ABSTRACT
Salmonella enterica Typhimurium is a rod-shaped Gram-negative bacterium that mostly enters the human body through contaminated food. It causes a gastrointestinal disorder called salmonellosis in humans and typhoid-like systemic disease in mice. OmpV, an outer membrane protein of S. Typhimurium, helps in adhesion and invasion of bacteria to intestinal epithelial cells and thus plays a vital role in the pathogenesis of S. Typhimurium. In this study, we have shown that intraperitoneal immunization with OmpV is able to induce high IgG production and protection against systemic disease. Further, oral immunization with OmpV-incorporated proteoliposome (OmpV-proteoliposome [PL]) induces production of high IgA antibody levels and protection against gastrointestinal infection. Furthermore, we have shown that OmpV induces Th1 bias in systemic immunization with purified OmpV, but both Th1 and Th2 polarization in oral immunization with OmpV-proteoliposome (PL). Additionally, we have shown that OmpV activates innate immune cells, such as monocytes, macrophages, and intestinal epithelial cells, in a Toll-like receptor 2 (TLR2)-dependent manner. Interestingly, OmpV is recognized by the TLR1/2 heterodimer in monocytes, but by both TLR1/2 and TLR2/6 heterodimers in macrophages and intestinal epithelial cells. Further, downstream signaling involves MyD88, interleukin-1 receptor-associated kinase (IRAK)-1, mitogen-activated protein kinase (MAPK) (both p38 and Jun N-terminal protein kinase (JNK)), and transcription factors NF-κB and AP-1. Due to its ability to efficiently activate both the innate and adaptive immune systems and protective efficacy, OmpV can be a potential vaccine candidate against S. Typhimurium infection. Further, the fact that OmpV can be recognized by both TLR1/2 and TLR2/6 heterodimers increases its potential to act as good adjuvant in other vaccine formulations.
KEYWORDS: OmpV, Salmonella, immune mechanisms
INTRODUCTION
Salmonella enterica Typhimurium and Salmonella enterica Enteritidis are two major nontyphoidal serovars of Salmonella that cause a gastroenteritis disorder called salmonellosis (1, 2). Salmonellosis is a major problem in developing countries, as it severely affects elderly people, younger children, and immunocompromised individuals, such as HIV patients (3). It is reported that S. Typhimurium contributes to 50% of salmonellosis cases in humans (4). The Global Burden of Diseases, Injuries, and Risk Factors Study (GBD) 2017 reported 95.1 million cases and 50,771 deaths worldwide due to salmonellosis in 2017 (5).
To date, no vaccine is available for S. Typhimurium or S. Enteritidis, and, thus, antibiotics are the only mode of treatment available for salmonellosis; but, the emergence of multidrug-resistant strains has complicated the treatment of this disease, leading to frequent outbreaks. Recently, a highly invasive strain of S. Typhimurium, ST313, has emerged that shows resistance to multiple antimicrobial agents, including those recommended as first-line treatment (6). Also, in Malawi, around 90% of strains causing salmonellosis are multidrug resistant (7). Thus, new vaccines against S. Typhimurium can potentially impact global health.
Outer membrane proteins (Omp) are considered the major candidates for vaccines or adjuvants, as they can be recognized by the host immune system to initiate innate and adaptive immune responses (8). Some outer membrane proteins of S. Typhimurium were evaluated for vaccine potential against salmonellosis. One such protein is OmpL, a transmembrane beta-barrel protein that showed protection against Salmonella infection in a mouse model (9). Apart from this, OmpF and OmpC from S. Typhi are also reported as potential candidates for a subunit vaccine against S. Typhimurium (3, 10). Another outer membrane protein, OmpA from S. Typhimurium, is known to activate dendritic cells (DCs) and induces Th1 polarization (11).
OmpV belongs to the MipA family of proteins and is documented as an outer membrane protein in various Gram-negative bacteria, such as S. Typhimurium (NP_460252.1 from the NCBI protein database), Vibrio cholerae (12), Vibrio parahaemolyticus (13), S. Paratyphi (14), Pasteurella multo ci da (15), Photobacterium damselae (16), etc. In our laboratory we wanted to characterize the role of S. Typhimurium OmpV toward pathogenesis and host immunomodulation.
S. Typhimurium enters the human body through contaminated food and water. Upon reaching the intestine, it adheres and invades intestinal epithelial cells (IECs). In an earlier study, we reported that OmpV, an outer membrane protein of S. Typhimurium, is a crucial adhesin and helps in adhesion of the bacteria to intestinal epithelial cells. Further, we have shown that deletion of OmpV can significantly affect the pathogenesis of S. Typhimurium (17).
As OmpV plays a key role in the virulence of S. Typhimurium, in this report, we did immunological characterization and investigated the protective efficacy of OmpV with the notion that OmpV might act as a potential vaccine candidate.
Our observations indicated that OmpV can trigger innate as well as adaptive immune responses, leading to T cell activation and antibody production. Further, we observed that OmpV-induced immune responses can protect mice from S. Typhimurium infection. Thus, OmpV can play an important role in subunit vaccine development for S. Typhimurium infection along with other candidates.
RESULTS
OmpV induces protective immunity against S. Typhimurium infection.
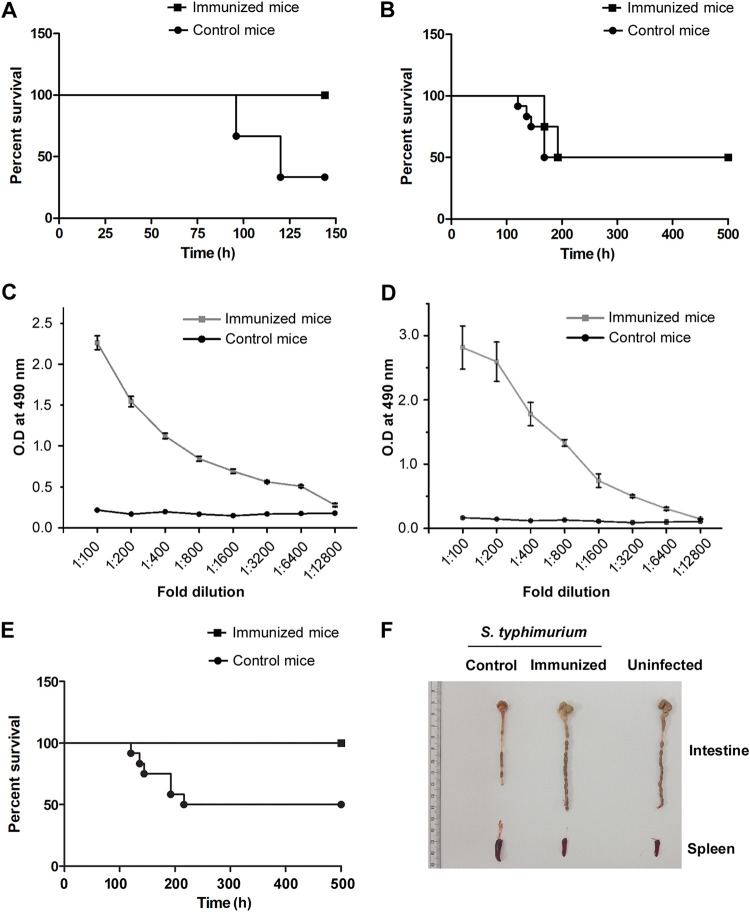
To probe whether immunization with OmpV, a crucial adhesion factor of S. Typhimurium, can generate protection against S. Typhimurium infection in mice, we first immunized mice by intraperitoneal administration of 4 doses of OmpV (25 μg/dose/mouse) at an interval of 7 days each. Two weeks following completion of the immunization schedule, mice were challenged with virulent S. Typhimurium through the intraperitoneal route and oral route to check protection against systemic as well as gastrointestinal infection. In cases of systemic infection with S. Typhimurium, we observed 100% survival (Fig. 1A), but no protection was obtained in cases of oral challenge (Fig. 1B). Further, we assessed the IgG in serum and IgA in stools of immunized mice and detected high IgG titers (Fig. 1C) but could not detect any IgA.
FIG 1.
OmpV induces protective immunity against S. Typhimurium infection. (A, B) Intraperitoneal immunization of mice with purified OmpV protects against systemic infection (A) but does not protect against gastrointestinal infection (B) of S. Typhimurium. Mice were immunized intraperitoneally at an interval of 7 days. Buffer was used for the control mice. Following 14 days after the last dose, mice were challenged with S. Typhimurium SL1344 intraperitoneally (A) or through the oral route (B), and survival was monitored until day 7 (A) or day 21 (B). (C, D) A high titer of IgG (C) and IgA (D) was observed in the serum of mice immunized intraperitoneally with OmpV (C) and in the stools of mice immunized orally with OmpV-proteoliposome (PL) (D). Mice were immunized intraperitoneally with four doses of OmpV, and buffer was used for the control mice (C). Mice were immunized with four doses of OmpV-proteoliposome (PL), and liposomes were used for control mice (D). Following 14 days after the last dose of immunization, serum and stool samples were obtained from the immunized and control mice (n = 3 mice/group, 3 biological replicates). (E) Oral immunization of mice with OmpV-proteoliposome (PL) protects against gastrointestinal infection of S. Typhimurium. Mice were immunized with OmpV-proteoliposome (PL). Following the last dose, mice were given gastrointestinal infection with oral challenge of S. Typhimurium SL1344, and survival was monitored. Mice immunized with liposomes were taken as controls. For panels A, B, and E, n = 12 mice/group (4 mice per group per experiment were taken, and 3 biological replicates were done). A Kaplan-Meier plot of cumulative mortality was prepared to compare the survival rate. (F) Similar intestinal length was observed in the naive noninfected mice as well as the infected mice immunized with OmpV-proteoliposome (PL). Mice were immunized with OmpV-proteoliposome (PL), and, after the last dose, mice were given gastrointestinal infection with S. Typhimurium SL1344. After 5 days of infection, the intestines and spleens of these mice were removed, observed, and matched with noninfected naive mice (n = 3 mice/group).
Further, to probe whether oral immunization can induce IgA production against OmpV, as well as protection against oral challenge with virulent bacteria, 4 doses of purified OmpV protein (25 μg/dose/mouse) were orally administered a week apart. Following the oral immunization schedule, we assessed the stool for IgA production and checked for protection against oral challenge with the virulent bacteria. However, even in cases of oral immunization with purified OmpV, we could not detect any IgA, and no protection was observed against oral challenge. This led us to believe that the acidic pH of the stomach and proteolytic enzymes in the gut might degrade the purified protein. Also, degradation of purified OmpV protein was observed following treatment with proteinase K, whereas the protein becomes resistant to proteinase K and remains intact when present in the outer membrane of S. Typhimurium. Therefore, to avoid degradation of the protein during oral immunization, we prepared proteoliposomes by incorporating OmpV into liposomes (OmpV-proteoliposome [PL]) with the idea that, in addition to protection, the liposome could act as an adjuvant (18). Following immunization with OmpV-proteoliposome (PL) (25 μg/dose/mouse) for four doses at an interval of 7 days, we observed a high IgA titer in stools (Fig. 1D). Further, we observed 100% survival of the immunized mice against oral challenge with S. Typhimurium (Fig. 1E). Furthermore, when we measured the intestinal length, 5 days following oral infection, we observed a significant shrinkage in intestinal length in liposome-immunized mice (control) indicating a higher level of infection, whereas the proteoliposome (PL)-immunized mice showed similar intestinal length as the uninfected mice (Fig. 1F).
The above data suggested that immunization of mice with OmpV and OmpV-proteoliposome (PL) can induce IgG and IgA antibody production, respectively, and thus provide protection against systemic and oral challenge.
OmpV induces dendritic cell maturation and Th1 differentiation.
