Summary
Apoptotic cells are quickly and efficiently engulfed and removed via the process of efferocytosis by either professional phagocytes such as macrophages or non-professional phagocytes including epithelial cells 1,2. In addition to debris removal, a key benefit of efferocytosis is that phagocytes engulfing apoptotic cells releases anti-inflammatory mediators3,4 that help reduce local tissue inflammation5; conversely, accumulation of uncleared apoptotic cells predisposes to a pro-inflammatory tissue milieu6–8. Due to their high proliferative capacity, intestinal epithelial cells (iEC) are sensitive to inflammation, irradiation, and chemotherapy induced DNA damage leading to apoptosis. Mechanisms of iEC death in the context of irradiation has been studied 9,10, but phagocytosis of dying iECs is poorly understood. Here, we identify an unexpected efferocytic role for Paneth cells, which reside in intestinal crypts and are linked to innate immunity and maintenance of the stem cell niche in the crypt11,12. Through a series of studies spanning in vitro efferocytosis, ex vivo intestinal organoids (‘enteroids’), and in vivo Cre-mediated deletion of Paneth cells, we show that Paneth cells mediate apoptotic cell uptake of dying neighbors. The relevance of Paneth cell-mediated efferocytosis was revealed ex vivo, and in mice after low dose Cesium-137 (137Cs) irradiation, mimicking radiation therapies given to cancer patients often causing significant apoptosis of iECs. These data advance a new concept that Paneth cells can act as phagocytes and identify another way in which Paneth cells contribute to the overall health of the intestine. These observations also have implications for individuals undergoing chemotherapy, or chronic inflammatory bowel disease.
In Brief
Shankman et al. uncover a new function for Paneth cells in phagocytic clearance of apoptotic intestinal epithelial cells. After demonstrating that Paneth cells engulf dying cells in enteroids ex vivo, they use mice with selective depletion of Paneth cells to show functional relevance for efferocytosis by Paneth cells during radiation treatments.
Results and Discussion
Apoptosis detection in enteroids and mouse models of Cesium-137 irradiation.
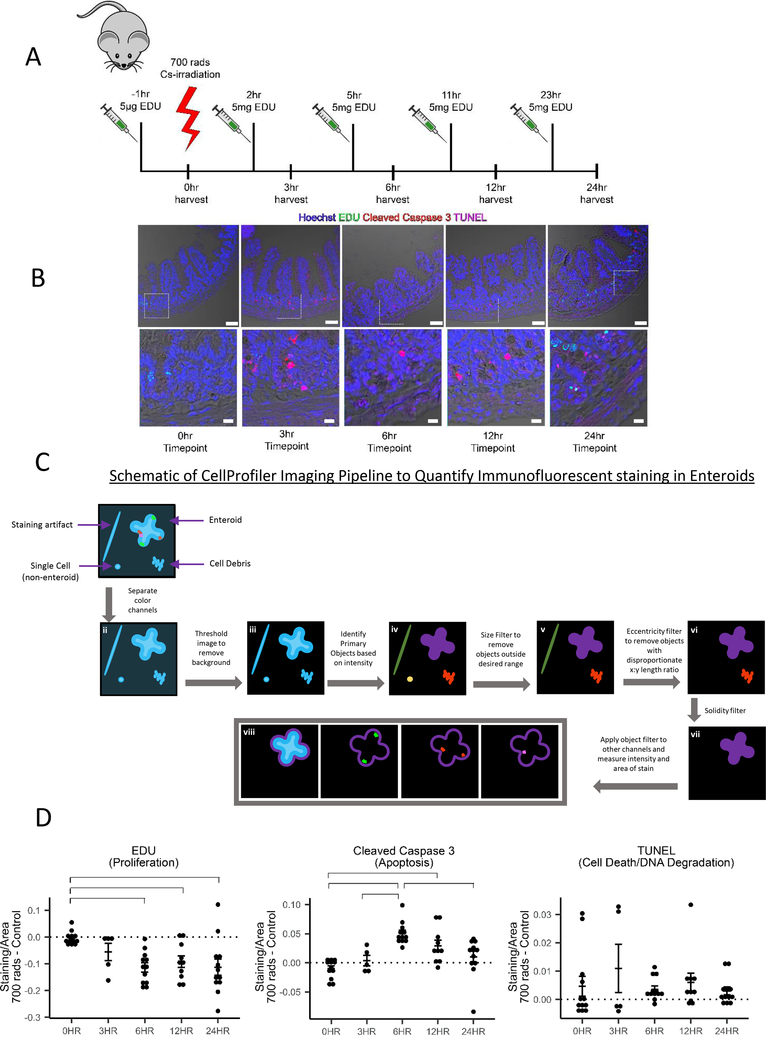
Patients undergoing irradiation receive multiple treatments of low dose irradiation in an attempt to kill rapidly proliferating cancer cells, yet this induces significant cell death of intestinal epithelial cells leading to mucositis. To model low dose irradiation in mice, we treated C57BL/6J mice with 700 rads of ionizing 137Cs irradiation (Figure 1A) and stained jejunal tissue for markers of proliferation (EDU), apoptosis (cleaved caspase-3), and late stage cell death (TUNEL), in addition to counterstaining with Hoechst (to mark the nuclei). We noted that peak apoptosis occurred between 3 and 6 hours post-irradiation and the proliferation began ~24h post-irradiation (Figure 1B).
Figure 1: Enteroids as a model for the progression of death and proliferation in the jejunum.
(A) Diagram of low dose irradiation treatment time course of C57BL/6J mice.
(B) Immunofluorescence staining of 4% PFA fixed paraffin embedded 5μm sections from mice 0, 3, 6, 12, and 24 hours post 700 rad 137Cs-irradiation. Hoechst (nuclei), EDU (proliferating cells), Cleaved Caspase-3 (apoptosis), TUNEL (late stage apoptosis/necrosis). Imaged on a Zeiss LSM880 with a 20X air objective. Scale bar = 50 μm. Below are digitally zoomed in images of the crypts using photoshop, scale bar = 10 μm.
(C) Graphical representation of the Cell Profiler pipeline used to analyze 137Cs-irradiated enteroids.
(D) Quantification of fluorescence intensity of EDU, Cleaved Caspase-3, and TUNEL staining in 700 rad 137Cs-irradiated enteroids. Irradiated samples were normalized to non-irradiated samples and as a function of the size of the enteroid. Data acquired from 12 independent mouse preparations of enteroids on 3 separate days. Significance was determined by 2-way ANOVA with Tukey post-test (p < 0.05) and is indicated by the brackets. (See also Figure S1)
We next sought to create a three-dimensional ex vivo tissue culture system of the small intestine (enteroids) to model the in vivo irradiation conditions. Four-day old enteroids derived from C57BL/6J mice were treated with 700 rads of ionizing irradiation and stained for EDU, cleaved caspase-3 and TUNEL. We created an analysis pipeline in Cell profiler13 to segment images from 137Cs-irradiated enteroids. Briefly, the cell profiler pipeline separated the fluorescence image channels and utilized the nuclear counterstain to filter out debris, and identify intact enteroids (Primary Objects) (see schematic in Figure 1C). Next, the pipeline applied the Primary Object mask to the EDU, cleaved caspase-3, and TUNEL channels, and the amount of fluorescence intensity was quantified (Figure 1D). To minimize variation due to enteroid preparation differences (Figure S1), we normalized the total amount of staining per area in the irradiated enteroids to non-irradiated enteroids (Figure 1D). Using this analysis pipeline, we found that the timeline of cleaved caspase-3 staining in the enteroid model closely followed that observed in the intact tissue of irradiated mice, peaking ~6 hours post 137Cs-irradiation and dissipating by 24 hours. Notably, if we do not normalize the EDU from the 137Cs-irradiated group to the control group, a clear increase in proliferation (i.e. resumed proliferation after 137Cs-irradiation) is seen at the 24-hour timepoint that is masked by the continued proliferation in the control group (Figure S1D, Figure S1E). Although TUNEL staining was absent in non-irradiated mice and enteroids, the TUNEL staining was variable in 137Cs-irradiated samples. Thus, cleaved caspase-3 (rather than TUNEL) appears to be a better indicator of injury and recovery in this model.
