Abstract
The ability to reproduce is essential in all branches of life. In metazoans, this process is initiated by formation of the germline, a group of cells that are destined to form the future gonads, the tissue that will produce the gametes. The molecular mechanisms underlying germline formation differs between species. In zebrafish, development of the germline is dependent on the specification, migration and proliferation of progenitors called the primordial germ cells (PGCs). PGC specification is dependent on a maternally provided cytoplasmic complex of ribonucleoproteins (RNPs), the germplasm. Here, we show that the conserved RNA-binding protein (RBP), Igf2bp3, has an essential role during early embryonic development and germline development. Loss of Igf2bp3 leads to an expanded yolk syncytial layer (YSL) in early embryos, reduced germline RNA expression, and mis-regulated germline development. We show that loss of maternal Igf2bp3 function results in translational de-regulation of a Nodal reporter during the mid-blastula transition. Furthermore, maternal igf2bp3 mutants exhibit reduced expression of germplasm transcripts, defects in chemokine guidance, abnormal PGC behavior and germ cell death. Consistently, adult igf2bp3 mutants show a strong male bias. Our findings suggest that Igf2bp3 is essential for normal embryonic and germline development, and acts as a key regulator of sexual development.
Author summary
Animals develop from a single egg cell that is fertilised by sperm, to form various tissues and organs. How the different tissues form is still not fully understood. In particular, we don’t know precisely how organisms develop to be males or females, or how balanced sex ratios are maintained in populations. However, if these processes are disrupted, either too many males or too many females could arise in populations, which can lead to failure to produce the next generation, eventually resulting in extinction of the species. Therefore, identifying the factors that allow these processes to happen properly and deciphering how they function is crucial. We have identified a protein called Igf2bp3 in the tropical fish, Zebrafish, which if disabled, leads to most of the zebrafish developing as males. In addition, the few females that do make it to adulthood produce eggs that are defective, leading to abnormal progeny that generally do not survive. Our work identifies an important factor in fish that determines normal development and balanced sex ratios. Igf2bp3 has counterparts in other animals and therefore, our work can shed light on how these processes are controlled across organisms.
Introduction
In eukaryotes, nascent RNA transcripts are bound in complexes of RNA-binding proteins (RBPs) known as heterogeneous nuclear ribonucleoproteins (hnRNPs) for further processing [1]. The purpose of these complexes are to direct the fate of its cargo RNAs, such as splicing [2,3], capping activity [4], polyadenylation [5] and export [6]. The composition of the hnRNPs contributes to the fate of cargo molecules. For example, hnRNPs containing the RNA-binding protein Igf2bp1 impart increased stability of bound RNAs, such as c-myc [7–9]. Mis-regulation of RNAs through abnormal RBP functions often lead to developmental consequences. For instance, knockout of mouse Igf2bp1 leads to gross defects in development [10] and knockout of the RBP Zar1 in zebrafish leads to abnormalities in sex determination, resulting in male-only fish [11]. Several questions remain regarding how individual RNA-binding proteins contribute to development and the mechanism for their actions in specific cell types.
The germline is one of the earliest cell lineages to be specified across many multicellular organisms [12,13]. The precursor of the germline is the primordial germ cell (PGC). In mammals, the germline forms in the developing embryo, a process which is induced by signals from neighbouring cells [14]. In other animals, including fruit flies and zebrafish, a substance termed ‘germplasm’ is inherited by the egg from the mother, and contains factors that promote the formation of germline cells and development of the gonads.
Primordial germ cells receiving germplasm maintain their fate independently of the surrounding soma, whilst proliferating and migrating to the gonadal ridges [15]. Therefore, germplasm components are tightly regulated, and key components, including ribonucleoprotein complexes, are thought to be essential for correct PGC behaviour [16–18]. Germplasm RNAs are post-transcriptionally regulated by factors including microRNAs and RNA-binding proteins. RNA-binding proteins have a wide range of roles in processes such as oocyte maturation [11,19] and axis formation [20,21].
In addition to the functions that RNA-binding proteins have when bound to their target RNAs, the mechanism of the interactions is also important. For example, Ybx1 protein acts as a translational repressor and localisation factor for RNAs during early zebrafish embryogenesis. Ybx1 interacts with a stem-loop element found in the 3’ UTR of its target transcripts such as sqt/ndr1 [21–24]. The DAZ family of proteins, which are critical for germline formation [25], interact with target RNAs via a specific sequence motif, and acts as translational activators [26,27]. More recently, post-transcriptional modifications to RNA are proposed to act as another marker for recognition by RNA-binding proteins. The modifications include N6-methyladenosine (m6A), which is recognised by a host of proteins such as Ythdc2 and Ythdf2, which have been proposed to have a role in germline development [28–31].
Another group of proteins that are now known to recognise m6A modifications is the Insulin-growth factor 2 mRNA binding protein (Igf2bp) family [32]. Igf2bp family members are nucleocytoplasmic proteins containing two RNA-recognition motifs (RRMs) and two K-Homology (KH) didomains [33] that are highly conserved. Igf2bp proteins have many diverse functions. In addition to acting as RNA stabilisers [32,34], Igf2bp proteins also have roles in localisation. For instance, Xenopus Igf2bp3 binds to a vg1 3’ UTR element to localise vg1 transcripts to the vegetal cortex of oocytes [35–37]. Subsequent work has shown Igf2bp proteins to play a role in development in Xenopus [38], mice [10] and zebrafish [34]. However, whether Igf2bp proteins have a role in vertebrate germline development remains unexplored, although many lines of evidence point towards this possibility. In zebrafish oocytes, zebrafish igf2bp3 transcript is enriched in the germplasm-containing structure, the Balbiani body [20], and Drosophila igf2bp has been shown to regulate germ cell maintenance [39].
To determine the role of Igf2bp3 in zebrafish development, we generated and analysed zebrafish mutants affecting igf2bp3, and find that maternal Igf2bp3 is essential for proper germline development. Loss of zygotic igf2bp3 is sufficient to induce a male-sex bias in adult zebrafish. Loss of Igf2bp3 leads to a reduction in canonical germline markers, and maternal igf2bp3 mutants have a mis-regulated germline, with depletion of primordial germ cells and aberrant PGC behaviour. These results highlight a new function for this family of proteins in germline development and identify a key maternal factor that is essential for germline development, maintenance and PGC migration.
Methods
Ethics statement
Adult zebrafish were kept at their ambient temperature [40] in the University of Warwick aquatics facility, in compliance with the University of Warwick animal welfare and ethical review board (AWERB) and the UK home office animal welfare regulations, covered by the UK Home office licenses PPL 70/7836 and P782A73C4 to KS, and PEL 30/2308 and X59628BFC to the University of Warwick.
Animal husbandry
Embryos were collected from pair-wise or pooled mating of adult wild type or mutant zebrafish and incubated at 28.5°C in 0.3X Danieau’s solution with methylene blue (17 mM NaCl, 2 mM KCl, 0.12 mM MgSO4, 1.8 mM, Ca(NO3)2, 1.5 mM HEPES, pH 7.6). Embryos were manually dechorionated with a pair of Dumont #5 tweezers.
