Abstract
Lung cancer is one of the main causes of tumor lethality worldwide. Circular RNAs (circRNAs) have essential roles in tumor progression. However, in non-small cell lung carcinoma (NSCLC), the role of circRNAs remains unknown. In the present study, the expression, function and molecular mechanisms of a new circRNA, circRNA_103615, were investigated in NSCLC. Reverse transcription-quantitative PCR (RT-qPCR) was used to detect circRNA_103615 expression levels in NSCLC and normal tissues, as well as in NSCLC cell lines. MTT assay, flow cytometric assay, colony formation assay, and cell migration and invasion assays were used to examine the function of circRNA_103615 in NSCLC cells. MTT assay, colony formation assay, RT-qPCR, and western blotting were used to detect the effect of circRNA_103615 on cisplatin resistance. The results demonstrated that NSCLC cell lines and tissues had increased levels of circRNA_103615 compared with normal cells and normal tissues, respectively. Functionally, silencing of circRNA_103615 by small interfering RNA resulted in suppression of cell growth, migration, and invasion, but promotion of cell apoptosis. In addition, the cisplatin resistance of NSCLC was reversed by the silencing of circRNA_103615. Notably, ATP binding cassette subfamily Bmember 1 (ABCB1) expression was significantly decreased following circRNA_103615 knockdown, and ABCB1 overexpression reversed the effects of circRNA_103615 silencing on NSCLC cisplatin resistance. Thus, the present study indicated that circRNA_103615 may serve as a critical oncogene and potential novel biomarker in NSCLC, as well as a potential cisplatin resistance promoter, by regulating ABCB1 expression.
Keywords: circRNA_103615, cisplatin resistance, ATP binding cassette subfamily B member 1, non-small cell lung carcinoma
Introduction
Lung cancer is one of the predominant causes of tumor lethality worldwide, with a 5-year survival rate of only 19.3%. Additionally, ~85% of lung cancers are non-small cell lung cancers (NSCLCs) that are resistant to radiotherapy and chemotherapy (1-5). Therefore, the present study explored the molecular mechanisms involved in the resistance of chemotherapy drugs in NSCLC.
Circular RNAs (circRNAs), a newly appreciated class of RNA expressed across diverse phyla, have covalently closed-loop structures without a 5 ‘cap or a 3’ poly A tail (6,7). In human cancers, the abnormal expression of circRNAs has been observed (8-12). A recent study has shown a significant role of the circRNA circRNA_100876 in chemotherapy resistance, development, and migration, as well as tumor invasion of NSCLC (13). circRNA circ_0067934 regulates the miR-1324/FZD5/Wnt/β-catenin axis in hepatocellular carcinoma promoting tumor growth and metastasis (14). In esophageal squamous cell carcinoma, circ_0067934 is upregulated and promotes proliferation (15). In gastric cancer, cisplatin resistance is enhanced by suppression of miR-198 by PIK3R1 that is upregulated by circular RNA AKT3(16). Although these circRNAs have important roles in the development and metastasis of NSCLC, the chemoresistance regulatory mechanisms of circRNA in NSCLC remain unclear.
In the present study, the differentially expressed circRNAs between NSCLC tissues and matched adjacent normal tissues were investigated by retrieving the microarray data in the GSE112214 dataset, normalizing the data and analyzing them by using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) after applying log2 transformation. The results showed that several circRNAs were significantly increased in NSCLC tissues. circRNA_103615 is a newly identified circRNA, and was one of the significantly overexpressed circRNAs in NSCLC in microarray analysis. However, the function and mechanism of circRNA_103615 have not been reported in cancer. Therefore, the present study investigated the circRNA_103615 expression levels in NSCLC cell lines and tissues and explored its function and mechanism in NSCLC cells.
