exome of patients would allow identification of rare variants. Early studies using exon sequencing to identify rare variants in other infectious diseases, which were focused on the TLR4 gene in meningococcal disease and on five TLR genes in tuberculosis, showed an excess of rare (and some more frequent) coding changes in patients compared to controls (98, 99). Therefore, WES can potentially open up new avenues for discovering rare variants that predispose individuals to fungal infections.
However, the majority of low-frequency and/or rare variants that have been associated with infectious diseases, including systemic Candida infections, are noncoding variants (intronic or intergenic) (100, 101). To explore the role of common noncoding variants, follow-up studies on the genetics of fungal diseases made use of genomic tools, such as genotype imputation, custom genotyping arrays, and whole-genome sequencing, to reveal novel associations between phenotypes and variants. For example, a pilot association study performed a screen of ∼120,000 SNPs across 186 genetic loci related to immune function among hospitalized patients with candidemia compared to those in healthy and patient-matched controls and revealed significant associations between novel SNPs in the CD58, TAGAP, and LCE4A-C1orf68 genes and candidemia susceptibility (101). Of note, the presence of two or more high-risk SNPs within these loci resulted in an ∼20-fold increased risk of developing candidemia, indicating a possible synergistic effect on increasing the infection risk (101). A large GWAS of volunteers contributing DNA from the 23andMe database identified three significant associations between yeast infection and variants downstream of the PRKCH gene, within DSG1, and C14orf177 (102). Another pilot GWAS, which was performed in children with dermatophytosis caused by the fungal species Trichophyton tonsurans, identified SNPs in eight genes involved in leukocyte activation, melanocyte function, and extracellular matrix remodeling that have been significantly associated with increased infection rate (103). All these studies indicated the role of common variants in contributing to variability in the susceptibility to fungal diseases. Despite significant progress over the last few years in identifying susceptibility genes for fungal infections, there is still much genetic information unexplored, and the molecular mechanisms underlying susceptibility are not fully understood due to challenges that are discussed below.
(ii) Limitations of studying the genetics of fungal diseases. GWASs for identifying genetic risk factors in fungal infections have not been as successful as those in other complex diseases, such as autoimmune diseases (104), because of several limitations. One of the major limitations in studying the genetics of fungal infections is the lack of power due to relatively small patient cohorts. Large sample sizes are required in order to obtain sufficient statistical power to detect true disease associations (105). The collection of a patient cohort is also complicated by the possible presence of asymptomatic infections or different ethnicities. Patient cohorts must be ethnically homogeneous and well phenotyped in order to identify phenotype- and population-specific associations. Considering the genetic substructure of human populations, it is crucial to consider that the allele frequency differs substantially among ethnic groups, and in certain cases, for example, in the African ancestry, it is possible to find larger variation and a lower linkage disequilibrium (106). The admixture of ethnic groups (107) as well as subtle differences in the ethnic compositions of cases and controls (108) can lead to false-positive results. While careful matching of demographic factors can reduce the number of false-positive results, statistical methods (nicely reviewed in reference 107) can now be applied to address this issue and mitigate these caveats.
Another limitation is that most GWASs have been focused on identifying only common variants whose minor allele frequency (MAF) is >5% (105, 109, 110), missing low-frequency or rare variants. To identify rare variants, next-generation sequencing (NGS) studies with relatively small cohorts followed by testing of associated variants in larger cohorts might be a promising complementary strategy (110). After validating the SNP in a validation cohort, a “wet-lab” functional validation of the disease-causing effect of this genetic variant is critical and required to confirm the causal relationship between genetics and phenotype.
Another limitation is that a GWAS alone, while providing significant associations between a genetic variant and a disease, cannot explain the biological consequence or pinpoint the causal gene, especially when noncoding genetic variants are discovered (111). A possible approach for exploring the link between a GWAS genetic variant and its effect is to statistically correlate variants with measured biological quantitative data by performing quantitative trait loci (QTL) analysis. For example, a statistical correlation between a genetic variant to gene expression is called expression-QTL (eQTL) analysis (112), to cytokine production is cytokine-QTL (cQTL) analysis (113), to DNA methylation is methylation-QTL (meQTL) analysis (114), among others (115). Of note, eQTL and cQTL analyses have already been implemented for studying interindividual variability in cytokine production in response to fungal pathogens (113, 116). In particular, the GOLM1 gene was associated with C. albicans-induced IL-6 production, and a genetic variant within this locus was also associated with increased susceptibility to candidemia (113).
In addition, it is becoming increasingly clear that the outcome of an infectious disease reflects the dynamic interaction between humans, pathogen genotypes, and the environment (117). The host-fungal interaction exhibits features of a dynamic system that may exert genetic effects known as genotype-by-genotype (GxG) interactions (16). Those GxG interactions led to a slow host-pathogen coevolution (especially in cases of a commensal fungi such as C. albicans); this phenomenon might justify the host heterogeneity in the frequency of polymorphisms and haplotypes among populations (118). At the same time, the fungal pathogen can rapidly acquire mutations to adapt to host polymorphisms in a specific population, resulting in considerable genomic variation across fungi from different geographic regions (119–121). In turn, rapid pathogen evolution or host-pathogen coevolution might have caused a fluctuation over time of the disease susceptibility genes across populations, as mathematically modeled by Lambrechts et al. (122), and may have directly played a role in the limited success of GWASs on fungal (or, in general, infectious) disease susceptibility. Last but not least, classical GWASs can detect only the genetic component of the three-way interplay between the host immune system, different pathogen morphotypes, and the environment. In particular, host and pathogen genetic variability interaction with environmental influences are even more challenging to model, and they can be collectively defined as gene-environment (GxE) interactions (123). For example, environmental factors such as pH and/or an imbalanced microbiome influence the susceptibility to develop recurrent vulvovaginal candidiasis (RVVC) (124). Specific interactions between commensal bacteria and fungi could play an important role in the development of invasive candidiasis (125). Therefore, a more integrative and multilevel analysis of host, pathogen, and environmental variation is required to account for all these interactions while studying the pathogenesis of a fungal disease.
(iii) Overcoming limitations: the introduction of functional genomics approaches. Given the complexity of host-pathogen interactions, conventional experimental approaches that study only individual molecular components (either of the host or pathogen) cannot provide a comprehensive picture of these interactions. The development of high-throughput data acquisition technologies and the possibility to integrate multi-omics data sets have laid the foundations for a new discipline: systems biology (126, 127). The increasing use of systems biology is tightly intertwined with that of functional genomics, which represents a novel, more powerful multilevel manner for studying the genetics of complex diseases (128, 129) (Fig. 2). Thus, the integration of high-throughput multi-omics data (transcriptomics, proteomics, metabolomics, lipidomics, etc.) with genetics can be used to prioritize genes for follow-up functional experiments to better understand their role in host immune defense and identify molecular pathways that underlie disease pathogenesis (Table 2). Several studies have applied a functional genomics approach to understand host genetic susceptibility to fungal infections, where genome-wide data (also called “static biomarkers” [129]) were integrated, validated, or complemented with other multi-omics data sets in the context of the disease, where host-pathogen interactions are dynamically changing. Table 3 lists the studies in the last 5 years that identified a genetic variant associated with fungal infections using a systems genomics approach.
TABLE 2.
Selected high-throughput methods for studying host-pathogen interactions
| Methoda | Purpose | Reference(s) |
|---|---|---|
| RNA-seq | Transcript analysis | 145 |
| Dual RNA-seq | Transcript analysis of both the host and the pathogen | 146, 147 |
| scRNA-seq | Transcript analysis | 136 |
| GRO-seq | Transcription | 148 |
| PRO-seq | Genome-wide map of transcriptionally engaged Pol II | 149 |
| Nascent-seq | Transcription | 150 |
| ChIA-PET | Chromatin conformation | 151 |
| Hi-C | Chromatin conformation | 152, 153 |
| 5-C-seq | Chromatin conformation | 154 |
| DNase-seq | Open chromatin | 155 |
| ATAC-seq | Open chromatin | 156 |
| ChIP-seq | Mapping DNA regulatory elements | 157 |
| BS-seq | Genome methylation | 158 |
| RRBS-seq | Genome methylation | 159 |
| ITS1-seq | Fungi detection | 160 |
| Nano LC-MS/MS | Host and fungal quantitative proteome analysis without isolation | 161 |
seq, sequencing; scRNA, single cell RNA; GRO, global run-on; PRO, precision nuclear run-on; ChIA-PET, chromatin interaction analysis by paired-end tag sequencing; 5-C, chromosome conformation capture carbon copy; ATAC, assay for transposase-accessible chromatin; ChIP, chromatin immunoprecipitation; BS, bisulfite; RRBS, reduced representation bisulfite sequencing; ITS1, internal transcribed spacer 1; Nano LC-MS/MS, nanoscale liquid chromatography tandem mass spectrometry.