Affinity purification, mass spectrometric analysis and identification of Igf2bp3
NHS (N-hydroxysuccinimide) activated Sepharose 4 fastflow (GE Healthcare) beads coupled with an RNA aptamer alone control or aptamer fusion (zebrafish ndr1 3’UTR sequences fused to a Tobramycin aptamer; S1 Methods) was incubated with 10 mg of 20 min post-fertilisation embryonic lysates, washed and eluted with Tobramycin antibiotic [41,42]. The eluates were run on 5–16% gradient polyacrylamide gel, washed with deionized water and stained with Coomassie blue for 15 minutes. Protein bands were cut into cubes of ∼1mm. Gel pieces were de-stained twice using 50% ethanol (Thermo Fisher Scientific) in 50 mM ammonium bicarbonate (ABC, Fluka) at 22°C for 15 min and dehydrated with 100% ethanol for 5 min with shaking (650 rpm). Dehydrated gel pieces were reduced with 10 mM DTT (Sigma) at 55°C for 30 min and then alkylated with 55 mM iodoacetamide (Sigma) for 20 min in the dark at 22°C. Samples were then washed with 50% ethanol in 50 mM ABC at 22°C for 15 min and then dehydrated with 100% ethanol for 5 min. The gel pieces were hydrated and incubated with 2.5 ng/μl of trypsin (Promega) in 50 mM ammonium bicarbonate (ABC) overnight at 37°C. Peptides were extracted from gel pieces three times with 5% formic acid in 25% acetonitrile (ACN; Thermo Fisher Scientific) with 5-minute sonications in a water bath. The supernatants were combined in a fresh vial, dried using a vacuum centrifuge at 40°C, resuspended in 55 μl of 2.5% acetonitrile containing 0.05% trifluoroacetic acid and sonicated for 30 mins. 20μl of the sample was used for liquid chromatography-mass spectrometry analysis as performed previously [43]. Samples were analysed using reversed phase chromatography with two columns, an Acclaim PepMap μ-precolumn cartridge 300 μm i.d. x 5 mm, 5 μm, 100 Å and an Acclaim PepMap RSLC 75 μm i.d. x 50 cm, 2 μm, 100 Å (Thermo Scientific) was used to separate tryptic peptides prior to mass spectrometric analysis. The columns were installed on an Ultimate 3000 RSLCnano system (Dionex) at 40°C. Mobile phase buffer A composed of 0.1% formic acid and mobile phase B composed of acetonitrile containing 0.1% formic acid. Samples were loaded onto the μ-precolumn equilibrated in 2% aqueous acetonitrile containing 0.1% trifluoroacetic acid for 5 min at 10 μL min-1, after which peptides were eluted onto the analytical column at 250 nL min-1 by increasing the mobile phase B concentration from 6% B to 37% over 100 min, followed by a 3 minute wash at 80% B and a 10 min re-equilibration at 4% B. Eluting peptides were converted to gas-phase ions by means of electrospray ionization and analysed on a Thermo Orbitrap Fusion (Thermo Scientific). Survey scans of peptide precursors from 375 to 1500 m/z were performed at 120K resolution (at 200 m/z) with a 2x105 ion count target. The maximum injection time was set to 150 ms. Tandem MS was performed by isolation at 1.2 Th using the quadrupole, HCD fragmentation with normalized collision energy of 33, and rapid scan MS analysis in the ion trap. The MS2 ion count target was set to 5x103. The maximum injection time was 200 ms. Precursors with charge state 2–6 were selected and sampled for MS2. The dynamic exclusion duration was set to 40 s with a 10 ppm tolerance around the selected precursor and its isotopes. Monoisotopic precursor selection was turned on. The instrument was run in top speed mode. The acquired tandem mass spectra, as Xcalibur (version 2.2) raw files were analysed using Max-Quant software v1.6.0.16 [44,45] against UniProtK Danio rerio (UP000000437). Trypsin was specified as the digestion enzyme, with up to 2 missed cleavages, and a parent ion mass tolerance of 4.5 ppm for the initial search, with recalibration enabled. Oxidation of methionine was set as a variable modification and carbamidomethyl of cysteine as a fixed modification for all searches. The MS-MS data were further collated using Scaffold software. Fold change in proteins between the control aptamer and zebrafish ndr1 fusion RNA samples were calculated using emPAI in scaffold. The emPAI is a label-free, relative quantitation of the proteins in a mixture based on protein coverage by the peptide matches in a database search result. Igf2bp3 showed > 4-fold enrichment in ndr1-bound eluates.
Generation and establishment of igf2bp3 mutants
Cas9 mutants were generated in the TU background. Target sequences were verified by PCR sequencing and the conserved igf2bp3 cDNA sequence was used as a target in CHOPCHOP [46] to produce the target site “GGCTCCCTTCCTCGTAAAAAG” in exon 1. Embryos were injected with capped 150 pg Cas9 RNA and 50 pg sgRNA at the 1-cell stage, and raised to adulthood as described [47]. F0s were outcrossed to identify heterozygous F1s, which were outcrossed again to expand the line, followed by mating of heterozygous sibling to retrieve homozygous mutants. The igf2bp3 transgenic insertion alleles were obtained from the Lin/Burgess retroviral insertion collection, outcrossed over two generations and subsequently, heterozygous carriers were inter-crossed to homozygosity. Experiments were typically carried out with at least two females, and results from one representative clutch are shown.
Genotyping mutants
DNA was isolated from fin clips by incubating with lysis buffer (10 mM Tris pH 8.3, 50 mM KCl) with proteinase K (200 μg/mL) at 55°C overnight, followed by inactivation at 95°C for 10 minutes and lysates were used directly for PCR (primer sequences provided in S1 Methods). PCR products were visualised on a 2–3% agarose gel. The igf2bp3la020659Tg insertion mutant fish were genotyped by the use of three primers in a single PCR reaction, a forward/reverse primer flanking the insertion and a second reverse primer specific to the long terminal repeats in the retroviral construct. This generates a 250 bp WT product and a ~800 bp product for the insertion allele. The igf2bp3la010361Tg insertion allele was genotyped by the use of three primers in a single PCR reaction, a forward/reverse primer flanking the insertion and a second forward primer specific to the long terminal repeats in the retroviral construct. This generates a 350 bp WT product and a ~900 bp product for the insertion allele. The igf2bp3Δ7 deletion allele contains a continuous 7 bp deletion in exon 1. This mutation generates a BssaI (NEB) restriction enzyme site.
Synthesis of RNAs
Synthetic capped mRNAs were transcribed using the SP6 mMessage mMachine, following the manufacturer’s instructions. Cas9 sgRNA was transcribed using the T7 HiScribe High Yield RNA Synthesis kit, following the manufacturer’s instructions. RNAs were purified with phenol-chloroform extraction. The pSP64-mmGFP5-nos1-3’UTR [48] and pSP64-eGFP-F-nos1-3’UTR [49] constructs were linearised with SacII and NotI restriction enzymes (NEB) respectively, following the manufacturer’s instructions. These were used to label the cytoplasm and cell membranes of the PGCs. Probes for whole mount in situ hybridisation were produced with Promega SP6/T7/T3 polymerases with DIG-labelling mix (Roche) and purified with lithium chloride extraction.
Whole mount in situ hybridisation
Embryos were fixed in 4% paraformaldehyde in PBS (phosphate buffered saline), and processed for whole mount in situ hybridization (WISH) to detect gene expression as described previously [50]. Experiments were typically carried out with at least two females and we analysed independent clutches from each female. Results from one female are shown for gdf3 and wnt8a, whereas the dazl WISH represents two clutches from two independent females, which showed 12/35 (34%) and 15/29 (52%), respectively.
Total RNA extraction and qRT-PCR
Embryos were collected at various stages and lysed in TRIzol, followed by RNA extraction using the Monarch Total RNA Miniprep kit according to the manufacturer’s instructions. For the one cell stage, maternal, maternal/zygotic igf2bp3 mutants in Tg(buc:buc-egfp); buc106+/- background or maternal/zygotic igf2bp3 mutants were used. For 1K and 50% epiboly stages, maternal/zygotic igf2bp3 mutants in Tg(buc:buc-egfp); buc106+/- background or maternal/zygotic igf2bp3 mutants alone were used. Embryos were collected and incubated at 28.5°C until the appropriate stage for RNA extraction. First strand cDNA was synthesised using the qPCRBIO cDNA Synthesis kit and qPCR performed (primer sequences provided in S1 Methods) using the qPCRBIO SyGreen Blue Mix Lo-ROX or Luna Universal qPCR Master Mix. Analysis was performed using the Stratagene Mx3005P.