Materials and methods
Patients and tissue specimens
The samples used in the present study were NSCLC tissue samples and adjacent non-cancerous tissue samples, which were obtained from 60 patients who underwent primary surgical resection of NSCLC at the Fuda Cancer Hospital (Guangzhou, China) from May 2010 to May 2015. Patients were included in the current study if they did not receive any therapy other than surgery. Written informed consent was obtained from all the participants before enrollment. No preoperative therapy was received by any of the patients. All procedures involving human samples were approved and performed in accordance with the ethical standards of the Human Resources Ethics Committee of the Fuda Cancer Hospital.
Cell culture
The human NSCLC cell lines A549, NCI-H1299, H1650 and H358, and the normal lung cell line Beas-2B were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.).
Bioinformatics analysis
Microarray data from the Gene Expression Omnibus dataset GSE112214 (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE112214) were retrieved to examine the expression of circRNAs in NSCLCs. GEO2R was used to analyze normalized microarray data after applying log2 transformation.
Cell transfection
All cells were maintained at 37˚C with 5% CO2. Cells (2x105 cells/well) were plated in 6-well plates at 24 h and then transfected with control, small interfering (si) RNA si-circRNA_103615 (100 nM/well in 6-well plates) or the overexpression plasmid pcDNA3.1-ABCB1 (4.0 µg/well in 6-well plates; Genepharm, Inc.) at 80% confluence using Lipofectamine 3000 (Life Technologies; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The transfection assay was performed at 37°C for 48 h. After 48 h of transfection, subsequent experiments were performed. The sequence of si-circRNA_103615 was 5'-CTGCTGACCGCTTCCGGGAGG-3'. The sequence of si-NC was 5'-GCAAGCTGACCCTGAAGTT-3'. A negative control oligo (si-NC) or the empty vector pcDNA3.1 were used as controls.
Cell proliferation assay
A549 cells transfected with either si-circRNA_103615 or si-NC were seeded into 96-well plates at 5,000 cells/well and examined after 24, 48, 72 and 96 h. Methyl thiazolyl tetrazolium (MTT; Sigma-Aldrich; Merck KGaA) assay was used to examine cell proliferation as per the manufacturer's protocol (Sigma-Aldrich; Merck KGaA). A microplate reader was used to detect the optical density at 490 nm.
Cell migration and invasion
For cell migration assays, uncoated 24-well Transwell chambers (8 µm pores; BD Biosciences) were used. For cell invasion assays, the 24-well Transwell chambers were coated with Matrigel (BD Biosciences) for cell invasion. A549 cells transfected with either si-circRNA_103615 or si-NC (1x105 cell/well) suspended in 200 µl DMEM (with 1% FBS) were added into each chamber. DMEM medium with 20% FBS was added to the bottom wells. Subsequently, the cells were allowed to migrate or invade for 48 h at 37˚C with 5% CO2. Migratory or invasive cells were stained with DAPI. The stained cells in four random visual fields were counted under a fluorescence microscope (IX71-F22FL/PH; Olympus Corporation).
Cell apoptosis and cell cycle
Cell apoptosis was detect using an Annexin V-FITC Apoptosis kit (cat. no. 556547; BD Biosciences), as per the manufacturer's protocol. Cell cycle distribution was detected using a propidium iodide (PI)/RNase Staining buffer (cat. no. 550825; BD Biosciences), as per manufacturer's protocol. The percentage of apoptotic cells was determined by flow cytometry (C6; BD Biosciences), and the data were analyzed using FlowJo software (V10.7.1; FlowJo LLC)
Colony formation
Cells were plated in 6-well plates at 1x103 per well and incubated for 14 days. Subsequently, cells were fixed with 4% paraformaldehyde at 37˚C, and stained with crystal violet (Sigma-Aldrich; Merck KGaA). The number of colonies were counted with ImageJ (1.52v; National Institutes of Health).