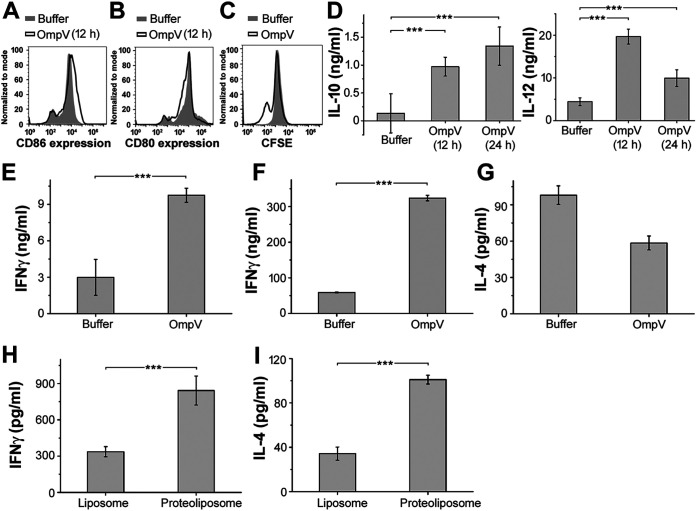
CD4+ T cells or T helper (Th1 and Th2) cells are instrumental to high-affinity antibody production and B cell class switching (19). Dendritic cells (DCs) act as the professional antigen-presenting cell for T cell activation and Th polarization. For T cell activation and differentiation, DCs mainly provide 3 signals. Out of these, signal 1 is provided by interaction of peptide-major histocompatibility complex (MHC) and T cell receptor, and signal 2 is provided by the interaction of costimulatory molecules (CD80, CD86) with CD28 on the T cell. Signal 3 is provided by the cytokines produced by the activated DCs or cytokines present in the milieu secreted by other immune cells in the microenvironment (20). With activation via its innate receptor, a DC undergoes maturation and becomes efficient in terms of antigen presentation. One of the markers for DC maturation is increased expression of costimulatory molecules. To assess, whether OmpV can induce DC maturation, we treated bone marrow-derived dendritic cells (BMDCs) with OmpV and checked for surface expression of CD80 and CD86. We observed an increase in the expression of CD86 on the surface of OmpV-treated BMDCs (Fig. 2A), but no increase was observed in CD80 expression, probably due to the high basal level of CD80 expression on the surface of BMDCs (Fig. 2B). Further, we wanted to check whether OmpV-primed DCs can lead to proliferation of T cells. Towards this, we performed a T cell proliferation assay. We observed dilution in carboxyfluorescein diacetate succinimidyl ester (CFSE) fluorescence, indicating proliferation of T cells (Fig. 2C).
FIG 2.
OmpV induces dendritic cell maturation and Th cell differentiation. (A) Increased expression of CD86 in BMDCs activated with OmpV compared to the control. (B) No increase in CD80 expression in OmpV-treated BMDCs compared to the control. (A, B) BMDCs were pretreated with polymyxin B (PmB) followed by treatment with OmpV. Following incubations, the surface expression of costimulatory molecules CD86 and CD80 were analyzed by flow cytometry. (C) OmpV induces proliferation of Th cells as indicated by a decrease in CFSE fluorescence. CFSE-labeled CD4+ T cells were cocultured with splenic DCs targeted with OmpV. At day 3, CFSE fluorescence was detected using flow cytometry. CFSE-labeled CD4+ T cells treated with buffer were used as controls. (A to C) Histograms are representative of three independent experiments. (D) OmpV leads to the production of a high IL-12/IL-10 ratio. BMDCs were treated with PmB followed by OmpV. Following incubations, the supernatants were collected and estimated for IL-10 and IL-12 cytokines using an ELISA. (E) OmpV leads to the production of high levels of IFN-γ. CD4+ T cells were cocultured with OmpV-targeted splenic DCs for 3 days. The collected supernatant was probed for the presence of IFN-γ using an ELISA. (D, E) Bar graphs are expressed as mean ± standard error of the mean (SEM) from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 versus the buffer-treated cells). (F, G) Treatment with purified OmpV leads to the production of high levels of IFN-γ in vivo. (H, I) OmpV-proteoliposome (PL) treatment leads to the production of both IFN-γ and IL-4 in vivo. OmpV or buffer was administered to mice intraperitoneally (4 doses a week apart) (F, G). Mice were immunized orally with four doses of OmpV-proteoliposome (PL)/liposome (H, I). Following 14 days after the last dose, cells were isolated from peripheral lymph nodes (F, G) or mesenteric lymph nodes (H, I) and activated with PMA and ionomycin. The supernatants were collected, and IFN-γ and IL-4 were quantified by ELISA. Bar graphs are expressed as mean ± SEM from three independent experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 versus the buffer-treated mice (F, G) or mice immunized with liposome (H, I).
As previously mentioned, T cell differentiation depends on cytokines secreted by antigen-presenting cells or the presence of cytokines in the microenvironment (21). Towards this, we wanted to check whether OmpV-activated DCs can produce interleukin-12 (IL-12) or IL-10, which will impact Th1, Th2, or regulatory T cell (Treg) differentiation. Towards this, we observed that OmpV-primed DCs secrete high levels of IL-12 compared to IL-10 (IL-12/IL-10 ratio around 20:1), indicating differentiation of T helper cells might happen toward the Th1 phenotype (Fig. 2D). To further confirm whether OmpV induces polarization of T helper cells toward the Th1 phenotype, we analyzed cytokines released by T cells cocultured with OmpV-targeted DCs. We observed a significant production of interferon gamma (IFN-γ) in the supernatant, confirming polarization of activated T cells toward Th1 (Fig. 2E).
Because other cell types also contribute to cytokine production in the milieu in vivo, we wanted to check whether under in vivo conditions OmpV induces Th polarization toward Th1 as well. Towards this, we administered 4 doses of purified OmpV in mice intraperitoneally. Following immunization, T cells were isolated from inguinal, popliteal, and mesenteric lymph nodes and stimulated using phorbol myristate acetate (PMA) and ionomycin. We observed high production of IFN-γ, confirming polarization of Th cells toward the Th1 phenotype (Fig. 2F and G). Further, after oral administration of OmpV-proteoliposome (PL), T cells were isolated from mesenteric lymph nodes and assessed for the production of IFN-γ and IL-4 upon activation with PMA and ionomycin. We observed production of more IFN-γ and less IL-4, suggesting Th differentiation toward both Th1 and Th2 phenotypes with more Th1 bias (Fig. 2H and I).
OmpV can activate innate immune cells, leading to the production of proinflammatory mediators.
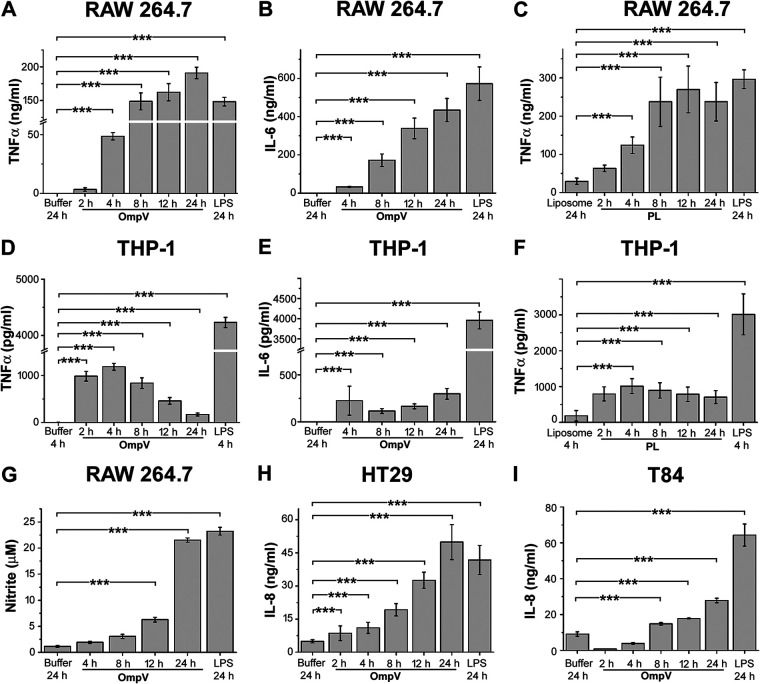
As innate immune responses are crucial in shaping adaptive immunity, we further investigated the innate immune responses involved in OmpV signaling. Towards this, we at first treated RAW 264.7 (murine macrophages), THP-1 (human monocytes), HT29, and T84 (human intestinal epithelial cells or IECs) cells with different doses of OmpV or OmpV-proteoliposome (PL) and incubated for 24 h. We observed significant production of various proinflammatory cytokines and chemokines in response to different doses of OmpV or OmpV-proteoliposome (PL) (see Fig. S4A to I in the supplemental material). In both RAW 264.7 macrophages (Fig. S4A to C) and THP-1 monocytes (Fig. S4D to F), considerable production of tumor necrosis factor alpha (TNF-α) and IL-6 was observed in response to 2 μg/ml of OmpV or OmpV-proteoliposome (PL), and a plateau was observed in proinflammatory cytokine release in macrophages and monocytes with an increase in OmpV/OmpV-proteoliposome (PL) doses beyond 2 μg/ml (Fig. S4A to D and F) except for IL-6 production by THP-1 cells (Fig. S4E). Further, we observed that IL-8 was significantly produced by HT29 cells (Fig. S4G) and T84 IECs (Fig. S4H) in response to different doses of OmpV, and there was significant release of IL-8 at a dose of 2 μg/ml, beyond which there was either a decrease (Fig. S4G) or a plateau (Fig. S4H) in IL-8 production. Apart from cytokines, another major sign of inflammatory responses by macrophages is nitric oxide (NO) production (22). So, we checked NO production in response to different doses of OmpV. Maximum NO production was observed at a concentration of 2 μg/ml OmpV (Fig. S4I). Further, we assessed cell viability by MTT (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) assay with different doses of OmpV following incubation for 24 h (Fig. S4J to M). We observed a minimum effect on cell viability with an increase in dosage in all the different cell types used, except in RAW 264.7 macrophages where a dose-dependent decrease in cell viability was observed (Fig. S4J). As significant production of proinflammatory responses with no significant decrease in cell viability was observed in all the cell types in response to 2 μg/ml OmpV, we therefore further used this concentration for time course studies and other studies.
For time course studies, we treated RAW 264.7 macrophages, THP-1 monocytes, and T84 and HT29 IECs with 2 μg/ml of OmpV or OmpV-proteoliposome (PL) and incubated them for different time periods. After respective incubations, the supernatants were collected and analyzed for the presence of proinflammatory mediators (Fig. 3A to I). We assessed TNF-α and IL-6 production by RAW 264.7 macrophages and THP-1 monocytes and observed a time-dependent increase of both TNF-α and IL-6 in response to OmpV until 24 h in RAW 264.7 macrophages (Fig. 3A and B), whereas OmpV-proteoliposome (PL) treatment led to maximum TNF-α production at 12 h (Fig. 3C). In THP-1 monocytes, maximum release of TNF-α was observed at 4 h (Fig. 3D and F) in response to both OmpV and OmpV-proteoliposome (PL), and IL-6 was maximally produced at 24 h (Fig. 3E) in response to OmpV treatment. Further, nitrite production in RAW 264.7 macrophages was assessed at different time points upon treatment of OmpV, and maximum generation was seen at 24 h (Fig. 3G). Furthermore, we assessed IL-8 production in both HT29 and T84 cells in response to OmpV at different time points, and maximum release was observed at 24 h (Fig. 3H and I). Lipopolysaccharide (LPS; 1 μg/ml) was used as a positive control for these experiments.
FIG 3.
Time-dependent analysis for the production of proinflammatory mediators in RAW 264.7 macrophages, THP-1 monocytes, and IECs. (A to C) Time-dependent increase in TNF-α and IL-6 production was observed in RAW 264.7 macrophages. (D to F) In THP-1 monocytes, TNF-α was maximally produced at 4 h and IL-6 at 24 h. (G) A time-dependent increase in nitric oxide production was observed in RAW 264.7 macrophages. (H, I) A time-dependent increase in IL-8 production was observed in HT29 (H) and T84 cells (I). (A to I) Cells were treated with PmB followed by OmpV or OmpV-proteoliposome (PL) and incubated for different time periods as indicated. Following respective incubations, supernatants were collected and analyzed for cytokines (A to F and H and I) or nitric oxide (G). LPS was used as a positive control. Bar graphs are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 versus the buffer-treated or liposome-treated cells).
