Abstract
Gastric cancer is a type of cancer that is characterized by high morbidity and mortality rates. Long non-coding RNA (lncRNA) β-1,3-galactosyltransferase 5-AS1 (B3GALT5-AS1) was previously found to be highly expressed in the serum of patients with gastric cancer. However, the regulatory effects of B3GALT5-AS1 in gastric cancer remain poorly understood. The present study aimed to investigate the effects of B3GALT5-AS1 in gastric cancer cell lines. The expression levels of B3GALT5-AS1 were determined in different gastric cancer cell lines (AGS, HGC-27 and MKN-45) using reverse transcription-quantitative PCR. The potential interaction between B3GALT5-AS1 and casein kinase 2 a1 (CSNK2A1) was evaluated using an RNA binding protein immunoprecipitation and RNA pull down assays. Western blot analysis was performed to measure protein expression levels. Cell Counting Kit-8 assay was utilized to determine cell viability, whilst cell invasion and migration were assessed using Transwell and wound healing assays, respectively. Apoptotic cells were evaluated using TUNEL assays. The results showed that B3GALT5-AS1 expression was upregulated in MKN-45 cells compared with the control group. In addition, B3GALT5-AS1 could bind to CSNK2A1 to regulate its expression. B3GALT5-AS1 knockdown attenuated cell viability, invasion and migration, whilst promoting cell apoptosis. These effects were partly reversed by CSNK2A1 overexpression. Overall, results of the present study revealed that interference with B3GALT5-AS1 impeded gastric cancer cell migration and invasion whilst promoting apoptosis by regulating CSNK2A1 expression. These findings suggested that B3GALT5-AS1 and CSNK2A1 may serve a tumorigenic role in the progression of gastric cancer and serve as therapeutic targets for this type of cancer.
Keywords: β-1, 3-galactosyltransferase 5-AS1, casein kinase 2 a1, gastric cancer, apoptosis
Introduction
Gastric cancer is one of the most common and incurable types of cancer (1,2). Gastric cancer is associated with high rates of incidence and mortality, which imposes a considerable economic burden on society with 1 million new cases annually in the United States (1,2). However, the overall survival rate of patients with early stages of gastric cancer is significantly higher compared with that in patients with advanced stages of gastric cancer (3). Therefore, diagnosis of gastric cancer in the early stages is critical for improving the survival rate. In addition to upper gastrointestinal endoscopy and the detection of common tumor biomarkers, including carcinoembryonic antigen (CEA), cancer antigen (CA)72-4 and CA19-9 in the serum (4-6), long non-coding RNAs (lncRNAs) is garnering interest for reported their role in the early diagnosis, therapy and prognosis of gastric cancer (7,8).
The effects of lncRNAs in gastric cancer are gradually becoming unraveled. Wei and Wang (9) showed that lncRNA maternally expressed 3 was highly expressed in gastric cancer tissues compared with that in adjacent normal tissues (9). In addition, other studies revealed that lncRNA small nucleolar host gene (SNHG)1 and SNHG7 overexpression could promote gastric cancer cell proliferation (10,11). A study demonstrated that B3GALT5-AS1 expression was markedly increased in patients with gastric cancer compared with those with normal colonic epithelia (12). Moreover, high serum B3GALT5-AS1 levels were found to be associated with TNM stage and lymph node metastasis (13), suggesting a role of B3GALT5-AS1 in gastric cancer occurrence and progression. Another study suggested that serum levels of B3GALT5-AS1 could also serve as a diagnostic biomarker of colorectal cancer (14). Indeed, B3GALT5-AS1 was demonstrated to be localized predominantly in the nucleus, where it could directly bind to the promoter of microRNA (miR)-203 to upregulate the expression of the target genes of miR-203, which in turn suppresses colon cancer metastasis to the liver (12). However, to date, the regulatory mechanism of B3GALT5-AS1 in gastric cancer remains elusive.
It has been previously demonstrated that casein kinase 2 a1 (CSNK2A1) is associated with cell invasion and migration in several types of cancer, including lung (15) and breast cancer (16). Additionally, overexpression of CSNK2A1 in gastric cancer cells enhanced cell proliferation, invasion and migration (17), suggesting a regulatory role of CSNK2A1 in gastric cancer.
Therefore, in the present study, the expression profile of B3GALT5-AS1 was determined in gastric cancer cell lines. Additionally, whether B3GALT5-AS1 served a role in gastric cancer cell proliferation, migration, invasion and apoptosis and its potential relationship with CSNK2A1 was assessed.