Protein gel electrophoresis and Western blot
SDS-PAGE gels were prepared using the Bio-Rad protein electrophoresis systems. Zebrafish embryos were homogenised in RIPA (50 mM Tris-HCl, 150 mM NaCl, 1% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate, 1 mM EDTA, 0.1% (w/v) SDS) lysis buffer supplemented with protease inhibitor cocktail (Sigma-Aldrich) using a syringe and needle. Supernatants were collected after a brief centrifugation and boiled in 4X loading buffer (200 mM Tris-HCl (pH 6.8), 400 mM DTT, 8% SDS, 0.4% bromophenol blue and 40% glycerol) before loading. After transfer of proteins, membranes were rinsed with TBSTw (Tris Buffered Saline, 0.1% Tween-20) once and blocked in 5% skimmed-milk powder in TBSTw for 1 hour before incubation with primary antibody overnight. After incubation, membranes were rinsed 4X in TBSTw for 5 minutes and transferred to secondary antibody for 4 hours at room temperature, excess antibody subsequently removed by a further 4 washes in TBSTw for 5 minutes before detection with ECL Western blotting reagent (Bio-Rad) following manufacturer’s instructions. Signal detection was performed using a ChemiDoc MP Imaging system (Bio-Rad) or with CL-XPosure Film (34089, Thermo Fisher Scientific).
Image acquisition
For live tracking of PGCs, embryos were mounted in 0.8% LMPA on a heated stage maintained at 28.5°C and imaged with an Andor Revolution Spinning Disk system, based on a Nikon Ni-E PFS inverted microscope equipped with a Yokogawa CSU-X1 spinning disk unit, fitted with a 488 nm laser and captured with an iXon Ultra 888 EMCCD camera. Images were captured using either a Nikon Plan Apochromat 20X/0.75 NA or the Nikon Apochromat 60X/1.49 NA oil immersion objectives. Images were acquired with the Andor iQ3 software.
For time-lapse imaging of migrating PGCs, images were acquired using a 20X objective and z-stacks were generated with 1μm step-sizes and at 1-minute intervals for 60 minutes. Maximum intensity projections were generated using ImageJ/Fiji, and the MTrackJ plugin was used to derive displacement, speed and straightness of PGCs [51].
For analysing PGC filopodia dynamics, z-stacks were generated with the 60X objective with 0.5 μm step-sizes at 10 second intervals for 2–10 minutes. Using ImageJ/Fiji, maximum intensity projections were generated and the filopodia numbers per PGC were recorded by counting the filopodia for a single timepoint. Persistence of the filopodia was calculated by counting the number of consecutive frames that a single filopodium was present. The length of a filopodium was calculated as the average length over its observable lifetime. The frequency of filopodia projections was calculated by measuring the direction of the projections relative to the embryonic midline. To image PGCs in vivo during segmentation, the Zeiss LSM 880 scanning confocal microscope was used.
Immunofluorescence and TUNEL assay
To detect the YSL, the cell membranes were labelled with an anti-β-Catenin antibody (C2206, Sigma Aldrich) and nuclei were labelled with DAPI stain as described [21]. The embryos were washed in PBS, mounted in 1% low melting agarose in PBS, and imaged with a Zeiss LSM 880 scanning confocal microscope, using 25X Zeiss Plan-Neofluar 25X/0.8 NA and 40X Plan-Neofluar 40X/1.3 NA objective lenses.
For TUNEL labelling of PGCs, embryos were fixed and processed for immunofluorescence using an anti-Vasa antibody (ab209710, Abcam) as described [52], and subsequently processed for TUNEL labelling following the manufacturer’s instructions (Sigma Aldrich).
Nodal/Squint translation assay
MZigf2bp3la659Tg mutant and control embryos were obtained from crosses of adult mutants and either heterozygous carriers or wild type siblings, respectively. For detection of Sqt-GFP reporter at high stage, embryos derived from crosses of heterozygous siblings were used as controls. TU WT embryos served as controls for detection of ΔΥΒΕ-Sqt-GFP at high stage. Synthetic mRNAs for microinjections were synthesized using the mMessage mMachine SP6 kit with Not I linearised pCS2+ plasmids containing either sqt-gfp:sqt3’UTR or a sqt-gfp:ΔYBE 3’UTR control construct lacking the YBE sequence. The fusion constructs harbour a 3xFLAG:enterokinase cleavage site cassette [53,54] fused to EGFP one amino acid after the pro-protein convertase cleavage site RRHRR, so that EGFP is fused to mature Nodal peptide after processing. Embryos were microinjected at the 1-cell stage with 100–150 pg of either sqt-GFP or ΔYBE sqt-GFP RNA [21,24]. Rhodamine dextran (0.5%, 70 kDa) was co-injected as an injection control for all experiments except high stage sqt-gfp injections. Injected embryos were dechorionated by Pronase treatment (2 mg/ml for 2 minutes) and incubated at 28.5°C until mounting and imaging. Embryos were mounted for lateral views in agarose-coated glass-bottom dishes and imaged on an Andor Revolution XD microscope equipped with an Andor iXon 888 camera. Z-stacks of 50 μm in 0.56–0.59 μm slices were acquired using a 488 nm laser at 40% and a 561 nm laser at 10%. Intensity measurements were carried out using Fiji software. Maximum intensity projections of the Z-stacks were generated. Threshold-segmentation in the rhodamine channel followed by “Analyse Particles” produced a single ROI corresponding to the area of the blastoderm that has been imaged. Mean pixel values in the ROI were measured in both channels. To normalize GFP signal, the mean GFP intensity was divided by the mean rhodamine intensity. Bar plots were produced in R-studio.
Injections with igf2bp3 mRNA
To rescue the MZigf2bp3la659Tg and MZigf2bp3Δ7 phenotypes, we injected igf2bp3 mRNA made from a pCS2+ igf2bp3 cDNA plasmid that was linearised with NotI. Capped mRNA was generated using the mMessage mMachine SP6 kit, and 1-cell stage mutant or control embryos were injected with 100 or 200 pg igf2bp3 mRNA. Similarly, igf2bp1 RNA was generated and injected. Injected embryos were scored for YSL expansion at the 1K cell stage, or fixed at prim5 for WISH to detect ddx4/vasa expression.
Statistical analysis
Data analysis was typically performed with two-tailed t-tests, except for analysis of sex ratios in igf2bp3 mutants, which was performed with a Binomial test [55].
Results
Mutations in igf2bp3 result in biased sex ratios in adult zebrafish
Igf2bp3 was identified from a proteomic screen to identify RNA-binding proteins in early zebrafish embryonic lysates that bind an RNA aptamer. Igf2bp3 was enriched 4-fold compared to controls. Analysis of igf2bp3 expression by WISH and available RNAseq datasets shows that igf2bp3 is ubiquitously expressed and at high levels during early development (S1A and S1B Fig). Igf2bp3 protein is expressed maternally and zygotically (S2C Fig), precluding the use of morpholinos to uncover the role of igf2bp3 in development.
To identify zebrafish lines with genomic lesions in igf2bp3, we searched a mutant collection generated by the Lin and Burgess laboratories [56]. We identified several retroviral transgenic insertion alleles that harbour integration of the Tg(nLacZ-GTvirus) in intron one of the igf2bp3 locus: igf2bp3la010361Tg and igf2bp3la020659Tg, hereafter referred to as igf2bp3la361Tg and igf2bp3la659Tg (Fig 1B). The igf2bp3 insertion mutant alleles are predicted to result in loss-of-function by preventing splicing of exons one and two in igf2bp3 transcripts, leading to premature termination codons. RT-PCRs of exon-spanning junctions of igf2bp3 showed a reduction of RNA levels (S2B Fig). Western blots performed using an antibody directed against a central region of Igf2bp3 downstream of the insertion site did not reveal any detectable Igf2bp3 in mutant embryo lysates (S2C Fig).