OmpV induces activation of innate immune cells via Toll-like receptor 2.
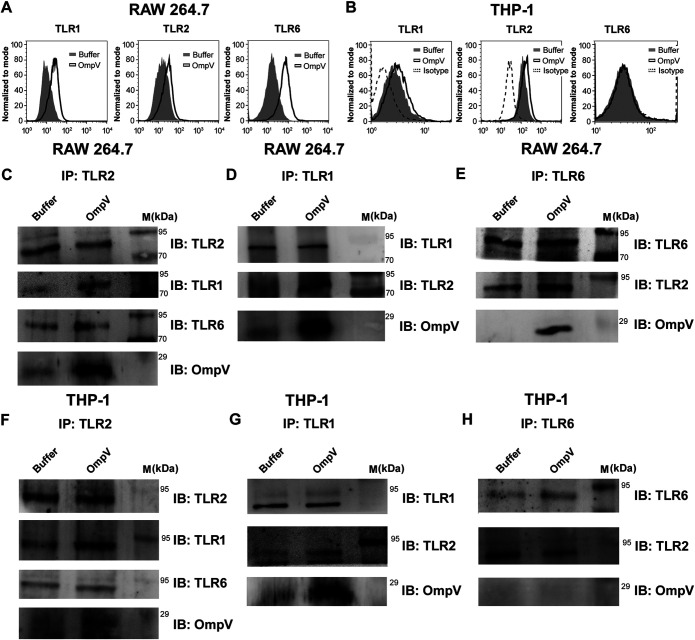
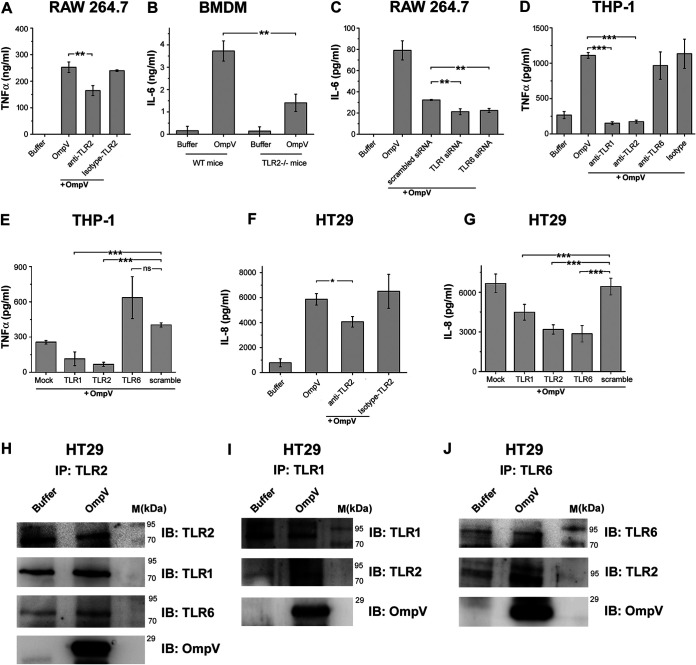
Generally, for the production of proinflammatory mediators, pathogen-associated molecular patterns (PAMPs) are recognized by pattern recognition receptors (PRRs) present on immune cells. As OmpV induces proinflammatory cytokine production, then certainly it acted as a PAMP and was recognized by PRRs. Therefore, we wanted to check which PRR is involved in recognition of OmpV. Being an outer membrane protein, OmpV would probably come into contact with surface PRRs at first. The majority of surface PRRs include Toll-like receptors (TLRs), out of which TLR1, TLR2, TLR4, and TLR6 are mostly known to recognize bacterial ligands (23). Therefore, we first checked whether there was any increase in surface expression of these TLRs in response to OmpV treatment. We observed an increase in surface expression of TLR1, TLR2, and TLR6 in OmpV-activated RAW 264.7 macrophages (Fig. 4A and Fig. S5), whereas in OmpV-activated THP-1 monocytes, we observed an increase in surface expression of only TLR1 and TLR2 (Fig. 4B and Fig. S5). Therefore, our observations indicated that TLR1, TLR2, and TLR6 in RAW 264.7 macrophages and TLR1 and TLR2 in THP-1 monocytes could be involved in the recognition of OmpV. Also, we observed involvement of both TLR2 and TLR6 in OmpV recognition in macrophages differentiated from THP-1 monocytes (data not shown).
FIG 4.
OmpV induces increases in surface expression of TLRs and coimmunoprecipitates with TLR2, TLR1, and TLR6 in macrophages and TLR1 and TLR2 in monocytes. (A, B) OmpV induces increased surface expression of TLR1, TLR2, and TLR6 on RAW 264.7 macrophages (A) and TLR1 and TLR2 on THP-1 monocytes (B). Cells were treated with PmB followed by OmpV. Following incubations, the surface expression of TLRs was analyzed using flow cytometry. Histograms are representatives of three independent experiments. (C to H) OmpV coimmunoprecipitates with TLR2, TLR1, and TLR6 in RAW 264.7 macrophages (C to E), whereas it only coimmunoprecipitates with TLR1 and TLR2 but not with TLR6 in THP-1 monocytes (F to H). Cells were treated with PmB followed by OmpV or buffer, and whole-cell lysates were prepared following incubation. Further, lysates were immunoprecipitated with anti-TLR2/anti-TLR-1/anti-TLR6 antibody and checked for the presence of TLR1, TLR2, TLR6, and OmpV. Buffer-treated cells were used as controls for coimmunoprecipitation experiments. IP indicates the antibody used for immunoprecipitation, whereas IB indicates the antibody used for immunoblotting. Western blots are representative of three independent experiments.
Further, we observed that in RAW 264.7 macrophages, OmpV coimmunoprecipitated with TLR2, TLR1, and TLR6 (Fig. 4C to E), whereas in OmpV-activated THP-1 monocytes, OmpV coimmunoprecipitated only with TLR2 and TLR1 but not with TLR6 (Fig. 4F to H). To check any aggregation of protein during immunoprecipitation, isotype controls were used (data not shown). Similar to the surface expression data, the coimmunoprecipitation study strongly suggested that TLR2 might be involved in OmpV recognition by innate immune cells. Moreover, as it was known from the literature that TLR2 generally heterodimerizes either with TLR1 or TLR6, our study indicated that in response to OmpV, TLR2 probably heterodimerizes with both TLR1 and TLR6 in macrophages but only with TLR1 in monocytes.
For further confirmation of the role of TLR2 in macrophages, we used neutralizing antibody against TLR2 in RAW 264.7 cells and bone marrow-derived macrophages (BMDM) from TLR2−/− mice. In both cases, we observed a significant decrease in cytokine production (Fig. 5A and B), confirming involvement of TLR2 in OmpV recognition in macrophages. Further, to confirm the dimerizing partners of TLR2 in recognition of OmpV in macrophages, we knocked down TLR1 and TLR6 with small interfering RNA (siRNA) in RAW 264.7 cells and activated with OmpV. Following activation, we observed a significant decrease in the proinflammatory signal in both TLR1 as well as TLR6 knockdown cells compared to signal observed in the controls, confirming involvement of both TLR1 and TLR6 in the recognition of OmpV along with TLR2 in macrophages (Fig. 5C). Knockdown of TLR1 and TLR6 was confirmed by checking surface expression using flow cytometry (Fig. S1). These data suggested that both TLR1/TLR2 and TLR2/TLR6 heterodimers are involved in recognition of OmpV in macrophages.
FIG 5.
In macrophages and IECs, OmpV is recognized by both TLR1/TLR2 and TLR2/TLR6 heterodimers, whereas in monocytes, OmpV is recognized only by TLR1/TLR2 heterodimers. (A) Neutralization of TLR2 indicated its involvement in recognition of OmpV in RAW 264.7 macrophages. Cells were pretreated with anti-TLR2 neutralizing antibody followed by PmB and OmpV. Following incubation, supernatants were analyzed for TNF-α production. (B) BMDMs from mice deficient in TLR2 (TLR2−/− mice) confirmed involvement of TLR2 in recognition of OmpV in macrophages. BMDMs from wild-type mice and TLR2−/− mice were treated with PmB followed by OmpV for 24 h. Supernatants were collected and analyzed for IL-6 production. (C) A significant decrease in production of IL-6 was observed upon knockdown of TLR1 and TLR6 in macrophages. RAW 264.7 cells were transfected with siRNA followed by treatment with PmB and OmpV. Following incubation, supernatants were collected and quantified for IL-6 by ELISA. Nontargeted (scrambled) siRNA was used as a negative control. (D) A neutralization experiment indicated the involvement of TLR1 and TLR2 in recognition of OmpV in monocytes. THP-1 cells were pretreated with neutralizing antibodies followed by PmB and OmpV. Supernatants were collected following incubation and analyzed for TNF-α production. (E) A significant decrease in production of TNF-α was observed upon knockdown of TLR1 and TLR2 but not upon knockdown of TLR6 in monocytes. THP-1 cells were transfected with shRNA followed by treatment with PmB and OmpV, and supernatants were collected after incubation and checked for the presence of TNF-α. Nontargeted (scrambled) shRNA was used as a negative control. (F) Neutralization of TLR2 indicates its involvement in recognition of OmpV in IECs. HT29 cells were pretreated with neutralizing antibody for 1 h followed by PmB and OmpV. The supernatants were collected and analyzed for IL-8 production. (G) A significant decrease in IL-8 production was observed upon knockdown of TLR1, TLR2, and TLR6 in IECs. HT29 cells were transfected with shRNA and were then treated with PmB followed by OmpV. Following incubation, supernatants were collected, and IL-8 was quantified by ELISA. Nontargeted (scrambled) shRNA was used as a negative control. (A to G) Bar graphs are expressed as mean ± SEM from three independent experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 versus only OmpV-treated cells (A, D, F), versus the OmpV-treated BMDMs from wild-type mice (B), versus scrambled siRNA-transfected OmpV-treated cells (C), or versus scrambled shRNA-transfected OmpV-treated cells (E, G). (H to J) OmpV coimmunoprecipitates with TLR1, TLR2, and TLR6 in IECs. HT29 cells were treated with PmB followed by OmpV. Following incubation, whole-cell lysates were prepared. These lysates were immunoprecipitated with anti-TLR2 (H), anti-TLR1 (I), or anti-TLR6 (J) antibody and checked for the presence of TLR1, TLR2, TLR6, and OmpV by immunoblotting. Buffer-treated cells were used as controls for coimmunoprecipitation experiments. IP indicates the antibody used for immunoprecipitation, whereas IB indicates the antibody used for immunoblotting. Western blots are representatives of three independent experiments.
Similarly, using neutralizing antibodies against TLR1, TLR2, and TLR6 in THP-1 monocytes, we observed that, along with TLR2, only TLR1 is involved in recognition of OmpV, and TLR6 is not involved (Fig. 5D). These data, along with surface expression (Fig. 4B) and coimmunoprecipitation data (Fig. 4F to H), indicated that in monocytes, OmpV is recognized probably by TLR1/TLR2 heterodimers, and there is no involvement of TLR2/TLR6 heterodimers. Further confirmation was achieved using short hairpin RNA (shRNA)-mediated knockdown of TLR1, TLR2, and TLR6 in THP-1 monocytes (Fig. 5E). Knockdown of TLRs was observed and quantified using Western blotting and densitometry (Fig. S2).
In neutralization studies in IECs, shRNA-mediated knockdown and coimmunoprecipitation experiments confirmed involvement of TLR2 in recognition of OmpV (Fig. 5F to J). Knockdown of TLRs was quantified using semiquantitative real-time PCR (RT-PCR) (Fig. S3). Further, coimmunoprecipitation studies suggested that in IECs, TLR2 probably forms heterodimers with both TLR1 and TLR6, which is similar to macrophages.
The above observations in macrophages, monocytes, and intestinal epithelial cells confirmed that both TLR1 and TLR2 are mainly responsible for activation of innate immunity against OmpV.