Materials and methods
Cell culture
Gastric cancer cell lines, AGS, HGC-27 and MKN-45 were purchased from the China Infrastructure of Cell Line Resources, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. The human gastric mucosa cell line GES-1 (cat. no. CL-0563) was obtained from Procell Life Science & Technology Co., Ltd.. All cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and maintained in an incubator with 5% CO2 at 37˚C.
Plasmid transfection
For B3GALT5-AS1 knockdown, two short hairpin RNAs (shRNA/sh), namely sh-B3GALT5-AS1-1 (5'-GCATAAGAGAGACCAACTTGG-3') and sh-B3GALT5-AS1-2 (5'-GCAAGACAGCGCATTGATTGG-3'), were constructed using the pGPU6/Neo plasmid (Shanghai GenePharma Co., Ltd.). Scrambled shRNA [shRNA-negative control (NC)] served as the NC (5'-ACTTGCGCTTGCGAAAATCTATATAGC-3'). The pcDNA-MFHAS1 plasmid (Shanghai GenePharma Co., Ltd.) was used for CSNK2A1 overexpression (ov-CSNK2A1). The empty vector, ov-NC, served as the negative control for the overexpression experiments. Cells were seeded into six-well plates at a density of 5x105 cells/well. When they reached 80% confluence, cells were transfected with 3 µg shRNA-encoding plasmids or 5 µg overexpression plasmids using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Following incubation for 48 h at 37˚C, transfected cells were used for subsequent experiments.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the four cell lines (GES-1, AGS, HGC-27 and MKN-45) using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Following reverse transcription using AMV Reverse Transcription kit (cat. no. A3500; Promega Corporation), this reaction was performed at 25˚C for 5 min, 50˚C for 20 min, and then 75˚C for 5 min. The FastStart Universal SYBR Green Master kit (Roche Diagnostics GmbH) was used for qPCR in an ABI PRISM 7900 HT system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer sequences used were as follows: B3GALT5-AS1 forward, 5'-GATCCACGTCCAGGCTCACT-3' and reverse, 5'-GTGCTGGCTGTCAGGATGAG-3'; CSNK2A1 forward, 5'-GAACGCTTTGTCCACAGTGA-3' and reverse, 5'-TATCGCAGCAGTTTGTCCAG-3' and GAPDH forward, 5'-AACAGCGACACCCACTCCTC-3' and reverse, 5'-GGAGGGGAGATTCAGTGTGGT-3'. The thermocycling conditions for PCR reaction was as follows: Initial denaturation for 5 min at 95˚C, followed by 40 cycles of 30 sec at 95˚C and 45 sec at 65˚C. The relative mRNA expression was calculated using the 2-∆∆Cq method (18) and GAPDH served as the internal control.
Bioinformatics analysis
The starBase 2.0 database (http://starbase.sysu.edu.cn/) was used to predict the binding sites between B3GALT5-AS1 and CSNK2A1.
RNA immunoprecipitation (RIP) assay
RIP assay was performed using the Millipore Magna RIP™ RNA kit (EMD Millipore). Briefly, MKN-45 cells were resuspended in RIP lysis buffer (EMD Millipore) and incubated on ice for 5 min. Subsequently, the cell lysate (100 µl per antibody per RIP) was incubated with an anti-CSNK2A1 antibody (1:1,000; cat. no. ab70774; Abcam) or IgG antibody (0.2 µg/ml; cat. no. ab190475; Abcam) and 40 µl protein A/G magnetic beads (EMD Millipore) at 4˚C overnight. The protein-RNA complexes were digested with 2 µl proteinase K buffer at 55˚C for 30 min. The beads buffer was spun down at 2,000 x g for 30 sec, and the supernatant was transferred to a fresh tube. Finally, the extracted RNA was subjected to RT-qPCR.
RNA pull-down assay
RNA pull-down assay was performed using Pierce™ Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Inc.). Briefly, MKN-45 cells were lysed by RIPA lysis buffer (Sigma-Aldrich; Merck KGaA). B3GALT5-AS1 primers were 5'-CCTTGAGAGACGAAGCAC-3' (sense) and 5'-ATTTCACGGATGAGACGAC-3' (antisense). B3GALT5-AS1 was labeled with biotin using Pierce RNA 3' End Desthiobiotinylation Kit (Thermo Fisher Scientific, Inc.) and then bound to streptavidin magnetic beads (~10 µg), before this complex was incubated with 1 ml cell lysates at 4˚C for 1 h. RNA-protein complexes were eluted by adding 30 µl SDS sample buffer to the beads and heating at 95˚C for 5 min. The samples were cooled on ice for 1 min, 2 µl Benzonase added, and then incubated for 15 min at room temperature. The sample buffer was heated again at 95˚C for 5 min. The magnetic beads were spun down at room temperature at 12,000 x g for 1 min. The presence of CSNK2A1 protein in the RNA-protein complexes was analyzed by western blotting.