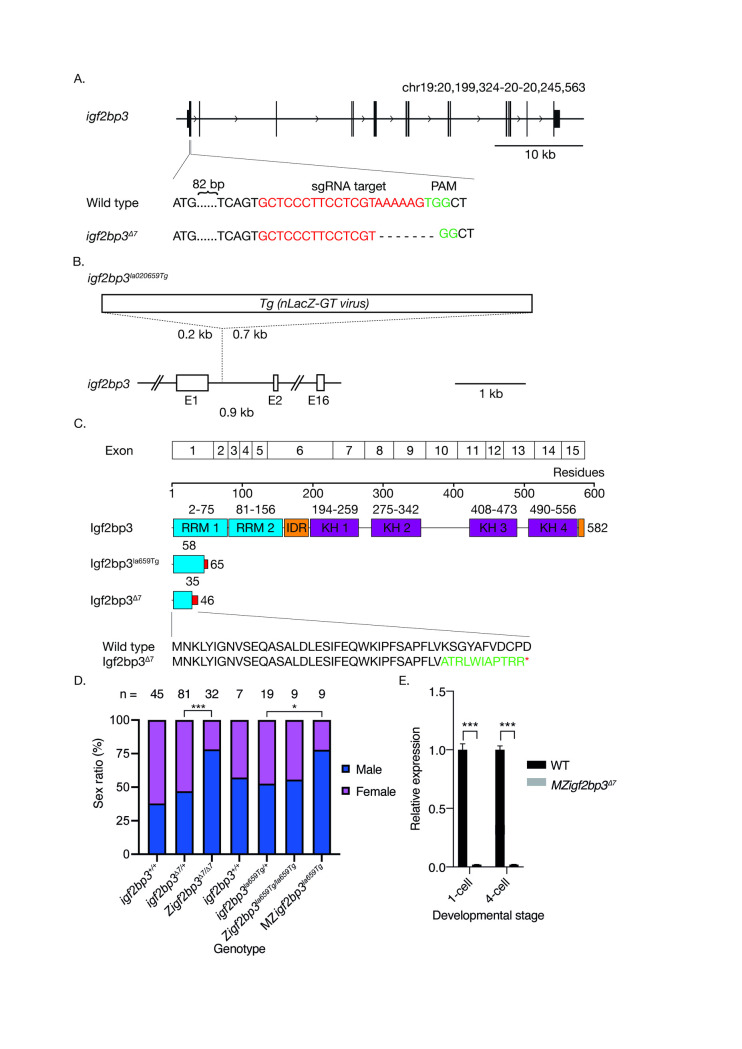
Fig 1. Loss of igf2bp3 results in skewed male:female sex ratios.
A. Generation of the igf2bp3 mutant allele by Cas9. Schematic showing the igf2bp3 locus on chromosome 19. Exon 1 of igf2bp3 was targeted by CRISPR-Cas9, resulting in a 7 bp deletion and frameshift. B. Schematic of the igf2bp3la659Tg with a 6 kb Tg(nLacZ-GTvirus) retroviral insertion in intron 1. C. The igf2bp3Δ7 mutant is predicted to be protein null. The igf2bp3Δ7 mutation generates a premature stop codon at residue 46 whereas the igf2bp3la659Tg results in a predicted truncated protein of 65 amino acids. D. Loss of igf2bp3 results in skewed sex ratios. Adult zygotic igf2bp3Δ7 and MZigf2bp3la659Tg mutant fish show a significant male bias compared to wild type, heterozygous igf2bp3Δ7 as well as igf2bp3la659Tg, and Zigf2bp3la659Tg fish. Asterisks indicate level of significance (p *<0.05, **<0.01, ***<0.001). E. The igf2bp3Δ7 mutation results in reduced igf2bp3 levels. qRT-PCRs at the 1-cell and 4-cell stages show significantly reduced igf2bp3 transcript levels in igf2bp3Δ7 mutant embryos (p<0.001).
To generate igf2bp3 null mutants, we used CRISPR-Cas9 mutagenesis, and identified mutants harbouring a 7 bp deletion within igf2bp3 exon one, hereafter referred to as igf2bp3Δ7 (Fig 1A). The deletion is predicted to result in a frameshift at residue 35 that leads to a truncated Igf2bp3 peptide of 47 residues which lacks the RNA-binding domains and the predicted intrinsic disordered regions (Fig 1C), and results in a significant reduction in igf2bp3 RNA (Fig 1E).
Zygotic igf2bp3la659Tg embryos appear morphologically normal and grow to adults at expected Mendelian ratios (Figs 1D and S2E and S2F). However, progeny of igf2bp3la659Tg adults (i.e., MZigf2bp3la659Tg) show biased sex ratios as adults (Fig 1D), with an approximately 3:1 male-female ratio. We also observed a similar male sex bias (3:1) in zygotic igf2bp3Δ7 deletion mutant adults (Figs 1D and S1C), suggesting that Igf2bp3 might have important roles in normal germline, gonad and/or sexual development.
Maternal Igf2bp3 plays a critical role in early development
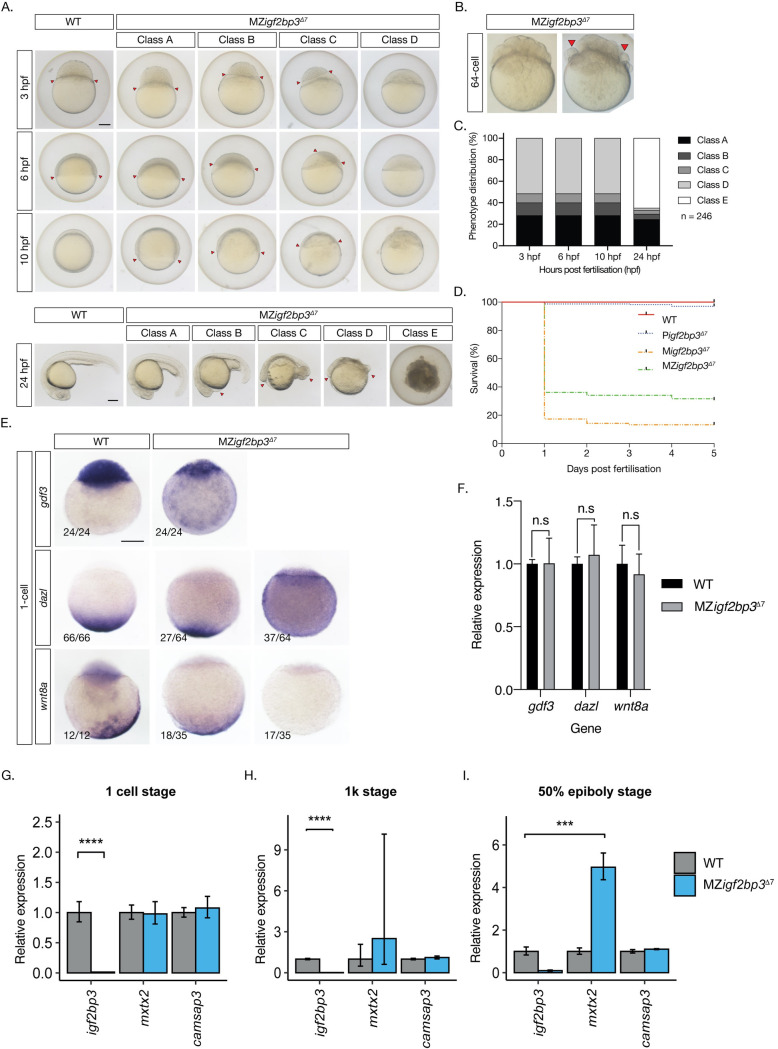
Although zygotic igf2bp3 mutants are viable up to adulthood, we observed defects in the progeny of homozygous igf2bp3 mutant females, i.e., maternal igf2bp3Δ7 embryos. Early cell divisions and cleavage are similar to wild type embryos in the majority of maternal igf2bp3 mutants (Fig 2B). However, a small proportion of deletion mutant embryos (10–15% from different females) show defects in cell adhesion at 32–64 cell stage, with some blastoderm cells rounding up and detaching from the embryo (red arrowheads, Fig 2B). These embryos later manifest severe phenotypes (described below). We observed gross defects across the blastoderm from the 1K-cell stage, resulting in delayed progression through gastrula and lethality in the majority of embryos by 24 hpf (Fig 2A–2D). We have categorised the embryos as class A (delayed progression of gastrulation), class B (abnormal blastoderm and expanded YSL), class C (pockets of blastomeres and massively expanded YSL), class D (severe). All class C and D embryos are lethal by 24 hpf (class E). The majority of these embryos are not viable and fail to progress beyond 24 hpf.
Fig 2. Maternal igf2bp3 is required for normal embryonic development.
