Abstract
Human cytomegalovirus (HCMV) manipulates multiple cellular processes to facilitate virus replication, including the control of mRNA translation. We previously showed that the HCMV TRS1 protein (pTRS1) promotes cap-dependent mRNA translation independent of its ability to antagonize the antiviral protein PKR. Here we find that pTRS1 enhances internal ribosome entry site (IRES) activity using a novel circular RNA reporter that lacks an mRNA cap and poly(A) tail. Additionally, pTRS1 expression increases the activity of cellular IRESs that control the expression of proteins needed for efficient HCMV replication. We find that the ability of pTRS1 to enhance cap-independent translation is separable from its ability to antagonize PKR, but requires the pTRS1 RNA binding domain. Together these data show that pTRS1 stimulates cap-independent translation and suggest a role for pTRS1 in alternative translation initiation pathways during HCMV infection.
Keywords: human herpesvirus, human cytomegalovirus, HCMV, mRNA translation, protein synthesis, cap-independent translation, internal ribosome entry site, IRES
1. Introduction
Control and regulation of the cellular protein synthesis machinery is critical for virus replication (1–3) as viral mRNAs are translated by cellular ribosomes. Thus viral protein synthesis relies exclusively on the cellular protein synthesis machinery. To ensure efficient viral mRNA translation, viruses often encode proteins that subvert cellular defenses that limit protein synthesis in infected cells, and direct the cellular protein synthesis machinery to viral mRNAs. Viral factors that interface with the translation machinery are therefore important determinants of virus replication, and viral pathogenesis.
The rate-limiting step in mRNA translation is translation initiation (4, 5). In the standard model, initiation begins with binding of the eIF4F complex (composed of eIF4A, eIF4E and eIF4G) to the 7-methylguanosine (m7G) mRNA cap (6–8). The bound eIF4F complex recruits the 43S preinitation complex (43S PIC), consisting of eIF3, eIF2-GTP, an initiator methionyl tRNA (Met-tRNAi) and the 40S ribosomal subunit. Together with the mRNA these factors comprise the 48S complex (9–11), which scans the mRNA 5’ untranslated region (5’UTR) until the translation start codon is recognized, at which time the 60S ribosomal subunit is recruited and translation elongation begins (12).
While most translation requires the mRNA cap for initiation, translation initiation can also occur through cap-independent mechanisms. Internal ribosome entry sites, or IRESs, are RNA structures that directly recruit ribosomal subunits and/or translation initiation factors to allow for translation initiation independent of the m7G cap (5, 13–15). The presence of an IRES allows for efficient mRNA translation under conditions where eIF4F abundance is limited, and cap-dependent translation is inhibited, as happens during periods of cell stress (16, 17). In fact, most mRNAs containing IRES elements encode proteins that are involved in stress response and cell death pathways (17, 18). Thus IRES-mediated translation provides an alternative to cap-dependent translation that allows for ongoing protein synthesis during cellular stressors that reduce overall levels of protein synthesis.
Viruses commonly inhibit cap-dependent translation initiation as a means to usurp the cellular translation machinery (13). IRES elements within viral mRNAs allow for ongoing viral protein synthesis despite inhibition of cap-dependent cellular mRNA translation. The poliovirus IRES provides an excellent example (19–21). Poliovirus encodes a viral protease that cleaves eIF4G during infection, thus inhibiting cap-dependent translation. However, the carboxyl-terminus of cleaved eIF4G binds stem loop structures in the poliovirus IRES (located in the 5’UTR) and helps recruit eIF4A and the 43S PIC. The 48S complex then scans through additional RNA structures in the poliovirus 5’UTR and initiates translation at the proper start codon (22–26). eIF4E is not necessary for the translation of poliovirus RNA, as the presence of an mRNA cap is not necessary for eIF4G recruitment, or viral RNA translation. Similarly, rhinovirus, picornavirus, flavivirus, and pestivirus RNAs contain IRES elements to ensure the synthesis of viral proteins despite inactivation of eIF4F-dependent translation during infection (27–30).
While cap-independent translation has been well characterized in the context of RNA virus infections, its role in the translation of DNA virus mRNAs has been studied in less detail. One of the best characterized DNA virus IRES elements is from the Kaposi’s sarcoma-associated herpesvirus (KSHV) v-cyclin transcript. The majority of KSHV transcripts are translated in a cap-dependent manner, however, synthesis of the KSHV vFLIP protein is initiated by an IRES element within the v-cyclin transcript (31), increasing the coding capacity of the mRNA. Another herpesvirus, human cytomegalovirus (HCMV), transcribes its m7G capped mRNAs in the nucleus, which are then transported to the cytoplasm and translated by host ribosomes (32, 33). Unlike most viruses, HCMV infection does not inhibit host protein synthesis, and both cellular and viral mRNAs are translated throughout infection (34, 35). The ongoing translation of host mRNAs encoding proteins required for HCMV replication is due in part to increased eIF4F levels in infected cells (36, 37). However, the translation of HCMV mRNAs becomes increasingly resistant to eIF4F disruption as infection progresses (38), suggesting alternative translation initiation mechanisms exist in infected cells. While only a single HCMV IRES has been described in detail, ~100 HCMV 5’UTRs have been shown to promote cap-independent translation in a bicistronic mRNA reporter screen (39, 40). These data suggest that HCMV uses an eIF4F-independent mechanism to initiate translation on viral mRNAs and facilitate virus replication.
The HCMV TRS1 protein (pTRS1) regulates translation during HCMV infection. We previously showed that pTRS1 increases overall levels of protein synthesis and stimulates cap-dependent translation of reporter mRNAs containing both cellular and HCMV 5’UTRs, though pTRS1 preferentially enhances the translation of mRNAs containing viral 5’UTRs (41). pTRS1 binds double-stranded (dsRNA) and the m7G cap, and also inhibits activation of the antiviral kinase PKR, to maintain protein synthesis during infection (41–46). Additionally, pTRS1 stimulates mRNA translation in cells lacking PKR (41) showing that pTRS1 enhances cap-dependent translation independent of its function as a PKR antagonist.
Here we show that HCMV pTRS1 also enhances cap-independent translation. pTRS1 increased the activity of multiple viral IRESs in a bicistronic reporter assay. To circumvent technical issues associated with bicistronic reporters, we developed a new assay that uses circular RNAs to measure the effect of IRES trans-acting factors (ITAFs) on IRES activity. pTRS1 enhances translation of circular mRNAs containing viral IRESs and increases the activity of two cellular IRES that control the expression of cellular proteins required for HCMV replication. pTRS1 also stimulates cap-independent translation of bicistronic and circular mRNAs in cells lacking PKR, showing that the ability of pTRS1 to stimulate cap-independent translation is separable from its ability to antagonize PKR. The ability of pTRS1 to bind RNA was necessary to stimulate cap-independent translation, as a pTRS1 mutant that cannot bind dsRNA does not enhance IRES activity. Together these data show that pTRS1 stimulates cap-independent translation, and may play a role in the eIF4F-independent translation of viral transcripts previously reported during HCMV infection(38).