Western blotting
Total proteins were isolated from cell lysates using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) and then quantified using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). Subsequently, 20 µg protein extracts per lane were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes (EMD Millipore). Following blocking with 5% skim milk at room temperature for 2 h, membranes were incubated with antibodies against CSNK2A1 (1:1,000; cat. no. ab70774; Abcam), matrix metalloproteinase (MMP)2 (1:2,000; cat. no. AF0577; Affinity Biosciences), MMP9 (1:2,000; cat. no. AF5228; Affinity Biosciences), phosphorylated (p)-c-Met (1:1,000; cat. no. AF3128; Affinity Biosciences), c-Met (1:1,000; cat. no. AF6128; Affinity Biosciences), Bcl-2 (1:5,000; cat. no. AF6139; Affinity Biosciences), Bax (1:5,000; cat. no. AF0120; Affinity Biosciences), cleaved caspase-3 (1:2,000; cat. no. AF7022; Affinity Biosciences), caspase-3 (1:2,000; cat. no. AF6311; Affinity Biosciences) or GAPDH (1:5,000; cat. no. AF7021; Affinity Biosciences) at 4˚C overnight. The next day, the membranes were incubated with corresponding HRP-conjugated secondary antibodies (1:5,000; cat. no. S0001; Affinity Biosciences) at room temperature for 2 h. The protein bands were visualized using the Immobilon Western Chemilum HRP substrate (EMD Millipore) and analyzed using Image Lab Software 3.0 (Bio-Rad Laboratories, Inc.).
Cell Counting Kit 8 (CCK-8) assay
For CCK-8 assays, MKN-45 cells were seeded into a 96-well plates at a density of 5,000 cells per well and incubated at 37˚C overnight. The next day, 10 µl CCK-8 reagent (Dojindo Molecular Technologies, Inc.) was added into each well and cells were incubated for 2 h at 37˚C. Finally, the optical density (OD) in each well was measured using a microplate reader (Bio-Rad Laboratories, Inc.) at a wavelength of 450 nm.
Cell invasion assay
The upper chamber of the Transwell chamber (Corning, Inc.) was coated with 100 µl Matrigel (1 mg/ml; BD Biosciences) and incubated at 37˚C for 4 h. The cell density was then adjusted to 2x105 cells/ml and 100 µl cell suspension in serum-free RPMI-1640 medium was added to the upper chamber and cultured at 37˚C in the presence of 5% CO2 for 24 h. The lower chamber was supplemented with 600 µl RPMI-1640 medium containing 20% FBS. At 24 h post-incubation, invasive cells were fixed with 4% paraformaldehyde at room temperature for 0.5 h, stained with 0.1% crystal violet at 37˚C for 10 min and photographed in randomly selected nine fields of view under a light microscope (magnification, x100).
Cell migration assay
MKN-45 cells were seeded into a six-well plate (5x105 cells/well) and when they reached 100% confluence, scratches were created using a 20-µl pipette tip. The medium was then replaced with fresh medium containing 1% FBS and cells were cultured at 37˚C for 24 h (19). Finally, images of the migrated cells were captured at 0 and 24 h in randomly selected fields of view under a light microscope (magnification, x100). The migration distance was calculated as the width of the scratch at 24 h minus the width of the scratch at 0 h. The relative migration rate was calculated by normalizing to the control group.
Cell apoptosis assay
Apoptotic cells were assessed using an One Step TUNEL Apoptosis Assay kit (Beyotime Institute of Biotechnology). Briefly, MKN-45 cells (3x105 cells/ml) were fixed with 4% paraformaldehyde at room temperature for 30 min and then incubated with 0.3% Triton X-100 at room temperature for 5 min. Subsequently, each well of the 24-well plate was supplemented with 50 µl TUNEL detection reagent for 60 min at 37˚C in the dark. DAPI staining solution (10 µg/ml) was used to visualize all nuclei for 5 min at 37˚C. The coverslips were then washed with PBS and mounted on slides with anti-fading solution. The apoptotic cells (green fluorescence) in three random fields were observed under a fluorescence microscope (magnification, x100).
Statistical analysis
All data in the study are expressed as the mean ± SD and analyzed using an independent unpaired t-test or one-way ANOVA followed by a Tukey's post hoc test. Each experiment was repeated ≥ three times. P<0.05 was considered to indicate a statistically significant difference.