A. Maternal igf2bp3Δ7 mutants show severe defects by blastula and gastrula stages. B. At the 64-cell stage, most maternal igf2bp3Δ7 mutants are similar to wild type embryos (left), but a proportion of embryos (right; <15%) show rounded cells (red arrowheads). C-D. The majority of igf2bp3 mutants do not survive beyond 1 dpf. E. Animal-vegetal markers (gdf3, dazl and wnt8a) show altered distribution in igf2bp3Δ7 embryos by whole mount in situ hybridization (WISH). F. qRT-PCRs show that expression levels of gdf3, dazl and wnt8a transcripts is not significantly different from wild type control embryos at the 1-cell stage. G-I. qRT-PCR with maternal igf2bp3Δ7 embryos show that expression of igf2bp3 transcripts is significantly reduced at all stages examined (1 cell, 1K, and 50% epiboly), whereas expression of the yolk syncytial layer (YSL) transcript mxtx2 is initially unchanged (G), but shows variable increase at the 1K stage (F), with substantially higher expression at 50% epiboly compared to controls (I), and the microtubule and spectrin-associated camsap3 transcript is largely unchanged at all stages examined. p ***<0.01 or ****<0.001 from 3 biological replicates. Scale bar in A, 200 μm.
In maternal igf2bp3la659Tg mutant embryos, early cell divisions are not affected (S2F Fig) and lethality is not observed, although mutant embryos are delayed briefly at late blastula stages. There is no visible morphological defect in MZigf2bp3la659Tg mutants, with expression of myoD in the myotome and shh in the midline comparable to wild type embryos and the swim bladder inflates normally in MZigf2bp3la659Tg embryos (S2D and S2E Fig). To determine if paralogue redundancy underlies the viability of the igf2bp3la659Tg mutant embryos, we examined the expression of other igf2bp transcripts. Whereas igf2bp3 expression is not detected in igf2bp3la659Tg mutant embryos at all stages, we do not find an increase in the expression levels of igf2bp1, igf2bp2a, or igf2bp2b transcripts (S2G Fig). Therefore, paralogue gene expression is not altered in maternal igf2bp3la659Tg mutant embryos.
Distribution of maternal RNAs is disrupted in maternal igf2bp3 mutant embryos
As Igf2bp3 is an RNA-binding protein that is involved in the regulation of vg1 (now known as gdf3) in Xenopus and enriched in the Balbiani body, we examined maternal-zygotic igf2bp3Δ7 embryos at the 1-cell stage for expression of the animal pole marker gdf3 and vegetal markers, dazl and wnt8a. MZigf2bp3Δ7 embryos showed markedly reduced gdf3 expression in the blastoderm, with some expression detected in the yolk of mutant embryos. Expression of dazl and wnt8a transcripts is either reduced or not detectable in the vegetal cortex, and a proportion of maternal mutant embryos show diffuse staining for dazl (Fig 2E). Quantitative real-time PCR (qRT-PCR) in 1-cell stage embryos show that the expression levels of gdf3, dazl and wnt8a in MZigf2bp3Δ7 mutant embryos is not different from controls (Fig 2F). Therefore, it is likely that maternal Igf2bp3 plays a key role in the distribution, anchoring and/or stability of a subset of maternal RNAs.
The extra-embryonic yolk syncytial layer (YSL) is expanded in maternal igf2bp3 mutants
In maternal igf2bp3Δ7 as well as MZigf2bp3la659Tg mutant embryos at 3 hpf, we observed an expansion of the extra-embryonic yolk syncytial layer (YSL; class B,C in Figs 2A and S3A and S3B). Remarkably, this expansion is not observed in MZigf2bp3la659Tg at 4.5 hpf, and MZigf2bp3la659Tg mutant embryos recover to complete gastrulation and survive (S3A and S3B Fig). By contrast, the majority of maternal igf2bp3Δ7 embryos show an expanded YSL, a small blastoderm, defects during gastrulation and do not survive beyond 24 hpf (Fig 2A–2D). Consistent with these observations, quantitative real-time PCR (qRT-PCR) in early embryos show that the YSL transcript mxtx2 is increased from the 1K-cell stage and is significantly increased at gastrula stages in MZigf2bp3Δ7 embryos but in MZigf2bp3la659Tg mxtx2 levels are either unchanged or slightly reduced (Figs 2G–2I and S3C–S3E). The microtubule and spectrin-associated camsap3 transcript is unchanged in both mutants at all stages examined. Injection of igf2bp3 mRNA into MZigf2bp3Δ7 mutant embryos fails to rescue the YSL expansion (S4A Fig). Therefore, maternal Igf2bp3 plays an important role in the allocation of embryonic versus extra-embryonic cell fates and embryonic survival.
Maternal Igf2bp3 mutant embryos show translational deregulation
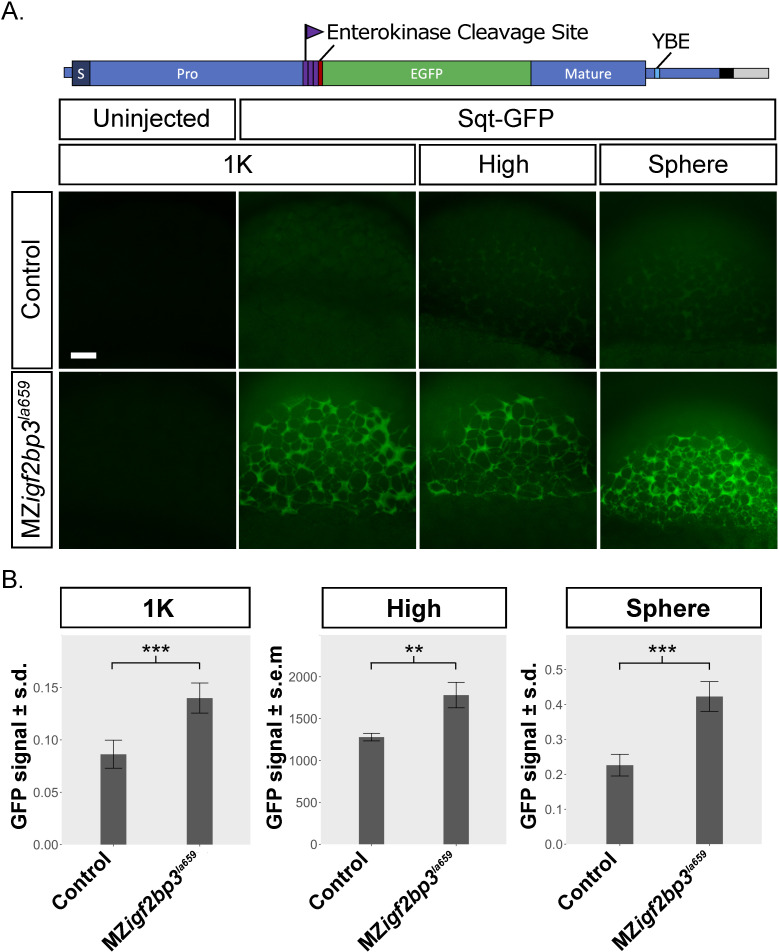
YSL expansion could potentially arise from cleavage or cytoplasmic segregation defects [57–59]. However, we did not see any defects in cell divisions or cleavage in mutant embryos for MZigf2bp3la659Tg or MZigf2bp3Δ7 (Figs 2B and S2F). We identified Igf2bp3 as a component of an RNA-binding complex that recognizes the 3’ untranslated (3’UTR) region of nodal/squint RNA. Defects in another protein in the same RNP complex, Ybx1, also result in YSL expansion, which we have shown to arise from translational de-regulation and premature Nodal signaling in maternal ybx1 mutant embryos [21]. We therefore investigated if maternal Igf2bp3 functions in translational control of nodal/sqt RNA. In MZigf2bp3la659Tg embryos injected with sqt-gfp fusion reporter RNA (Fig 3A), we found high levels of Sqt-GFP protein expression in the blastoderm at 1K, high and sphere stages. By contrast, control (wild type or heterozygous igf2bp3la659Tg/+) embryos showed weak expression only from sphere stages (Fig 3A and 3B). Similarly, a sqt-gfp reporter lacking the binding motif for Ybx1 (ΔYBE Sqt-GFP) also showed elevated expression in MZigf2bp3la659Tg embryos (S5A and S5B Fig). Therefore, maternal Igf2bp3 likely functions in translational control of Nodal signaling in early zebrafish embryos.