2. Materials and methods
2.1. Cells and viruses.
HeLa cells and HEK293T cells were maintained in Dulbecco’s Modified Eagle Medium (Sigma) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin solution (Sigma). HeLa cells expressing a Cas9 vector and HeLa cells lacking PKR expression (Control and PKR KO, respectively (45)) were maintained in media as above supplemented with 1 ug/ml puromycin.
2.2. Generation of recombinant plasmids.
The poliovirus and KSHV vFLIP bicistronic reporters were a gift from Dr. Felicia Goodrum at the University of Arizona (39). The circular GFP reporter (TR-circGFP) was a gift from Dr. Aravind Asokan at the University of North Carolina at Chapel Hill (47). To clone IRES elements into the circular RNA reporter, the circular GFP reporter plasmid was amplified using circGFP F and circGFP R (Table 1). The poliovirus IRES element was amplified from the bicistronic reporter above using poliocirc F and poliocirc R (Table 1). The KSHV vFLIP IRES was amplified from the bicistronic reporter above using KSHVcirc F and KSHVcirc R (Table 1). The IRES elements were ligated into the amplified circular GFP reporter using Gibson cloning (NEB). The BiP and c-myc IRES elements were ordered as gBlock gene fragments (IDT; Table 1) and ligated to the amplified circular GFP reporter using Gibson cloning. The sequences of all circular GFP constructs were confirmed by Sanger sequencing. The pTRS1 triple mutant has been described previously (46).
Table 1.
Primer and gBlock Gene Fragment Sequences
| Primer Name | Sequence (5’ → 3’) |
|---|---|
| circGFP F | ACCATGGTGAGCAAGGGCGA |
| circGFP R | GTCGACTGCAGAATTCAGAT |
| poliocirc F | ATCTGAATTCTGCAGTCGACTTAAAACAGCTCTGGGGTTGTTCC |
| poliocirc R | TCGCCCTTGCTCACCATGGTATATCTTAACAATGAGGTAATTCC |
| KSHVcirc F | ATCTGAATTCTGCAGTCGACTTGGACAGACTCCTACTT |
| KSHVcirc R | TCGCCCTTGCTCACCATGGTTTGTATATGTGAAGGCACCG |
| BiP IRES gBlock | ATCTGAATTCTGCAGTCGACAGGTCGACGCCGGCCAAGACAGCACAGA CAGATTGACCTATTGGGGTGTTTCGCGAGTGTGAGAGGGAAGCGCCGC GGCCTGTATTTCTAGACCTGCCCTTCGCCTGGTTCGTGGCGCCTTGTGA CCCCGGGCCCCTGCCGCCTGCAAGTCGAAATTGCGCTGTGCTCCTGTG CTACGGCCTGTGGCTGGACTGCCTGCTGCTGCCCAACTGGCTGGCAAG ACCATGGTGAGCAAGGGCGA |
| c-myc IRES gBlock | ATCTGAATTCTGCAGTCGACTAATCCAGAACTGGATCGGGGTAAAGTGA CTTGTCAAGATGGGAGAGGAGAAGGCAGAGGGAAAACGGGAATGGTTT TTAAGACTACCCTTTCGAGATTTCTGCCTTATGAATATATTCACGCTGAC TCCCGGCCGGTCGGACATTCCTGCTTTATTGTGTTAATTGCTCTCTGGG TTTTGGGGGGCTGGGGGTTGCTTTGCGGTGGCAGAAAGCCCCTTGCAT CCTGAGCTCCTTGGAGTAGGGACCGCATATCGCCTGTGTGAGCCAGAT CGCTCCGCAGCCGCTGACTTGTCCCCGTCTCCGGGAGGGCATTTAAAT TTCGGCTCACCGCATTTCTGACAGCCGGAGACGGACACTCGGCGTCCC GCCCGCCTGTCCCCGCGGCGATTCCAACACCATGGTGAGCAAGGGCG A |
| GAPDH F | CTGTTGCTGTAGCCAAATTCGT |
| GAPDH R | ACCCACTCCTCCACCTTTGAC |
| RLUC F | TCGGTTGGCAGAAGCTATGA |
| RLUC R | CCGATAAATAACGCGCCCAA |
| FLUC F | ACAAAGGCTATCAGGTGGCT |
| FLUC R | CGTGCTCCAAAACAACAACG |
| qcircGFP F | GCAGTGCTTCAGCCGCTAC |
| qcircGFP R | GTGTCGCCCTCGAACTTCAC |
2.3. Luciferase assays.
HeLa cells were seeded into 12-well plates (150,000 cells/well) and transfected with the indicated plasmids using polyethylenimine (PEI, Polysciences). Twenty-four hours after transfection cells were washed in 1 ml 1X PBS (Sigma) and lysed in 150 μl 1X Passive Lysis Buffer (Promega) for 10 min with rocking at room temperature. After lysis supernatants were cleared of debris by centrifugation at 10,000 x g for 1 min. For firefly luciferase, 40 μl of luciferase reagent (Promega) was added to 8 μl of sample, and luciferase activity was measured using a luminometer (Molecular Devices). For experiments using bicistronic reporters, 40 μl of Stop and Glo reagent was added to each sample after measuring firefly luciferase levels. 40 μl of renilla luciferase reagent (Promega) was then added, and luciferase activity was again measured. The amount of luciferase activity was normalized to the amount of protein present in each sample as determined by Bradford assay (Amresco).
2.4. Western blot analysis.
Cells were washed once in 1X PBS before scraping, and cell pellets were stored at −80˚C until analysis. Cell pellets were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP40, 1% sodium deoxycholate) containing protease and phosphatase inhibitors (Roche) for 10 min on ice. Cell debris was removed by centrifugation at 10,000 x g for 10 min. The lysate protein concentration was determined by Bradford assay and equal amounts of protein were loaded onto SDS-PAGE gels. The resolved proteins were transferred to 0.45 μm nitrocellulose membranes (Amersham) and blocked in 5% non-fat milk in TBS-T (20 mM Tris-HCl, pH 7.6, 140 mM NaCl, 0.1% Tween 20) for 1 hour at room temperature. For mouse primary antibodies, membranes were incubated with the appropriate antibody diluted in 1% BSA in TBS-T for one hour at room temperature. For rabbit primary antibodies, membranes were incubated in the appropriate antibody diluted in 5% BSA in TBS-T overnight. Secondary antibodies were diluted 1:10,000 in 1% BSA in TBS-T and incubated with the membrane for 1 hour at room temperature. Proteins were visualized by chemiluminesence using WesternBlot ECL (Advansta) and an imager (ChemiDoc; BioRad). The following antibodies were used: anti-GFP (1:10,000; Sigma G1544), anti-tubulin (1:20,000; ThermoScientific A11126), anti-TRS1 (1:100; (48)), anti-His (1:1,000; CST #2365). Western blot densitometry was quantified using ImageJ software.