Results
Interaction between B3GALT5-AS1 and CSNK2A1
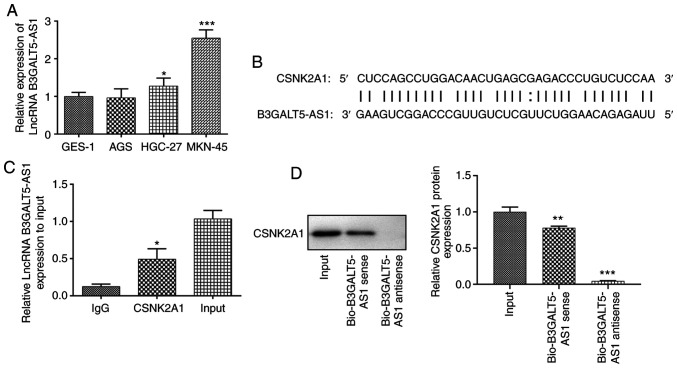
The expression levels of B3GALT5-AS1 were first determined in a range of gastric cell lines. The expression levels of B3GALT5-AS1 in HGC-27 cells were higher compared with that in the human gastric mucosa cell line GES-1 (Fig. 1A). Compared with that in GES-1 cells, B3GALT5-AS1 expression was significantly higher in MKN-45 cells (Fig. 1A). It has been previously suggested that B3GALT5-AS1 may originate from autocrine gastric tumor cells and is closely associated with gastric tumor metastasis (13). However, the specific regulatory mechanism underlying the effect of B3GALT5-AS1 on tumor metastasis remains unclear. Therefore, based on the expression of B3GALT5-AS1, the MKN-45 cell line was used for the subsequent experiments. mRNA of CSNK2A1 was located in Chr20:453618-453657. Bioinformatics analysis predicted that B3GALT5-AS1 could bind to the sequence of CSNK2A1 (Fig. 1B). Notably, bioinformatics analysis predicted binding sites between the sequences of B3GALT5-AS1 and CSNK2A1, suggesting that B3GALT5-AS1 may modulate CSNK2A1 expression by direct binding (12,20). As shown in Fig. 1C, the interaction between B3GALT5-AS1 and CSNK2A1 was evaluated using RIP assays. Further evidence of a CSNK2A1-complex interaction was found via a RNA pull down assay (Fig. 1D). Since B3GALT5-AS1 was found to be enriched in the CSNK2A1-complex, direct binding of CSNK2A1 to B3GALT5-AS1 was reproduced in vitro by a RNA pull-down assay.
Figure 1.
Interaction between B3GALT5-AS1 and CSNK2A1. (A) Expression levels of B3GALT5-AS1 in a range of gastric cell lines were determined by reverse transcription-quantitative PCR. *P<0.05 and ***P<0.001 vs. GES-1. (B) Potential binding sites between B3GALT5-AS1 and CSNK2A1 were predicted using the starBase 2.0 database. Interaction between B3GALT5-AS1 and CSNK2A1 was evaluated using (C) RNA immunoprecipitation. *P<0.05 vs. IgG. (D) RNA pull-down assays. **P<0.01 and ***P<0.001 vs. Input. B3GALT5-AS1, β-1,3-galactosyltransferase 5-AS1; CSNK2A1, casein kinase 2 a1; LncRNA, long non-coding RNA.
Efficiency of B3GALT5-AS1 knockdown and CSNK2A1 overexpression
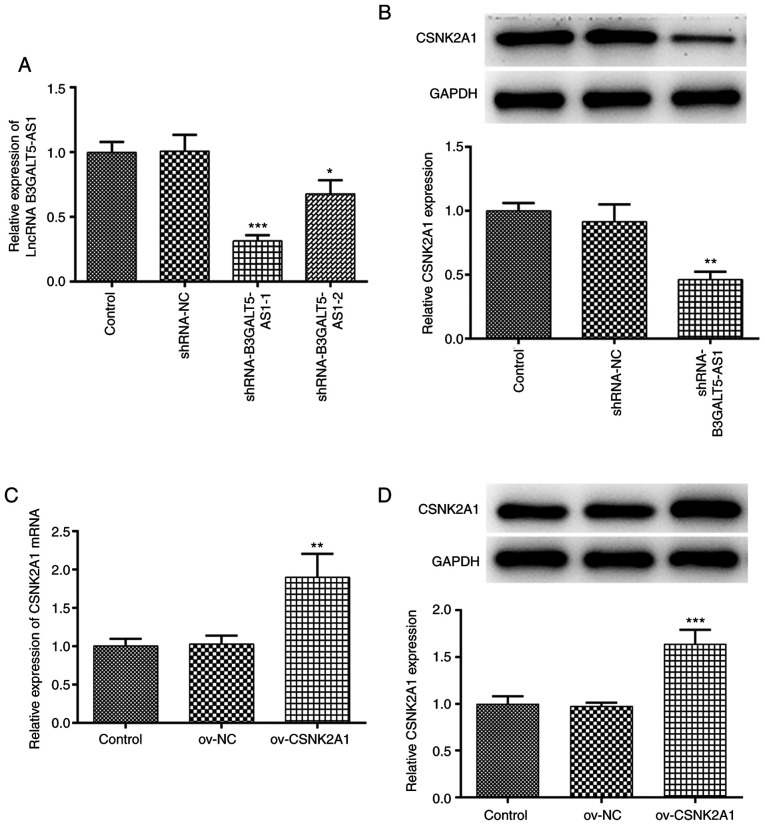
Since B3GALT5-AS1 expression was the highest in MKN-45 cells, these cells were transfected with plasmids encoding shRNAs to knockdown the expression B3GALT5-AS1. As shown in Fig. 2A, between the two shRNA clones, shRNA-B3GALT5-AS1-1 exhibited the more potent silencing effect compared with that mediated by shRNA-B3GALT5-AS1-2. The protein expression level of CSNK2A1 in MKN-45 cells transfected with shRNA-B3GALT5-AS1-1 was significantly lower compared with that in the shRNA-NC group. However, no significant differences were observed in the protein expression of CSNK2A1 between the control and shRNA-NC groups (Fig. 2B). Subsequently, MKN-45 cells were transfected with a CSNK2A1 overexpression plasmid. mRNA and protein expression of CSNK2A1 were both significantly upregulated following cell transfection with ov-CSNK2A1 compared with those transfected with ov-NC (Fig. 2C and D).