TABLE 3.
Genetic variants associated with fungal infections found in the last 5 years using a systems genomics approach
| Gene | Polymorphism(s) | Chromosome location | Reported association | Functional validation/evidence | Reference |
|---|---|---|---|---|---|
| GOLM1 | rs11141235 | 9q21 | Increased candidemia susceptibility | cQTL locus: lower C. albicans-induced IL-6 production | 113 |
| IFIH1 | rs1990760, rs3747517 | 2q24.2 | Increased candidemia susceptibility | Reduced C. albicans-induced IL-10 in PMCs | 132 |
| MAP3K8 | rs1360119 | 10p11.23 | Increased candidemia susceptibility | Reduced IL-6, IL-8 and IFN-γ in serum of candidemia patients | 100 |
| SPTBN5 (eQTL of PLA2G4B) | rs8028958 | 15q15.1 | Increased candidemia susceptibility | Lower C. albicans-induced IL-6 and ROS in PBMCs | 133 |
| LY86 | rs9405943 | 6p25.1 | Increased candidemia susceptibility | Lower migration towards MCP-1 of monocytes with knockdown of LY86 | 136 |
| SIGLEC15 | rs2919643 | 18q21.1 | Increased RVVC susceptibility | Increased C. albicans-induced IL-17A, IL-22 and IFN-γ | 137 |
| MFHAS1 | rs139408032 | 8p23.1 | NAa | cQTL locus: higher M. circinelloides-induced FGF-2 production | 138 |
| FRMD4A | rs61836093 | 10p13 | NA | cQTL locus: higher C. albicans-induced FGF-2 production | 138 |
| CSF1 | rs1999713 | 1p13 | Decreased cryptococcosis susceptibility in HIV patients | Upregulation of CSF1 upon C. neoformans stimulation of human PBMCs; higher phagocytosis and killing of C. neoformans in PBMCs from HIV patients pretreated with M-CSF | 134 |
NA, not applicable.
A specific role of the type I interferon pathway in anti-Candida host defense was supported by integrating transcriptional analysis and functional genomics (130) using Candida-stimulated human immune cells. Of note, the importance of this pathway was validated through immunological and genetic studies in both healthy volunteers and patients with systemic candidiasis or suffering from CMC. Moreover, polymorphisms in type I interferon genes modulated Candida-induced cytokine production, and they were correlated with susceptibility to systemic candidiasis (130). The first transcriptome-wide association study (TWAS) (131) of the fungal immunology field identified molecular pathways underlying candidemia susceptibility using unbiased transcriptomics data, which were then validated in a patient cohort. Significant associations between CCL8, STAT1, PSMB8, and SP110 polymorphisms and susceptibility to candidemia were identified by integrating transcriptomics data, and a candidemia GWAS followed up by functional in vitro validation in the context of Candida infection (130). Another study suggested that RIG-I-like receptor (RLR) MDA5 has a critical role in anti-Candida host immune defense by integrating genetic, transcriptomic, and immunological data generated from mouse and human studies (132). The additive value of integrating multiple molecular data sets became even more apparent by two follow-up studies where genes and pathways underlying candidemia susceptibility were prioritized. In the first study, suggestive genetic associations together with transcriptomic data prioritized novel pathways implicated in candidemia susceptibility, including the complement and hemostasis pathways (100). In the second study, integration of GWAS data with variants that affect cytokine levels (cytokine-QTLs) from different Candida-stimulated cell types prioritized lipid and arachidonic acid metabolism as potential mechanisms that affect monocyte-derived cytokines to influence susceptibility to candidemia (133).
Although African populations suffer the most from infectious diseases, they are still underrepresented in studies of disease susceptibility (117). The first genome-wide association study of susceptibility to cryptococcosis in HIV patients was carried out with genotype data from 524 patients of African descent. This study identified six loci upstream of the CSF1 gene (encoding macrophage colony-stimulating factor [M-CSF]) that were significantly associated with the disease susceptibility and validated in a separate cohort. Functional data from RNA sequencing (RNA-seq) of human peripheral blood mononuclear cells (PBMCs) stimulated with C. neoformans and in vitro experiments with HIV patients’ PBMCs confirmed the crucial role of M-CSF for anti-Cryptococcus host defense mechanisms (134).
Given that genetic variants significantly associated with a disease are often regulated in a context- and cell-specific way (135), with the development of single-cell RNA-seq, it has become possible to prioritize genes in a cell-type-specific fashion. For example, by combining bulk and single-cell transcriptome data in response to Candida stimulation with GWAS data on candidemia susceptibility, the LY86 antigen has been prioritized and further validated to exert a protective role against candidemia risk (136). Furthermore, genes and cellular processes that contribute to the pathogenesis of RVVC, including cellular morphogenesis and metabolism, and cellular adhesion were identified through the integration of genomic approaches and immunological studies in two independent cohorts of patients with RVVC and healthy individuals (137). In particular, the role of SIGLEC15, a lectin expressed by various immune cells that binds sialic acid, in Candida recognition and RVVC susceptibility was also validated in the same study with both in vitro and in vivo functional assays (137). Wang et al. in the Hi-HOST Phenome Project (H2P2) identified two SNPs significantly associated with basic fibroblast growth factor (FGF2) production in response to Mucor circinelloides and C. albicans, posing those allelic variants as potential candidates for the antifungal host immune response (138). However, they did not validate whether and how the presence of these SNPs are associated with an increased risk of fungal infections.
Overall, such an integrative functional approach is valuable in the context of not only fungal infections but also other infectious diseases, for which the limited size of patient cohorts limits the power of the GWAS. The reasons for using such an approach are 3-fold. First of all, this approach makes use of large-population-based cohort studies in the context of the disease that can be excellent models to get a powerful analysis to understand disease pathophysiology. Second, it is very versatile and provides independent layers of evidence intersecting with each other: from the multi-omics untargeted molecular candidate to the experimental or clinical evidence (top down) and vice versa (bottom up) in a multidisciplinary and collaborative way. Last but not least, the ultimate aim of functional genomic studies is to provide “actionable data” with translational potential. Since this can be a pathway-based targeted approach, it is possible to validate and clinically translate those findings to patients. Knowing the underlying pathways of human host defense allows, for example, for the identification of ways to prevent the disease, the development of novel diagnostic tools to be used in patient risk stratification, and the identification of new potential therapeutics. For a robust implementation of such a host-oriented therapy, it is particularly crucial to make sure that the results are validated in a physiologically relevant model, preferably relevant primary models of disease or appropriate patient samples or clinical strains. It has been shown that not always what has been validated in human cell lines (139), in mice (140), or in a laboratory pathogen strain (141, 142) holds true in patient’s cells or fungal clinical isolates.
FUTURE PERSPECTIVES
Over the last decades, the study of the genetics of infectious disease susceptibility has been revolutionized, and it has been developed more rapidly thanks to new technologies. This progress was important at multiple levels. First, it has made the research process more effective, comprehensive, and productive, providing valuable new findings on host-pathogen interactions. Such an evolution of the field combined with an interdisciplinary approach can be a useful tool to identify potential novel therapeutic drug targets. In this respect, a recent study has shown that the proportion of drug mechanisms with direct genetic support increases significantly across the drug development phases, indicating that prioritizing a genetically supported drug target could double the success rate in drug discovery (143). A stratification of patients based on genetic profiling would pinpoint the patients with high risk of disease and who will benefit most from the drug. Unless additional clinical trials provide evidence of a treatment effect based on genetic profiling, we should be aware and cautious of the benefits and harms of new drug targets. In addition, a host-directed therapeutic target may also result in a weaker selection pressure on pathogens, potentially making it more difficult for a pathogen to evolve beyond the control of the host immune response.