Fig 3. MZigf2bp3la659Tg mutant embryos show premature translation of a Nodal reporter.
A. Schematic of the sqt-gfp fusion reporter construct showing the Sqt pro-domain, the enterokinase cleavage site, 3xFLAG epitope tag, eGFP and mature Nodal peptide sequences. Expression of a Sqt-GFP reporter expression (green) in MZigf2bp3la659Tg embryos compared to wild type or control heterozygous embryos. Representative examples from three independent experiments are shown, imaged at the 1K, high or sphere stage, respectively. Scale bar, 50 μm. B. Bar charts represent mean GFP intensity in the blastoderm of imaged embryos. GFP signals were normalized to co-injected rhodamine dextran control. Number of embryos analysed: 1K (n = 9 WT and 10 MZigf2bp3la659Tg), high (n = 10 Control and 11 MZigf2bp3la659Tg), sphere (n = 9 WT and 9 MZigf2bp3la659Tg). Asterisks indicate level of significance from two-tailed t tests (p *<0.05, **<0.01, ***<0.001).
Igf2bp3 is essential for proper germline development
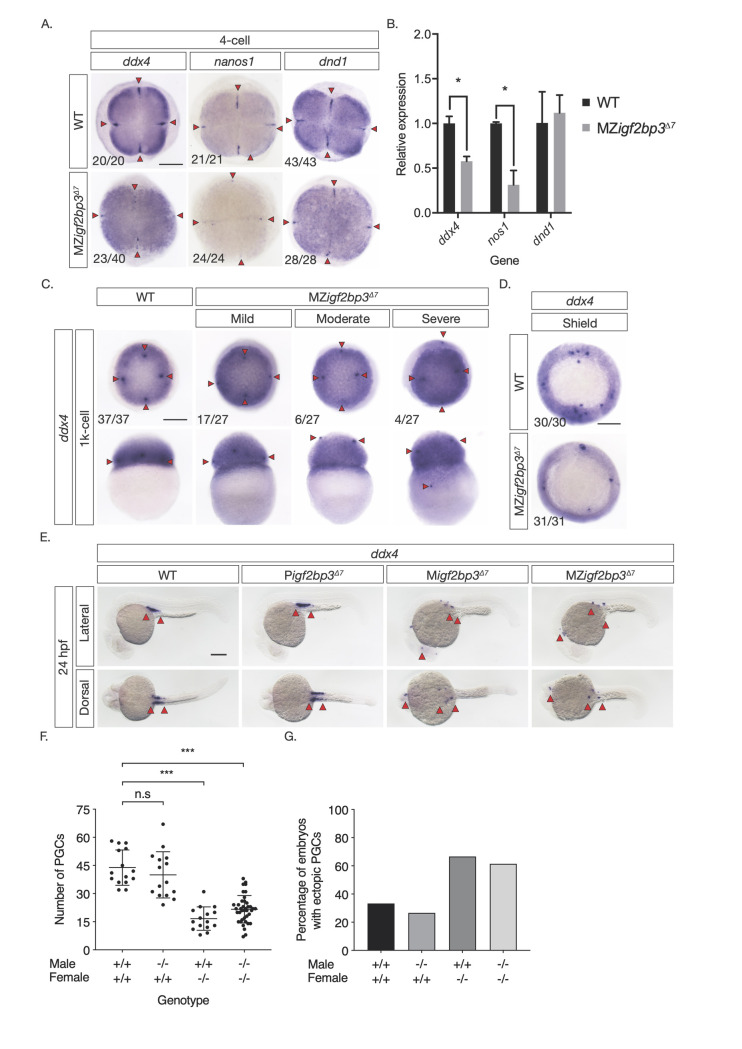
We found altered distribution of dazl RNA in MZigf2bp3Δ7 mutant embryos at the 1-cell stage, and altered sex ratios with a male bias in Zigf2bp3Δ7 as well as MZigf2bp3la659Tg adults. In zebrafish, male sex bias can arise from depletion of germplasm or primordial germ cells (PGCs). Therefore, we examined the expression of ddx4, nos1 and dnd1, in MZigf2bp3Δ7 embryos. WISH and qRT-PCRs show that germplasm markers are reduced in mutant embryos from the 4-cell stage (Fig 4A and 4B). PGCs are displaced and found in ectopic positions in the blastoderm from the 1K-cell stage and reduced during gastrulation (Fig 4C and 4D). Ectopic and reduced number of PGCs are observed in the gonadal ridge by 24 hpf (Fig 4E–4G) in maternal igf2bp3 mutants (progeny of wild-type males crossed with mutant females, Migf2bp3) and MZigf2bp3 mutants, but not in wild type (WT) and paternal igf2bp3 mutants (progeny of igf2bp3 males crossed with WT females, Pigf2bp3). These defects are also observed in MZigf2bp3la361Tg and MZigf2bp3la659Tg mutant embryos (S6A–S6G Fig). The reduction in PGCs is not rescued by injection of either igf2bp1 or igf2bp3 capped mRNA (S4B Fig). Therefore, maternal Igf2bp3 function is crucial for normal PGC development.
Fig 4. Igf2bp3 is required for normal germline development.
A. Whole mount in-situ hybridisation (WISH) in early embryos shows altered expression of germline markers ddx4, dnd1, and nanos1 in igf2bp3Δ7 mutant embryos. B. qRT-PCR to detect early germplasm markers shows reduced levels of ddx4 and nos1 expression in mutant embryos at the 4-cell stage, whereas expression levels of dnd1 is not significantly different from control wild type embryos. C. Primordial germ cells (PGCs; red arrowheads) in igf2bp3Δ7 embryos are ectopically located at the 1k-cell stage to varying extents ranging from mild or moderate to severe. D. Primordial germ cells are severely reduced or not detected in igf2bp3Δ7 mutants by gastrula stages. E,F. WISH (E) and quantitation (F) of ddx4-positive cells (red arrowheads) shows reduced and ectopic primordial germ cells in 24 hpf maternal igf2bp3Δ7 (Migf2bp3Δ7) and maternal-zygotic igf2bp3Δ7 mutants (MZigf2bp3Δ7) compared to WT siblings and paternal igf2bp3Δ7 (Pigf2bp3Δ7) mutants; p *<0.05, **<0.01, ***< 0.001. G. Loss of maternal igf2bp3 leads to some ectopic primordial germ cells across the trunk (red arrowheads) and occasionally in the hindbrain region. Bar graph shows the number of embryos with ectopic germ cells in WT, Pigf2bp3Δ7, Migf2bp3Δ7, and MZigf2bp3Δ7 mutants. Scale bar in A and C-E, 200 μm. N = 15 embryos each for WT, Pigf2bp3Δ7, and Migf2bp3Δ7 and 39 for MZigf2bp3Δ7 mutants.
Germline progenitor cells mis-migrate in MZigf2bp3 embryos
To understand the basis of the PGC defects in igf2bp3 mutant embryos, we performed live-imaging of PGCs using a GFP-nos1 3’UTR fluorescent reporter. We detected aberrant and convoluted PGC migration in MZigf2bp3la659Tg embryos from late-gastrula stage (S1 Movie), and PGCs are spread out along the animal-vegetal axis in mutant embryos. At the tailbud stage (10 hpf), both control and MZigf2bp3la659Tg embryos show trailing clusters of PGCs (S2 and S3 Movies). However, PGC numbers are reduced in MZigf2bp3la659Tg (Fig 5A and 5B) and igf2bp3Δ7 (S7A and S7B Fig) mutant embryos. Many PGCs are found at ectopic locations, with some even in the hindbrain region (Figs 4E and S6C and S6F) in mutant embryos and a number of PGCs significantly further away from the midline (S3 Movie). Some mutant PGCs form clusters that are not closely associated with other PGCs, indicating disruptions in collective migration of mutant germline cells.