Figure 2.
B3GALT5-AS1 silencing and CSNK2A1 overexpression on MKN-cells. (A) Expression levels of B3GALT5-AS1 in MKN-45 cells following transfection with shRNA-NC, shRNA-B3GALT5-AS1-1 or shRNA-B3GALT5-AS1-2 were determined by RT-qPCR. (B) Western blot analysis showing the protein expression levels of CSNK2A1. *P<0.05, **P<0.01 and ***P<0.001 vs. shRNA-NC. (C) mRNA expression levels of CSNK2A1 following transfection with ov-NC or ov-CSNK2A1 were assessed by RT-qPCR. (D) Western blot analysis showing the protein expression levels of CSNK2A1 following transfection of MKN-45 cells with ov-NC or ov-CSNK2A1. **P<0.01 and ***P<0.001 vs. ov-NC. B3GALT5-AS1, β-1,3-galactosyltransferase 5-AS1; CSNK2A1, casein kinase 2 a1; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; LncRNA, long non-coding RNA; Ov, expression; shRNA, short hairpin RNA.
B3GALT5-AS1/CSNK2A1 axis controls cell migration and invasion
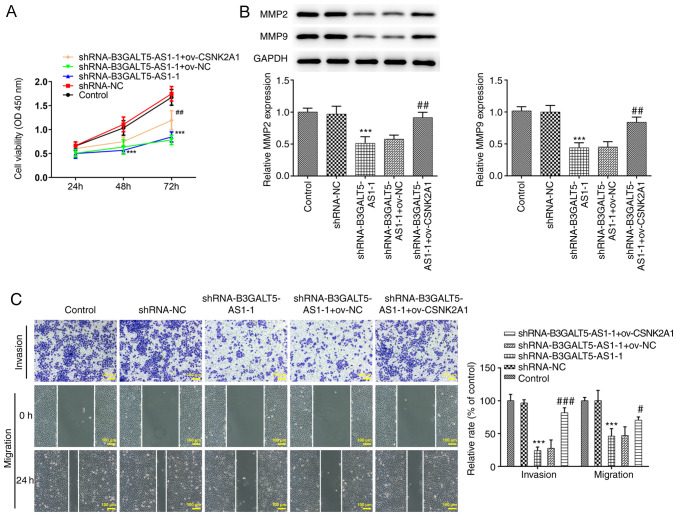
Subsequently, effects of gene knockdown or overexpression on the migratory and invasive behavior of MKN-45 cells were investigated. Cell viability was significantly decreased following cell transfection with shRNA-B3GALT5-AS1-1 compared with those transfected with shRNA-NC at 72 h. However, co-transfection with shRNA-B3GALT5-AS1-1 and ov-CSNK2A1 partially but significantly elevated cell viability compared with that in the shRNA-B3GALT5-AS1-1 + ov-NC group (Fig. 3A). MMPs are involved in the remodeling of the extracellular matrix and basement membrane, which is a key process in the invasion and metastasis of cancer cells (21,22). The protein expression levels of MMP2 and MMP9 were significantly reduced after cell transfection with shRNA-B3GALT5-AS1-1 compared with that in the shRNA-NC group, which was significantly reversed by CSNK2A1 plasmid co-transfection (Fig. 3B). Transwell assays showed that the invasive ability of MKN-45 cells was significantly decreased following B3GALT5-AS1 knockdown compared with that in the shRNA-NC group (Fig. 3C). B3GALT5-AS1 silencing also significantly attenuated the migratory ability of MKN-45 cells compared with that in cells transfected with shRNA-NC (Fig. 3C). For both migration and invasion, CSNK2A1 plasmid co-transfection significantly reversed the inhibitory effects of B3GALT5-AS1 knockdown (Fig. 3C).