2.5. Quantitative real-time PCR (qRT-PCR).
RNA was extracted from cells using Trizol essentially as described previously (41). Briefly, 100 μl of cell lysate was mixed with 1 ml of Trizol and incubated at room temperature for 10 min. 200 μl of chloroform was then added, and samples were shaken for 60 sec before centrifugation at 10,000 x g for 10 min. The aqueous layer was removed and an equal volume of isopropanol was added. Samples were incubated at −20°C overnight, and the RNA was pelleted by centrifugation at 10,000 x g for 30 min at 4°C. RNA pellets were washed once in 70% ethanol, resuspended in 1X DNase buffer containing 20U DNase (Turbo DNase free kit; Ambion), and incubated at 37°C for 30 min. DNase was inactivated according to the manufacturer’s directions. RNA concentrations were determined using a spectrophotometer (NanoDrop) and equal amounts of RNA were reverse transcribed to cDNA using the High Capacity cDNA Reverse Transcription Kit (ThermoFisher). 2 μl of cDNA was added to 1X SYBR Green Master Mix with the appropriate primers, and real-time PCR was performed using the following conditions: 95°C for 5 minutes, followed by 40 cycles of 95°C for 10 seconds, 55°C for 30 seconds, and 72°C for 90 seconds. Changes in RNA levels were determined by the ΔΔCt method as before using GAPDH as the reference gene (41). The following primer pairs were used (sequences listed in Table 1): GAPDH (GAPDH F and GAPDH R), renilla luciferase (RLUC F and RLUC R), firefly luciferase (FLUC F and FLUC R), circular GFP (qcircGFP F and qcircGFP R). All experiments were performed at least in triplicate and statistical significance was determined using a Student’s paired t-test.
3. Results
3.1. pTRS1 enhances the activity of the poliovirus IRES.
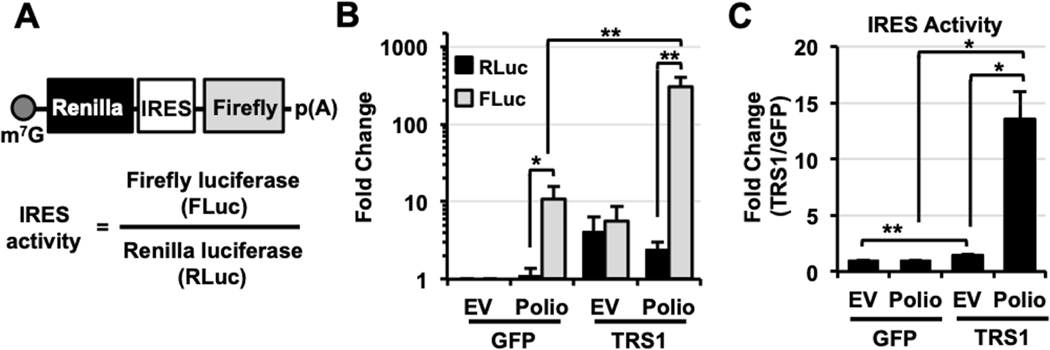
We previously found that pTRS1 increases the cap-dependent translation of monocistronic luciferase reporters (41). To determine if pTRS1 also enhances cap-independent translation we measured the effect of pTRS1 expression on the translation of a bicistronic mRNA reporter containing the poliovirus IRES (Polio) (19, 21, 49). Translation of the first cistron of the bicistronic mRNA (renilla luciferase) is cap-dependent, while translation of the downstream cistron (firefly luciferase) is driven by an IRES element in a cap-independent manner (Fig 1A). A bicistronic reporter containing no IRES element (empty vector (EV)) served as a control. Cells were co-transfected with either the EV or Polio bicistronic reporter with either a TRS1 expression vector or a control vector expressing GFP, and the levels of renilla and firefly luciferase were quantified (Fig 1B). In cells expressing GFP, the polio reporter showed a 10-fold increase in firefly luciferase levels compared to the EV, with no significant change in renilla luciferase levels, consistent with the known activity of the poliovirus IRES (39). pTRS1 expression increased renilla luciferase levels from both the EV and polio reporter by approximately 5-fold compared to cells expressing GFP, consistent with the ability of pTRS1 to enhance cap-dependent translation. As compared to cells expressing GFP, pTRS1 expression induced firefly luciferase to similar extent in cells transfected with EV reporter, but had a much greater effect on firefly activity driven by the polio IRES. The ratio of firefly to renilla luciferase activity serves as a measure of IRES activity (Fig 1A). Comparing the poliovirus IRES activity in cells transfected withy GFP or pTRS1, we found that pTRS1 expression did not affect IRES activity from the control reporter (Fig 1C), as levels of renilla and firefly luciferase were increased similarly (Fig 1B). However, pTRS1 expression did increase polio IRES activity by 13-fold compared to cells expressing the GFP (Fig 1C). These results suggest that pTRS1 enhances cap-independent translation driven by a viral IRES.
Figure 1. pTRS1 stimulates poliovirus IRES activity.
(A) Bicistronic mRNA reporter schematic. IRES activity is defined as the level of firefly luciferase (FLuc) compared to the level of renilla luciferase (RLuc). (B) HeLa cells were co-transfected with a bicistronic reporter containing no IRES element (EV; empty vector) or a reporter containing the poliovirus IRES (Polio) in the presence of GFP or TRS1. The average fold change of RLuc (black bars) and Fluc (grey bars) compared to EV co-transfected with GFP is shown (n=3). (C) As in (B), fold change in EV and Polio IRES activity in the presence of pTRS1 compared to GFP. *(p<0.05); **(p<0.01)