Integrating such a plethora of omics data would catalyze the identification of diagnostic markers that might be useful for severity stratification or eligibility for specific treatments. Considering the host variability in immune-related genes, personalized therapies based on an individual’s genetic profile, such as immunotherapy-based interventions or targeted antifungal prophylaxis in genetically susceptible individuals, are leading to increasingly more powerful precision medicine. Nonetheless, risk stratification approaches guiding the clinical decision-making process based on a patient’s individual susceptibility profile are expected to be promising. From a more basic science and biotechnological aspect, new technologies are gaining ground in the study of the genetics of infectious diseases, such as single-cell sequencing at the transcriptome level and whole-genome sequencing for the primary immunodeficiencies at the genomic level.
It is expected that in the coming years, novel technologies that will help dissect the interaction of host genetics and the metagenomic (microbiome and also mycobiome) make-up of an individual will be further integrated, as an increasing number of studies will investigate these complementary genomes of an individual. Some of the available technologies that can potentially be implemented and integrated with the genetic level are the organ-on-chips approach, which would allow us to better dissect the human-fungus-environment interaction in a more dynamic manner, which is more comparable to human physiology.
These novel tools in a systems genomic approach framework will also be used to decipher the pathophysiology of emerging fungal infections (e.g., Candida auris). In addition to this, such an approach needs to be employed more extensively in populations of non-European ancestry.
ACKNOWLEDGMENTS
We thank Diletta Rosati for the help with the figure realization.
F.L.V.D.V. was supported by a Vidi grant of the Netherlands Association for Scientific Research, the Europeans Union’s Horizon 2020 research and innovation program under grant agreement number 847507, HDM-FUN, and the La Caixa foundation (identifier [ID] 100010434). M.G.N. was supported by an ERC Advanced Grant (number 833247), a Spinoza Grant of the Netherlands Organization for Scientific Research, and a Competitiveness Operational Program Grant of the Romanian Ministry of European Funds (FUSE).
Biographies

Mariolina Bruno obtained her M.D. from Sapienza University of Rome (Italy) and received a degree in Life Sciences from the Sapienza School from Advances Studies (SSAS). She is currently a final-year Ph.D. candidate in the Department of Internal Medicine of Radboudumc in Nijmegen (The Netherlands). She has been working on a project aimed at defining how C. auris is recognized by immune cells. She is currently investigating host susceptibility factors for Aspergillus infection in patients with chronic pulmonary aspergillosis (CPA) and chronic granulomatous disease (CGD), with a particular focus on immunometabolism.

Mihai G. Netea was born and studied medicine in Cluj-Napoca (Romania). He completed his Ph.D. at the Radboud University Nijmegen (The Netherlands) on studies investigating the cytokine network in sepsis. After working as a postdoc at the University of Colorado, he returned to Nijmegen where he finished his clinical training as an infectious diseases specialist and where he currently heads the division of Experimental Medicine, Department of Internal Medicine, Nijmegen University Nijmegen Medical Center. His main research interests are pattern recognition of fungal pathogens and the induction of antifungal immunity, primary immunodeficiencies in innate immune system, and the study of the memory traits of innate immunity.
Contributor Information
Mihai G. Netea, Email: mihai.netea@radboudumc.nl.
Karen M. Ottemann, University of California, Santa Cruz
REFERENCES
- 1.GBD 2017 Causes of Death Collaborators. 2018. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392:1736–1788. 10.1016/S0140-6736(18)32203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathogens 13:e1006290. 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison AC. 1954. Protection afforded by sickle-cell trait against subtertian malarial infection. Br Med J 1:290–294. 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herndon CN, Jennings RG. 1951. A twin-family study of susceptibility to poliomyelitis. Am J Hum Genet 3:17–46. [PMC free article] [PubMed] [Google Scholar]
- 6.Kallmann FJ, Reisner D. 1943. Twin studies on the significance of genetic factors in tuberculosis. Am Rev Tuberc 47:549–574. [Google Scholar]
- 7.Fine PE. 1981. Immunogenetics of susceptibility to leprosy, tuberculosis, and leishmaniasis. an epidemiological perspective. Int J Lepr Other Mycobact Dis 49:437–454. [PubMed] [Google Scholar]
- 8.Malaty HM, Engstrand L, Pedersen NL, Graham DY. 1994. Helicobacter pylori infection: genetic and environmental influences. A study of twins. Ann Intern Med 120:982–986. 10.7326/0003-4819-120-12-199406150-00002. [DOI] [PubMed] [Google Scholar]
- 9.Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, Kwang TY, Hsu ST, Lin SY, Hsu LC. 1989. Hepatitis B virus markers in Chinese twins. Anticancer Res 9:737–741. [PubMed] [Google Scholar]
- 10.Sørensen TI, Nielsen GG, Andersen PK, Teasdale TW. 1988. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med 318:727–732. 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 11.Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES. 2002. Detecting recent positive selection in the human genome from haplotype structure. 6909. Nature 419:832–837. 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 12.Domínguez-Andrés J, Netea MG. 2019. Impact of historic migrations and evolutionary processes on human immunity. Trends Immunol 40:1105–1119. 10.1016/j.it.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shultz AJ, Sackton TB. 2019. Immune genes are hotspots of shared positive selection across birds and mammals. Elife 8:e41815. 10.7554/eLife.41815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abel L, Dessein AJ. 1998. Genetic epidemiology of infectious diseases in humans: design of population-based studies. Emerg Infect Dis 4:593–603. 10.3201/eid0404.980409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matzaraki V, Kumar V, Wijmenga C, Zhernakova A. 2017. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol 18:76. 10.1186/s13059-017-1207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacPherson A, Otto SP, Nuismer SL. 2018. Keeping pace with the red queen: identifying the genetic basis of susceptibility to infectious disease. Genetics 208:779–789. 10.1534/genetics.117.300481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Templeton SP, Rivera A, Hube B, Jacobsen ID. 2018. Editorial: Immunity to human fungal pathogens: mechanisms of host recognition, protection, pathology, and fungal interference. Front Immunol 9:2337. 10.3389/fimmu.2018.02337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eades CP, Armstrong-James DPH. 2019. Invasive fungal infections in the immunocompromised host: mechanistic insights in an era of changing immunotherapeutics. Med Mycol 57:S307–S317. 10.1093/mmy/myy136. [DOI] [PubMed] [Google Scholar]
- 19.Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi (Basel) 3:57. 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LIFE. 2020. The burden of fungal disease. http://www.life-worldwide.org/awareness-advocacy.