Figure 3.
B3GALT5-AS1/CSNK2A1 axis regulates MKN-45 cell migration and invasion. (A) Cell viability in the different transfection groups was evaluated by Cell Counting Kit-8 assay. (B) Western blot analysis showing the protein expression levels of MMP2 and MMP9 in different groups. (C) Cell invasion and migration in different groups of cells after transfection. Scale bar, 100μm, ***P<0.001 vs. shRNA-NC; #P<0.05, ##P<0.01 and ###P<0.001 vs. shRNA-B3GALT5-AS1-1 + ov-NC. B3GALT5-AS1, β-1,3-galactosyltransferase 5-AS1; CSNK2A1, casein kinase 2 a1; MMP, matrix metalloproteinase; LncRNA, long non-coding RNA; Ov, expression; shRNA, short hairpin RNA; OD, optical density.
B3GALT5-AS1/CSNK2A1 axis modulates cell apoptosis
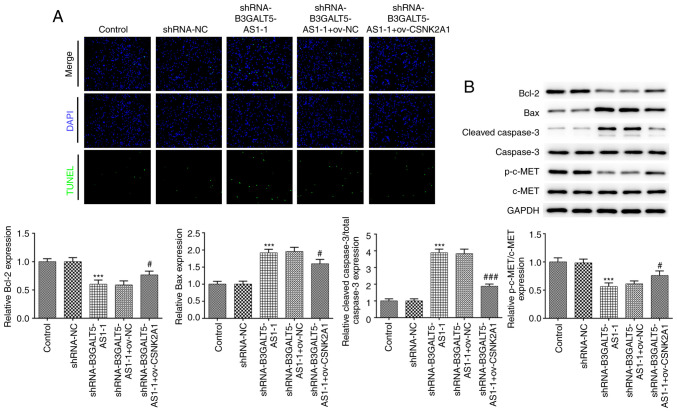
Furthermore, a TUNEL assay was performed to measure cell apoptosis. The number of apoptotic cells in the shRNA-B3GALT5-AS1-1 group was markedly higher compared with that in the shRNA-NC or control group, which was reversed by CSNK2A1 plasmid co-transfection (Fig. 4A). The levels of the pro-apoptotic proteins Bax and cleaved caspase-3 were both significantly upregulated following B3GALT5-AS1 knockdown, but their protein levels were significantly restored after CSNK2A1 overexpression in addition to B3GALT5-AS1 knockdown (Fig. 4B). By contrast, the expression levels of the anti-apoptotic proteins Bcl-2 and p-c-Met were decreased in the shRNA-B3GALT5-AS1-1 group compared with those in the shRNA-NC group. However, co-transfection of MKN-45 cells with shRNA-B3GALT5-AS1-1 and ov-CSNK2A1 upregulated Bcl-2 and p-c-Met compared with cells transfected with shRNA-B3GALT5-AS1-1 and ov-NC (Fig. 4B). Dysregulation in c-Met has been implicated in the pathogenesis and development of gastric cancer, where its downregulation can induce apoptosis (23,24). These results aforementioned suggest that B3GALT5-AS1 knockdown promotes gastric cancer cell apoptosis by binding to CSNK2A1.
Figure 4.
B3GALT5-AS1/CSNK2A1 axis regulates cell apoptosis. (A) Cell apoptosis in different transfection groups was assessed using a TUNEL assay. Magnification, x100. (B) Western blot analysis showing the protein levels of Bcl-2, Bax, cleaved caspase-3, caspase-3, p-c-Met and c-Met in different groups. ***P<0.001 vs. shRNA-NC; #P<0.05 and ###P<0.001 vs. shRNA-B3GALT5-AS1-1 + ov-NC. B3GALT5-AS1, β-1,3-galactosyltransferase 5-AS1; CSNK2A1, casein kinase 2 a1; p-, phosphorylated; Ov, expression; shRNA, short hairpin RNA; NC, negative control.