Paneth cells are phagocytic and can ingest apoptotic intestinal epithelial cells
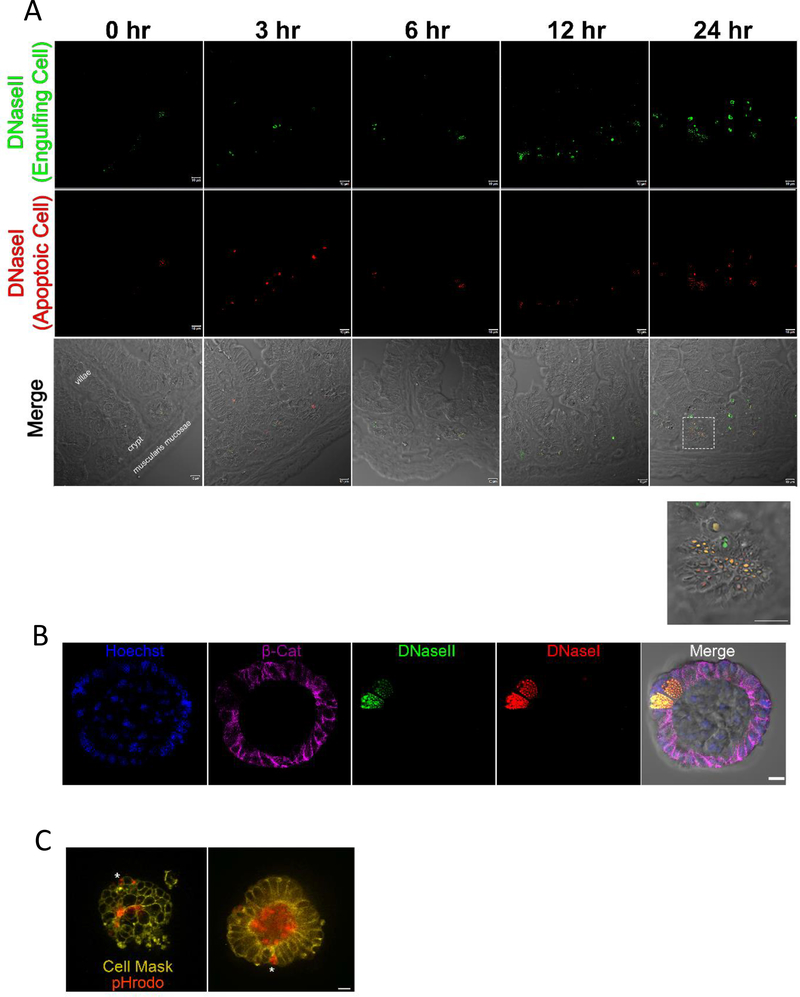
Apoptotic intestinal epithelial cells (iECs) are generally thought to be shed into the lumen and excreted14, at least as part of the routine turnover of the intestinal epithelial cells that occurs every 3–5 days. However, the irradiated mice had cleaved caspase-3 staining clustered in the crypt of small intestine, not the apex, and did not appear by morphology to be in the process of extrusion (Figure 1B). To further address the appearance of apoptotic cells and their efferocytosis by the neighboring cells, we stained sections from irradiated mice for both apoptotic cells and the engulfing phagocytes with Dual ISOL Apoptag (Figure 2A). The apoptag kit differentially stains the DNA break points created by DNaseI in the apoptotic cells versus DNaseII-mediated cleavage in the lysosome of efferocytic phagocytes that cleaves the DNA of the engulfed apoptotic cells15. DNaseII activity was found almost exclusively in the crypts compared to the villi (Figure 2A, Figure S2), consistent with the notion that apoptotic iECs are being engulfed by neighboring iECs. Interestingly, closer inspection showed puncta of DNaseII activity in the crypt correlated with cells having secretory granules similar to Paneth cells11. To further investigate this, we stained 4-day old irradiated enteroids with Dual ISOL Apoptag and β-Catenin (to visualize the iEC boundaries), which revealed that Paneth cells exclusively accounted for the DNaseII activity in the enteroids (Figure 2B). Paneth cells are central players in innate immunity in the intestine through their secretion of antimicrobials such as α-defensins, secretory phospholipase, and lysozyme11. Work from Hans Clevers’ laboratory has also delineated a key role for Paneth cells in the maintenance of the intestinal stem cell niche, in part through Wnt and Notch signaling pathways16,17. Paneth cells are also highly autophagic18, but the ability of Paneth cells to prune apoptotic cells in the crypt and the implications of their ability to engulf dying neighbors have not been investigated.
Figure 2: Paneth Cells can act as phagocytes within the small intestine.
(A) Immunofluorescence staining of 4% PFA fixed paraffin embedded 5μm sections from mice 0, 3, 6, 12, and 24 hours post 700 rad 137Cs-irradiation. Apoptag staining shows DNaseI (red) and DNaseII (green) staining. Imaged on a Zeiss LSM880 with a 63X oil objective. Scale bar = 10 μm. (See also Figure S2).
(B) Apoptag staining of 4 day old enteroids irradiated with 700 rads 137Cs-irradiated 6 hours prior to fixation with Cytofix/Cytoperm. Enteroids were also stained with nuclear counterstain, Hoechst (blue), and beta catenin (magenta). Imaged on a Zeiss LSM880 63X oil objective. Scale bar = 10 μm.
(C) Frame from live imaging of membrane labeled UV irradiated 4 day old enteroids (CellMask, yellow) co-stained with pHrodo. Asterix indicates engulfed cargo within an intact intestinal epithelial cell. Imaged on a Leica DMi8 inverted motorized microscope with temperature and CO2 controlled OKO chamber. Images were captured with a 63X oil objective. Scale bar = 10 μm. (See also Video S1 and Video S2)
In the enteroid system, we noted that all Paneth cells have DNaseII activity (data not shown). This was likely because the enteroids, unlike the intestine, cannot expel cells into the lumen to be eliminated by fecal excretion, and the Paneth cells remain in contact with apoptotic cells even in healthy enteroids. Given that engulfment of apoptotic cells and autophagy can share many of the same intracellular machinery19,20 we performed live imaging in the enteroid model using CellMask as a membrane marker, and propidium iodide (PI) as a dead cell marker. We then recorded cell death and subsequent engulfment of the apoptotic corpse (Video S1, Figure S3). Next, we stained enteroids with CellMask and the pH sensitive dye pHrodo, to demonstrate that acidification occurs within the iECs, indicative of acidic phagosome maturation of the engulfed apoptotic corpse (Figure 2C, Video S2).