- 21.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almeida F, Rodrigues ML, Coelho C. 2019. The still underestimated problem of fungal diseases worldwide. Front Microbiol 10:214. 10.3389/fmicb.2019.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodrigues ML, Nosanchuk JD. 2020. Fungal diseases as neglected pathogens: a wake-up call to public health officials. PLoS Negl Trop Dis 14:e0007964. 10.1371/journal.pntd.0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denham ST, Wambaugh MA, Brown JCS. 2019. How environmental fungi cause a range of clinical outcomes in susceptible hosts. J Mol Biol 431:2982–3009. 10.1016/j.jmb.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Underhill DM, Iliev ID. 2014. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 14:405–416. 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falci DR, Stadnik CMB, Pasqualotto AC. 2017. A review of diagnostic methods for invasive fungal diseases: challenges and perspectives. Infect Dis Ther 6:213–223. 10.1007/s40121-017-0154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lass-Flörl C. 2017. Current challenges in the diagnosis of fungal infections. Methods Mol Biol 1508:3–15. 10.1007/978-1-4939-6515-1_1. [DOI] [PubMed] [Google Scholar]
- 28.Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. 2018. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360:739–742. 10.1126/science.aap7999. [DOI] [PubMed] [Google Scholar]
- 29.Lionakis MS, Netea MG, Holland SM. 2014. Mendelian genetics of human susceptibility to fungal infection. Cold Spring Harb Perspect Med 4:a019638. 10.1101/cshperspect.a019638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muskett H, Shahin J, Eyres G, Harvey S, Rowan K, Harrison D. 2011. Risk factors for invasive fungal disease in critically ill adult patients: a systematic review. Crit Care 15:R287. 10.1186/cc10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erjavec Z, Kluin-Nelemans H, Verweij PE. 2009. Trends in invasive fungal infections, with emphasis on invasive aspergillosis. Clin Microbiol Infect 15:625–633. 10.1111/j.1469-0691.2009.02929.x. [DOI] [PubMed] [Google Scholar]
- 32.Richardson M, Lass-Flörl C. 2008. Changing epidemiology of systemic fungal infections. Clin Microbiol Infect 14:5–24. 10.1111/j.1469-0691.2008.01978.x. [DOI] [PubMed] [Google Scholar]
- 33.Cortés JA, Corrales IF. 2018. Invasive candidiasis: epidemiology and risk factors. In Silva de Loreto E, Moraes Tondolo JS. (ed), Fungal infection. IntechOpen, London, United Kingdom. 10.5772/intechopen.81813. [DOI] [Google Scholar]
- 34.Ahmed A, Azim A, Baronia AK, Marak KRSK, Gurjar M. 2014. Risk prediction for invasive candidiasis. Indian J Crit Care Med 18:682–688. 10.4103/0972-5229.142178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanzani M, Lewis R. 2018. Development and applications of prognostic risk models in the management of invasive mold disease. J Fungi 4:141. 10.3390/jof4040141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F, Zhou M, Zou Z, Li W, Huang C, He Z. 2018. A risk prediction model for invasive fungal disease in critically ill patients in the intensive care unit. Asian Nursing Res 12:299–303. 10.1016/j.anr.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 37.León C, Ruiz-Santana S, Saavedra P, Almirante B, Nolla-Salas J, Alvarez-Lerma F, Garnacho-Montero J, León MA, EPCAN Study Group. 2006. A bedside scoring system (“Candida score”) for early antifungal treatment in nonneutropenic critically ill patients with Candida colonization. Crit Care Med 34:730–737. 10.1097/01.CCM.0000202208.37364.7D. [DOI] [PubMed] [Google Scholar]
- 38.Lionakis MS. 2019. Genetic variation and fungal infection risk: state of the art. Curr Fungal Infect Rep 13:250–259. 10.1007/s12281-019-00362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cottier F, Pavelka N. 2012. Complexity and dynamics of host–fungal interactions. Immunol Res 53:127–135. 10.1007/s12026-012-8265-y. [DOI] [PubMed] [Google Scholar]
- 40.Salazar F, Brown GD. 2018. Antifungal innate immunity: a perspective from the last 10 years. J Innate Immun 10:373–397. 10.1159/000488539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roilides E, Dimitriadou-Georgiadou A, Sein T, Kadiltsoglou I, Walsh TJ. 1998. Tumor necrosis factor alpha enhances antifungal activities of polymorphonuclear and mononuclear phagocytes against Aspergillus fumigatus. Infect Immun 66:5999–6003. 10.1128/IAI.66.12.5999-6003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Netea MG, van Tits LJ, Curfs JH, Amiot F, Meis JF, van der Meer JW, Kullberg BJ. 1999. Increased susceptibility of TNF-alpha lymphotoxin-alpha double knockout mice to systemic candidiasis through impaired recruitment of neutrophils and phagocytosis of Candida albicans. J Immunol 163:1498–1505. [PubMed] [Google Scholar]
- 43.van de Veerdonk FL, Netea MG. 2010. T-cell subsets and antifungal host defenses. Curr Fungal Infect Rep 4:238–243. 10.1007/s12281-010-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verma A, Wüthrich M, Deepe G, Klein B. 2015. Adaptive immunity to fungi. Cold Spring Harb Perspect Med 5:a019612. 10.1101/cshperspect.a019612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dewi IMW, Van de Veerdonk FL, Gresnigt MS. 2017. The multifaceted role of T-Helper responses in host defense against Aspergillus fumigatus. J Fungi 3:55. 10.3390/jof3040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sparber F, LeibundGut-Landmann S. 2019. Interleukin-17 in antifungal immunity. Pathogens 8:54. 10.3390/pathogens8020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lanternier F, Cypowyj S, Picard C, Bustamante J, Lortholary O, Casanova J-L, Puel A. 2013. Primary immunodeficiencies underlying fungal infections. Curr Opin Pediatr 25:736–747. 10.1097/MOP.0000000000000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antachopoulos C, Walsh TJ, Roilides E. 2007. Fungal infections in primary immunodeficiencies. Eur J Pediatr 166:1099–1117. 10.1007/s00431-007-0527-7. [DOI] [PubMed] [Google Scholar]
- 49.Segal BH, Leto TL, Gallin JI, Malech HL, Holland SM. 2000. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore) 79:170–200. 10.1097/00005792-200005000-00004. [DOI] [PubMed] [Google Scholar]
- 50.Almyroudis NG, Holland SM, Segal BH. 2005. Invasive aspergillosis in primary immunodeficiencies. Med Mycol 43 Suppl 1:S247–259. 10.1080/13693780400025203. [DOI] [PubMed] [Google Scholar]
- 51.Blumental S, Mouy R, Mahlaoui N, Bougnoux M-E, Debré M, Beauté J, Lortholary O, Blanche S, Fischer A. 2011. Invasive mold infections in chronic granulomatous disease: a 25-year retrospective survey. Clin Infect Dis 53:e159–e169. 10.1093/cid/cir731. [DOI] [PubMed] [Google Scholar]
- 52.van de Geer A, Nieto-Patlán A, Kuhns DB, Tool ATJ, Arias AA, Bouaziz M, de Boer M, Franco JL, Gazendam RP, van Hamme JL, van Houdt M, van Leeuwen K, Verkuijlen PJH, van den Berg TK, Alzate JF, Arango-Franco CA, Batura V, Bernasconi AR, Boardman B, Booth C, Burns SO, Cabarcas F, Bensussan NC, Charbit-Henrion F, Corveleyn A, Deswarte C, Azcoiti ME, Foell D, Gallin JI, Garcés C, Guedes M, Hinze CH, Holland SM, Hughes SM, Ibañez P, Malech HL, Meyts I, Moncada-Velez M, Moriya K, Neves E, Oleastro M, Perez L, Rattina V, Oleaga-Quintas C, Warner N, Muise AM, López JS, Trindade E, Vasconcelos J, Vermeire S, et al. 2018. Inherited p40phox deficiency differs from classic chronic granulomatous disease. J Clin Invest 128:3957–3975. 10.1172/JCI97116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehrer RI, Cline MJ. 1969. Leukocyte myeloperoxidase deficiency and disseminated candidiasis: the role of myeloperoxidase in resistance to Candida infection. J Clin Invest 48:1478–1488. 10.1172/JCI106114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cech P, Papathanassiou A, Boreux G, Roth P, Miescher PA. 1979. Hereditary myeloperoxidase deficiency. Blood 53:403–411. 10.1182/blood.V53.3.403.403. [DOI] [PubMed] [Google Scholar]