Discussion
The incidence rate of gastric cancer is particularly high in developing countries, such as China and India (25). Data suggests that gastric cancer ranks second in terms of incidence among all malignancies in China (26,27). Gastric cancer has long been considered to be a difficult challenge to treat clinically (28). Therapeutic interventions in patients with gastric cancer typically involve alterations in diet, psychological support and changes in daily life habits to optimize the therapeutic effects and improve the quality of life (29). The moderate diagnostic value of CEA and CA199 for the early detection of gastric cancer highlights the importance and urgency of developing novel biomarkers with high specificity and sensitivity (30). It has been reported that B3GALT5-AS1 can acts as a diagnostic and prognostic biomarker of gastric cancer (13).
The present study demonstrated that B3GALT5-AS1 was highly expressed in MKN-45 cells compared with that in normal gastric mucosa GES-1 cells. In addition, binding sites between the sequences of B3GALT5-AS1 and CSNK2A1 were identified. B3GALT5-AS1 silencing repressed the protein expression of CSNK2A1. The catalytic subunit of CK2α is encoded by CSNK2A1(31) and is associated with cell proliferation and invasion in colorectal cancer (32). A previous study showed that the knockdown of CSNK2A1 expression attenuated the proliferative and invasive capabilities of breast carcinoma cells (16). In the present study, B3GALT5-AS1 knockdown in MKN-45 cells reduced cell viability, which was reversed by CSNK2A1 overexpression. The reduced MKN-45 cell migration and invasion, mediated by shRNA-B3GALT5-AS1, were also restored following CSNK2A1 overexpression. Furthermore, silencing of B3GALT5-AS1 expression induced cell apoptosis in a manner that was partially counteracted by CSNK2A1 overexpression. However, there are some limitations in the present study. These findings were based on in vitro cell model, which require further validation using in vivo animal models or human tissues in future studies. In addition, the underlying mechanism of the B3GALT5-AS1/CSNK2A1 axis regulating gastric cancer physiology has not been studied in depth in the present study.
To conclude, the present study demonstrated that knocking down B3GALT5-AS1 expression, which was upregulated in gastric cancer cells, could reduce cell viability and inhibit migration whilst inducing cell apoptosis. These effects could be restored by increasing the expression of CSNK2A1.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PW and GBS acquired the data. GXD and BQW contributed to the study design and analysis of the data. PW drafted the manuscript and BQW revised it critically for important intellectual content. All authors read and approved the final manuscript. BQW and PW are responsible for confirming the authenticity if the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol. 2020;18:534–542. doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki T, Kitagawa Y, Nankinzan R, Yamaguchi T. Early gastric cancer diagnostic ability of ultrathin endoscope loaded with laser light source. World J Gastroenterol. 2019;25:1378–1386. doi: 10.3748/wjg.v25.i11.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J, Zheng W. An alternative method for screening gastric cancer based on serum levels of CEA, CA19-9, and CA72-4. J Gastrointest Cancer. 2018;49:57–62. doi: 10.1007/s12029-016-9912-7. [DOI] [PubMed] [Google Scholar]
- 6.Mori A, Ohashi N, Maruyama T, Tatebe H, Sakai K, Shibuya T, Inoue H, Okuno M. Cardiovascular tolerance in upper gastrointestinal endoscopy using an ultrathin scope: Prospective randomized comparison between transnasal and transoral procedures. Dig Endosc. 2008;20:79–83. [Google Scholar]
- 7.Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, Bleotu C, Diaconu CC, Chivu-Economescu M. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25:2029–2044. doi: 10.3748/wjg.v25.i17.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song P, Jiang B, Liu Z, Ding J, Liu S, Guan W. A three-lncRNA expression signature associated with the prognosis of gastric cancer patients. Cancer Med. 2017;6:1154–1164. doi: 10.1002/cam4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3850–3856. [PubMed] [Google Scholar]
- 10.Hu Y, Ma Z, He Y, Liu W, Su Y, Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491:926–931. doi: 10.1016/j.bbrc.2017.07.137. [DOI] [PubMed] [Google Scholar]