Further, we also checked for the presence of contaminants like peptidoglycan, lipoproteins, and LPS in OmpV protein as they can activate TLR2. For this, we used a chemical inhibitor against the peptidoglycan-activated nucleotide-binding oligomerization domain-like (NOD) signaling pathway (i.e., Gefitinib). The results indicated an absence of peptidoglycan contamination, as no decrease in cytokine production was observed on pretreatment with inhibitor (Fig. S9). Furthermore, intact mass spectra of protein indicated an absence of any lipoprotein contamination (data not shown). We also checked protein for LPS contamination using a Limulus amebocyte lysate (LAL) assay, and only an insignificant amount of LPS was observed.
Downstream signaling of TLR activation by OmpV in innate immune cells involves MyD88 and IRAK-1.
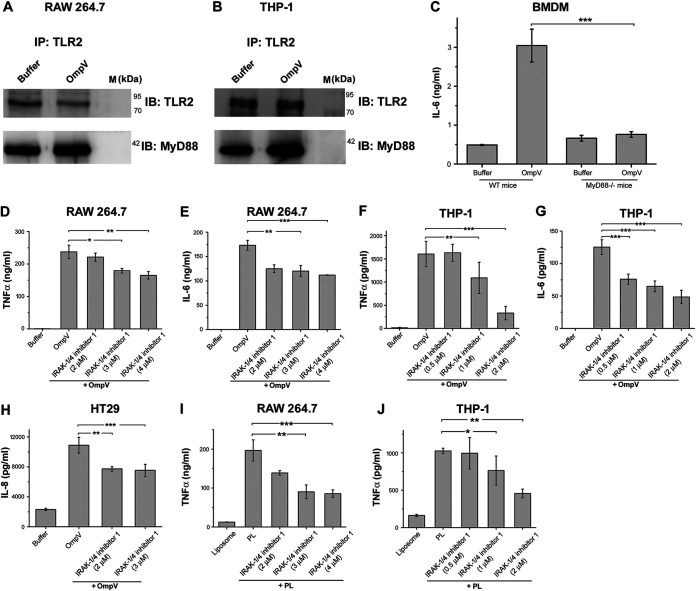
MyD88 is one of the major adaptor molecules recruited to the cytoplasmic tail of TLR2 (24). However, TLR2 could induce its signaling via MyD88-independent pathways as well (25, 26); therefore, we checked whether MyD88 is involved in OmpV-mediated signaling. We observed that MyD88 coimmunoprecipitated with TLR2 in both OmpV-activated macrophages and monocytes (Fig. 6A and B). Further, the involvement of MyD88 was confirmed using bone marrow-derived macrophages (BMDMs) from MyD88−/− mice (Fig. 6C). For further signaling, MyD88 recruits the IRAK-1/4 complex, and activated IRAK-1 carries forward downstream signaling (27). Therefore, to investigate the involvement of IRAK-1, we used an IRAK-1/4 inhibitor and observed a significant decrease in TNF-α and IL-6 production in both macrophages and monocytes following OmpV activation in cells pretreated with the IRAK-1 inhibitor (Fig. 6D to G) compared with levels observed in the control-treated cells. Furthermore, in intestinal epithelial HT29 cells, we observed a significant decrease in IL-8 production with prior inhibition of IRAK-1 compared to the control cells (Fig. 6H). These results confirmed the involvement of both MyD88 and IRAK-1 in OmpV-mediated induction of innate immune responses. We observed a similar pattern upon OmpV-proteoliposome (PL) treatment (Fig. 6I and J).
FIG 6.
MyD88 and IRAK are involved in OmpV-mediated signaling in macrophages, monocytes, and IECs. (A, B) Increased association of MyD88 with TLR2 in OmpV-activated macrophages and monocytes. Cell lysates of OmpV-treated RAW 264.7 macrophages (A) and THP-1 monocytes (B) were immunoprecipitated with anti-TLR2 antibody and checked for the presence of MyD88. Buffer-treated cells were used as controls. IP indicates the antibody used for immunoprecipitation, whereas IB indicates the antibody used for immunoblotting. Western blots are representatives of three independent experiments. (C) A significant decrease in proinflammatory cytokine production was observed in macrophages under MyD88-deficient conditions. BMDMs from wild-type and MyD88−/− mice were treated with PmB followed by OmpV. Following incubation, supernatants were collected and analyzed for IL-6 production. (D to G) A significant decrease in proinflammatory cytokine production was observed with inhibition of IRAK-1/4 in macrophages and monocytes. (H) A significant decrease in IL-8 was observed upon inhibition of IRAK-1/4 in IECs. (I, J) IRAK is involved in OmpV-proteoliposome (PL)-mediated signaling. (D to J) RAW 264.7 macrophages, THP-1 monocytes, or HT29 cells were pretreated with IRAK-1/4 inhibitor followed by treatment with PmB and OmpV or OmpV-proteoliposome (PL). Following incubations, supernatants were collected and analyzed for cytokine production by ELISA. Bar graphs are expressed as mean ± SEM from three independent experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 versus the OmpV-activated BMDMs from wild-type mice (C), versus only OmpV-treated cells (D to H), or versus only OmpV-proteoliposome (PL)-treated cells (I, J).
Both NF-κB and AP-1 transcription factors are involved in OmpV-mediated activation of innate immune responses.
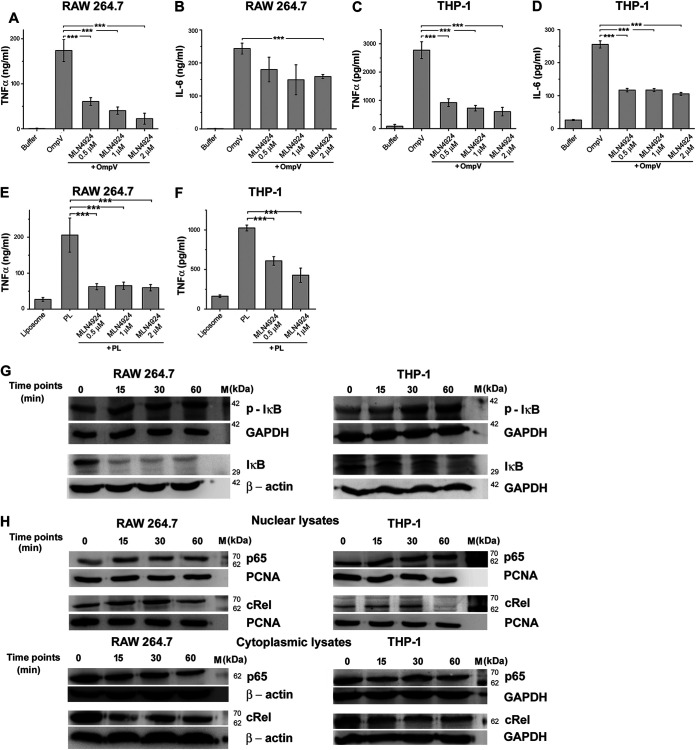
Downstream signaling of PAMP-PRR interaction culminates in the activation of transcription factors. Primarily NF-κB and AP-1 transcription factors are involved in triggering inflammatory responses (28). Therefore, we checked whether these transcription factors are involved in OmpV-mediated activation of innate immune responses. Using chemical inhibitors against NF-κB, we observed a significant decrease in production TNF-α and IL-6 in both macrophages and monocytes (Fig. 7A to D). These results indicated the involvement of NF-κB in OmpV-mediated proinflammatory signaling.
FIG 7.
NF-κB is involved in OmpV-mediated proinflammatory responses. (A to D) A significant decrease in TNF-α and IL-6 production was observed upon pretreatment with NF-κB inhibitor in OmpV-activated RAW 264.7 macrophages (A, B) and THP-1 cells (C, D). (E, F) OmpV-proteoliposome (PL)-mediated proinflammatory signaling involves NF-κB. (A to F) Cells were pretreated with NF-κB inhibitor followed by treatment with PmB and OmpV or OmpV-proteoliposome (PL). Following incubations, supernatants were collected and analyzed for cytokine production. Bar graphs are expressed as mean ± SEM from three independent experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 versus only OmpV-treated cells (A to D) or versus only OmpV-proteoliposome (PL)-treated cells (E, F). (G) Phosphorylation of IκB was observed in OmpV-treated macrophages and monocytes. (H) Translocation of NF-κB subunits p65 and cRel from the cytoplasm to the nucleus was observed in OmpV-treated macrophages and monocytes. (G, H) RAW 264.7 and THP-1 cells were treated with PmB followed by OmpV. Following incubation, whole-cell lysates were prepared at different time points, and levels of p-IκB (phosphorylated IκB) and IκB were assessed by Western blotting (G), or nuclear and cytoplasmic fractions were prepared, and levels of p65 and cRel were checked by Western blotting (H). GAPDH and β-actin were used as loading controls for whole-cell and cytoplasmic lysates (G, H) and PCNA was used as a loading control for nuclear lysates (H). Western blots are representatives of three independent experiments.
It is known that NF-κB remains present in the cytosol in an inactivated state bound to the inhibitor IκB. When IκB gets phosphorylated and degraded, NF-κB moves to the nucleus, leading to transcriptional upregulation of cytokine genes (29). Therefore, to further confirm the involvement of NF-κB in response to OmpV, we checked phosphorylation of IκB using Western blotting. We observed increased phosphorylation (p-IκB) in both OmpV-activated RAW 264.7 macrophages and THP-1 monocytes (Fig. 7G). Further, it is known that among the five NF-κB family members, p65 (REL-A), Rel-B, or cRel are mainly involved in proinflammatory responses (30). Therefore, we checked the translocation of p65 and cRel subunits to the nucleus using Western blotting. We observed an increase in translocation of both p65 and cRel subunits in nuclear lysates with a corresponding decrease in the cytoplasmic lysate of both OmpV-activated RAW 264.7 macrophages and THP-1 monocytes (Fig. 7H and Fig. S6).
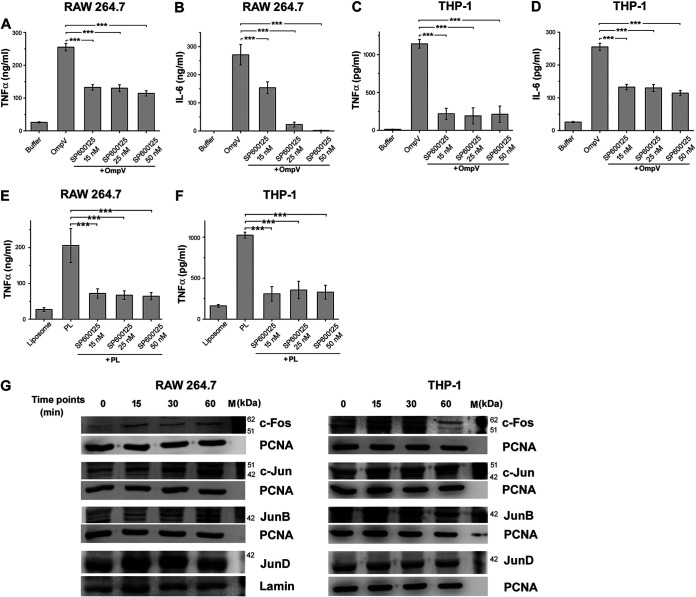
Similar to NF-κB, using a chemical inhibitor, we observed that OmpV-mediated innate immune responses could involve AP-1 activation as well (Fig. 8A to D). Further, AP-1 is also composed of different family members. Out of these, Jun and Fos family members mainly form heterodimers and are involved in transcriptional upregulation of proinflammatory cytokine genes. We checked the presence of these family members in the nucleus of OmpV-activated macrophages and monocytes. We observed increased expression of c-Fos, c-Jun, and JunD subunits in the nuclear lysate of OmpV-activated RAW 264.7 macrophages and c-Fos and c-Jun in THP-1 monocytes (Fig. 8G and Fig. S7).
FIG 8.