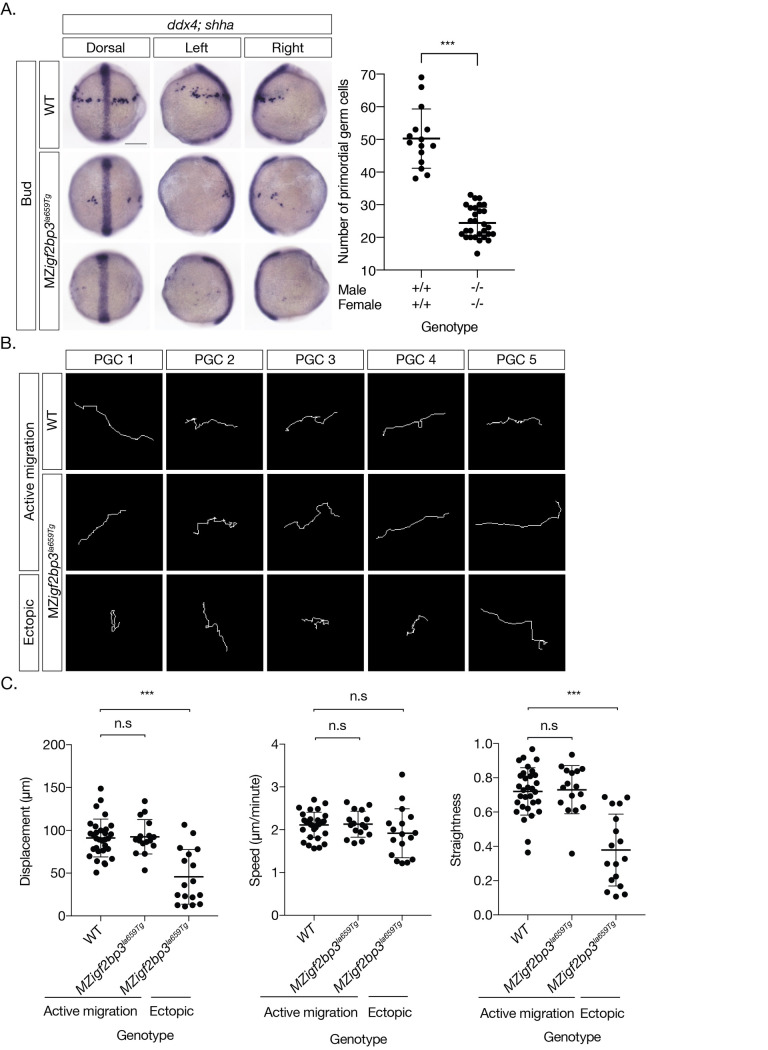
Fig 5. Primordial germ cell migration is aberrant in igf2bp3 mutant embryos.
A. WISH to detect ddx4 positive PGCs and shh in the midline at Bud stage. PGCs are reduced (scatter plot, right) and ectopically located in MZigf2bp3la659Tg embryos compared to wild type (WT) embryos. B. Migration tracks of PGCs in bud stage embryos, labelled with GFP-nos1 reporter. MZigf2bp3la659Tg embryos show aberrant PGC migration compared to WT controls. C. PGC track analysis showing displacement, speed and straightness index. Ectopic PGCs in MZigf2bp3la659Tg mutants show significantly reduced total displacement and straightness, although speed of individual PGCs is not altered in mutant embryos compared to WT controls. Scale bars, 200 μm; p*<0.05, **<0.01, ***<0.001; Number of PGCs analysed = 31, 16 and 17 for WT, actively migrating and ectopic MZigf2bp3la659Tg PGCs, respectively.
We compared the behaviour of actively migrating cells and ectopic PGCs in WT and igf2bp3la659Tg embryos. PGC migration tracks were traced and movement dynamics (speed, displacement and track straightness) were measured. Actively migrating PGCs exhibit similar dynamics in mutant and wild type embryos and produce linear tracks (Fig 5B). By contrast, ectopic PGCs in igf2bp3 mutant embryos show significantly convoluted paths, resulting in reduced net displacement over the course of migration (Fig 5C and S4 Movie).
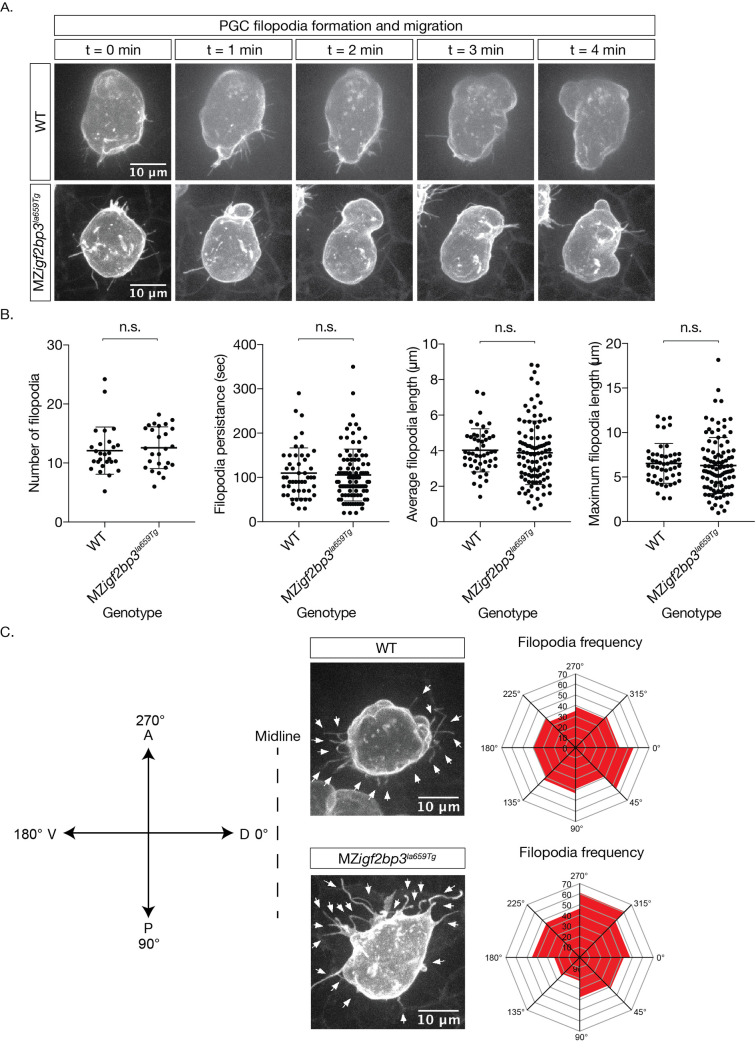
In order to investigate further the aberrant migration in igf2bp3 mutants, we analysed the behaviour of individual PGCs and imaged the actin-based protrusions, filopodia. We injected farnesylated eGFP mRNA to label the PGCs, and imaged embryos from 10 hpf. The behaviour of individual filopodia was measured over their lifetime: the number of filopodia per PGC, the persistence of each filopodium, the average and maximum length. The overall morphology of the PGCs appeared to be consistent (Fig 6A), and the number and length of the filopodia were comparable through the duration of time-lapse imaging (Fig 6B). In WT PGCs, the projections are biased towards the midline, shown as an increase of filopodia with projections towards the right (S5 Movie). However, in MZigf2bp3la659Tg PGCs, the filopodial projections are not directed towards the midline. (Fig 6C and S6 Movie). We also observed extensive blebbing, but without any particular orientation in mutant PGCs (S6 Movie). Therefore, PGC behaviour is perturbed in igf2bp3la659Tg mutant embryos.
Fig 6. PGC membrane projections show altered behaviour in MZigf2bp3la659Tg mutants.
A. Images of cell membranes of PGCs labelled with a Farnesylated-GFP-nos1 live reporter in bud stage mutant or wild type embryos. B. Quantitation of filopodia and behaviour (persistence, length) in WT and MZigf2bp3la659Tg PGCs. C. Projection angle relative to their destination, showing an altered skew in maternal igf2bp3la659Tg PGCs, although the projection parameters, namely numbers, persistence, average and maximum lengths did not appear to be changed. p *<0.05, **<0.01, ***<0.001. N = 27 WT and 26 MZigf2bp3la659Tg PGCs; number of filopodia = 50 WT and 100 mutant filopodia. Number of PGCs analysed for projection frequency = 25 per genotype; Scale bars, 10 μm.