- 55.Corvilain E, Casanova J-L, Puel A. 2018. Inherited CARD9 deficiency: invasive disease caused by ascomycete fungi in previously healthy children and adults. J Clin Immunol 38:656–693. 10.1007/s10875-018-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drummond RA, Franco LM, Lionakis MS. 2018. Human CARD9: a critical molecule of fungal immune surveillance. Front Immunol 9:1836. 10.3389/fimmu.2018.01836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drummond RA, Swamydas M, Oikonomou V, Zhai B, Dambuza IM, Schaefer BC, Bohrer AC, Mayer-Barber KD, Lira SA, Iwakura Y, Filler SG, Brown GD, Hube B, Naglik JR, Hohl TM, Lionakis MS. 2019. CARD9+ microglia promote antifungal immunity via IL-1β- and CXCL1-mediated neutrophil recruitment. Nat Immunol 20:559–570. 10.1038/s41590-019-0377-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vinh DC, Patel SY, Uzel G, Anderson VL, Freeman AF, Olivier KN, Spalding C, Hughes S, Pittaluga S, Raffeld M, Sorbara LR, Elloumi HZ, Kuhns DB, Turner ML, Cowen EW, Fink D, Long-Priel D, Hsu AP, Ding L, Paulson ML, Whitney AR, Sampaio EP, Frucht DM, DeLeo FR, Holland SM. 2010. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood 115:1519–1529. 10.1182/blood-2009-03-208629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sampaio EP, Hsu AP, Pechacek J, Bax HI, Dias DL, Paulson ML, Chandrasekaran P, Rosen LB, Carvalho DS, Ding L, Vinh DC, Browne SK, Datta S, Milner JD, Kuhns DB, Long Priel DA, Sadat MA, Shiloh M, De Marco B, Alvares M, Gillman JW, Ramarathnam V, de la Morena M, Bezrodnik L, Moreira I, Uzel G, Johnson D, Spalding C, Zerbe CS, Wiley H, Greenberg DE, Hoover SE, Rosenzweig SD, Galgiani JN, Holland SM. 2013. Signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations and disseminated coccidioidomycosis and histoplasmosis. J Allergy Clin Immunol 131:1624–1634. 10.1016/j.jaci.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niehues H, Rösler B, van der Krieken DA, van Vlijmen-Willems IMJJ, Rodijk-Olthuis D, Peppelman M, Schalkwijk J, van den Bogaard EHJ, Zeeuwen PLJM, van de Veerdonk FL. 2019. STAT1 gain-of-function compromises skin host defense in the context of IFN-γ signaling. J Allergy Clin Immunol 143:1626–1629.e5. 10.1016/j.jaci.2018.11.033. [DOI] [PubMed] [Google Scholar]
- 61.Boisson B, Wang C, Pedergnana V, Wu L, Cypowyj S, Rybojad M, Belkadi A, Picard C, Abel L, Fieschi C, Puel A, Li X, Casanova J-L. 2013. An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity 39:676–686. 10.1016/j.immuni.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M, Israel L, Chrabieh M, Audry M, Gumbleton M, Toulon A, Bodemer C, El-Baghdadi J, Whitters M, Paradis T, Brooks J, Collins M, Wolfman NM, Al-Muhsen S, Galicchio M, Abel L, Picard C, Casanova J-L. 2011. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 332:65–68. 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ling Y, Cypowyj S, Aytekin C, Galicchio M, Camcioglu Y, Nepesov S, Ikinciogullari A, Dogu F, Belkadi A, Levy R, Migaud M, Boisson B, Bolze A, Itan Y, Goudin N, Cottineau J, Picard C, Abel L, Bustamante J, Casanova J-L, Puel A. 2015. Inherited IL-17RC deficiency in patients with chronic mucocutaneous candidiasis. J Exp Med 212:619–631. 10.1084/jem.20141065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lévy R, Okada S, Béziat V, Moriya K, Liu C, Chai LYA, Migaud M, Hauck F, Al Ali A, Cyrus C, Vatte C, Patiroglu T, Unal E, Ferneiny M, Hyakuna N, Nepesov S, Oleastro M, Ikinciogullari A, Dogu F, Asano T, Ohara O, Yun L, Della Mina E, Bronnimann D, Itan Y, Gothe F, Bustamante J, Boisson-Dupuis S, Tahuil N, Aytekin C, Salhi A, Al Muhsen S, Kobayashi M, Toubiana J, Abel L, Li X, Camcioglu Y, Celmeli F, Klein C, AlKhater SA, Casanova J-L, Puel A. 2016. Genetic, immunological, and clinical features of patients with bacterial and fungal infections due to inherited IL-17RA deficiency. Proc Natl Acad Sci U S A 113:E8277–E8285. 10.1073/pnas.1618300114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, Davis J, Hsu A, Asher AI, O’Shea J, Holland SM, Paul WE, Douek DC. 2008. Impaired TH17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452:773–776. 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, Alzahrani M, Al-Muhsen S, Halwani R, Ma CS, Wong N, Soudais C, Henderson LA, Marzouqa H, Shamma J, Gonzalez M, Martinez-Barricarte R, Okada C, Avery DT, Latorre D, Deswarte C, Jabot-Hanin F, Torrado E, Fountain J, Belkadi A, Itan Y, Boisson B, Migaud M, Arlehamn CSL, Sette A, Breton S, McCluskey J, Rossjohn J, de Villartay J-P, Moshous D, Hambleton S, Latour S, Arkwright PD, Picard C, Lantz O, Engelhard D, Kobayashi M, Abel L, Cooper AM, Notarangelo LD, Boisson-Dupuis S, Puel A, Sallusto F, Bustamante J, Tangye SG, Casanova J-L. 2015. Immunodeficiencies. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science 349:606–613. 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng J, van de Veerdonk FL, Crossland KL, Smeekens SP, Chan CM, Shehri TA, Abinun M, Gennery AR, Mann J, Lendrem DW, Netea MG, Rowan AD, Lilic D. 2015. Gain-of-function STAT1 mutations impair STAT3 activity in patients with chronic mucocutaneous candidiasis (CMC). Eur J Immunol 45:2834–2846. 10.1002/eji.201445344. [DOI] [PubMed] [Google Scholar]
- 68.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LAB, Gilissen C, Arts P, Rosentul DC, Carmichael AJ, Smits-van der Graaf CAA, Kullberg BJ, van der Meer JWM, Lilic D, Veltman JA, Netea MG. 2011. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med 365:54–61. 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Q, Davis JC, Lamborn IT, Freeman AF, Jing H, Favreau AJ, Matthews HF, Davis J, Turner ML, Uzel G, Holland SM, Su HC. 2009. Combined Immunodeficiency Associated with DOCK8 Mutations. N Engl J Med 361:2046–2055. 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lionakis MS, Levitz SM. 2018. Host control of fungal infections: lessons from basic studies and human cohorts. Annu Rev Immunol 36:157–191. 10.1146/annurev-immunol-042617-053318. [DOI] [PubMed] [Google Scholar]
- 71.Puel A, Döffinger R, Natividad A, Chrabieh M, Barcenas-Morales G, Picard C, Cobat A, Ouachée-Chardin M, Toulon A, Bustamante J, Al-Muhsen S, Al-Owain M, Arkwright PD, Costigan C, McConnell V, Cant AJ, Abinun M, Polak M, Bougnères P-F, Kumararatne D, Marodi L, Nahum A, Roifman C, Blanche S, Fischer A, Bodemer C, Abel L, Lilic D, Casanova J-L. 2010. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med 207:291–297. 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas DC, Witte JS. 2002. Point: population stratification: a problem for case-control studies of candidate-gene associations? Cancer Epidemiol Biomarkers Prev 11:505–512. [PubMed] [Google Scholar]
- 73.Netea MG, Wijmenga C, O'Neill LAJ. 2012. Genetic variation in Toll-like receptors and disease susceptibility. Nat Immunol 13:535–542. 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 74.Pana Z-D, Farmaki E, Roilides E. 2014. Host genetics and opportunistic fungal infections. Clin Microbiol Infect 20:1254–1264. 10.1111/1469-0691.12800. [DOI] [PubMed] [Google Scholar]
- 75.Khanna N, Stuehler C, Lünemann A, Wójtowicz A, Bochud P-Y, Leibundgut-Landmann S. 2016. Host response to fungal infections – how immunology and host genetics could help to identify and treat patients at risk. Swiss Med Wkly 146:w14350. 10.4414/smw.2016.14350. [DOI] [PubMed] [Google Scholar]
- 76.Campos CF, van de Veerdonk FL, Gonçalves SM, Cunha C, Netea MG, Carvalho A. 2019. Host genetic signatures of susceptibility to fungal disease. Curr Top Microbiol Immunol 422:237–263. 10.1007/82_2018_113. [DOI] [PubMed] [Google Scholar]
- 77.Plantinga TS, Johnson MD, Scott WK, van de Vosse E, Velez Edwards DR, Smith PB, Alexander BD, Yang JC, Kremer D, Laird GM, Oosting M, Joosten LAB, van der Meer JWM, van Dissel JT, Walsh TJ, Perfect JR, Kullberg BJ, Netea MG. 2012. Toll-like receptor 1 polymorphisms increase susceptibility to candidemia. J Infect Dis 205:934–943. 10.1093/infdis/jir867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koldehoff M, Beelen DW, Elmaagacli AH. 2013. Increased susceptibility for aspergillosis and post-transplant immune deficiency in patients with gene variants of TLR4 after stem cell transplantation. Transpl Infect Dis 15:533–539. 10.1111/tid.12115. [DOI] [PubMed] [Google Scholar]