3.2. KSHV vFLIP IRES activity is enhanced in the presence of pTRS1.
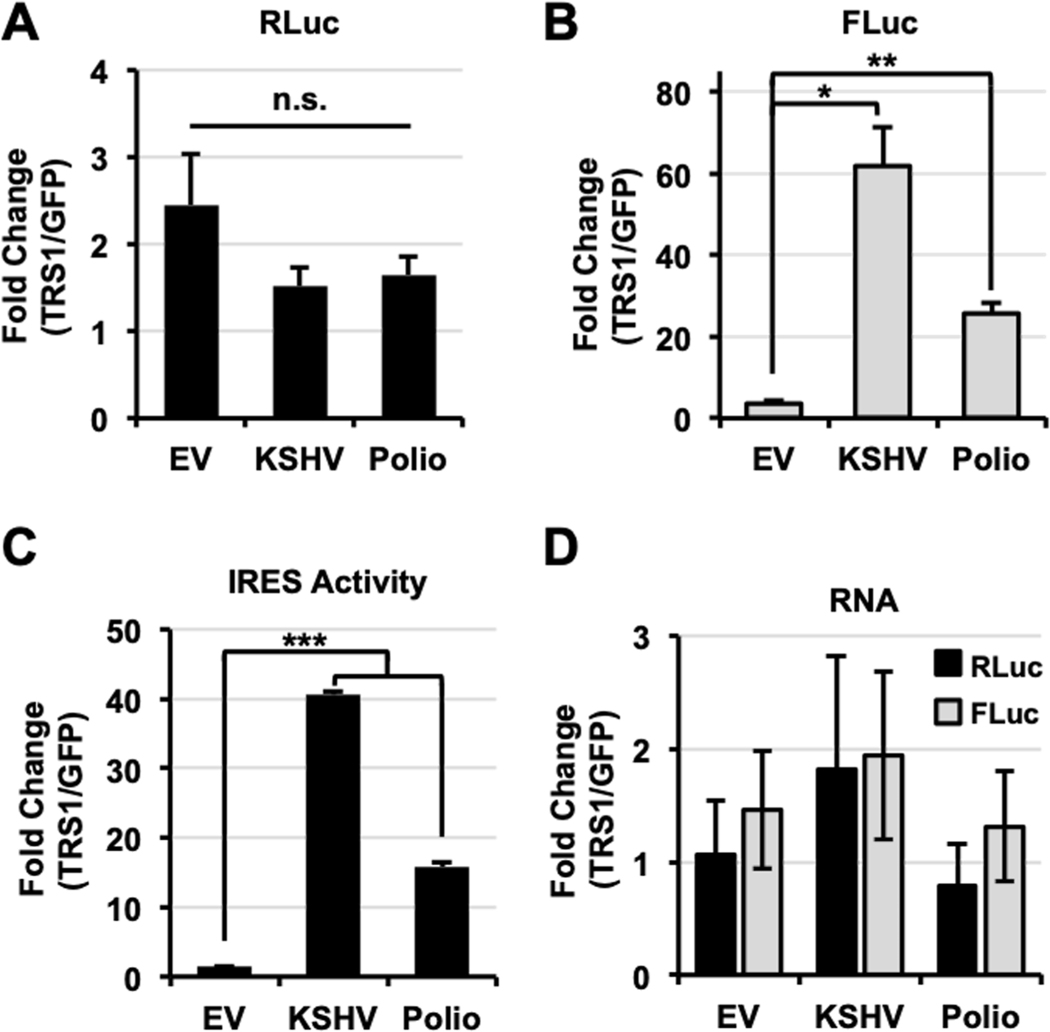
To determine if the effect of pTRS1 on cap-independent translation was specific to the polio IRES, we examined the effect of pTRS1 on the activity of an additional viral IRES from the Kaposi’s sarcoma-associated herpersvirus (KSHV) vFLIP mRNA (31). pTRS1 did not change renilla luciferase activity (Fig 2A), but significantly increased firefly luciferase activity (Fig 2B) from the vFLIP reporter. In fact, pTRS1 expression had an even greater effect on vFLIP IRES activity than poliovirus IRES activity (Fig 2C). An important caveat of bicistronic reporter assays is the potential for cryptic promoter activity or cryptic splicing to alter expression of the downstream cistron. However we found that pTRS1 expression did not lead to a significant change in either firefly or renilla RNA levels as compared to cells expressing GFP (Fig 2D). Thus the increase in poliovirus and KSHV vFLIP IRES activity in the presence of pTRS1 was not due to cryptic promoter activity or cryptic RNA splicing. These data demonstrate that pTRS1 stimulates viral IRES activity, providing further support for the conclusion that pTRS1 enhances cap-independent translation.
Figure 2. pTRS1 enhances virus IRES activity.
HeLa cells were co-transfected with a bicistronic reporter containing no IRES element (EV), the KSHV vFLIP IRES (KSHV) or the poliovirus IRES (Polio) and either GFP or TRS1. Average fold change in RLuc (A), FLuc (B) and IRES activity (C) in cells transfected with TRS1 compared to cells transfected with GFP is shown for each reporter (n=3). (D) RNA was extracted from cells transfected as in (A). RLuc (black bars) and Fluc (grey bars) RNA levels were determined by RT-qPCR. Average fold change in RLuc and FLuc RNA in cells transfected with TRS1 compared to cells transfected with GFP is shown for each bicistronic reporter (n=2). *(p<0.05); **(p<0.01); ***(p<0.005)
3.3. pTRS1 RNA binding is necessary to promote translation of a circular mRNA.
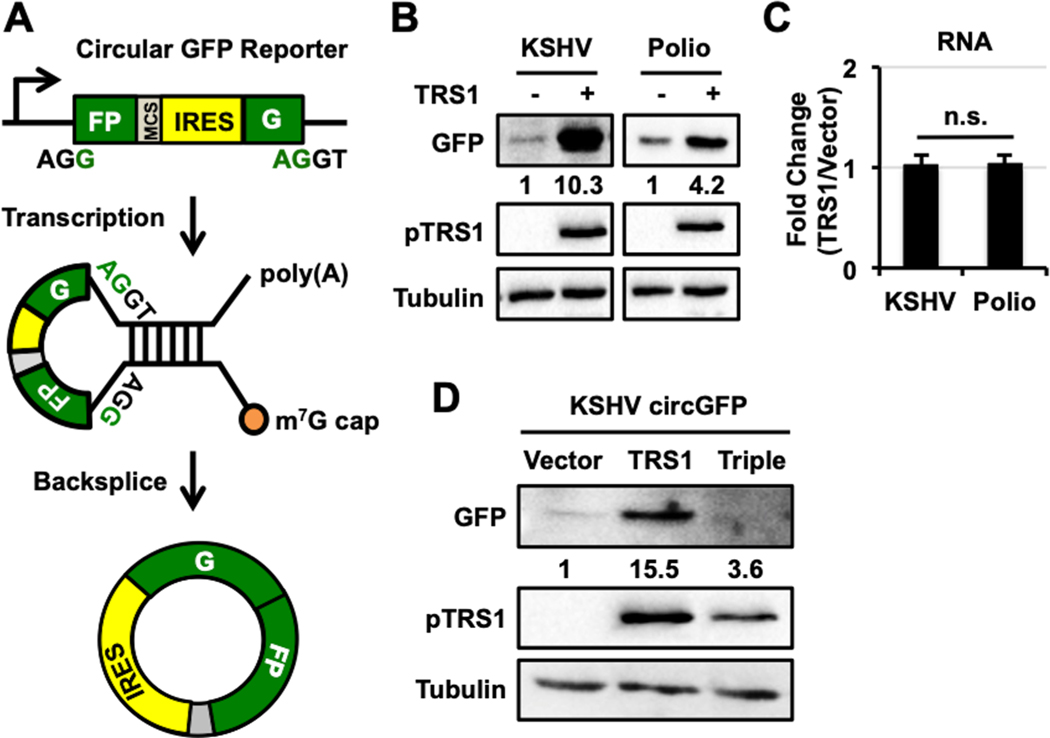
Translation of the firefly open reading frame (ORF) in the bicistronic reporter requires an IRES element. However, the bicistronic RNA encoding the firefly ORF has an m7G mRNA cap, which pTRS1 can bind (41). Thus pTRS1 could promote translation of the firefly ORF by bridging an interaction with cap-associated proteins and the IRES element. If so, the ability of pTRS1 to stimulate IRES activity would not be truly independent of the m7G mRNA cap. To test this possibility, we determined the effect of pTRS1 expression on the translation of a circular reporter RNA. The circular RNA reporter consists of a single exon minigene containing split GFP in the reverse order (Fig 3A). Backsplicing of the primary transcript produces a circular RNA (circRNA), which lacks both a m7G RNA cap and poly(A) tail, and contains the GFP coding sequence in the correct order. The inclusion of an IRES upstream of the GFP start codon allows the circRNA to be translated (47, 50). The lack of both an m7G mRNA cap and poly(A) tail on the mature mRNA encoding GFP makes this system extremely useful for measuring the effect of IRES trans-activating-factors (ITAFs) on IRES activity.