OmpV-mediated proinflammatory signaling involves transcription factor AP-1. (A to D) A significant decrease in TNF-α and IL-6 was observed upon pretreatment with AP-1 inhibitor in OmpV-activated macrophages (A, B) and monocytes (C, D). (E, F) AP-1 is involved in OmpV-proteoliposome (PL)-mediated proinflammatory signaling. (A to F) RAW 264.7 and THP-1 cells were pretreated with AP-1 inhibitor followed by treatment with PmB and OmpV or OmpV-proteoliposome (PL). Following respective incubations, supernatants were collected and analyzed for cytokines by ELISA. Bar graphs are expressed as mean ± SEM from three independent experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 versus only OmpV-treated cells or OmpV-proteoliposome (PL)-treated cells. (G) Translocation of AP-1 subunits to the nucleus was observed in OmpV-targeted cells. RAW 264.7 and THP-1 cells were treated with PmB followed by OmpV and incubated for different time points as indicated. Following incubations, nuclear fractions were extracted and probed for AP-1 subunits. PCNA and lamin were used as loading controls for nuclear lysates. Western blots are representatives of three independent experiments.
All of the above results suggested the involvement of NF-κB and AP-1 in OmpV-mediated proinflammatory responses.
Further, with proteoliposome treatment, we observed that, similar to purified OmpV, downstream signaling in proteoliposome-activated innate immune cells also involves NF-κB (Fig. 7E and F) and AP-1 (Fig. 8E and F).
Involvement of p38 and Jun N-terminal protein kinase in OmpV-mediated signaling.
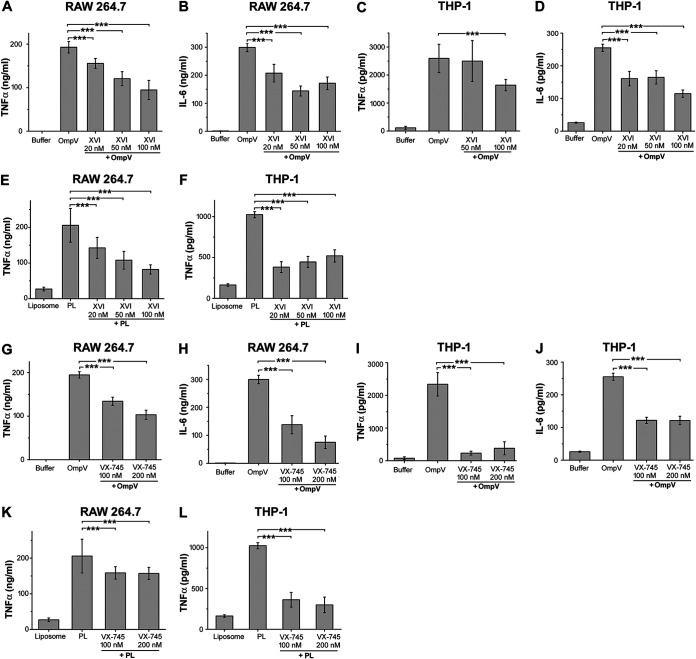
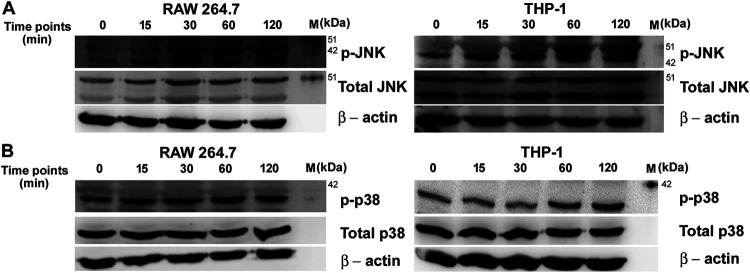
Activation of the AP-1 transcription factor happens through the activation of upstream mitogen-activated protein kinases (MAPKs) (31). Therefore, OmpV-mediated activation of AP-1 suggested that OmpV can activate MAPK cascades in innate immune cells. Towards this, we used pharmacological inhibitors against p38 and Jun N-terminal protein kinase (JNK) MAPKs. We observed a significant decrease in TNF-α and IL-6 production in macrophages (Fig. 9A, B, E, G, H and K) and monocytes (Fig. 9C, D, F, I, J and L) in the presence of JNK and p38 inhibitors upon activation with both purified OmpV and OmpV-proteoliposome (PL), suggesting their involvement in OmpV-activated innate immune cells. Further, we checked the phosphorylation status, which is indicative of the activation status of p38 and JNK MAPKs. We observed a time-dependent increase in phosphorylation of both JNK and p38 (Fig. 10A and B and Fig. S8) upon OmpV treatment.
FIG 9.
p38 and JNK are involved OmpV-mediated signaling. (A to D) A significant decrease in TNF-α and IL-6 production was observed upon pretreatment with JNK inhibitor in OmpV-activated macrophages (A, B) and monocytes (C, D). (E, F) JNK is involved in OmpV-proteoliposome (PL)-mediated proinflammatory signaling. (A to F) RAW 264.7 macrophages (A, B, E) and THP-1 monocytes (C, D, F) were pretreated with JNK inhibitor followed by treatment with PmB and OmpV (A to D) or OmpV-proteoliposome (PL) (E, F). Following incubations, supernatants were analyzed for proinflammatory cytokines by ELISA. Bar graphs are expressed as mean ± SEM from three independent experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 versus only OmpV-treated cells (A to D) or OmpV-proteoliposome (PL)-treated cells (E, F). (G to J) A significant decrease in TNF-α and IL-6 was observed upon pretreatment with p38 inhibitor in OmpV-activated macrophages (G, H) and monocytes (I, J). (K, L) p38 is involved in OmpV-proteoliposome (PL)-mediated signaling in macrophages and monocytes. RAW 264.7 (G, H, K) and THP-1 cells (I, J, L) were pretreated with p38 inhibitor followed by treatment with PmB and OmpV or OmpV-proteoliposome (PL). Following incubations, supernatants were collected and analyzed for proinflammatory cytokines by ELISA. Bar graphs are expressed as mean ± SEM from three independent experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 versus only OmpV-treated cells (G to J) or OmpV-proteoliposome (PL)-treated cells (K, L).
FIG 10.
OmpV induces phosphorylation of JNK and p38 in macrophages, monocytes. (A, B) OmpV treatment leads to the phosphorylation of JNK and p38 in macrophages and monocytes. RAW 264.7 macrophages and THP-1 monocytes were treated with PmB followed by OmpV (2 μg/ml) for different time points. Following incubations, whole-cell lysates were prepared and probed for phosphorylated and total JNK (A) and p38 (B). β-Actin was used as a loading control. Western blots are representatives of three independent experiments.
The above results suggested that induction of OmpV-mediated innate immune responses involves MAPK activation.
DISCUSSION
OmpV belongs to a very large family of proteins known as the MipA (MltA-interacting protein) family. MltA is a membrane-bound lytic transglycosylase. In Escherichia coli, MipA is considered a structural protein, and it probably acts as a scaffold to form a complex between MltA and penicillin-binding protein 1 (PBP1) to help in enlargement and septation of the murein sacculus (32). However, the sequence of OmpV varies in different organisms, and a range of functions are so far attributed to OmpV. In E. coli, it was documented that OmpV helps in antibiotic resistance (33).
In V. parahaemolyticu s (34), V. alginolyticus (35), and P. damselae (16), OmpV is required for NaCl regulation. Apart from this, another MipA protein from Salmonella Paratyphi A provides protection against the bacterium (36). Also, immunization with MipA antigen is known to confer protection against the enterotoxigenic E. coli strains (37).
In our earlier study, we found that OmpV acts as an important adhesin of S. Typhimurium (17). Deletion of the ompV gene leads to a significant decrease in adhesion of S. Typhimurium to the host intestinal epithelial cells. Apart from this, OmpV can modulate actin filaments by activating integrin signaling cascades critical in the invasion process. Deletion of the ompV gene leads to a significant decrease in S. Typhimurium pathogenesis in mice. Hence, OmpV plays a crucial role in the pathogenesis of S. Typhimurium (17). Further, in the present study, we focused on the potential of OmpV to act as a vaccine candidate.
Activation of the immune system, antibody production and protection against disease are major roles of an ideal vaccine candidate (38). Therefore, we first probed whether OmpV immunization can evoke the immune system and give protection against S. Typhimurium infection in mice. As S. Typhimurium causes gastrointestinal infection and salmonellosis in humans and typhoid-like systemic disease in mice, we evaluated protection in cases of both gastrointestinal infection and systemic disease. We observed that intraperitoneal immunization of mice with OmpV generates a high IgG titer as well as protection against systemic infection of S. Typhimurium, but no protection was observed against gastrointestinal infection, and no detectable level of IgA was observed (Fig. 1). Further, we found that oral immunization with purified OmpV protein could not evoke any antibody response or protection against Salmonella infection probably due to degradation of protein in the acidic environment of the stomach. To protect the protein from degradation, we incorporated the protein into liposomes to form OmpV-proteoliposome (PL). Further, liposomes are already known to act as an adjuvant (18). Upon oral immunization of mice with OmpV-proteoliposome (PL), we observed an induction of high IgA titer as well as protection against gastrointestinal infection (Fig. 1).
OmpV can induce activation of B cells as observed in terms of production of IgG and IgA (Fig. 1). However, antibody production by B cells requires T helper cells (Th1 or Th2), which in turn are activated by antigen-presenting cells, such as dendritic cells (DCs). Therefore, we checked the modulation of dendritic cell activation as well as T cell activation and polarization in response to OmpV. Towards that, we observed maturation of OmpV-activated DCs in terms of increased surface expression of the costimulatory molecule CD86 (Fig. 2). In addition to that, we detected T cell proliferation and Th1 polarization in vitro induced by OmpV-activated DCs (Fig. 2). Th1 polarization was also observed in vivo in response to purified OmpV, but both Th1 and Th2 polarization were observed in vivo with more Th1 bias in response to OmpV-proteoliposome (PL) (Fig. 2). It is known that Th2 helps in IgA production (39); therefore, Th2 polarization along with Th1 by OmpV-proteoliposome (PL) treatment probably helped in IgA production and protection in gastrointestinal infection.
Furthermore, as adaptive responses are shaped by the innate immune system, we next studied the immunomodulatory effect of OmpV on innate immune cells, such as monocytes and macrophages. Being a gut microbe, S. Typhimurium first comes into contact with intestinal epithelial cells (IECs), which also have characteristics of innate immune cells. Therefore, we also used IECs for our immunomodulatory study. During this study, we found significant production of proinflammmatory mediators following activation with OmpV (Fig. 3). During the investigation, OmpV and OmpV-proteoliposome (PL) were found to elicit proinflammatory responses through TLR1 and TLR2 (Fig. 4 and 5). Interestingly, it has been reported in the literature that mice deficient in TLR2 and TLR4 are more susceptible to S. Typhimurium infection (40). Our observations underlined the necessity of TLR2 in OmpV recognition. Further, downstream signaling involved MyD88, IRAK-1, MAPKs, and transcription factors NF-κB and AP-1 in OmpV-activated monocytes and macrophages (Fig. 6 to 10).
The above results established that OmpV can activate innate and adaptive responses, leading to IgG and IgA antibody production and protection, revealing efficacy of OmpV as a vaccine candidate.
Further, interestingly, we have observed that in monocytes, OmpV signals through TLR2/6 heterodimers, whereas in macrophages and IECs, both TLR1/2 and TLR2/6 heterodimers were found to be involved (Fig. 4 and 5). A similar observation was reported by us with Vibrio parahaemolyticus OmpU in macrophages (41). Therefore, it seems that S. Typhimurium OmpV is another naturally found ligand having the ability to recognize via both TLR1/2 and TLR2/6 heterodimers. In the case of VpOmpU, the preferred dimer was TLR2/6, but in Salmonella, OmpV signaling via both TLR1/2 and TLR2/6 heterodimers were found to participate equally. This particular characteristic highlights the usage of OmpV as a good adjuvant in vaccine formulations, as adjuvants are desired to evoke good innate immune responses to add/increase the immune potential of antigens.
The results altogether indicated that OmpV might have potential as a vaccine against S. Typhimurium infection and also as an adjuvant in other vaccine formulations.