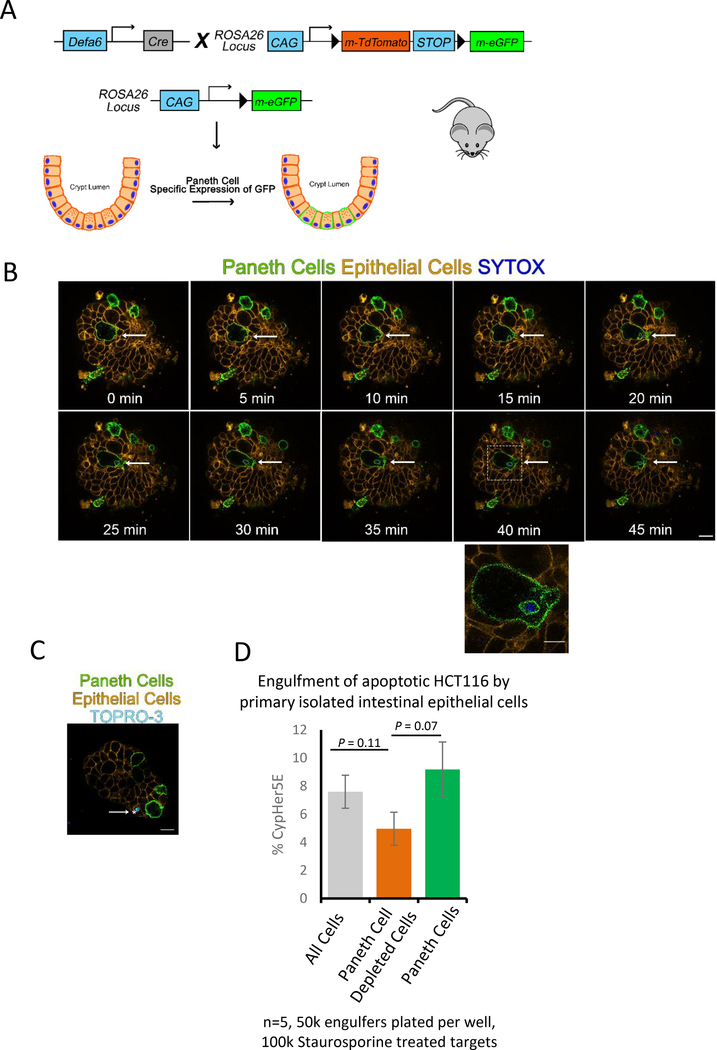
To definitively identify whether Paneth cells engulf dying iEC, we developed a mouse strain capable of tracking Paneth cells, denoted Defa6-cremTmG. To generate Defa6-cremTmG, we crossed Paneth cell specific Defa6-cre mice21 to the mT/mG mouse22 strain that basally expresses membrane targeted tdTomato in all cells, but irreversibly switches to membrane targeted GFP in Paneth cells after a Cre mediated excision event (Figure 3A). Therefore, in the Defa6-cremTmG mice, Paneth cell membranes are green, while all non-Paneth cell membranes are red. Next, we induced apoptotic death of specific cells within enteroids derived from Defa6-cremTmG mice by laser ablation. We confirmed cell death by testing the uptake of TO-PRO-3 and SYTOX BLUE, DNA staining dyes that are normally impermeable in live cells but permeable in apoptotic cells (Figure S4A, Video S3). Next, enteroids were live imaged where we discovered Paneth cells that engulf nearby dying cells (Figure 3B). We could also visualize TO-PRO-3 positive engulfed vesicles in other epithelial cells (Figure 3C). It is important to note that the presence of TO-PRO-3 or SYTOX BLUE indicates that the portions of apoptotic cells ingested by the Paneth cells must contain DNA from a dying cell.
Figure 3: Paneth cells can mediate uptake of apoptotic cells.
(A) Diagram of Paneth cell tracing mouse (Defa6-cremTmG) generation.
(B) Frames from live imaging of enteroids from Defa6-cremTmG. Arrow indicates cell that was killed using laser ablation with a 405nm laser and partially engulfed by neighboring Paneth cell (Green). Enteroid was scanned once every 5min by a LSM780 scanning confocal microscope with temperature and CO2 controlled environmental chamber. 63X oil objective. Scale bar = 10 μm. Zoom in region Scale bar = 5 μm. (See also Figure S3 and Video S3).
(C) Defa6-cremTmG (iECs = orange, Paneth Cells = green) co-stained with TO-PRO-3. Arrows indicate cells that have ingested DNA from a dying cell. Imaged on a Zeiss LSM780 with a 63X oil objective. Scale bar = 10 μm.
(D) Percentage of mouse iECs from Defa6-cremTmG mice that engulfed apoptotic HCT116 cells. Two tailed t-tests were used to compare groups. Bars are standard error of five independent experiments. (See also Figure S4).
To complement the image analysis above, we developed an ex vivo flow cytometry-based engulfment model using the Defa6-cremTmG mice to determine the efferocytosis by Paneth cells. Fluorescence activated cell sorting was performed on intestinal tissue of Defa6-cremTmG mice to create single cell suspensions of Paneth cells, non-Paneth cells, or both, which were then plated on a matrigel coated 96-well plate. These cells were fed CypHer5E-labeled apoptotic human colonic epithelial cells (HCT116 cells) (Figure S4B). CypHer5E has been reliably used to study phagocytosis23–25 as its fluorescence increases during acidification, allowing us to distinguish between corpse binding versus internalization and processing of the ingested apoptotic cells. We found that purified Paneth cells could ingest apoptotic cells in this assay, further confirming that Paneth cells can function as phagocytes. When we used the total iEC, approximately 8% of the iECs were capable of phagocytosing apoptotic HCT116 cells. Interestingly, when the Paneth cells were removed from the total iEC, the engulfment capacity of this population dropped by a third (Figure 3D).