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract RNA from NSCLC tissues and cells, as per the manufacturer's instructions. PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.) was used to synthesized cDNA for analysis of circRNA_103615, multidrug resistance-associated protein 1 (MRP1), breast cancer resistance protein (BCRP), glutathione S-transferase π (GST-π), high mobility group box 1 (HMGB1), and ATP binding cassette subfamily Bmember 1 (ABCB1) expression levels. SYBR Green kit (Qiagen GmbH) was used to run qPCR on an ABI Prism 7500 Sequence Detector (Applied Biosystems; Thermo Fisher Scientific, Inc.). The temperature protocol for the reaction was as follows: 95˚C for 10 min; 40 cycles at 95˚C for 15 sec and 60˚C for 1 min. GAPDH was used as an internal reference gene. Relative fold changes in expression were calculated using the formula 2-ΔΔCq. The primers for the circRNA_103615 and GAPDH are listed in Table I.
Table I.
Primer sequences used in the present study.
| Gene | Primer | Sequence (5'-3') |
|---|---|---|
| circRNA_103615 | Forward | CTGACCCTTGACAGCATACGGATG |
| Reverse | GAAGTTACGGTCGGTGTATGGACGAG | |
| GAPDH | Forward | GCACCGTCAAGGCTGAGAAC |
| Reverse | ATGGTGGTGAAGACGCCAGT | |
| MRP1 | Forward | GCACTGGCTTCTAACTATTGG |
| Reverse | TCTCATTGAAGTGTGAGTACAC | |
| BCRP | Forward | TGGCTTAGACTCAAGCACAGC |
| Reverse | TCGTCCCTGCTTAGACATCC | |
| GST-π | Forward | AATGGATCCTCCACCATGCCGCCCTACACCGTGGT |
| Reverse | GACCTCGAGCTACTGTTTCCCGTTGCCAT | |
| HMGB1 | Forward | ATATGGCAAAAGCGGACAAG |
| Reverse | GCAACATCACCAATGGACAG | |
| ABCB1 | Forward | TGGTTCAGGTGGCTCTGGAT |
| Reverse | CTGTAGACAAACGATGAGCTAT |
circRNA, circular RNA; MRP1, multidrug resistance-associated protein 1; BCRP, breast cancer resistance protein; GST-π, glutathione S-transferase π; HMGB1, high mobility group box 1; ABCB1, ATP binding cassette subfamily Bmember 1.
Half-maximal inhibitory concentration (IC50) value
A549 cells transfected with either si-circRNA_103615 or si-NC were seeded into 96-well plates at 5,000 cells/well. The culture medium was subsequently replaced with medium containing cisplatin (DDP) (32, 16, 8, 4, 2, 1, 0.5, 0.25 and 0.125 mg/ml) for 48 h, and cell viability was evaluated with an MTT assay (Sigma-Aldrich; Merck KGaA), as per the manufacturer's protocol (Sigma-Aldrich; Merck KGaA). A microplate reader was used to detect the optical density at 490 nm.
Western blot analysis
RIPA lysis extraction buffer (Thermo Fisher Scientific, Inc.) was used for total protein extraction from NSCLC cells and protein concentration was determined by performing a bicinchoninic acid assay (Thermo Fisher Scientific, Inc.). Subsequently, 8-12% SDS-PAGE was used to separate protein samples (25 µg), and then the samples were transferred on PVDF membranes (EMD Millipore). The membrane was blocked in PBS containing 0.1% Tween-20 (Beyotime Institute of Biotechnology) and 5% non-fat dry milk at room temperature for 2 h. The membranes were first incubated with the following primary antibodies: Anti-ABCB1 (1:1,000; cat. no. 13342; Cell Signaling Technology, Inc.) or anti-GAPDH (1:1,000; cat. no. 5174; Cell Signaling Technology, Inc.) for overnight incubation at 4˚C. The membranes were then incubated with horseradish peroxidase-anti-rabbit secondary antibody (1:2,000; cat. no. 14708; Cell Signaling Technology, Inc.) for 1 h at room temperature. Finally, the protein bands were detected using ECL solution (Pierce; Thermo Fisher Scientific, Inc.) and images were captured using a FluorChem imaging system (ProteinSimple) GADPH was used as a loading control.