Further, the fact that there is an outer membrane protein in S. Typhi with 99.46% homologous sequence to OmpV opens up the possibility that OmpV could give protection against typhoid. Furthermore, OmpV can also be used as a part of subunit vaccines along with other potential vaccine candidates.
MATERIALS AND METHODS
Cell lines, bacterial and mouse strains, and chemicals.
Cell lines: THP-1, a human monocytic cell line (NCCS Pune, India), RAW 264.7, a murine macrophage cell line (ATCC, VA, USA), T84, a human carcinoma cell line (NCCS Pune, India), and HT29, a human colon cancer cell line (NCCS Pune, India), were used for this study. RPMI 1640 medium (for RAW 264.7 and THP-1 cells) and Dulbecco’s minimal essential medium (DMEM; for HT29 and T84 cells) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen Life Technologies, CA, USA) were used to culture cells. Cells were maintained at 37°C in 5% CO2.
Bacterial strain: S. Typhimurium SL1344, a gift from Mahak Sharma (IISER Mohali, Punjab, India), were grown and maintained in Luria-Bertani agar (LA) plates at 37°C and Luria-Bertani (LB) broth in the presence of 50 μg/ml streptomycin (HiMedia, Maharashtra, India). E. coli BL21 (DE3) were obtained from Microbial Type Culture Collection (MTCC) and maintained in LA plates and LB broth at 37°C.
Mice: BALB/c mice (6 to 8 weeks old) were used for in vivo immunization experiments and isolation of BMDCs and splenic DCs. For isolation of BMDMs, TLR2−/−, MyD88−/−, and wild-type C57BL/6 mice (6 to 8 weeks old) were used.
All the antibiotics were obtained from HiMedia (Maharashtra, India); oligonucleotides were custom synthesized from Integrated DNA Technologies (IDT, CA, USA) and DNA-modifying enzymes were obtained from New England Biolabs (MA, USA). Nickel-nitrilotriacetic acid (Ni-NTA) agarose, plasmid, and DNA isolation kits were obtained from Qiagen (Hilden, Germany).
Purification and refolding of recombinant OmpV.
S. Typhimurium OmpV was cloned in E. coli BL21(DE3) and purified as described earlier (17). Briefly, the amplified ompV gene (accession no. NC_003197.2 [1,364,189 to 1,364,935 bp]) was cloned into the pET-14b vector obtained from Novagen (Irene, South Africa) and transformed into chemically competent E. coli BL21(DE3) cells. The cells were lysed, and protein was purified from inclusion bodies using Ni-NTA affinity chromatography. The purified protein was then refolded by rapid dilution method with constant stirring using refolding buffer (10% glycerol, 0.5% lauryldimethylamine oxide [LDAO], and 0.01% Triton X-100 in phosphate-buffered saline [PBS]). Further, buffer was exchanged (0.5% LDAO and 0.01% Triton X-100 in 1× PBS) using a size-exclusion chromatography column (Sephacryl S-200). Fractions were collected and run on an SDS-PAGE gel, which was then visualized using Coomassie brilliant blue staining.
Preparation of proteoliposome.
Proteoliposome was prepared by dissolving cholesterol (Sigma, MO, USA) and asolectin (Sigma, MO, USA) in a 1:1 ratio in chloroform (Sigma, MO, USA). This mixture was poured into a round-bottom flask, and a film was prepared on the surface of the flask. The flask was then dried in a vacuum desiccator to remove the remaining chloroform content. After 3 to 4 h, 1 ml of recombinantly purified Salmonella OmpV (500 μg) dissolved in 1× PBS was added to this flask. The film of lipids was thus dissolved and further washed three times by ultracentrifuging at 350,000 × g for 15 min at 4°C. Similarly, the control liposome was prepared by suspending the film made on the surface of the flask in 1× PBS.
Preparation of bone marrow-derived macrophages from wild-type, TLR2−/− and MyD88−/− mice.
Femur and tibia bones were extracted from 6- to 8-week-old mice (C57BL/6, TLR2−/−, or MyD88−/−), and then any muscle tissue was cleaned off and washed with ice-cold PBS. Further, the bones were dipped in 70% alcohol for 2 min and transferred to RPMI complete medium. Further, with the help of sterile scissors, the epiphyses of the bones were cut, and the bones were flushed with medium, releasing the bone marrow cells into the medium. Cells were washed with complete RPMI medium and treated with 2 to 3 ml of ammonium-chloride-potassium (ACK) lysis buffer (Life Technologies, CA, USA) for 5 min to remove erythrocytes. Further, these cells were resuspended in bone marrow differentiation medium (RPMI 1640 supplemented with 10% FBS, 100 U/ml of penicillin and streptomycin each, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids [NEAA], 1% β-mercaptoethanol, and 20 ng/ml of macrophage colony-stimulating factor [M-CSF]) and seeded in 24-well plates. Cells were incubated at 37°C with 5% CO2. The medium was changed every 2 days, and fresh bone marrow differentiation medium was added. On day 7, cells were put in medium without M-CSF and treated with 10 μg/ml of polymyxin B followed by 2 μg/ml of OmpV and incubated for 24 h. The supernatants were collected and analyzed for cytokines by enzyme-linked immunosorbent assay (ELISA). M-CSF was obtained from Peprotech (NJ, USA).
Preparation of bone marrow-derived dendritic cells.
Bone marrow cells were isolated as mentioned above and treated with 2 to 3 ml of ACK lysis buffer (Life Technologies, CA, USA) for 5 min to remove erythrocytes. Further, these cells were resuspended in BMDC differentiation medium (RPMI 1640 supplemented with 10% FBS, 100 U/ml of penicillin and streptomycin each, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids [NEAA], 1% β-mercaptoethanol, and 10 ng/ml of granulocyte-macrophage colony-stimulating factor [GM-CSF]) and seeded in 6-well plates. Cells were incubated at 37°C with 5% CO2. The medium was changed every alternate day for 7 days, and fresh BMDC differentiation medium was added. On day 7, cells were treated according to the experiment. GM-CSF was obtained from Peprotech (NJ, USA).
Isolation of splenic DCs.
Mouse spleen was ballooned using 5 ml of calcium-magnesium-free Hanks balanced salt solution (HBSS; 0.137 M NaCl, 5.4 mM KCl, 0.25 mM Na2HPO4, 0.44 mM KH2PO4, 4.2 mM NaHCO3, and 0.1% glucose [Sigma, MO, USA]) containing 400 U/ml collagenase D (Roche, Basel, Switzerland) for 25 min at 37°C and 5% CO2. Further, 0.5 M EDTA was added for 5 min to stop the reaction. The cell suspension was then filtered through a 0.45-μm filter into RPMI complete medium. Cells were washed with PBS and resuspended in 3 ml of 30% bovine serum albumin (BSA; Sigma, MO, USA). Further 1 ml of PBS was layered on top of 30% BSA and centrifuged at 620 × g for 30 min at 12°C with slow acceleration and deceleration. Cells from the interface were collected, having a CD11c-enriched population. These cells were further used in the experiments.
Isolation of T cells.
The mouse spleen was squashed and filtered through a 0.45-μm filter to obtain a single-cell suspension. Further, these cells were treated with ACK lysis buffer for 5 min to remove erythrocytes. Cells were washed and purified using a mouse CD4+ T-cell isolation kit II (Miltenyi Biotec, Bergisch Gladbach, Germany) by the negative selection method.
Briefly, 107 cells were suspended in 40 μl of magnetically activated cell sorting (MACS) buffer and 5 μl of biotin-conjugated antibody cocktail and incubated for 10 min at 4°C. Further, 30 μl of MACS buffer and 10 μl of microbead cocktail were added for 15 min at 4°C. Cells were then washed with MACS buffer and resuspended in 500 μl of MACS buffer. Cells were then loaded onto the preequilibrated mass spectrometry (MS) MACS column and washed three times with 3 ml of MACS buffer. Flowthrough containing T cells was collected, washed with MACS buffer, and finally resuspended in complete RPMI medium. These cells were then used for experiments.
Coculture of splenic DCs and T cells.
Carboxyfluorescein succinimidyl ester (CFSE)-labeled T cells were plated at a density of 0.5 × 106 cells/ml. For CFSE labeling, 5 μM CellTrace CFSE (ThermoFisher, MA, USA) was added per 2 million T cells and incubated for 10 min at room temperature. CFSE was then quenched by adding an equal amount of FBS for 10 min. These cells were then washed with PBS and plated accordingly. Further, OmpV (2 μg/ml for 24 h)-treated splenic DCs were diluted and added to the CFSE-labeled T cells in 1:20, 1:40, 1:80, and 1:160 DC:T cell ratios/100 μl/well (day 1). At day 3, CFSE fluorescence was detected using a BD FACS Accuri C6 (BD Biosciences, CA, USA).
Isolation of lymph nodes.
Mice were immunized with four doses of OmpV (25 μg/dose) intraperitoneally or with OmpV-proteoliposome (PL) (25 μg/dose) orally at an interval of 7 days. Buffer or liposome was used, respectively, for the control mouse. Following 14 days of the last dose, mice were sacrificed, and inguinal, mesenteric, popliteal lymph nodes (for intraperitoneal immunization), and mesenteric lymph nodes (for oral immunization) were extracted. These lymph nodes were squashed and filtered through 0.45-μm filters to collect single-cell suspensions. These cells were further stimulated with phorbol myristate acetate (PMA; 5 ng/ml) and ionomycin (500 μg/ml) for 24 h, and the presence of cytokines was detected in the supernatant by ELISA.
Survival assay.
Mice were immunized with four doses of OmpV (25 μg/dose) for intraperitoneal immunization and OmpV-proteoliposome (PL) (25 μg/dose) for oral immunization at an interval of 7 days. Buffer, in the case of intraperitoneal immunization, and liposomes, in the case of oral immunization, were used for control mice. Following 14 days after the last dose, mice were challenged with S. Typhimurium SL1344 either intraperitoneally (1 × 103 bacteria per mouse) or orally (7.5 × 103 bacteria per mouse), and survival was monitored. A Kaplan-Meier plot of cumulative mortality was prepared to compare the survival rate.
Detection of antibody titer.
Blood and stool were collected from the immunized (intraperitoneally and orally) mice and checked for the antibody titer. To check IgG titer, blood was taken from the retro-orbital route of immunized mice with the help of capillary action. The serum was isolated by centrifuging the blood at 1,000 × g for 10 min. The collected serum was then serially diluted for detection of OmpV by ELISA and for estimating antibody titer. Similarly, to check IgA titer, stools were collected from immunized mice and dissolved in 1 ml of PBS containing 0.01% sodium azide and 1% protease inhibitor per 100 mg of sample. Further, the dissolved stool sample was centrifuged at 16,000 × g for 10 min at 4°C. Serial dilutions of the supernatant were used to detect OmpV by ELISA to calculate antibody titer.
MTT cell viability assay.
Cell viability was determined using an MTT (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) assay using an MTT kit (Sigma, MO, USA) as per the manufacturer’s protocol. Briefly, THP-1, RAW 264.7, HT29, or T84 cells were plated at a density of 1 × 105 cells/100 μl using complete RPMI 1640 or DMEM complete medium. Cells were treated with 10 μg/ml polymyxin B for 30 min followed by treatments with different concentrations of OmpV and incubated at 37°C. Buffer-treated cells were used as controls. Following incubations, 10 μl of MTT was added and incubated for 4 h. The precipitated MTT formazan crystals were formed, which were then dissolved in acidified propanol (0.1 M HCl in propan-2-ol). The optical density at 570 nm (OD570) was taken on an iMark microplate absorbance reader (Bio-Rad Laboratories, CA, USA). Untreated cells represent 100% cell viability.
Quantification of nitric oxide.