- 79.Bochud P-Y, Chien JW, Marr KA, Leisenring WM, Upton A, Janer M, Rodrigues SD, Li S, Hansen JA, Zhao LP, Aderem A, Boeckh M. 2008. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med 359:1766–1777. 10.1056/NEJMoa0802629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van der Graaf CAA, Netea MG, Morré SA, Den Heijer M, Verweij PE, Van der Meer JWM, Kullberg BJ. 2006. Toll-like receptor 4 Asp299Gly/Thr399Ile polymorphisms are a risk factor for Candida bloodstream infection. Eur Cytokine Netw 17:29–34. [PubMed] [Google Scholar]
- 81.Cunha C, Di Ianni M, Bozza S, Giovannini G, Zagarella S, Zelante T, D’Angelo C, Pierini A, Pitzurra L, Falzetti F, Carotti A, Perruccio K, Latgé J-P, Rodrigues F, Velardi A, Aversa F, Romani L, Carvalho A. 2010. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood 116:5394–5402. 10.1182/blood-2010-04-279307. [DOI] [PubMed] [Google Scholar]
- 82.Plantinga TS, van der Velden WJFM, Ferwerda B, van Spriel AB, Adema G, Feuth T, Donnelly JP, Brown GD, Kullberg B-J, Blijlevens NMA, Netea MG. 2009. Early stop polymorphism in human DECTIN-1 is associated with increased Candida colonization in hematopoietic stem cell transplant recipients. Clin Infect Dis 49:724–732. 10.1086/604714. [DOI] [PubMed] [Google Scholar]
- 83.Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, Elbers CC, Johnson MD, Cambi A, Huysamen C, Jacobs L, Jansen T, Verheijen K, Masthoff L, Morré SA, Vriend G, Williams DL, Perfect JR, Joosten LAB, Wijmenga C, van der Meer JWM, Adema GJ, Kullberg BJ, Brown GD, Netea MG. 2009. Human dectin-1 deficiency and mucocutaneous fungal infections. N Engl J Med 361:1760–1767. 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cunha C, Aversa F, Lacerda JF, Busca A, Kurzai O, Grube M, Löffler J, Maertens JA, Bell AS, Inforzato A, Barbati E, Almeida B, Santos e Sousa P, Barbui A, Potenza L, Caira M, Rodrigues F, Salvatori G, Pagano L, Luppi M, Mantovani A, Velardi A, Romani L, Carvalho A. 2014. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med 370:421–432. 10.1056/NEJMoa1211161. [DOI] [PubMed] [Google Scholar]
- 85.Wójtowicz A, Lecompte TD, Bibert S, Manuel O, Rüeger S, Berger C, Boggian K, Cusini A, Garzoni C, Hirsch H, Khanna N, Mueller NJ, Meylan PR, Pascual M, van Delden C, Bochud P-Y, Swiss Transplant Cohort Study. 2015. PTX3 polymorphisms and invasive mold infections after solid organ transplant. Clin Infect Dis 61:619–622. 10.1093/cid/civ386. [DOI] [PubMed] [Google Scholar]
- 86.Gresnigt MS, Cunha C, Jaeger M, Gonçalves SM, Malireddi RKS, Ammerdorffer A, Lubbers R, Oosting M, Rasid O, Jouvion G, Fitting C, de Jong DJ, Lacerda JF, Campos A, Melchers WJG, Lagrou K, Maertens J, Kanneganti T-D, Carvalho A, Ibrahim-Granet O, van de Veerdonk FL. 2018. Genetic deficiency of NOD2 confers resistance to invasive aspergillosis. Nat Commun 9:2636. 10.1038/s41467-018-04912-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Break TJ, Jaeger M, Solis NV, Filler SG, Rodriguez CA, Lim JK, Lee C-CR, Sobel JD, Netea MG, Lionakis MS. 2015. CX3CR1 is dispensable for control of mucosal Candida albicans infections in mice and humans. Infect Immun 83:958–965. 10.1128/IAI.02604-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lionakis MS, Swamydas M, Fischer BG, Plantinga TS, Johnson MD, Jaeger M, Green NM, Masedunskas A, Weigert R, Mikelis C, Wan W, Lee C-CR, Lim JK, Rivollier A, Yang JC, Laird GM, Wheeler RT, Alexander BD, Perfect JR, Gao J-L, Kullberg B-J, Netea MG, Murphy PM. 2013. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J Clin Invest 123:5035–5051. 10.1172/JCI71307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Collar AL, Swamydas M, O’Hayre M, Sajib MS, Hoffman KW, Singh SP, Mourad A, Johnson MD, Ferre EMN, Farber JM, Lim JK, Mikelis CM, Gutkind JS, Lionakis MS. 2018. The homozygous CX3CR1-M280 mutation impairs human monocyte survival. JCI Insight 3:e95417. 10.1172/jci.insight.95417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Swamydas M, Gao J-L, Break TJ, Johnson MD, Jaeger M, Rodriguez CA, Lim JK, Green NM, Collar AL, Fischer BG, Lee C-CR, Perfect JR, Alexander BD, Kullberg B-J, Netea MG, Murphy PM, Lionakis MS. 2016. CXCR1-mediated neutrophil degranulation and fungal killing promote Candida clearance and host survival. Sci Transl Med 8:322ra10. 10.1126/scitranslmed.aac7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wójtowicz A, Gresnigt MS, Lecompte T, Bibert S, Manuel O, Joosten LAB, Rüeger S, Berger C, Boggian K, Cusini A, Garzoni C, Hirsch HH, Weisser M, Mueller NJ, Meylan PR, Steiger J, Kutalik Z, Pascual M, van Delden C, van de Veerdonk FL, Bochud P-Y, Swiss Transplant Cohort Study (STCS). 2015. IL1B and DEFB1 polymorphisms increase susceptibility to invasive mold infection after solid-organ transplantation. J Infect Dis 211:1646–1657. 10.1093/infdis/jiu636. [DOI] [PubMed] [Google Scholar]
- 92.Sainz J, Pérez E, Gómez-Lopera S, Jurado M. 2008. IL1 gene cluster polymorphisms and its haplotypes may predict the risk to develop invasive pulmonary aspergillosis and modulate C-reactive protein level. J Clin Immunol 28:473–485. 10.1007/s10875-008-9197-0. [DOI] [PubMed] [Google Scholar]
- 93.The International HapMap Consortium. 2003. The international HapMap project. Nature 426:789–796. 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 94.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. 2015. A global reference for human genetic variation. Nature 526:68–74. 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu L, Okada S, Kong X-F, Kreins AY, Cypowyj S, Abhyankar A, Toubiana J, Itan Y, Audry M, Nitschke P, Masson C, Toth B, Flatot J, Migaud M, Chrabieh M, Kochetkov T, Bolze A, Borghesi A, Toulon A, Hiller J, Eyerich S, Eyerich K, Gulácsy V, Chernyshova L, Chernyshov V, Bondarenko A, María Cortés Grimaldo R, Blancas-Galicia L, Madrigal Beas IM, Roesler J, Magdorf K, Engelhard D, Thumerelle C, Burgel P-R, Hoernes M, Drexel B, Seger R, Kusuma T, Jansson AF, Sawalle-Belohradsky J, Belohradsky B, Jouanguy E, Bustamante J, Bué M, Karin N, Wildbaum G, Bodemer C, Lortholary O, Fischer A, Blanche S, et al. 2011. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med 208:1635–1648. 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shelburne SA, Ajami NJ, Chibucos MC, Beird HC, Tarrand J, Galloway-Peña J, Albert N, Chemaly RF, Ghantoji SS, Marsh L, Pemmaraju N, Andreeff M, Shpall EJ, Wargo JA, Rezvani K, Alousi A, Bruno VM, Futreal PA, Petrosino JF, Kontoyiannis DP. 2015. Implementation of a pan-genomic approach to investigate holobiont-infecting microbe interaction: a case report of a leukemic patient with invasive mucormycosis. PLoS One 10:e0139851. 10.1371/journal.pone.0139851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arts P, Simons A, AlZahrani MS, Yilmaz E, AlIdrissi E, van Aerde KJ, Alenezi N, AlGhamdi HA, AlJubab HA, Al-Hussaini AA, AlManjomi F, Alsaad AB, Alsaleem B, Andijani AA, Asery A, Ballourah W, Bleeker-Rovers CP, van Deuren M, van der Flier M, Gerkes EH, Gilissen C, Habazi MK, Hehir-Kwa JY, Henriet SS, Hoppenreijs EP, Hortillosa S, Kerkhofs CH, Keski-Filppula R, Lelieveld SH, Lone K, MacKenzie MA, Mensenkamp AR, Moilanen J, Nelen M, ten Oever J, Potjewijd J, van Paassen P, Schuurs-Hoeijmakers JHM, Simon A, Stokowy T, van de Vorst M, Vreeburg M, Wagner A, van Well GTJ, Zafeiropoulou D, Zonneveld-Huijssoon E, Veltman JA, van Zelst-Stams WAG, Faqeih EA, van de Veerdonk FL, Netea MG, Hoischen A. 2019. Exome sequencing in routine diagnostics: a generic test for 254 patients with primary immunodeficiencies. Genome Med 11:38. 10.1186/s13073-019-0649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smirnova I, Mann N, Dols A, Derkx HH, Hibberd ML, Levin M, Beutler B. 2003. Assay of locus-specific genetic load implicates rare Toll-like receptor 4 mutations in meningococcal susceptibility. Proc Natl Acad Sci U S A 100:6075–6080. 10.1073/pnas.1031605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. 2007. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One 2:e1318. 