- 11.Yang S, Liu Y, Li MY, Ng CSH, Yang SL, Wang S, Zou C, Dong Y, Du J, Long X, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16(124) doi: 10.1186/s12943-017-0700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Wei Z, Wu K, Dai W, Zhang C, Peng J, He Y. Long noncoding RNA B3GALT5-AS1 suppresses colon cancer liver metastasis via repressing microRNA-203. Aging (Albany NY) 2018;10:3662–3682. doi: 10.18632/aging.101628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng W, Zong W, Li Y, Shen X, Cui X, Ju S. Abnormally expressed long noncoding RNA B3GALT5-AS1 may serve as a biomarker for the diagnostic and prognostic of gastric cancer. J Cell Biochem. 2020;121:557–565. doi: 10.1002/jcb.29296. [DOI] [PubMed] [Google Scholar]
- 14.Ding Y, Feng W, Ge JK, Dai L, Liu TT, Hua XY, Lu X, Ju SQ, Yu J. Serum level of long noncoding RNA B3GALT5-AS1 as a diagnostic biomarker of colorectal cancer. Future Oncol. 2020;16:827–835. doi: 10.2217/fon-2019-0820. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Ham S, Yang K, Kim K. Protein kinase CK2 activation is required for transforming growth factor β-induced epithelial-mesenchymal transition. Mol Oncol. 2018;12:1811–1826. doi: 10.1002/1878-0261.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae JS, Park SH, Jamiyandorj U, Kim KM, Noh SJ, Kim JR, Park HJ, Kwon KS, Jung SH, Park HS, et al. CK2α/CSNK2A1 phosphorylates SIRT6 and is involved in the progression of breast carcinoma and predicts shorter survival of diagnosed patients. Am J Pathol. 2016;186:3297–3315. doi: 10.1016/j.ajpath.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Jiang C, Ma Z, Zhang G, Yang X, Du Q, Wang W. CSNK2A1 promotes gastric cancer invasion through the PI3K-Akt-mTOR signaling pathway. Cancer Manag Res. 2019;11:10135–10143. doi: 10.2147/CMAR.S222620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Kramer N, Walzl A, Unger C, Rosner M, Krupitza G, Hengstschläger M, Dolznig H. In vitro cell migration and invasion assays. Mutat Res. 2013;752:10–24. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Jiang HM, Dong JK, Song K, Wang TD, Huang WK, Zhang JM, Yang XY, Shen Y, Zhang J. A novel allosteric site in casein kinase 2α discovered using combining bioinformatics and biochemistry methods. Acta Pharmacol Sin. 2017;38:1691–1698. doi: 10.1038/aps.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H, Jin H, Zhao H, Wang J, Li X, Yan H, Wang S, Guo X, Xue L, Li J, et al. RhoGDIβ promotes Sp1/MMP-2 expression and bladder cancer invasion through perturbing miR-200c-targeted JNK2 protein translation. Mol Oncol. 2017;11:1579–1594. doi: 10.1002/1878-0261.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H. Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: Recent advances. Sensors (Basel) 2018;18(3249) doi: 10.3390/s18103249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu ML, Zhou XX, Ren MT, Shi KD, Yu MS, Jiao WR, Wang YM, Zhong WX, Ji F. Blockage of ETS homologous factor inhibits the proliferation and invasion of gastric cancer cells through the c-Met pathway. World J Gastroenterol. 2020;26:7497–7512. doi: 10.3748/wjg.v26.i47.7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, Zhang H, Ning T, Liu D, Deng T, Liu R, Bai M, Zhu K, Li J, Fan Q, et al. Exosome-delivered c-Met siRNA could reverse chemoresistance to cisplatin in gastric cancer. Int J Nanomedicine. 2020;15:2323–2335. doi: 10.2147/IJN.S231214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Sun K, Zheng R, Zeng H, Zhang S, Xia C, Yang Z, Li H, Zou X, He J. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng R, Zeng H, Zhang S, Chen W. Estimates of cancer incidence and mortality in China, 2013. Chin J Cancer. 2017;36(66) doi: 10.1186/s40880-017-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shishida M, Toyota K, Ikeda M, Karakuchi N, Inoue M, Saito Y, Takahashi T. Clinical complete response after chemotherapy and palliative surgery for unresectable gastric cancer. Case Rep Oncol. 2020;13:689–695. doi: 10.1159/000507781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(1010428317714626) doi: 10.1177/1010428317714626. [DOI] [PubMed] [Google Scholar]
- 30.Lamerz R. Role of tumour markers, cytogenetics. Ann Oncol. 1999;10 (Suppl 4):S145–S149. [PubMed] [Google Scholar]
- 31.Bodenbach L, Fauss J, Robitzki A, Krehan A, Lorenz P, Lozeman FJ, Pyerin W. Recombinant human casein kinase II. A study with the complete set of subunits (alpha, alpha' and beta), site-directed autophosphorylation mutants and a bicistronically expressed holoenzyme. Eur J Biochem. 1994;220:263–273. doi: 10.1111/j.1432-1033.1994.tb18622.x. [DOI] [PubMed] [Google Scholar]
- 32.Zou J, Luo H, Zeng Q, Dong Z, Wu D, Liu L. Protein kinase CK2α is overexpressed in colorectal cancer and modulates cell proliferation and invasion via regulating EMT-related genes. J Transl Med. 2011;9(97) doi: 10.1186/1479-5876-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.