Statistical analysis
All experiments were repeated three times, and quantitative data were presented as the mean ± SD. Student's t-test was used for statistical analysis (two groups that were not matched/paired were compared using an unpaired t-test, and two groups that were matched/paired were compared using a paired t-test). Comparisons among >2 groups were performed using analysis of variance followed by Dunnett's or Tukey's test. Correlation analysis was conducted using Pearson's correlation coefficient. The association between circRNA_103615 expression levels and the patient clinicopathological characteristics was determined by χ2 test. Kaplan-Meier survival analysis was used to calculate the survival curve, and the log-rank test was employed to determine statistical significance. P<0.05 was considered to indicate a statistically significant difference.
Results
circRNA_103615 expression is increased in NSCLC clinical specimens and cell lines
Microarray data in the Gene Expression Omnibus dataset GSE112214 were retrieved to examine the expression of circRNAs in NSCLC, and GEO2R was used to analyze normalized microarray data after applying log2 transformation. The data showed that circRNA_103615 expression was increased in NSCLC tissues compared with normal tissues (Fig. 1A). Therefore, circRNA_103615 was selected for further investigation in the present study. RT-qPCR was used to detect the circRNA_103615 expression levels in 60 NSCLC tissue samples and matched normal tissues. The results demonstrated that circRNA_103615 expression levels were significantly increased in NSCLC tissues compared with normal tissues (Fig. 1B). Next, the patient cohort was separated into high circRNA 103615 (n=30) and low circRNA_103615 groups (n=30) according to the median circRNA_103615 expression (15.1585), and Kaplan-Meier survival analysis revealed that low circRNA_103615 expression levels in NSCLC patients were associated with a higher survival rate (Fig. 1C). Furthermore, the circRNA_103615 expression levels of NSCLC cell lines A549, NCI-H1299, H1650 and H358 and a normal lung cell line Beas-2B were examined by RT-qPCR assay. The results revealed that the circRNA_103615 expression levels were significantly increased in NSCLC cell lines compared with the normal Beas-2B cells (Fig. 1D). The present findings demonstrated that circRNA_103615 was significantly upregulated in NSCLC clinical samples and cell lines, suggesting that its dysregulation may promote NSCLC progression.
Figure 1.
circRNA_103615 is upregulated in NSCLC clinical specimens and cells. (A) Analysis of the GSE112214 dataset revealed that circRNA_103615 was upregulated in NSCLC tissues compared with that in normal samples. (B) circRNA_103615 levels were examined by RT-qPCR in 60 NSCLC tissues and their adjacent normal tissues collected in the present study. ***P<0.001. (C) Kaplan-Meier survival analysis. (D) circRNA_103615 levels were detected by RT-qPCR in human NSCLC cell lines A549, NCI-H1299, H1650 and H358 and a normal lung cell line Beas-2B. ***P<0.001 vs. Beas-2B. Data are presented as the mean ± SD (n=3). circRNA, circular RNA; NSCLC, non-small cell lung carcinoma; RT-qPCR, reverse transcription-quantitative PCR.
Silencing of circRNA_103615 attenuates tumor progression in NSCLC
To investigate the function of circRNA_103615 in NSCLC, an siRNA that specifically targeted the circRNA_103615 junction sites was constructed (si-circRNA_103615) and transfected into the A549 cells. RT-qPCR was used to examine the transfection and silencing efficiency of si-circRNA_103615. The results confirmed that circRNA_103615 expression levels were significantly decreased in A549 cells transfected with the si-circRNA_103615 compared with the control (Fig. 2A). The effect of circRNA_103615 silencing in cell proliferation was analyzed by MTT assay. The results showed that the numbers of viable A549 cells were suppressed at 48-96 h following si-circRNA_103615 transfection (Fig. 2B), suggesting that knockdown of circRNA_103615 attenuated NSCLC cell proliferation in vitro. In addition, G1 arrest was induced by si-circRNA_103615 transfection (Fig. 2C), and cell apoptosis was promoted (Fig. 2D) compared with the NC-transfected A549 cells. Finally, the clone formation ability of A549 cell was significantly decreased following silencing of circRNA_103615 (Fig. 2E).