10.1371/journal.pone.0001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matzaraki V, Gresnigt MS, Jaeger M, Ricaño-Ponce I, Johnson MD, Oosting M, Franke L, Withoff S, Perfect JR, Joosten LAB, Kullberg BJ, van de Veerdonk FL, Jonkers I, Li Y, Wijmenga C, Netea MG, Kumar V. 2017. An integrative genomics approach identifies novel pathways that influence candidaemia susceptibility. PLoS One 12:e0180824. 10.1371/journal.pone.0180824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kumar V, Cheng S-C, Johnson MD, Smeekens SP, Wojtowicz A, Giamarellos-Bourboulis E, Karjalainen J, Franke L, Withoff S, Plantinga TS, van de Veerdonk FL, van der Meer JWM, Joosten LAB, Sokol H, Bauer H, Herrmann BG, Bochud P-Y, Marchetti O, Perfect JR, Xavier RJ, Kullberg BJ, Wijmenga C, Netea MG. 2014. Immunochip SNP array identifies novel genetic variants conferring susceptibility to candidaemia. Nat Commun 5:4675. 10.1038/ncomms5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tian C, Hromatka BS, Kiefer AK, Eriksson N, Noble SM, Tung JY, Hinds DA. 2017. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat Commun 8:599. 10.1038/s41467-017-00257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abdel-Rahman SM, Preuett BL. 2012. Genetic predictors of susceptibility to cutaneous fungal infections: a pilot genome wide association study to refine a candidate gene search. J Dermatol Sci 67:147–152. 10.1016/j.jdermsci.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maskarinec SA, Johnson MD, Perfect JR. 2016. Genetic susceptibility to fungal infections: what is in the genes? Curr Clin Microbiol Rep 3:81–91. 10.1007/s40588-016-0037-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN. 2008. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9:356–369. 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 106.Goddard KA, Hopkins PJ, Hall JM, Witte JS. 2000. Linkage disequilibrium and allele-frequency distributions for 114 single-nucleotide polymorphisms in five populations. Am J Hum Genet 66:216–234. 10.1086/302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tian C, Gregersen PK, Seldin MF. 2008. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet 17:R143–R150. 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bulik-Sullivan BK, Loh P-R, Finucane H, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM. 2015. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47:291–295. 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gloyn AL, McCarthy MI. 2010. Variation across the allele frequency spectrum. Nat Genet 42:648–650. 10.1038/ng0810-648. [DOI] [PubMed] [Google Scholar]
- 110.Cirulli ET, Goldstein DB. 2010. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 11:415–425. 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 111.Ko DC, Urban TJ. 2013. Understanding human variation in infectious disease susceptibility through clinical and cellular GWAS. PLoS Pathog 9:e1003424. 10.1371/journal.ppat.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Croteau-Chonka DC, Rogers AJ, Raj T, McGeachie MJ, Qiu W, Ziniti JP, Stubbs BJ, Liang L, Martinez FD, Strunk RC, Lemanske RF, Liu AH, Stranger BE, Carey VJ, Raby BA. 2015. Expression quantitative trait loci information improves predictive modeling of disease relevance of non-coding genetic variation. PLoS One 10:e0140758. 10.1371/journal.pone.0140758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li Y, Oosting M, Deelen P, Ricaño-Ponce I, Smeekens S, Jaeger M, Matzaraki V, Swertz MA, Xavier RJ, Franke L, Wijmenga C, Joosten LAB, Kumar V, Netea MG. 2016. Inter-individual variability and genetic influences on cytokine responses against bacterial and fungal pathogens. Nat Med 22:952–960. 10.1038/nm.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huan T, Joehanes R, Song C, Peng F, Guo Y, Mendelson M, Yao C, Liu C, Ma J, Richard M, Agha G, Guan W, Almli LM, Conneely KN, Keefe J, Hwang S-J, Johnson AD, Fornage M, Liang L, Levy D. 2019. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun 10:1–14. 10.1038/s41467-019-12228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vandiedonck C. 2018. Genetic association of molecular traits: a help to identify causative variants in complex diseases. Clin Genet 93:520–532. 10.1111/cge.13187. [DOI] [PubMed] [Google Scholar]
- 116.Li Y, Oosting M, Smeekens SP, Jaeger M, Aguirre-Gamboa R, Le KTT, Deelen P, Ricaño-Ponce I, Schoffelen T, Jansen AFM, Swertz MA, Withoff S, van de Vosse E, van Deuren M, van de Veerdonk F, Zhernakova A, van der Meer JWM, Xavier RJ, Franke L, Joosten LAB, Wijmenga C, Kumar V, Netea MG. 2016. A functional genomics approach to understand variation in cytokine production in humans. Cell 167:1099.e14–1110.e14. 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 117.Chapman SJ, Hill AVS. 2012. Human genetic susceptibility to infectious disease. Nat Rev Genet 13:175–188. 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 118.Karlsson EK, Kwiatkowski DP, Sabeti PC. 2014. Natural selection and infectious disease in human populations. Nat Rev Genet 15:379–393. 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gabaldón T, Fairhead C. 2019. Genomes shed light on the secret life of Candida glabrata: not so asexual, not so commensal. Curr Genet 65:93–98. 10.1007/s00294-018-0867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ashu EE, Hagen F, Chowdhary A, Meis JF, Xu J. 2017. Global population genetic analysis of Aspergillus fumigatus. mSphere 2:e00019-17. 10.1128/mSphere.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ropars J, Maufrais C, Diogo D, Marcet-Houben M, Perin A, Sertour N, Mosca K, Permal E, Laval G, Bouchier C, Ma L, Schwartz K, Voelz K, May RC, Poulain J, Battail C, Wincker P, Borman AM, Chowdhary A, Fan S, Kim SH, Le Pape P, Romeo O, Shin JH, Gabaldon T, Sherlock G, Bougnoux M-E, d’Enfert C. 2018. Gene flow contributes to diversification of the major fungal pathogen Candida albicans. Nat Commun 9:1–10. 10.1038/s41467-018-04787-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lambrechts L. 2010. Dissecting the genetic architecture of host–pathogen specificity. PLoS Pathogens 6:e1001019. 10.1371/journal.ppat.1001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Burgner D, Jamieson SE, Blackwell JM. 2006. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect Dis 6:653–663. 10.1016/S1473-3099(06)70601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rosati D, Bruno M, Jaeger M, ten Oever J, Netea MG. 2020. Recurrent vulvovaginal candidiasis: an immunological perspective. Microorganisms 8:144. 10.3390/microorganisms8020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kapitan M, Niemiec MJ, Steimle A, Frick JS, Jacobsen ID. 2019. Fungi as part of the microbiota and interactions with intestinal bacteria, p 265–301. In Rodrigues ML (ed), Fungal physiology and immunopathogenesis. Springer International Publishing, Cham, Switzerland. [DOI] [PubMed] [Google Scholar]