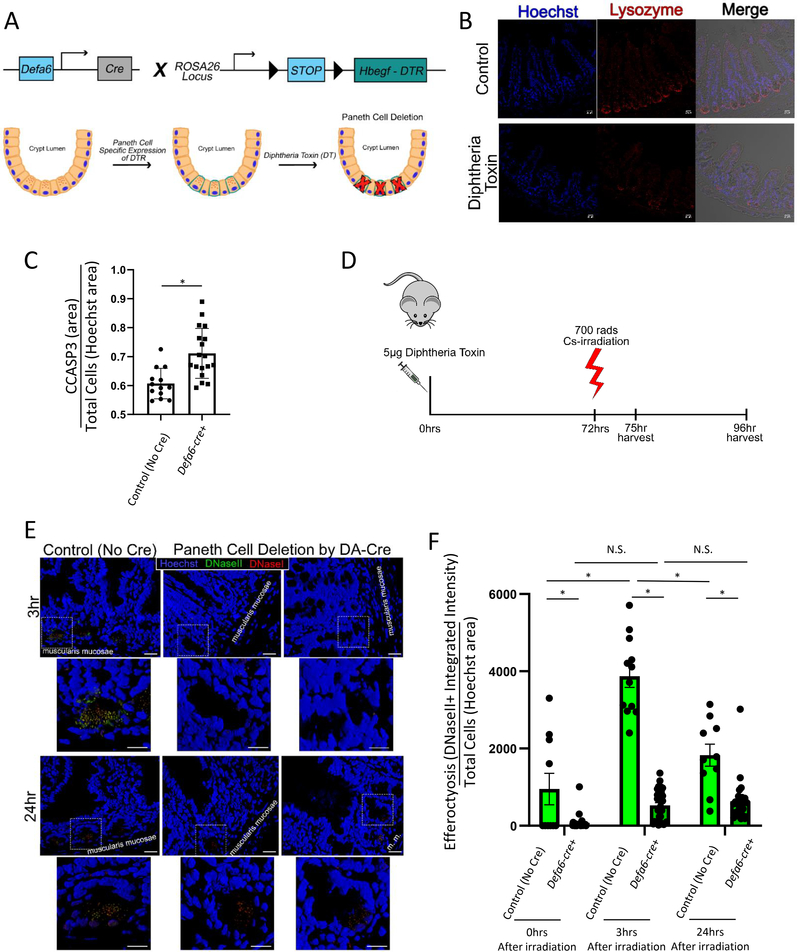
Loss of Paneth cells reduces efferocytosis during irradiation
To investigate the requirement of Paneth cell mediated phagocytosis in vivo, we sought to create an experimental system where we could inducibly cause the loss of Paneth cells. For this, we used the well-established diphtheria toxin (DT) model. Although mice normally lack the receptor for diphtheria toxin, transgenic expression of the human diphtheria toxin receptor (DTR) in the tissue of choice causes it to be susceptible to DT-induced cell death. Next, we crossed the Paneth cell specific Defa6-cre mice to an inducible diphtheria toxin receptor (ROSA26iDTR26) expressing mice to generate Defa6-cre ROSAiDTR/+ mice (Figure 4A). In Defa6-cre ROSAiDTR/+ mice, we expected the DT treatment to specifically induce death of Paneth cells. Administration of diphtheria toxin was able to selectively induce apoptosis of Paneth cells in vivo after 3 days27, as confirmed by the loss of Paneth cells (by lysozyme staining, Figure 4B). At this time we also noted that there was a significant increase in cleaved caspase 3 staining within the crypts of Defa6-cre ROSAiDTR/+ treated with diphtheria toxin, which is likely due to the apoptotic death of the Paneth cells, but might also be explained by the lack of Paneth cells to clear nearby apoptotic cells (Figure 4C). Next, we induced death of intestinal epithelial cells via irradiation in these mice (Figure 4D) and scored the intensity of signal for apoptotic (DNaseI) versus engulfed apoptotic (DNaseII) cells using the ApopTag kit. In the Defa6-cre ROSAiDTR/+ mice administered with diphtheria toxin and the resultant loss of Paneth cells, we see a notable decrease in the amount of DNaseII staining (Figure 4E), compared to control mice (ROSAiDTR/+ mice lacking Cre) given the diphtheria toxin. Of particular interest, there is a clear increase in efferocytosis (indicated by increased DNaseII staining cells) in the control mice at 3hr and 24hr post irradiation that is absent in the Defa6-cre ROSAiDTR/+ mice treated with DT (Figure 4F). These data suggested that the Paneth cells contribute to the in vivo clearance of dying cells in a model of irradiation-induced cell death.
Figure 4: Specific ablation of Paneth cells reduces cell clearance in vivo after irradiation.
(A) Schematic of diphtheria toxin induced Paneth cell depletion approach.
(B) Immunofluorescence staining of 4% PFA fixed paraffin embedded 5μm sections from Paneth cell depleted, irradiated mice stained for nuclei (Hoechst = blue), and Paneth cells (lysozyme = red). Imaged on a Zeiss LSM880 with a 20X air objective. Scale bar = 20 μm.
(C) Quantification of CCASP3 staining area as a function of Hoechst staining area in intestinal crypts (jejunum) prior to irradiation. Unpaired two tailed t-test was used to determine statistical significance p < 0.05.
(D) Timeline of diphtheria toxin induced Paneth cell depletion and analysis.
(E) Three dimensional rendering of Paneth cell depleted, irradiated mice stained for Apoptag (DNaseI = red, DNaseII = green) with nuclear counterstaining (Hoechst = blue). Imaged on a Zeiss LSM780 with a 63X oil objective. Three dimensional rendering created using Zen Black. Original images scale bar = 20 μm. Zoom-in images scale bar = 10 μm.
(F) Quantification of DNaseII activity (integrated intensity) in intestinal crypts (in jejunum) after 700 rads 137Cs-irradiation normalized to cell density (Hoechst nuclear counterstaining area) using FIJI. 0hr: Defa6-cre- (control) n = 13, Defa6-cre+ n = 30; 3hr: Defa6-cre- (control) n = 12, Defa6-cre+ n = 33; 24hr: Defa6-cre- (control) n = 10, Defa6-cre+ n = 27. 2-way ANOVA with Tukey post tests were used to determine statistical significance. p < 0.05.
Collectively, these data advance a new concept and has several implications. First, our data identify Paneth cells as previously unrecognized phagocytes within the intestinal tissue. Through a combination of in vitro, ex vivo, and in vivo approaches, we show that Paneth cells can engulf neighboring apoptotic cells in the small intestine after 137Cs-irradiation induced injury. Paneth cells are important in the maintenance of intestinal health in homeostatic conditions by maintaining the Lgr5+ intestinal stem cell niche and preventing bacterial infections11. While recent studies show that other iEC cell types such as Tuft cells and enteroendocrine cells can help compensate (at least in part) for the loss of Paneth cells in homeostatic conditions27, Paneth cells are necessary to help clear dying cells after irradiation. While our study reveals the efferocytic role of Paneth cells in clearing apoptotic cells in the intestine, the recently identified ‘gMacs’ that reside near the intestinal crypt might also contribute to apoptotic cell clearance28. Though infrequent, we noticed rare instances of DNaseII activity within Paneth cells in vivo in non-irradiated conditions (Figure 2A), suggesting that Paneth cells have an innate ability to act as phagocytes that is enhanced in disease contexts. Work by Yu et al. explore the capacity of Paneth cells to enter a multipotent state after lethal irradiation29. While this may partially explain how Paneth cells are able to take on a phagocytic role in our sublethal irradiation model, we show in Figure 3B and Figure 3D that Paneth cells can also engulf neighboring cells in the absence of direct irradiation/damage. Little is known about apoptotic recognition receptors and their role in mucositis, in part because cellular engulfment in the intestine by iECs is a nascent field1,15, and represents an exciting new line of research.
The new efferocytic role of Paneth cells identified here has possible implications for diseases such as Crohn’s disease and inflammatory bowel disease, where Paneth cell dysfunction plays a major role in disease progression 18,21. These data also have implications for cancer patients as many individuals with late stage cancer receive aggressive therapies that include some amount of radiation therapy. Recent reports state that approximately 40% of all cancer therapy patients experience gastrointestinal (GI) mucositis during their treatment, with the number jumping to 80% in patients receiving abdominal or pelvic irradiation30. Mucositis is the most common, painful, and multi-symptom side effect limiting the effective therapeutic dosage required to eradicate cancer31, and is caused by a massive surge of apoptotic epithelial cells in the gastrointestinal tract32–34. The discovery of Paneth cells as functionally relevant phagocytes in the intestine may represent a unique target to help alleviate intestinal inflammation in patients suffering from mucositis, by boosting Paneth cell-mediated efferocytosis. In this work, we also present a new type of CellProfiler pipeline, capable of segmenting enteroids based on their nuclear counterstain with low resolution images (both basally and after Cs irradiation). Whereas other modeling systems focus on gross death of enteroids, our semi-automated method to study a specific insult and subsequent recovery in enteroids has advantages that may be useful to other investigators.