Figure 2.
Silencing of circRNA_103615 attenuates cell proliferation, migration and invasion in NSCLC cells. A549 cells were transfected with si-circRNA_0023642 or with the control si-NC. (A) Knockdown of circRNA_103615 was confirmed by reverse transcription-quantitative PCR. (B) Cell proliferation was examined by MTT assay at different timepoints. (C) Flow cytometry assays were performed to examine the cell cycle progression. (D) Apoptotic changes were detected by flow cytometry after annexin V-FITC staining. (E) Colony formation assays. (F) Cell migration was examined by uncoated Transwell chambers assays. (G) Cell invasion was examined by Matrigel-coated Transwell chambers assays. Data are presented as the mean ± SD (n=3). ***P<0.001 vs. NC. circRNA, circular RNA; NSCLC, non-small cell lung carcinoma; si, small interfering; NC, negative control.
The association analysis of circRNA_103615 expression with the clinicopathological features of the patients enrolled in the present study revealed that increased expression of circRNA_103615 was significantly associated with increased tumor size, advanced TNM stage and the presence of metastases (Table II). Because cell migration and invasion are processes associated with tumor progression and metastasis, the present study explored the effect of circRNA_103615 on NSCLC cell migration and invasion. Transwell assays were utilized to examine migration and invasion of A549 cells transfected with either si-circRNA_103615 or si-NC in vitro. The results revealed that cell migration (Fig. 2F) and invasion (Fig. 2G) of A549 cells significantly decreased following silencing of circRNA_103615 expression. These findings suggested that the downregulation of circRNA_103615 may attenuate tumor progression in NSCLC.
Table II.
Association between clinical features and circRNA_103615 expression in 60 patients with non-small cell lung carcinoma.
| circRNA_103615 expression | ||||
|---|---|---|---|---|
| Clinical feature | No. of cases | Low | High | P-value |
| Age (years) | 0.452 | |||
| <55 | 34 | 18 | 16 | |
| ≥55 | 26 | 12 | 14 | |
| Sex | 0.845 | |||
| Male | 30 | 16 | 14 | |
| Female | 30 | 14 | 16 | |
| TNM stage | <0.01 | |||
| I-II | 24 | 20 | 4 | |
| III-IV | 36 | 10 | 26 | |
| Tumor size | <0.01 | |||
| <5 cm | 27 | 19 | 8 | |
| ≥5 cm | 33 | 11 | 22 | |
| Metastasis | <0.01 | |||
| No | 27 | 22 | 5 | |
| Yes | 33 | 8 | 25 | |
circRNA, circular RNA; TNM, tumor-node-metastasis.
Silencing of circRNA_103615 reverses the cisplatin resistance of NSCLC cells in vitro
The most commonly used drug for the treatment of NSCLC is cisplatin. MTT assay was used to explore the potential role of circRNA_103615 in cisplatin resistance in NSCLC. The results demonstrated that, after 48 h of treatment, the IC50 values of cisplatin in A549 cells transfected with si-circRNA_103615 (1.18 µg/ml) were decreased compared with those in control A549 cells (3.29 µg/ml) (Fig. 3A). In addition, when A549 cells were treated with 2 µg/ml cisplatin for a week in colony formation assays, the cell growth rate was significantly inhibited in A549 cells following si-circRNA_103615 transfection compared with control A549 cells (Fig. 3B). Therefore, downregulation of circRNA_103615 reversed the cisplatin resistance of NSCLC cells in vitro.
Figure 3.