- 126.Kitano H. 2002. Systems biology: a brief overview. Science 295:1662–1664. 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 127.Yan J, Risacher SL, Shen L, Saykin AJ. 2018. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief Bioinform 19:1370–1381. 10.1093/bib/bbx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bunnik EM, Le Roch KG. 2013. An introduction to functional genomics and systems biology. Adv Wound Care (New Rochelle) 2:490–498. 10.1089/wound.2012.0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dix A, Vlaic S, Guthke R, Linde J. 2016. Use of systems biology to decipher host–pathogen interaction networks and predict biomarkers. Clin Microbiol Infect 22:600–606. 10.1016/j.cmi.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 130.Smeekens SP, Ng A, Kumar V, Johnson MD, Plantinga TS, van Diemen C, Arts P, Verwiel ETP, Gresnigt MS, Fransen K, van Sommeren S, Oosting M, Cheng S-C, Joosten LAB, Hoischen A, Kullberg B-J, Scott WK, Perfect JR, van der Meer JWM, Wijmenga C, Netea MG, Xavier RJ. 2013. Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nat Commun 4:1342. 10.1038/ncomms2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, Ermel R, Ruusalepp A, Quertermous T, Hao K, Björkegren JLM, Im HK, Pasaniuc B, Rivas MA, Kundaje A. 2019. Opportunities and challenges for transcriptome-wide association studies. Nat Genet 51:592–599. 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jaeger M, van der Lee R, Cheng S-C, Johnson MD, Kumar V, Ng A, Plantinga TS, Smeekens SP, Oosting M, Wang X, Barchet W, Fitzgerald K, Joosten LAB, Perfect JR, Wijmenga C, van de Veerdonk FL, Huynen MA, Xavier RJ, Kullberg BJ, Netea MG. 2015. The RIG-I-like helicase receptor MDA5 (IFIH1) is involved in the host defense against Candida infections. Eur J Clin Microbiol Infect Dise 34:963–974. 10.1007/s10096-014-2309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jaeger M, Matzaraki V, Aguirre-Gamboa R, Gresnigt MS, Chu X, Johnson MD, Oosting M, Smeekens SP, Withoff S, Jonkers I, Perfect JR, van de Veerdonk FL, Kullberg B-J, Joosten LAB, Li Y, Wijmenga C, Netea MG, Kumar V. 2019. A genome-wide functional genomics approach identifies susceptibility pathways to fungal bloodstream infection in humans. J Infect Dis 220:862–872. 10.1093/infdis/jiz206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kannambath S, Jarvis JN, Wake RM, Longley N, Loyse A, Matzaraki V, Aguirre-Gamboa R, Wijmenga C, Doyle R, Paximadis M, Tiemessen CT, Kumar V, Pittman A, Meintjes G, Harrison TS, Netea MG, Bicanic T. 2020. Genome-wide association study identifies novel colony stimulating factor 1 locus conferring susceptibility to cryptococcosis in human immunodeficiency virus-infected South Africans. Open Forum Infect Dis 7:ofaa489. 10.1093/ofid/ofaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518:317–330. 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.de Vries DH, Matzaraki V, Bakker OB, Brugge H, Westra H-J, Netea MG, Franke L, Kumar V, van der Wijst MGP. 2020. Integrating GWAS with bulk and single-cell RNA-sequencing reveals a role for LY86 in the anti-Candida host response. PLoS Pathog 16:e1008408. 10.1371/journal.ppat.1008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jaeger M, Pinelli M, Borghi M, Constantini C, Dindo M, van Emst L, Puccetti M, Pariano M, Ricaño-Ponce I, Büll C, Gresnigt MS, Wang X, Gutierrez Achury J, Jacobs CWM, Xu N, Oosting M, Arts P, Joosten LAB, van de Veerdonk FL, Veltman JA, ten Oever J, Kullberg BJ, Feng M, Adema GJ, Wijmenga C, Kumar V, Sobel J, Gilissen C, Romani L, Netea MG. 2019. A systems genomics approach identifies SIGLEC15 as a susceptibility factor in recurrent vulvovaginal candidiasis. Sci Transl Med 11:eaar3558. 10.1126/scitranslmed.aar3558. [DOI] [PubMed] [Google Scholar]
- 138.Wang L, Pittman KJ, Barker JR, Salinas RE, Stanaway IB, Williams GD, Carroll RJ, Balmat T, Ingham A, Gopalakrishnan AM, Gibbs KD, Antonia AL, Heitman J, Lee SC, Jarvik GP, Denny JC, Horner SM, DeLong MR, Valdivia RH, Crosslin DR, Ko DC. 2018. An atlas of genetic variation linking pathogen-induced cellular traits to human disease. Cell Host Microbe 24:308.e6–323.e6. 10.1016/j.chom.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yu K, Chen B, Aran D, Charalel J, Yau C, Wolf DM, van ‘t Veer LJ, Butte AJ, Goldstein T, Sirota M. 2019. Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat Commun 10:3574. 10.1038/s41467-019-11415-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Radaelli E, Santagostino SF, Sellers RS, Brayton CF. 2018. Immune relevant and immune deficient mice: options and opportunities in translational research. ILAR J 59:211–246. 10.1093/ilar/ily026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Alvarez-Rueda N, Rouges C, Touahri A, Misme-Aucouturier B, Albassier M, Pape PL. 2020. In vitro immune responses of human PBMCs against Candida albicans reveals fungal and leucocyte phenotypes associated with fungal persistence. Sci Rep 10:6211. 10.1038/s41598-020-63344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rizzetto L, Giovannini G, Bromley M, Bowyer P, Romani L, Cavalieri D. 2013. Strain dependent variation of immune responses to A. fumigatus: definition of pathogenic species. PLoS One 8:e56651. 10.1371/journal.pone.0056651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Whittaker JC, Sanseau P. 2015. The support of human genetic evidence for approved drug indications. Nat Genet 47:856–860. 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 144.Lupiañez CB, Canet LM, Carvalho A, Alcazar-Fuoli L, Springer J, Lackner M, Segura-Catena J, Comino A, Olmedo C, Ríos R, Fernández-Montoya A, Cuenca-Estrella M, Solano C, López-Nevot MÁ, Cunha C, Oliveira-Coelho A, Villaescusa T, Fianchi L, Aguado JM, Pagano L, López-Fernández E, Potenza L, Luppi M, Lass-Flörl C, Loeffler J, Einsele H, Vazquez L, PCRAGA Study Group, Jurado M, Sainz J. 2015. Polymorphisms in host immunity-modulating genes and risk of invasive aspergillosis: results from the AspBIOmics Consortium. Infect Immun 84:643–657. 10.1128/IAI.01359-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349. 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chibucos MC, Soliman S, Gebremariam T, Lee H, Daugherty S, Orvis J, Shetty AC, Crabtree J, Hazen TH, Etienne KA, Kumari P, O’Connor TD, Rasko DA, Filler SG, Fraser CM, Lockhart SR, Skory CD, Ibrahim AS, Bruno VM. 2016. An integrated genomic and transcriptomic survey of mucormycosis-causing fungi. Nat Commun 7:12218. 10.1038/ncomms12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Muñoz JF, Delorey T, Ford CB, Li BY, Thompson DA, Rao RP, Cuomo CA. 2019. Coordinated host-pathogen transcriptional dynamics revealed using sorted subpopulations and single macrophages infected with Candida albicans. Nat Commun 10:1607. 10.1038/s41467-019-09599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845–1848. 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kwak H, Fuda NJ, Core LJ, Lis JT. 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339:950–953. 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Khodor YL, Rodriguez J, Abruzzi KC, Tang C-HA, Marr MT, Rosbash M. 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25:2502–2512. 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fullwood MJ, Liu MH, Pan YF, Liu J, Han X, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EGY, Huang PYH, Welboren W-J, Han Y, Ooi H-S, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KDSA, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RKM, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung W-K, Liu ET, Wei C-L, Cheung E, Ruan Y. 2009. An oestrogen receptor α-bound human chromatin interactome. Nature 462:58–64. 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. 2009. Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science 326:289–293. 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680. 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. 2006. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 16:1299–1309. 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, Zhou D, Luo S, Vasicek TJ, Daly MJ, Wolfsberg TG, Collins FS. 2006. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res 16:123–131. 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Buenrostro J, Wu B, Chang H, Greenleaf W. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109:21.29.1–21.29.9. 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316:1497–1502. 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 158.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452:215–219. 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770. 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Borman AM, Petch R, Linton CJ, Palmer MD, Bridge PD, Johnson EM. 2008. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J Clin Microbiol 46:933–938. 10.1128/JCM.02116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kitahara N, Morisaka H, Aoki W, Takeda Y, Shibasaki S, Kuroda K, Ueda M. 2015. Description of the interaction between Candida albicans and macrophages by mixed and quantitative proteome analysis without isolation. AMB Express 5:41. 10.1186/s13568-015-0127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]