STAR Methods:
RESOURCE AVAILABILTY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Kodi Ravichandran ravi@virginia.edu
Materials Availability
Defa6-cre mouse line was provided by Dr. Richard S. Blumberg from Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA, USA, and is subject to MTA restriction. All other mouse lines are commercially available and listed in the key resources table.
No other unique reagents were generated in this study.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| cleaved caspase-3 | Cell Signaling Technology | Cat# 9661 |
| lysozyme | BioGenex | Cat# AR024-10R |
| Donkey anti-Goat IgG (H+L) Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A-11056 |
| AnnexinV-FITC | Biolegend | Cat# 640905 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 5-ethynyl-2′-deoxyuridine (EDU) | GeneCopoeia | Cat# A003 |
| Diphtheria Toxin (DT) | Sigma Aldrich | Cat# 322326-1MG |
| Cytofix/Cytoperm | BD Biosciences | Cat# 554722 |
| ProLong Gold antifade reagent | Thermo Fisher Scientific | Cat# P36934 |
| 20% Paraformaldehyde | Electron Microscopy Sciences | Cat# 15713 |
| Antigen Unmasking Solution, Citrate-Based | Vector Laboratories | Cat# H-3300 |
| Staurosporine | Millipore Sigma | Cat# S5921 |
| 0.25% trypsin | Corning | Cat# 25-053Cl |
| Hoechst 33342 | Thermo Fisher Scientific | Catt# H3570 |
| propidium iodide | Thermo Fisher Scientific | Cat# P1304MP |
| 7-AAD | Biolegend | Cat# 420403 |
| TO-PRO-3 | Thermo Fisher Scientific | Cat# T3605 |
| CellMask Green | Thermo Fisher Scientific | Cat# C37608 |
| Cell trace violet | Thermo Fisher Scientific | Cat# C34571 |
| CypHer5E NHS Ester | GEHealhcare | Cat# PA15401 |
| EDTA (0.5 M), pH 8.0, RNase-free | Invitrogen | Cat# AM9260G |
| Critical Commercial Assays | ||
| CF640R TUNEL Assay | Biotium | Cat# 30074 |
| iClick EdU Andy Fluor 488 Imaging Kit | GeneCopoeia | Cat# A003 |
| ApopTag ISOL Dual Fluorescence Apoptosis Detection Kit | Milipore | Cat# APT1000 |
| Experimental Models: Cell Lines | ||
| HCT116 human colon cells (ATCC CCL-247) | American Type Culture Collection | https://www.atcc.org/products/all/CCL-247.aspx |
| Experimental Models: Organisms/Strains | ||
| Mouse: JAX C57BL/6J Mice | Jackson Laboratory | https://www.jax.org/strain/000664 |
| Mouse: Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratory | https://www.jax.org/strain/007676 |
| Mouse: C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J | Jackson Laboratory | https://www.jax.org/strain/007900 |
| Mouse: Defensin-6-cre (Defa6-cre) | Blumberg Laboratory | https://www.hms.harvard.edu/dms/immunology/fac/Blumberg.php |
| Software and Algorithms | ||
| CellProfiler v2.2 | Broad Institute of Harvard and MIT | https://cellprofiler.org/previous-releases |
| FIJI | Laboratory for Optical and Computation Instrumentation | https://imagej.net/Downloads |
| R | R Foundation | https://www.r-project.org/ |
| Prism 9 | GraphPad Software | www.graphpad.com |
| Enteroid Analysis Pipeline | This Paper | https://github.com/lsshankman/Paneth-Cells-as-novel-efferocytic-phagocytes-within-the-intestine |
| Other | ||
| MACS dead cell removal kit | Miltenyi Biotec | Cat# 130-090-101 |
| McCoy’s 5A Modified Medium | Thermo Fisher Scientific | Cat# 12330031 |
| intesticult media | Stem Cell Technologies | Cat# 06005 |
| Matrigel GFR Basement Membrane Matrix | Corning | Cat# 354230 |
| CellBIND 96-well Clear Flat Bottom Polystyrene Microplate | Corning | Cat# 3300 |
| L-Glutamine:Penicillin:Streptomycin (PSQ) | Thermo Fisher Scientific | Cat# 50-753-3016 |
| 70μm nylon filter | Corning | Cat# CLS431751 |
| 8 well glass chambered slides | Thermo Fisher Scientific | Cat# 154534 |
| 4 well coverslip bottom slides | Thermo Fisher Scientific | Cat# 155382 |
| pluriStrainer® 20 μm | pluriSelect | Cat# 43-50020-03 |
Data and Code Availability
The CellProfiler Pipeline used for this study is publicly available at https://github.com/lsshankman/Paneth-Cells-as-novel-efferocytic-phagocytes-within-the-intestine.
Since the time of its creation, CellProfiler has released several new software versions that are incompatible with the pipeline. User must also install CellProfiler v2.2 from https://cellprofiler.org/previous-releases.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice were given ad libitum access to food and water and housed on a 12–12 hour light-dark cycle. Sex matched 5–8 weeks old C57BL/6J mice (Jackson Laboratories) were used for the experiments in Figures 1 and 2. Defensin-6-cre (Defa6-cre)21 were provided by Dr. Richard S. Blumberg from Brigham and Women’s Hospital, and Harvard Medical School, and subsequently crossed with either Gt(ROSA)26Sortm4(ACTB-tdTomato, EGFP)Luo/J strain22 (Figure 3, mT/mG, Jackson laboratories Stock 007676) or C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J strain26 (ROSA26iDTR, Jackson laboratories stock 007900). For the experiments in Figure 4, mice were injected with diphtheria toxin 3 days prior to exposure to 700 rads137 Cs-irradiation. All mice were monitored by vivarium veterinary technicians for signs of distress and euthanized when recommended. Animals were maintained using standard husbandry and housing practices and treated according to an approved ACUC animal protocol (2992) on file at the University of Virginia.
Cell Culture
HCT116 human colon cells (ATCC CCL-247) were maintained by plating 1×104 cells in a 100mm plate in 5mL of McCoy’s 5A media with 10% BSA and 1% PSQ and passaged every three days. Cells were kept in a 37°C incubator with 5% CO2.
Enteroid Culture
Enteroids were harvested based on the protocol from Stem Cell Technologies with a few modifications. The entire small intestine was isolated in a biological safety cabinet and flushed with 10 mL of cold 1X PBS without Mg2+ Ca2+ (Corning, 21–040-CV) + 1% PSQ (Fisher scientific 50–753-3016) using a 25G needle. Tissue was then placed in cold 1XPBS + 1% PSQ, flayed, cut into 2mm segments, and transferred to 50mL conical tube containing cold 1XPBS + 1% PSQ. Tissue was rinsed 5 times by inverting the tube 6 times, allowing tissue to settle, aspirating supernatant, and adding 15mL cold 1XPBS + 1% PSQ. After the fifth wash, supernatant was aspirated and 25mL of chelation buffer (cold 1XPBS + 1% PSQ + 2mM EDTA) was added to the tissue. Tissue was allowed to rock for 1hr at 4°C. Next, the tissue was washed an additional 3 times with cold 1XPBS + 1% PSQ + 0.1%BSA, this time using a pipetman instead of tube inversion. On the third wash, the supernatant was strained through a 70μm nylon filter (Corning CLS431751–50EA) into a new 50mL conical tube and spun down at 200g x 5 min at 10°C. Supernatant was removed, tissue was resuspended in 10mL of cold 1XPBS + 1% PSQ + 0.1%BSA and transferred to a 15mL conical tube. 10μL of sample was removed to count the number of intact crypts for plating. Plating density was calculated and the sample was spun down and resuspended in media [1:1 ratio of intesticult media (Stem Cell Technologies #06005 + 1%PSQ) and growth factor depleted matrigel (corning # 354230)]. Plating density for imaging experiments was 200 crypts / 50 μL plated on 8 well glass chamber slides (Thermo Fisher Scientific #154534) or 4 well coverslip bottom slides (Thermo Fisher Scientific #155382). Media was changed daily.