Silencing of circRNA_103615 reverses the cisplatin resistance of NSCLC cells in vitro. (A) The IC50 values of cisplatin in A549 cells transfected with si-circRNA_103615 or si-NC were detected by MTT assay. (B) A549 cells transfected with si-circRNA_103615 or si-NC were subjected to colony formation assays in the presence of 2 µg/ml. Data are presented as the mean ± SD (n=3). ***P<0.001 vs. NC. circRNA, circular RNA; NSCLC, non-small cell lung carcinoma; si, small interfering; NC, negative control.
circRNA_103615 enhances cisplatin resistance by promoting ABCB1 expression in NSCLC cells
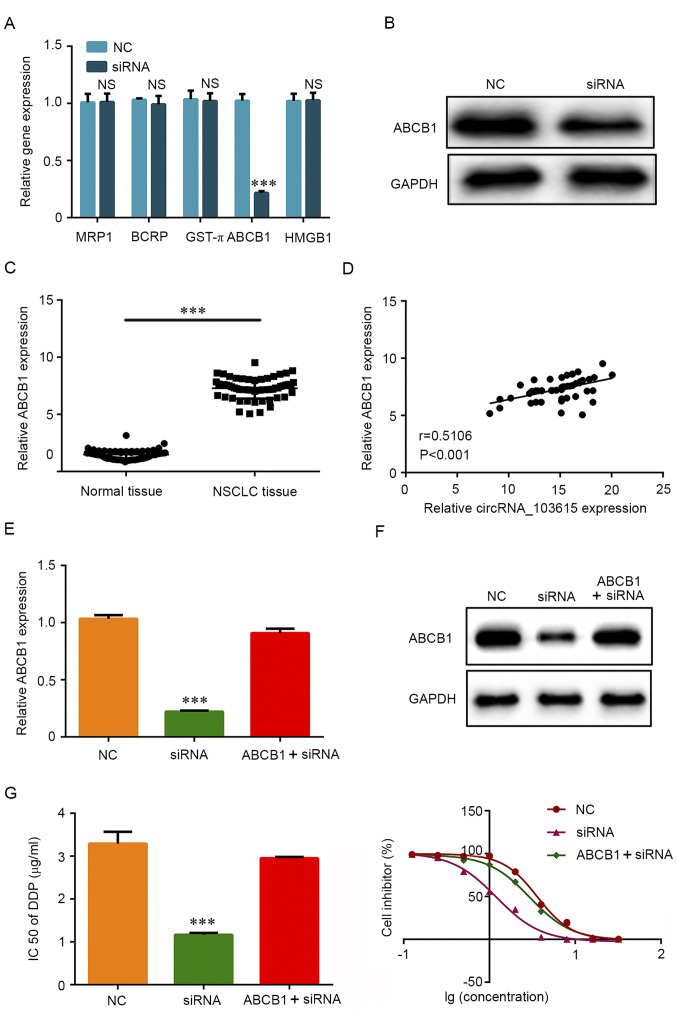
To explore the molecular mechanism underlying the role of circRNA_103615 in cisplatin resistance in NSCLC, RT-qPCR was used to examine the expression of a panel of classical multidrug resistance-related genes. The results reveled that the mRNA expression levels of ABCB1 were significantly decreased in A549 cells transfected with si-circRNA_103615 compared with NC-transfected A549 cells (Fig. 4A). No significant differences were observed in the expression levels of the classical multidrug resistance-related genes MRP1, BCRP, GST-π and HMGB1 (Fig. 4A). Furthermore, western blot analysis was used to detect the protein expression levels of ABCB1. The results demonstrated that ABCB1 protein expression levels were also markedly decreased in A549 cells following si-circRNA_103615 transfection compared with control A549 cells (Fig. 4B). In addition, ABCB1 mRNA expression levels were significantly increased in NSCLC tissues compared with normal tissues (Fig. 4C), and ABCB1 expression was positively correlated with circRNA_103615 expression in NSCLC tissues (Fig. 4D).