Figure 3. pTRS1 dsRNA binding is necessary to promote translation of a circular GFP mRNA.
(A) Circular GFP reporter schematic. A backsplicing event in the reporter mRNA results in the formation of a circular GFP mRNA containing no m7G cap or poly(A) tail. “G” and “FP” represent the split GFP exons. MCS = multiple cloning site. (B) HeLa cells were co-transfected with a circular GFP reporter containing either the KSHV vFLIP IRES (KSHV) or the poliovirus IRES (Polio) with either a vector control or TRS1, and GFP, pTRS1, and tubulin protein levels were measured by Western blot. Results of a representative experiment are shown (n=3). Numbers indicate the fold change in GFP expression compared to vector control, normalized to pTRS1 and tubulin levels. (C) RNA was extracted from cells transfected as in (B) and circular GFP mRNA levels were determined by RT-qPCR. The average fold change of GFP RNA in cells transfected with TRS1 compared to cells transfected with vector are shown for each reporter (n=3). The primers used span the GFP exon junctions, and thus only detect spliced mRNAs. (D) HeLa cells were co-transfected with a circular GFP reporter containing the KSHV vFLIP IRES and either a vector control (Vector), wild-type TRS1, or a TRS1 mutant that lacks the ability to bind dsRNA (Triple). A representative image of GFP and pTRS1 expression, as determined by Western blot (n=3), is shown. Fold change in GFP expression normalized to pTRS1 levels was determined as in (B).
We cloned the poliovirus and KSHV vFLIP IRESs into the circular GFP vector (polio circGFP and KSHV circGFP, respectively), and measured GFP expression from the circular RNA reporters. We previously showed that GFP was not expressed in the absence of an IRES element upstream of the GFP ORF, however the inclusion of either the poliovirus or KSHV vFLIP IRES allowed for efficient GFP expression (Fig 3B). We next determined how pTRS1 expression affected GFP expression from the polio circGFP and KSHV circGFP reporters. Both circGFP reporters expressed more GFP protein when co-transfected with pTRS1 as compared to an empty expression vector containing no ORF (pcDNA vector control) (Fig 3B), however pTRS1 expression did not affect spliced GFP RNA levels (Fig 3C). Together with the results from the bicistronic reporters assays above, these data suggest that pTRS1 promotes mRNA translation independent of the m7G mRNA cap or factors associated with the poly(A) tail.
pTRS1 has a non-canonical RNA binding domain that preferentially binds dsRNA (43, 44). While this function is dispensable for pTRS1 to antagonize PKR, we hypothesized it may be important to enhance cap-independent translation (46). To test this hypothesis we compared the ability of wild type pTRS1 and a pTRS1 dsRNA binding mutant (Triple) to increase GFP expression from the KSHV vFLIP circRNA (Fig 3D). pTRS1 Triple contains three arginine to alanine mutations in the RNA binding domain that abrogate its ability to bind dsRNA (43). As before, wild type pTRS1 increased GFP expression compared to the cells expressing GFP. However, significantly lower GFP expression was observed in cells transfected with the pTRS1 RNA binding mutant. These results show that the ability of pTRS1 to bind dsRNA is necessary for its ability to stimulate cap-independent translation.
3.4. pTRS1 stimulates the activity of cellular IRESs..
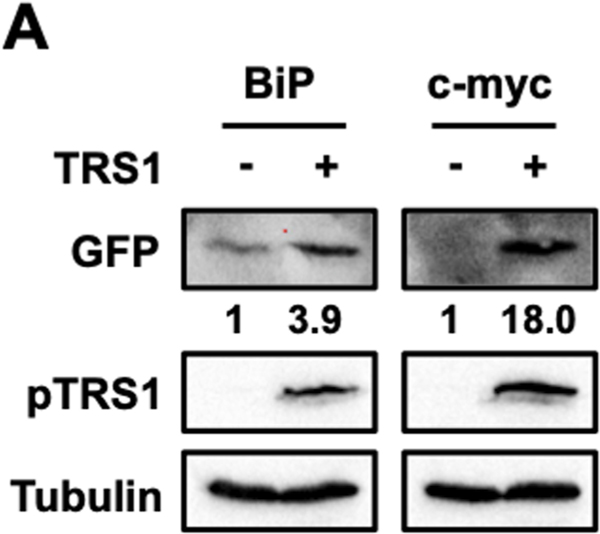
While most known IRESs are found in viral RNAs, some IRESs have also been reported in cellular RNAs. We examined the effect of pTRS1 expression on translation driven by two cellular IRESs, the binding immunoglobulin protein (BiP) and c-myc IRESs (15, 16, 18). Both BiP and c-myc are induced by HCMV infection and their expression is required for HCMV replication (51–56). Using our circular RNA reporter assay we confirmed that the BiP and c-myc IRES supported cap-independent translation, though the c-myc IRES had much lower basal activity (Fig 4). pTRS1 enhanced both BiP and c-myc IRES activity as shown by increased GFP protein expression compared to the empty pcDNA vector (vector control), though the effect was less pronounced than that observed with the poliovirus and vFLIP IRESs. Thus, in addition to increasing the activity of viral IRESs, pTRS1 also enhances translation driven by cellular IRES elements, including at least two IRESs that control the expression of cellular proteins needed for HCMV replication.
Figure 4. Cellular IRES activity is enhanced by pTRS1 expression.