Nitrite, a stable product formed by degradation of NO, was estimated in RAW 264.7 cells using Griess reagent (Sigma, MO, USA). RAW 264.7 cells (1 × 106 cells/ml) were plated using complete RPMI 1640 medium, and treatments were given for dose-response as well as time course studies. Polymyxin B (10 μg/ml) was added for 30 min before treatments to neutralize the effect of LPS. After respective treatment time points, supernatants were collected. Griess reagent was added to the supernatant in equal volumes and incubated for 15 min in the dark at room temperature. Then OD540 was taken using a microplate reader (Bio-Rad Laboratories, CA, USA), and nitrite concentration was estimated using the standard curve. The standard curve was made using sodium nitrite (Sigma, MO, USA) of known concentrations. E. coli LPS (1 μg/ml; Sigma, MO, USA) was used as a positive control in the experiments. Buffer volume equivalent to the highest dose of protein was used as negative control.
Quantification of cytokines using ELISA.
TNF-α, IL-6, IL-8, IL-10, IL-4, IFN-γ, and IL-12 were measured using BD OptEIA ELISA sets (BD Biosciences, CA, USA). RAW 264.7, THP-1, HT29, or BMDCs were plated at a density of 1 × 106 cells/ml, and OmpV treatments were given as indicated. Polymyxin B (10 μg/ml) was added for 30 min before treatments to neutralize the effect of LPS. Supernatants were collected at respective time points, and cytokine production was measured using ELISA. In the case of in vivo immunizations, lymph nodes were extracted, and cells obtained from these lymph nodes were plated at a density of 3 million cells/ml. These cells were activated using PMA (5 ng/ml) and ionomycin (500 μg/ml) for 24 h, and the supernatant was quantified for IL-4 and IFN-γ cytokine production.
Analysis of surface expression using flow cytometry.
To check the surface expression of TLRs in RAW 264.7 and THP-1 cells and CD80 and CD86 in BMDCs, cells were plated at a density of 1 × 106 cells/ml, and OmpV treatments were given following 10 μg/ml of polymyxin B for 30 min. After 24 h of OmpV treatment, cells were harvested and washed with 1× PBS and fluorescence-activated cell sorter (FACS) buffer (0.1% sodium azide and 1% FBS in 1× PBS). For THP-1 monocytes, TLR antibodies conjugated with fluorescein isothiocyanate (FITC)/allophycocyanin (APC)/phycoerythrin (PE) were added. On the other hand, in RAW 264.7 macrophages and BMDCs, Fc block was added and incubated for 20 min at 4°C before adding antibodies against TLRs, CD80, and CD86, respectively. Isotype controls were used in the case of THP-1 monocytes. Following incubations with respective antibodies, cells were washed twice and resuspended in FACS buffer and were acquired using a BD FACS Calibur flow cytometer (BD Biosciences, CA, USA) and analyzed with FlowJo software (http://www.flowjo.com).
PE-conjugated anti-mouse TLR1 antibody was obtained from eBiosciences (CA, USA); FITC-conjugated anti-human TLR1 was obtained from Abcam (Cambridge, UK).; FITC-conjugated anti-mouse/human CD282 (TLR2) and PE-conjugated anti-human CD286 (TLR6) antibodies were procured from BioLegend (CA, USA); APC-conjugated anti-mouse TLR6 antibody was acquired from R&D Systems (MN, USA); FITC-conjugated anti-mouse CD80 and PE-conjugated anti-mouse CD86 antibodies were obtained from BD Biosciences (CA, USA).
Neutralization of TLRs.
To find the TLRs involved in OmpV recognition, RAW 264.7/THP-1/HT29 cells were plated at a density of 1 × 105 cells/100 μl in a 96-well plate. After pretreatment with 5 μg/ml of anti-TLR neutralizing antibodies for 1 h (in RAW 264.7 cells and THP-1 cells) and 2 h (in HT29 cells), cells were treated with polymyxin B for 30 min followed by treatment with 2 μg/ml of OmpV. Supernatants were collected after 4 h for THP-1 cells and after 24 h for RAW 264.7 and HT29 cells. Further, TNF-α and IL-8 were quantified using an ELISA. Neutralizing antibody against TLR2 and its isotype were obtained from BioLegend (CA, USA), and neutralizing antibodies against human TLR1 and TLR6 and the isotypes were obtained from Invivogen (CA, USA).
Inhibitor studies.
To check the involvement of various signaling mediators, pharmacological inhibitors were used. For this, 1 × 106 cells/ml were plated and pretreated for 1 h with different pharmacological inhibitors. Following incubation, cells were treated with 2 μg/ml of OmpV for 24 h for RAW 264.7 cells and HT29 cells and 4 h for THP-1 cells. Following incubations, supernatants were collected and analyzed for TNF-α, IL-6, and IL-8 by ELISA. The inhibitors for IRAK-1 (IRAK-1/4 inhibitor) and AP-1 (SP600125) were obtained from Sigma (MO, USA). JNK (XVI) and p38 (VX745) inhibitors were purchased from Santa Cruz (TX, USA). Inhibitor for NF-κB (MLN4924) was obtained from R&D (MN, USA).
Preparation of whole-cell lysates and nuclear lysates.
Whole-cell lysates were prepared by using lysis buffer (50 mM Tris-Cl [pH 8], 150 mM NaCl, 0.1% Triton X-100, and 0.1% SDS). Briefly, cells were treated with OmpV or buffer following 10 μg/ml of polymyxin B treatments. Following incubations, cells were washed twice with ice-cold 1× PBS, and the pellet was resuspended in 200 μl of lysis buffer containing 1% mammalian protease inhibitor cocktail (Sigma, MO, USA). The cells were then sonicated three times at a 10 A pulse for 3 s each time. The lysed cells were centrifuged at 16,000 × g for 30 min at 4°C. The supernatants were collected and estimated using Bradford reagent. An equal amount of protein was loaded onto SDS-PAGE gels and were analyzed by Western blotting.
For nuclear lysates, treated cells were washed once with 1× PBS and then with hypotonic buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, and 0.5 M dithiothreitol [DTT]). Cells were pelleted at 1,850 × g for 5 min at 4°C. The pellet was resuspended in 150 μl of hypotonic buffer containing mammalian protease inhibitor cocktail. Cells were lysed using sonication twice at 10 A for 3 s each. After centrifugation at 3,300 × g for 15 min at 4°C, the supernatant was collected as a cytoplasmic fraction. The pellet was resuspended in 70 μl of low-salt buffer (20 mM HEPES [pH 7.9], 25% glycerol, 1.5 mM MgCl2, 0.02 M KCl, 0.2 mM EDTA, and 0.5 M DTT) along with mammalian protease inhibitor. Then, 30 μl of high-salt buffer (containing 0.8 M KCl) was added to the mixture and incubated for 10 min at 4°C. The mixture was then sonicated twice at 10 A for 3 s each and incubated for 30 min on ice. The suspension was then centrifuged at 24,000 × g for 30 min at 4°C. The supernatant was collected as nuclear lysate and analyzed by Western blotting.
Coimmunoprecipitation and immunoblots.
For immunoprecipitation experiments, RAW 264.7, THP-1, or HT29 cells were treated with buffer or OmpV (2 μg/ml), and whole-cell lysates were prepared. To these lysates (for approximately 500 μg of protein), 0.5 μg of antibody was added, and samples were incubated for 4 h at 4°C with continuous agitation. Further, protein A/G PLUS agarose beads (Santa Cruz Biotechnology, TX, USA) were added and incubated overnight at 4°C with constant agitation. The bead-protein complex was then washed three times with lysis buffer (50 mM Tris-Cl [pH 8], 150 mM NaCl, 0.1% Triton X-100, and 0.1% SDS), and then protein was eluted from beads using 5× loading buffer (250 mM Tris-Cl [pH 6.8], 10% SDS, 30% glycerol, 5% β-mercaptoethanol, and 0.02% bromophenol blue) and heated for 10 min at 100°C. Samples were loaded onto SDS-PAGE gels and probed for different proteins using immunoblotting.
Antibodies for TLR1, TLR2, p65, cRel, MyD88, JunD, IκB-α, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), β-actin, and lamin B1 were from Santa Cruz Biotechnology (TX, USA). Antibodies for c-Jun, c-Fos, TLR6, JunB, p-p38, total p38, phosphoSAPK/JNK (Thr183/Tyr185), and SAPK/JNK were from Cell Signaling Technology (MA, USA). Rabbit IgG isotype control was obtained from Santa Cruz Biotechnology (TX, USA). Antibody for phospho-IκB-α (pSer32/Ser36) and horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse IgG secondary antibodies were from Sigma (MO, USA).
Protein estimation by Bradford assay.
The protein concentration for recombinant protein, proteoliposomes, nuclear fractions, cytoplasmic fractions, and whole-cell fractions were measured using Bradford reagent (Sigma, MO, USA). BSA solution of different concentrations in the same buffer as that of the sample was used to make a standard curve. For purified protein and proteoliposome estimation, 95 μl of Bradford’s reagent was added to 5 μl of protein or proteoliposomes. For nuclear, cytoplasmic, or whole-cell lysates, 99 μl of Bradford’s reagent was added to 1 μl of sample. The samples were then mixed and kept in the dark for 15 min. The readings were taken in triplicate for each sample in a 96-well plate. The readings were taken at OD595 using an iMark microplate absorbance reader (Bio-Rad Laboratories, CA, USA), and concentration of the proteins was estimated using a BSA standard curve plot.
siRNA-mediated knockdown.
To check the involvement of TLRs in OmpV recognition by RAW 264.7 cells, 2.5 × 105 cells were plated in a 24-well plate and were transfected with ON-TARGETplus siRNA against mouse TLR1, mouse TLR6, or ON-TARGETplus nontargeted siRNA pool (Dharmacon, GE) using FuGENE HD (Promega) at a ratio of 1:5 (RNA:FuGENE) for 24 h. After incubation, knockdown was analyzed by FACS Calibur, and cells were treated with 2 μg/ml of OmpV for 12 h. Supernatants were collected, and cytokines were quantified by ELISA.
shRNA-mediated knockdown.
For knock down of TLR1, TLR2, and TLR6 in THP-1 and HT29 cells, an shRNA construct was cloned under the H1 promoter in green fluorescent protein (GFP)-expressing multipromoter vector as described by (42). GFP-expressing multipromoter vector, used for making the shRNA construct, was a kind gift from Samarjit Bhattacharyya (IISER Mohali, Punjab, India). Primer sequences used to make shRNA constructs are mentioned below. The scrambled sequence for TLR cloned in vector separately was used as a control.
Primers:
TLR1: ACAAATGTTCTTACTTCCT
TLR2: AGAGTTAAAAGAATCACAG
TLR6: AGACAAAGAACCTATTGTT
Scramble: GGAAGAACGATAAACTATA
The cloned plasmid was further transformed into E. coli Top10 cells, and colonies were selected using ampicillin resistance. The plasmids containing shRNA and scrambled sequence, respectively, were isolated from E. coli Top10 using a Qiagen miniprep kit and further transfected into THP-1 and HT29 cells using polyethylenimine (PEI) (1:3 ratio of DNA:PEI). The mixture was prepared by mixing 100 μl of Opti-MEM containing 1 μg of DNA and 100 μl of Opti-MEM containing 3 μl of PEI (1 mg/ml stock) and incubated at room temperature for 15 min. This mixture was subsequently added to the cells. After 48 h, whole-cell lysates were prepared, and Western blotting was performed in the case of THP-1 cells, and RNA was isolated and RT-PCR analysis was done in the case of HT29 cells to assess the knockdown. After knockdown, cells were treated with 2 μg/ml of OmpV for 12 h. Supernatants were then collected and quantified for TNF-α and IL-8 using an ELISA.
Ethical statement.
This study was carried out in strict accordance with the guidelines issued by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) (reg. no. 1842/GO/ReBiBt/S/15/CPCSEA). All protocols involving mice experiments were approved by the Institutional Animal Ethics Committee (IAEC) (IISERM/SAFE/PRT/2016-2018/005, 010, 015).
Statistical analysis.
Statistical significance was analyzed by the Student’s t test using GraphPad QuickCalcs software. The results were considered to be significant at a P value of <0.05 (*), a P value of <0.01 (**), and a P value of <0.001 (***) and were considered not significant (ns) at a P value of >0.05.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. This work was supported by funding from IISER Mohali.