Figure 4.
circRNA_103615 enhances cisplatin resistance by promoting ABCB1 expression in NSCLC cells. (A) The mRNA expression levels of classical multidrug resistance-related genes were detected by RT-qPCR in A549 cells transfected with si-circRNA_103615 or si-NC. (B) The protein expression levels of ABCB1 were detected by western blotting in A549 cells transfected with si-circRNA_103615 or si-NC. (C) ABCB1 expression levels were examined by RT-qPCR in 60 NSCLC tissues and their adjacent normal tissues collected in the present study. ***P<0.001. (D) The correlation between circRNA_103615 and ABCB1 expression in the 60 NSCLC tissues was evaluated using Pearson's correlation analysis (n=60, r=0.5106, P<0.001). (E) A549 cells were transfected with si-NC or si-circRNA_103615, or co-transfected with si-circRNA_103615 and an ABCB1-overexpression plasmid. Successful ABCB1 overexpression was confirmed by RT-PCR and (F) western blot analysis. ***P<0.001 vs. NC. (G) The IC50 value of cisplatin was detected by MTT assay. ***P<0.001 vs. NC. Data are presented as the mean ± SD (n=3). circRNA, circular RNA; ABCB1, ATP binding cassette subfamily Bmember 1; NSCLC, non-small cell lung carcinoma; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering; NC, negative control; IC50, half-maximal inhibitory concentration.
To further examine whether the effects of circRNA_103615 were mediated by ABCB1, an ABCB1-overexpressing plasmid was transfected in A549 cell lines to restore ABCB1 expression. Firstly, the efficiency of the ABCB1-overexpressing plasmid pcDNA3.1-ABCB1 was confirmed by RT-qPCR (Fig. S1). The results showed that pcDNA3.1 could be used as a transfection control. RT-qPCR results (Fig. 4E) and western blotting results (Fig. 4F) confirmed that the expression of ABCB1 was successfully increased in cells co-transfected with si-circRNA_103615 and pcDNA3.1-ABCB1 compared with cells transfected with si-circRNA_103615 alone. Of note, the IC50 values of cisplatin in the ABCB1-overexpressing, circRNA_103615-silenced A549 cells (2.95 µg/ml) were significantly increased compared with the cells transfected with si-circRNA_103615 alone (1.17 µg/ml) (Fig. 4G). The present findings indicated that circRNA_103615 promoted ABCB1 expression and subsequently enhanced cisplatin resistance in NSCLC cells.
Discussion
Highly efficient sequencing and bioinformatic analysis technologies have aided in the identification and functional study of circRNAs. Several circRNAs have been found to be associated with progression, migration and invasion of NSCLC. For example, the upregulation of circRNA hsa_circ_0007534 predicts an unfavorable prognosis for NSCLC and exerts oncogenic properties in vitro and in vivo (17). Overexpression of circRNA_100876 has been shown to have prognostic value in NSCLC (13). Microarray analysis has shown that circRNA hsa_circ_0007385 functions as an oncogene in NSCLC tumorigenesis (18). Notably, circRNA acts as a prognostic biomarker and inhibits tumor progression through the mechanism of sponge miRNA (19,20). For example, a novel circular RNA, hsa_circ_0043278, regulates miR-520, acts as a potential biomarker and promotes NSCLC proliferation and migration (21). A microarray profile of circRNAs has identified hsa_circ_0014130 as a new circRNA biomarker in NSCLC (22). CircFADS2 regulates the proliferation and invasion of lung cancer cells via the sponging of miR-498(23). circRNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p (24).
In the present study and for the first time, circRNA_103615 was investigated in tissues from patients with NSCLC, and circRNA_103615 expression levels were demonstrated to be significantly upregulated in NSCLC tissues and cell lines. In addition, functional assays revealed that silencing of circRNA_103615 attenuated the proliferation, migration, and invasion of NSCLC cells and promoted cell apoptosis. Further studies will be required in the future to explore the signaling pathways by which circRNA_103615 affects cell proliferation, migration and invasion.
Research in the chemoresistance mechanisms in NSCLC is rare. Thus, investigating the molecular mechanisms involved in the chemoresistance of NSCLC may provide an understanding of improved therapies for NSCLC. In the present study, silencing of circRNA_103615 significantly decreased the cisplatin IC50 values of A549 cells and reduced the expression levels of the classical multidrug resistance gene ABCB1, suggesting that circRNA_103615 promoted ABCB1 expression in NSCLC cells, thus, enhancing cisplatin resistance. It is worth noting that the expression of other classical multidrug resistance-related genes, including MRP1, BCRP, GST-π and HMGB1, was not affected. Therefore, ABCB1 appears to be the predominant protein mediating the role of circRNA_103615 in chemoresistance. Further studies will be needed in the future to explore the signaling pathways affected by circRNA_103615 and ABCB1 in chemoresistance.