HeLa cells were co-transfected with a circular GFP reporter containing either the BiP IRES (BiP) or the c-myc IRES (c-myc) with either a vector control or TRS1. A representative Western blot image showing GFP and pTRS1 (n=3). Numbers indicate the fold change in GFP expression compared to vector control, normalized to pTRS1 and tubulin levels.
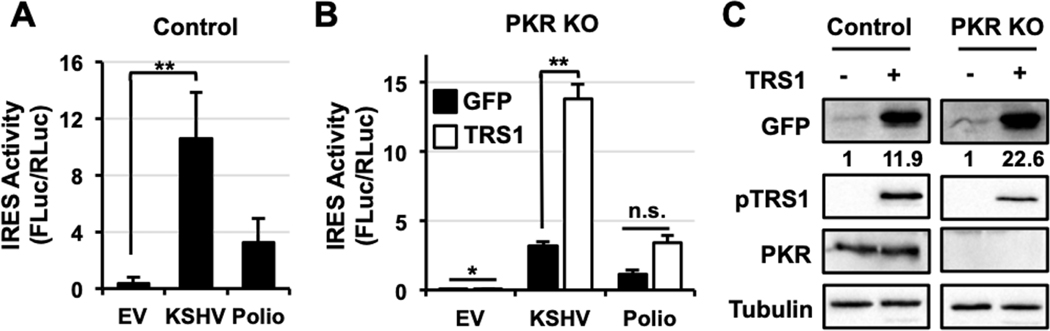
3.5. pTRS1 stimulates IRES activity independent of its ability to antagonize PKR.
pTRS1 enhances translation in part by blocking PKR activation, but also promotes cap-dependent translation independent of PKR antagonism (41, 45). To determine if the increase in IRES activity in the presence of pTRS1 was due to PKR inhibition, we measured the effect of pTRS1 on IRES activity in cells where CRISPR/Cas9 mutagenesis was used to ablate PKR expression (PKR KO cells, (45)), HeLa cells transfected with a guide RNA to the adeno-associated virus integration site (AAVS) served as a control. Using bicistronic reporters, we found that the polio and KSHV vFLIP IRESs had increased activity in PKR KO cells, demonstrating that PKR inhibits poliovirus and KSHV vFLIP IRES activity in this system (Fig 5A). However, we consistently saw a slight increase in polio IRES activity in PKR KO cells expressing pTRS1 compared to cells expressing GFP, though the increase was not statistically significant. In contrast, pTRS1 increased vFLIP IRES activity by 4-fold in PKR KO cells compared to cells expressing a control protein (GFP), a statistically significant increase in IRES activity (Fig 5B). We confirmed these results using the circGFP reporter containing the KSHV vFLIP IRES. pTRS1 expression increased GFP protein levels in PKR KO cells (Fig 5C) compared to an empty pcDNA vector control, showing that pTRS1 stimulates KSHV vFLIP IRES activity in a cap- and PKR-independent manner. Together our data show that pTRS1 stimulates cap-independent translation by promoting IRES activity, and that this function is independent of pTRS1 PKR inhibition, but dependent on the ability of pTRS1 to bind RNA.
Figure 5. pTRS1 stimulation of IRES activity is independent of PKR antagonism.
(A) Control HeLa or PKR KO HeLa cells were co-transfected with the a bicistronic reporter containing no IRES (EV), the KSHV vFLIP IRES (KSHV), or the poliovirus IRES (Polio) and GFP. Average IRES activity of each reporter (FLuc/RLuc) (n=3) is shown. (B) PKR KO HeLa cells were co-transfected with the a bicistronic reporter containing no IRES (EV), the KSHV vFLIP IRES (KSHV), or the poliovirus IRES (Polio) with either GFP (black bars) or TRS1 (white bars). Average IRES activity of each reporter (FLuc/RLuc) (n=3) is shown. (C) WT and PKR KO HeLa cells were co-transfected with a circular GFP reporter containing the KSHV vFLIP IRES and either a vector control or TRS1, anf GFP levels were meaureed by Western blot. A representivate image showing GFP and pTRS1 expression is shown (n=3). Numbers indicate the fold change in GFP expression compared to vector control, normalized to pTRS1 and tubulin levels. *(p<0.05); **(p<0.01)
4. Discussion
Our results raise the question of how pTRS1 enhances cap-independent translation. While pTRS1 binds the m7G mRNA cap (41), this function is dispensable to stimulate IRES activity as pTRS1 enhances translation of circRNA reporters lacking an mRNA cap and a poly(A) tail (Fig 3B). Here we show that the ability of pTRS1 to bind RNA is necessary for pTRS1 to enhance cap-independent translation (Fig 3D). Our group and others have shown that pTRS1 binds RNA, with a preference for binding to double-stranded RNA (dsRNA) (43, 57). The fact that pTRS1 stimulates the activity of multiple IRESs with minimal sequence homology suggests that pTRS1 might bind common IRES RNA structures. As pTRS1 also associates with 40S ribosomal subunits (41), these data suggest a model wherein pTRS1 binds to double-stranded RNA structures in IRESs and facilitates the recruitment of ribosomal subunits, resulting in enhanced cap-independent translation initiation.
pTRS1 potently antagonizes the antiviral kinase PKR, which inhibits both cap-dependent and -independent translation by limiting the availability of active ternary complexes (42, 45, 46, 58, 59). Our results show that PKR inhibition by pTRS1 contributes to increased cap-independent translation, as pTRS1 does not enhance IRES activity to the same magnitude in PKR KO cells (Fig 5B). However, pTRS1 maintains the ability to enhance cap-independent translation in PKR KO cells, demonstrating additional pTRS1 functions are also needed to enhance IRES activity. Multiple host RNA binding proteins recognize viral RNA elements and activate restriction pathways that limit viral protein synthesis (3). Therefore, it is possible that pTRS1 antagonizes additional restriction factors that recognize and inhibit the translation of viral RNA. If so, the requirement for the pTRS1 RNA binding domain suggests that pTRS1 competes with antiviral sensors to limit their detection of dsRNA structures.
Interestingly, in PKR KO cells, pTRS1 increased KSHV vFLIP IRES activity to a much greater extent than poliovirus IRES activity, which was not significantly enhanced by pTRS1 expression (Fig 5B). We found that the basal IRES activity for both viral IRESs increased in PKR KO cells compared to wild-type cells (Fig 5A), thus it could be that the majority of poliovirus IRES activity enhancement by pTRS1 is from PKR inhibition. Both viral IRESs require several initiation factors (e.g. eIF4A, eIF4G, eIF2) to initiation translation, however there are differences in the complement of proteins necessary for IRES activity between the two viral IRESs. For example, the KSHV vFLIP IRES requires eIF4E, while the poliovirus IRES requires PTB. Therefore, pTRS1 may interact with and nucleate a complex of initiation factors that promote KSHV vFLIP IRES activity more efficiently than poliovirus IRES activity. In either case, our data shows that the ability of pTRS1 to inhibit PKR activation and promote cap-independent translation are separable functions.