METHOD DETAILS
Mouse Irradiation and Diphtheria Toxin Treatment
For irradiation experiments, mice were subjected to 700 rads 137Cs-irradiation using a Shepperd Mark I irradiator. One hour prior to euthanasia, mice were injected with 5-ethynyl-2’-deoxyuridine (EDU : 5μg/ 20g mouse weight) to label actively proliferating cells. Defensin-6-cre (Defa6-cre)21 were crossed to Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J strain22 (mT/mG, Jackson Labs Stock 007676) to visualize Paneth cells from other epithelial cells within the mouse small intestine (Figure 3). Defa6-cre mice were also crossed to C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J strain26 (ROSA26iDTR mice) to determine the role of Paneth cell loss in vivo. At 6 weeks of age these mice were injected with 50 ng/μL diphtheria toxin (100 μL total volume, Sigma-Aldrich #322326) to eliminate the majority of Paneth cells within the small intestine. Seventy-two hours later, the mice were subjected to 700 rads 137Cs-irradiation. One hour prior to euthanasia mice were injected with 5-ethynyl-2’-deoxyuridine (EDU : 5μg/ 20g mouse) to label the actively proliferating cells. All mice were euthanized according to an approved ACUC animal protocol on file at the University of Virginia.
Irradiation of Enteroids
All preparations were allowed to grow for four days, media changed daily, prior to analysis. Enteroids were irradiated with either 700rads of 137Cs-irradiation (Shepperd Mark I) or 200mJ of UV (Stratagene, Stratalinker 1800). Plating density for imaging experiments was 200 crypts / 50 μL plated on 8 well glass chamber slides (Thermo Fisher Scientific #154534) or 4 well coverslip bottom slides (Thermo Fisher Scientific #155382).
Live Imaging of Enteroids
Four day old enteroids plated on 4 well coverslip bottom slides (Thermo Fisher Scientific #155382) were stained with TO-PRO-3 (Thermo Fisher Scientific #T3605, 1:1000) or CellMask Green (Thermo Fisher Scientific #C37608, 1:200 for 30min) and imaged at the University of Virginia’s Keck Imaging Center using a Zeiss 780 NLO scanning confocal microscope. Samples were kept in a temperature and CO2 controlled environmental chamber and imaged with an oil immersive 63X objective. Cells were targeted for death by overexposure to 405nm laser at 100% power for 20 iterations. Images were analyzed using Zen Black 2.3 SP1 software.
Enteroid Preparation for Flow Cytometry
For flow cytometry based analysis, after the chelation step (see STAR METHODS), supernatant was removed and tissue was forcefully dissociated by titrating six times with 10mL of cold 1XPBS + 1% PSQ + 0.1%BSA. Supernatant was transferred to a new tube and spun for 5 min x 100g at 10°C. Supernatant was removed and tissue is resuspended in warm media (Stem Cell Technologies #06005 + 1%PSQ) and incubated for 45min at 37°C. Next the tissue was titrated through a 20G needle five times and passed through a 20μm filter (pluriSelect #43–50020-03). After adding 5ml of 1XPBS + 1% PSQ + 0.1%BSA to the suspension and spinning for 5 min at 100g at 10°C, a MACS dead cell removal kit (Miltenyi Biotec #130–090-101) was used to remove some of the cellular debris prior to flow cytometry sorting.
Immunofluorescence
Four day old enteroids were treated with 2μM EDU 30min prior to irradiation of enteroids with 700rads of Cs-irradiation. Media was removed and replaced with fresh media immediately after irradiation. At the indicated timepoint, media was removed and enteroids were fixed for 15min with BD Cytofix/Cytoperm (#554722) and stored at 4°C in 1X PBS until ready for staining. One set of enteroids underwent apoptosis detection using the CF640R TUNEL Assay (Biotium #30074) allowing the reagent to sit for 2 hours at 37°C before EDU detection (GeneCopoeia #A003) for 30min. Finally, the slides were stained for cleaved caspase-3 (Cell Signaling Techology #9661, 1:200; #A-11056, 1:100), nuclear counterstained with Hoechst 33342 (ThermoFisher Scientific #H3570, 1:10000), and mounted using ProLong Gold antifade reagent (ThermoFisher Scientific #P36934). The second set of enteroids were stained with ApopTag ISOL Dual Fluorescence Apoptosis Detection Kit (Milipore #APT1000) to detect DNaseI and DNaseII activity and then nuclear counterstained with Hoechst before coverslipping. Samples stained for Hoechst, EDU, Cleaved Caspase 3, and TUNEL were imaged using a Zeiss inverted epifluorescence microscope. A 0.5cm2 area from each well was captured and analyzed using CellProfiler v2.2.
Tissue from mice irradiated with 700 rads Cs-irradiation were fixed with 4% paraformaldehyde in 1XPBS (Electron Microscopy Sciences #15713) for 24 hours, paraffin embedded, and sectioned at 5μm. Slides were rehydrated (5min 2X zylenes, 2X 100% EtOH, 2X 95% EtOH, 1X 80% EtOH, 2X dH2O) and underwent a citrate based antigen unmasking (Vector #H-3300) using a microwave to maintain a constant boil for 20min. Tissues were blocked for 30min at RT in 1X PBS + 3% BSA + 10% Normal goat serum (NGS) before staining. Tissue was stained with the same protocols listed above. One set of slides were stained with lysozyme diluted 1:2 with blocking solution (BioGenex #AR024–10R; ThermoFisher Scientific #A-11056, 1:100) and nuclear counterstained with Hoechst.
Fluorescence-activated Cell Sorting
Cells isolated from Defa6-cre Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J and C57BL/6J enteroids were stained with TO-PRO-3 (1:1000) and run on an Influx Cell Sorter run by the University of Virginia Flow Cytometry Core. TO-PRO-3 dead cells were gated out and cells were sorted based on GFP or tdTomato expression.
Apoptosis Induction in HCT116 Target Cells
1×104 Human colonic epithelial cells (HCT116) were plated on a 100mm plate in 5mL of McCoy’s 5A media with 10% BSA and 1% PSQ two days prior to experimentation. Twenty-four hours prior to experimentation media was replaced with new media containing 1μM Staurosporine (Millipore Sigma #S5921). On the day of the experiment, cells were dissociated using 0.25% trypsin (Corning #25–053Cl) for five minutes and the level of apoptosis was checked by flow cytometry of cells stained with AnnexinV-FITC (1:50, Biolegend #640905) and 7-AAD (1:1000, Biolegend #420403).