We thank the Small Animals Facility for Experimentation (SAFE)-IISER Mohali for animal maintenance. We also thank Samarjit Bhattacharyya (IISER Mohali) for help in shRNA-mediated knockdown studies.
Deepinder Kaur designed and performed the experiments, analyzed the data, and wrote the paper. Shraddha Gandhi performed experiments. Arunika Mukhopadhaya designed experiments, analyzed data, supervised the study, and wrote the paper.
We declare that we have no conflicts of interest in regard to the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Arunika Mukhopadhaya, Email: arunika@iisermohali.ac.in.
Igor E. Brodsky, University of Pennsylvania
REFERENCES
- 1.Graham SM. 2010. Nontyphoidal salmonellosis in Africa. Curr Opin Infect Dis 23:409–414. 10.1097/QCO.0b013e32833dd25d. [DOI] [PubMed] [Google Scholar]
- 2.Alghoribi MF, Doumith M, Alrodayyan M, Al Zayer M, Koster WL, Muhanna A, Aljohani SM, Balkhy HH, Desin TS. 2019. S. Enteritidis and S. Typhimurium harboring SPI-1 and SPI-2 are the predominant serotypes associated with human salmonellosis in Saudi Arabia. Front Cell Infect Microbiol 9:187. 10.3389/fcimb.2019.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacLennan CA, Martin LB, Micoli F. 2014. Vaccines against invasive Salmonella disease: current status and future directions. Hum Vaccin Immunother 10:1478–1493. 10.4161/hv.29054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sparham SJ, Kwong JC, Valcanis M, Easton M, Trott DJ, Seemann T, Stinear TP, Howden BP. 2017. Emergence of multidrug resistance in locally-acquired human infections with Salmonella Typhimurium in Australia owing to a new clade harbouring blaCTX-M-9. Int J Antimicrob Agents 50:101–105. 10.1016/j.ijantimicag.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 5.GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators. 2019. The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis 19:1312–1324. 10.1016/S1473-3099(19)30418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, Harris D, Clarke L, Whitehead S, Sangal V, Marsh K, Achtman M, Molyneux ME, Cormican M, Parkhill J, MacLennan CA, Heyderman RS, Dougan G. 2009. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res 19:2279–2287. 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon MA, Graham SM, Walsh AL, Wilson L, Phiri A, Molyneux E, Zijlstra EE, Heyderman RS, Hart CA, Molyneux ME. 2008. Epidemics of invasive Salmonella enterica serovar Enteritidis and S. enterica serovar Typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis 46:963–969. 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- 8.McConnell MJ, Dominguez-Herrera J, Smani Y, Lopez-Rojas R, Docobo-Perez F, Pachon J. 2011. Vaccination with outer membrane complexes elicits rapid protective immunity to multidrug-resistant Acinetobacter baumannii. Infect Immun 79:518–526. 10.1128/IAI.00741-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Wan C, Xu H, Wei H. 2013. Identification and characterization of OmpL as a potential vaccine candidate for immune-protection against salmonellosis in mice. Vaccine 31:2930–2936. 10.1016/j.vaccine.2013.04.044. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Toledo M, Valero-Pacheco N, Pastelin-Palacios R, Gil-Cruz C, Perez-Shibayama C, Moreno-Eutimio MA, Becker I, Perez-Tapia SM, Arriaga-Pizano L, Cunningham AF, Isibasi A, Bonifaz LC, Lopez-Macias C. 2017. Salmonella Typhi porins OmpC and OmpF are potent adjuvants for T-dependent and T-independent antigens. Front Immunol 8:230. 10.3389/fimmu.2017.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JS, Jung ID, Lee CM, Park JW, Chun SH, Jeong SK, Ha TK, Shin YK, Kim DJ, Park YM. 2010. Outer membrane protein a of Salmonella enterica serovar Typhimurium activates dendritic cells and enhances Th1 polarization. BMC Microbiol 10:263. 10.1186/1471-2180-10-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevenson G, Leavesley DI, Lagnado CA, Heuzenroeder MW, Manning PA. 1985. Purification of the 25-kDa Vibrio cholerae major outer-membrane protein and the molecular cloning of its gene: ompV. Eur J Biochem 148:385–390. 10.1111/j.1432-1033.1985.tb08850.x. [DOI] [PubMed] [Google Scholar]
- 13.Mao Z, Yu L, You Z, Wei Y, Liu Y. 2007. Cloning, expression and immunogenicty analysis of five outer membrane proteins of Vibrio parahaemolyticus zj2003. Fish Shellfish Immunol 23:567–575. 10.1016/j.fsi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Singh R, Shasany AK, Aggarwal A, Sinha S, Sisodia BS, Khanuja SP, Misra R. 2007. Low molecular weight proteins of outer membrane of Salmonella typhimurium are immunogenic in Salmonella induced reactive arthritis revealed by proteomics. Clin Exp Immunol 148:486–493. 10.1111/j.1365-2249.2007.03362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azama FM, Zamri-Saadc M, Rahima RA, Chumnanpoend P. 2020. Antigenic outer membrane proteins prediction of Pasteurella multocida serotype B: 2. AsPac J Mol Biol Biotechnol 28(4):102–116. 10.35118/apjmbb.2020.028.4.09. [DOI] [Google Scholar]
- 16.Wu L, Lin X, Wang F, Ye D, Xiao X, Wang S, Peng X. 2006. OmpW and OmpV are required for NaCl regulation in Photobacterium damsela. J Proteome Res 5:2250–2257. 10.1021/pr060046c. [DOI] [PubMed] [Google Scholar]
- 17.Kaur D, Mukhopadhaya A. 2020. Outer membrane protein OmpV mediates Salmonella enterica serovar typhimurium adhesion to intestinal epithelial cells via fibronectin and α1β1 integrin. Cell Microbiol 22:e13172. 10.1111/cmi.13172. [DOI] [PubMed] [Google Scholar]
- 18.Tandrup Schmidt S, Foged C, Korsholm KS, Rades T, Christensen D. 2016. Liposome-based adjuvants for subunit vaccines: formulation strategies for subunit antigens and immunostimulators. Pharmaceutics 8:7. 10.3390/pharmaceutics8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurieva RI, Chung Y. 2010. Understanding the development and function of T follicular helper cells. Cell Mol Immunol 7:190–197. 10.1038/cmi.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alloatti A, Kotsias F, Magalhaes JG, Amigorena S. 2016. Dendritic cell maturation and cross-presentation: timing matters! Immunol Rev 272:97–108. 10.1111/imr.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Garra A, Murphy KM. 2009. From IL-10 to IL-12: how pathogens and their products stimulate APCs to induce TH1 development. Nat Immunol 10:929–932. 10.1038/ni0909-929. [DOI] [PubMed] [Google Scholar]
- 22.Sharma JN, Al-Omran A, Parvathy SS. 2007. Role of nitric oxide in inflammatory diseases. Inflammopharmacology 15:252–259. 10.1007/s10787-007-0013-x. [DOI] [PubMed] [Google Scholar]
- 23.Janssens S, Beyaert R. 2003. Role of Toll-like receptors in pathogen recognition. Clin Microbiol Rev 16:637–646. 10.1128/cmr.16.4.637-646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warner N, Nunez G. 2013. MyD88: a critical adaptor protein in innate immunity signal transduction. J Immunol 190:3–4. 10.4049/jimmunol.1203103. [DOI] [PubMed] [Google Scholar]
- 25.Ding A, Yu H, Yang J, Shi S, Ehrt S. 2005. Induction of macrophage-derived SLPI by Mycobacterium tuberculosis depends on TLR2 but not MyD88. Immunology 116:381–389. 10.1111/j.1365-2567.2005.02238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Q, Qi L, Wu T, Wang J. 2012. Clostridium butyricum activates TLR2-mediated MyD88-independent signaling pathway in HT-29 cells. Mol Cell Biochem 361:31–37. 10.1007/s11010-011-1084-y. [DOI] [PubMed] [Google Scholar]
- 27.De Nardo D, Balka KR, Cardona Gloria Y, Rao VR, Latz E, Masters SL. 2018. Interleukin-1 receptor-associated kinase 4 (IRAK4) plays a dual role in myddosome formation and Toll-like receptor signaling. J Biol Chem 293:15195–15207. 10.1074/jbc.RA118.003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujioka S, Niu J, Schmidt C, Sclabas GM, Peng B, Uwagawa T, Li Z, Evans DB, Abbruzzese JL, Chiao PJ. 2004. NF-κB and AP-1 connection: mechanism of NF-κB-dependent regulation of AP-1 activity. Mol Cell Biol 24:7806–7819. 10.1128/MCB.24.17.7806-7819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanarek N, Ben-Neriah Y. 2012. Regulation of NF-κB by ubiquitination and degradation of the IκBs. Immunol Rev 246:77–94. 10.1111/j.1600-065X.2012.01098.x. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence T. 2009. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb Perspect Biol 1:a001651. 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gazon H, Barbeau B, Mesnard JM, Peloponese JM Jr, 2017. Hijacking of the AP-1 signaling pathway during development of ATL. Front Microbiol 8:2686. 10.3389/fmicb.2017.02686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vollmer W, von Rechenberg M, Holtje JV. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J Biol Chem 274:6726–6734. 10.1074/jbc.274.10.6726. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Zhang DF, Lin XM, Peng XX. 2015. Outer membrane proteomics of kanamycin-resistant Escherichia coli identified MipA as a novel antibiotic resistance-related protein. FEMS Microbiol Lett 362:fnv074. 10.1093/femsle/fnv074. [DOI] [PubMed] [Google Scholar]
- 34.Xu C, Ren H, Wang S, Peng X. 2004. Proteomic analysis of salt-sensitive outer membrane proteins of Vibrio parahaemolyticus. Res Microbiol 155:835–842. 10.1016/j.resmic.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Xu C, Wang S, Ren H, Lin X, Wu L, Peng X. 2005. Proteomic analysis on the expression of outer membrane proteins of Vibrio alginolyticus at different sodium concentrations. Proteomics 5:3142–3152. 10.1002/pmic.200401128. [DOI] [PubMed] [Google Scholar]
- 36.Yang TC, Ma XC, Liu F, Lin LR, Liu LL, Liu GL, Tong ML, Fu ZG, Zhou L. 2012. Screening of the Salmonella paratyphi A CMCC 50973 strain outer membrane proteins for the identification of potential vaccine targets. Mol Med Rep 5:78–83. 10.3892/mmr.2011.587. [DOI] [PubMed] [Google Scholar]
- 37.Hays MP, Kumar A, Martinez-Becerra FJ, Hardwidge PR. 2016. Immunization with the MipA, Skp, or ETEC_2479 antigens confers protection against enterotoxigenic E. coli strains expressing different colonization factors in a mouse pulmonary challenge model. Front Cell Infect Microbiol 6:181. 10.3389/fcimb.2016.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beverley PC. 2002. Immunology of vaccination. Br Med Bull 62:15–28. 10.1093/bmb/62.1.15. [DOI] [PubMed] [Google Scholar]
- 39.Wu Y, Kudsk KA, DeWitt RC, Tolley EA, Li J. 1999. Route and type of nutrition influence IgA-mediating intestinal cytokines. Ann Surg 229:662–667; discussion 667–8. 10.1097/00000658-199905000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, Jones MB, Dracheva T, Peterson SN, Monack DM, Barton GM. 2011. TLR signaling is required for Salmonella typhimurium virulence. Cell 144:675–688. 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gulati A, Kumar R, Mukhopadhaya A. 2019. Differential recognition of Vibrio parahaemolyticus OmpU by Toll-like receptors in monocytes and macrophages for the induction of proinflammatory responses. Infect Immun 87:e00809-18. 10.1128/IAI.00809-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma R, Gulia R, Bhattacharyya S. 2018. A critical role for sorting nexin 1 in the trafficking of metabotropic glutamate receptors. J Neurosci 38:8605–8620. 10.1523/JNEUROSCI.0454-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download IAI00121-21_Supp_1_seq12.pdf, PDF file, 1.3 MB (1.3MB, pdf)