How might the enhancement of cap-independent translation by pTRS1 impact HCMV replication? HCMV infection increases eIF4F abundance, and the eIF4F complex is required for the synthesis of several cellular proteins needed for virus replication (36–38). However, inhibiting the activity or formation of the eIF4F complex does not limit the association of HCMV mRNAs with polysomes, especially during the later stages of infection (38). Our results show that pTRS1 enhances the translation of a circRNA reporter, which lacks both an m7G cap and poly (A) tail, suggesting pTRS1 may help recruit ribosomes to mRNAs independent of the eIF4F complex. This could account for the ability of HCMV mRNAs to be efficiently translated despite disruption of the eIF4F complex. Recently, over 100 putative IRES elements within the HCMV genome have been identified (40). pTRS1 may stimulate the activity of these IRES elements during HCMV infection to promote viral protein synthesis. The increase in cap-independent translation caused by pTRS1 may also promote the expression of cellular proteins necessary for HCMV replication. BiP IRES activity and protein levels increase during HCMV infection, and BiP expression is necessary for efficient HCMV replication (51–54). Perhaps the stimulation of cap-independent translation by pTRS1 (Fig 4) allows for the efficient translation of BiP and other pro-viral cellular factors needed for successful HCMV replication.
5. Conclusions
In this study we find that HCMV pTRS1 enhances cap-independent translation. pTRS1 stimulates the activity of multiple IRESs in both a bicistronic reporter assay and a novel circular RNA assay of IRES activity. Additionally, pTRS1 stimulates cap-independent translation, independent of its ability to inhibit PKR, as it increases IRES activity in PKR KO cells. The ability of pTRS1 to bind RNA is necessary to stimulate cap-independent translation, suggesting that pTRS1 may bind specific mRNA sequences or structures to facilitate enhanced translation initiation.
Our results add to the growing list of functions ascribed to pTRS1 (41, 44–46, 57, 60, 61). While antagonism of PKR is an important role of pTRS1 during infection of primary fibroblasts, the enhancement of cap-independent translation may be more important in other cell types relevant to HCMV disease, such as CD34+ progenitor cells, epithelial cells or placental cells (62, 63). Further studies to define the mechanism by which pTRS1 stimulates both cap-dependent and cap-independent translation will therefore be critical to fully understand the role of pTRS1 in HCMV pathogenesis.
Acknowledgements
We would like to thank the Asokan and Marzluff labs for providing the circular GFP reporters and valuable conversations. We would also like to thank the Heise, Baric, Moody, and de Silva labs for their helpful comments and suggestions. Special thanks are given to Westchester Sanders and Anne Beall for their continued support.
Funding
This work was supported by NIH grant AI03311 to N.J.M. Additional support was provided by the North Carolina University Cancer Research Fund, and an award from the UNC Virology Training grant (T32 AI07419 to B.Z.).
List of Abbreviations
- HCMV
human cytomegalovirus
- IRES
internal ribosome entry site
- PKR
protein kinase R
- m7G
7-methylguanosine
- eIF
eukaryotic initiation factor
- PIC
preinitiation complex
- UTR
untranslated region
- ITAF
IRES trans-activating factor
- dsRNA
double-stranded RNA
- EV
empty vector
- KSHV
Kaposi’s sarcoma-associated herpesvirus
- ORF
open reading frame
- circRNA
circular RNA
- BiP
binding immunoglobin protein
- PKR KO
PKR knockout
Footnotes
Conflicts of Interest
The authors declare no competing financial interests regarding the publication of this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adeli K. 2011. Translational control mechanisms in metabolic regulation: critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. American Journal of Physiology - Endocrinology and Metabolism 301:E1051–E1064. [DOI] [PubMed] [Google Scholar]
- 2.Pyronnet S, Sonenberg N. 2001. Cell-cycle-dependent translational control. Current Opinion in Genetics & Development 11:13–18. [DOI] [PubMed] [Google Scholar]
- 3.Jan E, Mohr I, Walsh D. 2016. A Cap-to-Tail Guide to mRNA Translation Strategies in Virus-Infected Cells. Annual Review of Virology 3:283–307. [DOI] [PubMed] [Google Scholar]
- 4.Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annual Review of Biochemistry 83:779–812. [DOI] [PubMed] [Google Scholar]
- 5.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. 1983. New initiation factor activity required for globin mRNA translation. Journal of Biological Chemistry 258:5804–5810. [PubMed] [Google Scholar]
- 7.Tahara SM, Morgan MA, Shatkin AJ. 1981. Two forms of purified m7G-cap binding protein with different effects on capped mRNA translation in extracts of uninfected and poliovirus-infected HeLa cells. The Journal of Biological Chemistry 256:7691–7694. [PubMed] [Google Scholar]
- 8.Sonenberg N, Shatkin AJ. 1978. Nonspecific effect of m7GMP on protein-RNA interactions. The Journal of Biological Chemistry 253:6630–6632. [PubMed] [Google Scholar]
- 9.Shin B-S, Kim J-R, Walker SE, Dong J, Lorsch JR, Dever TE. 2011. Initiation Factor eIF2γ Promotes eIF2–GTP–Met-tRNAiMet Ternary Complex Binding to the 40S Ribosome. Nature structural & molecular biology 18:1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Méthot N, Song MS, Sonenberg N. 1996. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Molecular and Cellular Biology 16:5328–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asano K, Clayton J, Shalev A, Hinnebusch AG. 2000. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes & Development 14:2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valásek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. 2004. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Molecular and Cellular Biology 24:9437–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellen CUT, Sarnow P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Development 15:1593–1612. [DOI] [PubMed] [Google Scholar]
- 14.Le SY, Maizel JV. 1997. A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Research 25:362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Q, Sarnow P. 1997. Location of the internal ribosome entry site in the 5’ noncoding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Research 25:2800–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. 1998. C-Myc 5’ untranslated region contains an internal ribosome entry segment. Oncogene 16:423–428. [DOI] [PubMed] [Google Scholar]
- 17.Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proceedings of the National Academy of Sciences 96:13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johannes G, Sarnow P. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA (New York, NY) 4:1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325. [DOI] [PubMed] [Google Scholar]
- 20.Nomoto A, Kitamura N, Golini F, Wimmer E. 1977. The 5’-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proceedings of the National Academy of Sciences of the United States of America 74:5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelletier J, Flynn ME, Kaplan G, Racaniello V, Sonenberg N. 1988. Mutational analysis of upstream AUG codons of poliovirus RNA. Journal of Virology 62:4486–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hambidge SJ, Sarnow P. 1992. Translational enhancement of the poliovirus 5’ noncoding region mediated by virus-encoded polypeptide 2A. Proceedings of the National Academy of Sciences of the United States of America 89:10272–10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borman AM, Bailly JL, Girard M, Kean KM. 1995. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Research 23:3656–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer K, Petersen A, Niepmann M, Beck E. 1995. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. Journal of Virology 69:2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestova TV, Shatsky IN, Hellen CU. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Molecular and Cellular Biology 16:6870–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomakin IB, Hellen CU, Pestova TV. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Molecular and Cellular Biology 20:6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson EC, Hunt SL, Jackson RJ. 2007. Internal initiation of translation from the human rhinovirus-2 internal ribosome entry site requires the binding of Unr to two distinct sites on the 5’ untranslated region. The Journal of General Virology 88:3043–3052. [DOI] [PubMed] [Google Scholar]