Ex vivo Phagocytosis Assay
FACS sorted cells were plated 50,000 cells per well in a 3% matrigel coated (corning # 354230) 96 well plate (Corning #3300) and allowed to rest overnight at 37°C. Cells were then labeled with cell trace violet (ThermoFisher Scientific #C34571) for 30min at 37°C. Simultaneously, apoptotic HCT116 cells were labeled with CypHer5E (GE Healthcare, PA15401) as previously described15. The apoptotic population was purified by running the HCT116 cells over a MACS dead cell removal column (Miltenyi Biotec #130–090-101) and harvesting the bound population. Apoptotic HCT116 cells were added to the FACS sorted cells at a 2:1 ratio and incubated for 3 hours at 37°C before harvesting the cells by removing the supernatant and adding 0.25% trypsin for 5min. Cells were then run on an ATTUNE flow cytometer and analyzed for the amount of CypHer5E staining. Flow cytometry data was analyzed using FlowJo10.0.7.
Cell Profiler Pipeline for Enteroid Detection and Analysis
CellProfiler13 v2.2 DateRevision:20140723174500 was used to analyze images collected on a Zeiss inverted epifluorescence microscope (CellProfiler.org provided by the Broad Institute). In short, a nuclear counterstained image (Hoechst) was used to discriminate cellular debris and artifacts from intact enteroids using modules applyThreshold, IdentifyPrimaryObjects (Otsu 2-class global thresholding and Laplacian of Gaussian filters to distinguish clumped objects) MeasureObjectSizeShape, FilterObjects (filtered by area, solidity, and eccentricity). The primary objects were then used to measure the fluorescence intensity of the EDU, cleaved caspase-3, and TUNEL staining images. Data was exported to excel for further analysis. CellProfiler pipelines can be found at: https://github.com/lsshankman/Paneth-Cells-as-novel-efferocytic-phagocytes-within-the-intestine.
FIJI for Crypt Determination and Analysis
FIJI 35 was used to identify the intestinal crypts based on morphology in the bright field channel. Next, the z-stack fluorescent image was compressed using the extended depth of field plugin36 that locates the plane that is most in focus for each subregion of the image to convert a z-stack image into a 2D image. The crypt ROIs were then used to measure the area of positive signal and intensity of signal in each channel.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistics were performed in R and Prism 9 (GraphPad Software).
Analysis of enteroid staining
Twelve independent mice from three unique breeder pairs of C57B6/J mice were used to derive enteroid cultures for analysis. Each timepoint contained two replicate wells of enteroids, each well was seeded with 200 enteroids. Enteroids that met the requirements of the CellProfiler pipeline were analyzed for total area and integrated intensity of Hoechst (DNA), EDU (proliferation), CCASP3 (apoptosis), and TUNEL (late-stage apoptosis/necrosis). Data was plotted as the difference in integrated intensity between the 137Cs-irradiated sample compared to the non-irradiated control as a function of enteroid size. One-way ANOVA with Tukey post-hoc tests were performed and significance was determined by a p-value of less than 0.05.
Quantification of HCT116 engulfment by primary intestinal epithelial cells
Five independent experiments were conducted to determine the number of intestinal epithelial cells that were positive for CypHer5E (evidence of acidification in lysosomal compartments after engulfment of labeled, apoptotic HCT116 cells). In each experiment there were three replicates of the “all cells”, and “Paneth Cell Depleted Cells” groups and a single replicate of “Paneth Cells”. Significance was determined by a p-value of less than 0.05 using an unpaired 2-tailed t-test.
Quantification of cleaved caspase-3 staining in intestines
Crypts of intestines were drawn on blinded DIC images of mouse intestines. FIJI was then used to determine the area and integrated intensity of CCASP3 and Hoechst staining. Analysis included: Defa6-cre- (control) n = 13, Defa6-cre+ n = 18. Significance was determined by a p-value of less than 0.05 using an unpaired 2-tailed t-test.
Quantification of DNaseI/II staining in intestines
Crypts of intestines were drawn on blinded DIC images of mouse intestines. FIJI was then used to determine the area and integrated intensity of DNaseI, DNaseII, and Hoechst staining. Analysis included: 0hr: Defa6-cre- (control) n = 13, Defa6-cre+ n = 30; 3hr: Defa6-cre- (control) n = 12, Defa6-cre+ n = 33; 24hr: Defa6-cre- (control) n = 10, Defa6-cre+ n = 27. 2-way ANOVA with Tukey post hoc tests were used to determine statistical significance. p < 0.05
Supplementary Material
Video S1: Live imaging of enteroids reveals engulfment of dead cells by epithelial cells within the enteroid, Related to Figure 2. Three day old enteroids were irradiated with 200mJ UV light and stained with CellMask and propidium iodine (red). Images were acquired once every 5min using a Leica DMi8 inverted motorized microscope with temperature and CO2 controlled OKO chamber and a 63X oil objective. Scale bar = 5μm.
Video S2: Live imaging of enteroids revealed acidification of internalized cargo by epithelial cells within the enteroid, Related to Figure 2. Three day old enteroids were irradiated with 200mJ UV light and stained with CellMask and pHrodo red. Images were acquired once every 5min using a Leica DMi8 inverted motorized microscope with temperature and CO2 controlled OKO chamber and a 63X oil objective. Arrow indicates an engulfing cell that takes up corpse and becomes pHrodo positive.
Video S3: 405nm solid state laser induced cell death in selected cells of enteroids, Related to Figure 3 and Figure S4. Live imaging of a four-day old Defa6-cremTmG (iECs = orange, Paneth Cells = green) stained with TO-PRO-3 (red) imaged once every 5min immediately following laser ablation (Figure S4). Cell death occurred between 30min and 1 hour. A Zeiss 780 NLO scanning confocal equipped with a temperature and CO2 regulated environmental chamber and 63X oil objective was used to acquire these images.
Highlights.