In conclusion, the present study demonstrated that circRNA_103615 promoted proliferation, migration, invasion and cisplatin resistance by enhancing ABCB1 expression in NSCLC cells in vitro. Therefore, circRNA_103615 may serve as a potential biomarker in NSCLC, and circRNA_103615 suppression may serve as a potential therapeutic strategy for NSCLC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WH, HoL and ZL conceived the current study. HoL and WH conceived and designed the experiments. HoL and ZL performed the experiments. HuL and LZ analyzed statistical data. WH interpreted the data and revised the manuscript. HoL and WH confirmed the authenticity of all the raw data All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
All procedures involving human samples were approved by the Human Resources Ethics Committee of the Fuda Cancer Hospital. Written informed consent was obtained from all the participants before enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71:3–10. doi: 10.1016/j.lungcan.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 3.Jeremic B, Milicic B, Dagovic A, Aleksandrovic J, Nikolic N. Pretreatment clinical prognostic factors in patients with stage IV non-small cell lung cancer (NSCLC) treated with chemotherapy. J Cancer Res Clin Oncol. 2003;129:114–122. doi: 10.1007/s00432-002-0408-4. [DOI] [PubMed] [Google Scholar]
- 4.Yano T, Okamoto T, Fukuyama S, Maehara Y. Therapeutic strategy for postoperative recurrence in patients with non-small cell lung cancer. World J Clin Oncol. 2014;5:1048–1054. doi: 10.5306/wjco.v5.i5.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo S, Iwata S, Yamada T, Inoue Y, Ichihara H, Kichikawa Y, Katayose T, Souta-Kuribara A, Yamazaki H, Hosono O, et al. Impact of the integrin signaling adaptor protein NEDD9 on prognosis and metastatic behavior of human lung cancer. Clin Cancer Res. 2012;18:6326–6338. doi: 10.1158/1078-0432.CCR-11-2162. [DOI] [PubMed] [Google Scholar]
- 6.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. circRNA: A novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 8.Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, Zhang P, Xiong Z, He C, Huang Z, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32(e22281) doi: 10.1002/jcla.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C, Yuan W, Yang X, Li P, Wang J, Han J, Tao J, Li P, Yang H, Lv Q, Zhang W. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(19) doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L, Song R. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 2018;18(116) doi: 10.1186/s12935-018-0602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, Nan KJ. Over-expression of circRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D, Ni Y. circRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem Biophys Res Commun. 2018;497:626–632. doi: 10.1016/j.bbrc.2018.02.119. [DOI] [PubMed] [Google Scholar]
- 15.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, Yin R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6(35576) doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, Xu Z, Zeng A, Zhang X, Zhang X, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(71) doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi Y, Zhang B, Wang J, Yao M. Upregulation of circular RNA hsa_circ_0007534 predicts unfavorable prognosis for NSCLC and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:79–85. doi: 10.1016/j.gene.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 18.Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X, Sun BH, Di QG. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144:667–674. doi: 10.1007/s00432-017-2576-2. [DOI] [PubMed] [Google Scholar]
- 19.Jin X, Feng CY, Xiang Z, Chen YP, Li YM. circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7:66455–66467. doi: 10.18632/oncotarget.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Cui J, Li W, Liu G, Chen X, Gao X, Lu H, Lin D. A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif Cells Nanomed Biotechnol. 2019;47:810–821. doi: 10.1080/21691401.2019.1575847. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, Yuan H. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8(2878) doi: 10.1038/s41598-018-21300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep. 2018;38(BSR20180570) doi: 10.1042/BSR20180570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T, Song Z, Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun. 2018;503:1503–1509. doi: 10.1016/j.bbrc.2018.07.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.