- 28.Fletcher SP, Jackson RJ. 2002. Pestivirus Internal Ribosome Entry Site (IRES) Structure and Function: Elements in the 5′ Untranslated Region Important for IRES Function. Journal of Virology 76:5024–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaminski A, Howell MT, Jackson RJ. 1990. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. The EMBO journal 9:3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukavsky PJ. 2009. Structure and function of HCV IRES domains. Virus Research 139:166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bieleski L, Talbot SJ. 2001. Kaposi’s sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. Journal of Virology 75:1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mocarski ES Jr.. 2007. Betaherpes viral genes and their functions. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge University Press, Cambridge. [PubMed] [Google Scholar]
- 33.Stinski MF, Meier JL. 2007. Immediate–early viral gene regulation and function. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge University Press, Cambridge. [PubMed] [Google Scholar]
- 34.Tanaka S, Furukawa T, Plotkin SA. 1975. Human cytomegalovirus stimulates host cell RNA synthesis. Journal of Virology 15:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stinski MF. 1977. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. Journal of Virology 23:751–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh D, Perez C, Notary J, Mohr I. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. Journal of Virology 79:8057–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez C, McKinney C, Chulunbaatar U, Mohr I. 2011. Translational control of the abundance of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. Journal of Virology 85:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lenarcic EM, Ziehr B, De Leon G, Mitchell D, Moorman NJ. 2014. Differential role for host translation factors in host and viral protein synthesis during human cytomegalovirus infection. Journal of Virology 88:1473–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grainger L, Cicchini L, Rak M, Petrucelli A, Fitzgerald KD, Semler BL, Goodrum F. 2010. Stress-inducible alternative translation initiation of human cytomegalovirus latency protein pUL138. Journal of Virology 84:9472–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. 2016. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351:aad4939. [DOI] [PubMed] [Google Scholar]
- 41.Ziehr B, Lenarcic E, Vincent HA, Cecil C, Garcia B, Shenk T, Moorman NJ. 2015. Human cytomegalovirus TRS1 protein associates with the 7-methylguanosine mRNA cap and facilitates translation. Proteomics 15:1983–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braggin JE, Child SJ, Geballe AP. 2016. Essential role of protein kinase R antagonism by TRS1 in human cytomegalovirus replication. Virology 489:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bierle CJ, Semmens KM, Geballe AP. 2013. Double-stranded RNA binding by the human cytomegalovirus PKR antagonist TRS1. Virology 442:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakki M, Geballe AP. 2005. Double-stranded RNA binding by human cytomegalovirus pTRS1. Journal of Virology 79:7311–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziehr B, Vincent HA, Moorman NJ. 2016. Human Cytomegalovirus pTRS1 and pIRS1 Antagonize Protein Kinase R To Facilitate Virus Replication. Journal of Virology 90:3839–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent HA, Ziehr B, Moorman NJ. 2017. Mechanism of Protein Kinase R Inhibition by Human Cytomegalovirus pTRS1. Journal of Virology 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Wang Z. 2015. Efficient backsplicing produces translatable circular mRNAs. RNA (New York, NY) 21:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romanowski MJ, Shenk T. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. Journal of Virology 71:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balvay L, Rifo RS, Ricci EP, Decimo D, Ohlmann T. 2009. Structural and functional diversity of viral IRESes. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1789:542–557. [DOI] [PubMed] [Google Scholar]
- 50.Borchardt EK, Meganck RM, Vincent HA, Ball CB, Ramos SBV, Moorman NJ, Marzluff WF, Asokan A. 2017. Inducing circular RNA formation using the CRISPR endoribonuclease Csy4. RNA (New York, NY) 23:619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchkovich NJ, Yu Y, Pierciey FJ, Alwine JC. 2010. Human cytomegalovirus induces the endoplasmic reticulum chaperone BiP through increased transcription and activation of translation by using the BiP internal ribosome entry site. Journal of Virology 84:11479–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchkovich NJ, Maguire TG, Alwine JC. 2010. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. Journal of Virology 84:7005–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buchkovich NJ, Maguire TG, Paton AW, Paton JC, Alwine JC. 2009. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. Journal of Virology 83:11421–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buchkovich NJ, Maguire TG, Yu Y, Paton AW, Paton JC, Alwine JC. 2008. Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. Journal of Virology 82:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagemeier C, Walker SM, Sissons PJ, Sinclair JH. 1992. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. The Journal of General Virology 73 ( Pt 9):2385–2393. [DOI] [PubMed] [Google Scholar]
- 56.Monick MM, Geist LJ, Stinski MF, Hunninghake GW. 1992. The immediate early genes of human cytomegalovirus upregulate expression of the cellular genes myc and fos. American Journal of Respiratory Cell and Molecular Biology 7:251–256. [DOI] [PubMed] [Google Scholar]
- 57.Lenarcic EM, Ziehr BJ, Moorman NJ. 2015. An unbiased proteomics approach to identify human cytomegalovirus RNA-associated proteins. Virology 481:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. 2005. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell 122:901–913. [DOI] [PubMed] [Google Scholar]
- 59.Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG. 2001. Tight Binding of the Phosphorylated α Subunit of Initiation Factor 2 (eIF2α) to the Regulatory Subunits of Guanine Nucleotide Exchange Factor eIF2B Is Required for Inhibition of Translation Initiation. Molecular and Cellular Biology 21:5018–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaumorcel M, Lussignol M, Mouna L, Cavignac Y, Fahie K, Cotte-Laffitte J, Geballe A, Brune W, Beau I, Codogno P, Esclatine A. 2012. The Human Cytomegalovirus Protein TRS1 Inhibits Autophagy via Its Interaction with Beclin 1. Journal of Virology 86:2571–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Child SJ, Hakki M, De Niro KL, Geballe AP. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. Journal of Virology 78:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nogalski MT, Collins-McMillen D, Yurochko AD. 2014. Overview of Human Cytomegalovirus Pathogenesis, p 15–28, Human Cytomegaloviruses. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 63.Sinzger C, Digel M, Jahn G. 2008. Cytomegalovirus Cell Tropism, p 63–83, Human Cytomegalovirus. Springer, Berlin, Heidelberg. [DOI] [PubMed] [Google Scholar]