Paneth cells engulf dying cells in healthy enteroids
Dying epithelial neighbors promote Paneth cell engulfment activity
Targeted Paneth cell depletion decreases cell clearance in vivo after irradiation
Acknowledgements
The authors thank members of the Ravichandran laboratory at UVA for numerous discussions, and critical reading of the manuscript. This work is supported by grants to K.S.R. from NIGMS R35GM122542, and the Center for Cell Clearance/University of Virginia School of Medicine. Additional support was received through the American Cancer Society Roaring Fork Valley Postdoctoral Fellowship (130254-PF-17-098-01-CSM) awarded to L.S.S.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Arandjelovic S, Ravichandran KS. Phagocytosis of apoptotic cells in homeostasis. Nat Immunol 2015;16(9):907–17. (In eng). DOI: 10.1038/ni.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boada-Romero E, Martinez J, Heckmann BL, Green DR. The clearance of dead cells by efferocytosis. Nat Rev Mol Cell Biol 2020;21(7):398–414. (In eng). DOI: 10.1038/s41580-020-0232-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol 2014;14(3):166–80. DOI: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doran AC, Yurdagul A, Tabas I. Efferocytosis in health and disease. Nat Rev Immunol 2020;20(4):254–267. (In eng). DOI: 10.1038/s41577-019-0240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest 1998;101(4):890–8. DOI: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature 2015;517(7534):311–20. (In eng). DOI: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- 7.Vince JE, Silke J. The intersection of cell death and inflammasome activation. Cell Mol Life Sci 2016;73(11–12):2349–67. (In eng). DOI: 10.1007/s00018-016-2205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory CD, Ford CA, Voss JJ. Microenvironmental Effects of Cell Death in Malignant Disease. Adv Exp Med Biol 2016;930:51–88. (In eng). DOI: 10.1007/978-3-319-39406-0_3. [DOI] [PubMed] [Google Scholar]
- 9.Roche KC, Gracz AD, Liu XF, Newton V, Akiyama H, Magness ST. SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroenterology 2015;149(6):1553–1563.e10. (In eng). DOI: 10.1053/j.gastro.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He LX, Zhang ZF, Zhao J, et al. Ginseng oligopeptides protect against irradiation-induced immune dysfunction and intestinal injury. Sci Rep 2018;8(1):13916. (In eng). DOI: 10.1038/s41598-018-32188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol 2013;75:289–311. (In eng). DOI: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- 12.Bar-Ephraim YE, Kretzschmar K, Clevers H. Organoids in immunological research. Nat Rev Immunol 2020;20(5):279–293. (In eng). DOI: 10.1038/s41577-019-0248-y. [DOI] [PubMed] [Google Scholar]
- 13.McQuin C, Goodman A, Chernyshev V, et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol 2018;16(7):e2005970. DOI: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams JM, Duckworth CA, Burkitt MD, Watson AJ, Campbell BJ, Pritchard DM. Epithelial cell shedding and barrier function: a matter of life and death at the small intestinal villus tip. Vet Pathol 2015;52(3):445–55. (In eng). DOI: 10.1177/0300985814559404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CS, Penberthy KK, Wheeler KM, et al. Boosting Apoptotic Cell Clearance by Colonic Epithelial Cells Attenuates Inflammation In Vivo. Immunity 2016;44(4):807–20. DOI: 10.1016/j.immuni.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T, van Es JH, Snippert HJ, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011;469(7330):415–8. DOI: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durand A, Donahue B, Peignon G, et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc Natl Acad Sci U S A 2012;109(23):8965–70. (In eng). DOI: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008;456(7219):259–63. (In eng). DOI: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen EA, Baehrecke EH. Autophagy in animal development. Cell Death Differ 2020;27(3):903–918. (In eng). DOI: 10.1038/s41418-020-0497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doherty J, Baehrecke EH. Life, death and autophagy. Nat Cell Biol 2018;20(10):1110–1117. (In eng). DOI: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adolph TE, Tomczak MF, Niederreiter L, et al. Paneth cells as a site of origin for intestinal inflammation. Nature 2013;503(7475):272–6. (In eng). DOI: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis 2007;45(9):593–605. DOI: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 23.Han CZ, Juncadella IJ, Kinchen JM, et al. Macrophages redirect phagocytosis by non-professional phagocytes and influence inflammation. Nature 2016;539(7630):570–574. DOI: 10.1038/nature20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morioka S, Perry JSA, Raymond MH, et al. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 2018;563(7733):714–718. DOI: 10.1038/s41586-018-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry JSA, Morioka S, Medina CB, et al. Interpreting an apoptotic corpse as anti-inflammatory involves a chloride sensing pathway. Nat Cell Biol 2019;21(12):1532–1543. DOI: 10.1038/s41556-019-0431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buch T, Heppner FL, Tertilt C, et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods 2005;2(6):419–26. DOI: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 27.van Es JH, Wiebrands K, López-Iglesias C, et al. Enteroendocrine and tuft cells support Lgr5 stem cells on Paneth cell depletion. Proc Natl Acad Sci U S A 2019. (In eng). DOI: 10.1073/pnas.1801888117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Schepper S, Verheijden S, Aguilera-Lizarraga J, et al. Self-Maintaining Gut Macrophages Are Essential for Intestinal Homeostasis. Cell 2019;176(3):676. (In eng). DOI: 10.1016/j.cell.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Yu S, Tong K, Zhao Y, et al. Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell Stem Cell 2018;23(1):46–59.e5. (In eng). DOI: 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hauer-Jensen M, Denham JW, Andreyev HJ. Radiation enteropathy--pathogenesis, treatment and prevention. Nat Rev Gastroenterol Hepatol 2014;11(8):470–9. DOI: 10.1038/nrgastro.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elting LS, Cooksley C, Chambers M, Cantor SB, Manzullo E, Rubenstein EB. The burdens of cancer therapy. Clinical and economic outcomes of chemotherapy-induced mucositis. Cancer 2003;98(7):1531–9. DOI: 10.1002/cncr.11671. [DOI] [PubMed] [Google Scholar]
- 32.Gibson RJ, Keefe DM, Lalla RV, et al. Systematic review of agents for the management of gastrointestinal mucositis in cancer patients. Support Care Cancer 2013;21(1):313–26. DOI: 10.1007/s00520-012-1644-z. [DOI] [PubMed] [Google Scholar]
- 33.Boussios S, Pentheroudakis G, Katsanos K, Pavlidis N. Systemic treatment-induced gastrointestinal toxicity: incidence, clinical presentation and management. Ann Gastroenterol 2012;25(2):106–118. [PMC free article] [PubMed] [Google Scholar]
- 34.Sonis ST. The pathobiology of mucositis. Nat Rev Cancer 2004;4(4):277–84. DOI: 10.1038/nrc1318. [DOI] [PubMed] [Google Scholar]
- 35.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9(7):676–82. (In eng). DOI: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forster B, Van De Ville D, Berent J, Sage D, Unser M. Complex wavelets for extended depth-of-field: a new method for the fusion of multichannel microscopy images. Microsc Res Tech 2004;65(1–2):33–42. (In eng). DOI: 10.1002/jemt.20092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Live imaging of enteroids reveals engulfment of dead cells by epithelial cells within the enteroid, Related to Figure 2. Three day old enteroids were irradiated with 200mJ UV light and stained with CellMask and propidium iodine (red). Images were acquired once every 5min using a Leica DMi8 inverted motorized microscope with temperature and CO2 controlled OKO chamber and a 63X oil objective. Scale bar = 5μm.
Video S2: Live imaging of enteroids revealed acidification of internalized cargo by epithelial cells within the enteroid, Related to Figure 2. Three day old enteroids were irradiated with 200mJ UV light and stained with CellMask and pHrodo red. Images were acquired once every 5min using a Leica DMi8 inverted motorized microscope with temperature and CO2 controlled OKO chamber and a 63X oil objective. Arrow indicates an engulfing cell that takes up corpse and becomes pHrodo positive.
Video S3: 405nm solid state laser induced cell death in selected cells of enteroids, Related to Figure 3 and Figure S4. Live imaging of a four-day old Defa6-cremTmG (iECs = orange, Paneth Cells = green) stained with TO-PRO-3 (red) imaged once every 5min immediately following laser ablation (Figure S4). Cell death occurred between 30min and 1 hour. A Zeiss 780 NLO scanning confocal equipped with a temperature and CO2 regulated environmental chamber and 63X oil objective was used to acquire these images.
Data Availability Statement
The CellProfiler Pipeline used for this study is publicly available at https://github.com/lsshankman/Paneth-Cells-as-novel-efferocytic-phagocytes-within-the-intestine.
Since the time of its creation, CellProfiler has released several new software versions that are incompatible with the pipeline. User must also install CellProfiler v2.2 from https://cellprofiler.org/previous-releases.