MZigf2bp3 mutants have defects in the chemokine guidance system
PGC migration and cell behaviours depend on the chemokine guidance system, and disruptions in chemokine signalling can lead to aberrant PGC behaviours and mis-migration [60]. We examined the expression of chemokine guidance system components in MZigf2bp3la659Tg and MZigf2bp3Δ7 embryos. We find a significant increase in expression of the chemokine receptors cxcr4a and cxcr4b in both mutants at the 1K-cell stage, and during gastrulation, levels of the chemokine ligand cxcl12a are significantly reduced (S8 Fig). Therefore, the PGC migration defects and aberrant cell behaviours in maternal igf2bp3 mutant PGCs likely arise from defects in the chemokine guidance system.
Mis-migrating PGCs in MZigf2bp3 mutants undergo cell death
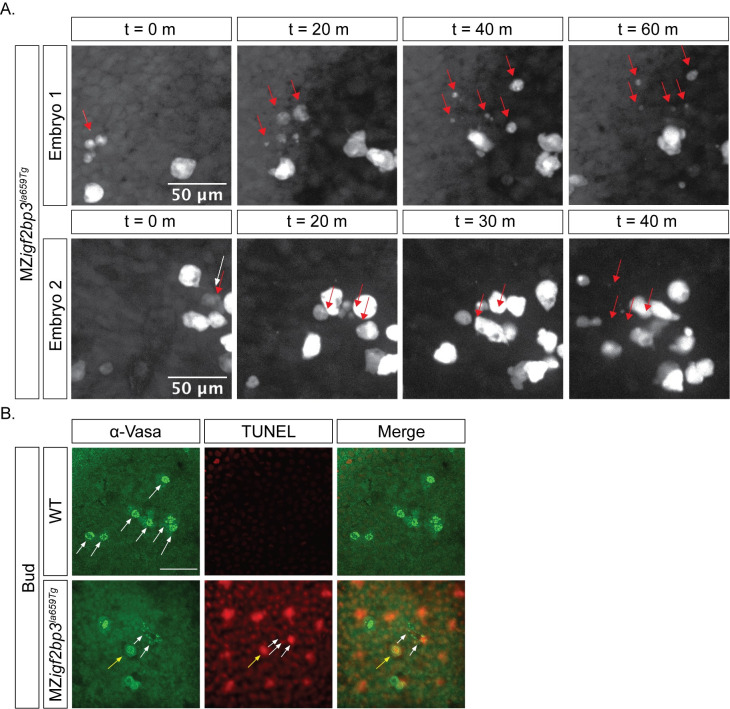
Finally, to investigate the reduction in PGC numbers in mutant embryos, we examined if cell proliferation and/or cell death are altered. Using live reporters, we did not observe any defects in PGC cell divisions in mutant embryos at gastrula or tailbud stages. However, some MZigf2bp3la659Tg PGCs fragment along their migration path from the tailbud stage (Fig 7A and S7 and S8 Movies). TUNEL labelling on fixed tailbud stage MZigf2bp3la659Tg mutant or wild type embryos together with immunostaining for Vasa protein shows Vasa-positive fragments, and co-localisation of the TUNEL label with Vasa-positive PGCs is observed in some cells (Fig 7B). Fragmentation is seen in 14% of mutant PGCs at tailbud (7/49 Vasa+ cells, n = 11 MZigf2bp3 embryos), whereas no fragmentation of Vasa-positive cells was found in wild type embryos (0/83 Vasa+ cells, n = 8 WT embryos). Therefore, PGC survival is compromised in the absence of Igf2bp3 function.
Fig 7. Loss of PGCs in MZigf2bp3la659Tg mutant embryos might arise from cell death.
A. Some PGCs are lost during migration in maternal igf2bp3la659Tg embryos. At bud stage, a subset of PGCs labelled with a GFP-nos1 live reporter display fragmentation during migration. B. Immunofluorescence with an α-Vasa antibody and TUNEL staining showed that compared to wild type embryos, in MZigf2bp3la659Tg embryos a subset of PGCs undergo cell death and fragmentation at the bud stage. Yellow arrow indicates co-localisation of Vasa and TUNEL labelling in a PGC, white arrows indicate Vasa+ fragments; Scale bar, 50 μm.
Discussion
In this study, we examined the role of the conserved RNA-binding protein, Igf2bp3, in zebrafish, and found that Igf2bp3 is essential for germline development and normal sex ratios in adults, suggesting that Igf2bp3 plays crucial roles in sex determination. We also uncovered a function for Igf2bp3 in early embryonic development, and specifically in the allocation of embryonic versus extra-embryonic cell fates, with maternal mutant embryos showing expansion of the YSL at the expense of the embryonic blastoderm. The YSL expansion, embryonic lethality and germline defects are not rescued by injection of igf2bp3 RNA into mutant embryos. Therefore, maternal Igf2bp3 is likely to play crucial roles during normal embryonic and germline development.
In a small number of igf2bp3 deletion mutant embryos, we also found cell adhesion defects and these embryos typically do not survive beyond gastrula stages. The cell adhesion defects are similar to the phenotypes described for a different igf2bp3 mutant allele [34]. The majority of our igf2bp3 deletion mutant embryos show expansion of the YSL and we found enhanced expression of the early YSL marker, mxtx2. By contrast, the expression of sqt/nodal and ntl/tbxta is variably increased in igf2bp3 mutant embryos at the 1K cell stage, and either reduced or comparable to control levels by early gastrula stages. Interestingly, we observed elevated levels of the mesendoderm gene gsc, which is a target of Nodal signalling. Consistent with this observation, we found premature and elevated levels of a Nodal reporter. This suggests premature translation and de-regulated Nodal signalling in maternal igf2bp3 mutant embryos. We previously showed that the RNA-binding protein Ybx1 (which was identified in an RNP complex together with Igf2bp3), also functions in translational repression of Nodal signalling in early zebrafish embryos [21]. Interestingly, a Nodal reporter lacking the binding motif for Ybx1 also showed elevated expression in MZigf2bp3 embryos, indicating that translational control by Igf2bp3 is likely independent of binding by Ybx1.
Previous studies in zebrafish and Xenopus found that igf2bp3 transcript is enriched in the Balbiani body [20], a transient structure in oocyte development, where germplasm is initially aggregated, and a recent report showed that Igf2bp3 protein interacts with the germplasm protein Bucky ball in yeast-two hybrid assays and through co-immunofluorescence [61]. The presence of Igf2bp3 as an RNA-binding protein in this structure suggests that it might regulate many RNAs involved in germline development. Accordingly, we find that germline transcripts including ddx4, nanos1 and dnd1, are reduced in maternal igf2bp3 mutant embryos at early stages. Recent studies examining the zebrafish transcriptome in igf2bp3 mutants reported that igf2bp3 acts as a stability factor for maternal and germplasm RNAs in zebrafish [34,61]. We have observed additional phenotypes that have not previously been reported, such as a mis-migration of germ cells and male sex bias in adult zygotic igf2bp3 mutants. Therefore, igf2bp3 might have crucial roles in further processes that are not strictly maternal.
Some germplasm RNAs are critical for the survival, proliferation or migration of PGCs. For example, dnd1 RNA acts as a survival and migration factor for zebrafish PGCs [16]. We found a reduction of germline RNAs, which is consistent with the reduced number of PGCs in igf2bp3 mutants at gastrula stages. In addition, we also observed ectopic germ cells at mid-late blastula stages. It is possible that the expanded YSL and reduced blastoderm results in altered positioning of germ cells in igf2bp3 mutant blastulae. This might be exacerbated by abnormal chemokine guidance system, leading to abnormal migratory behavior during gastrula and somitogenesis stages, resulting in ectopic localisation and depletion of PGCs. We also observed increased frequency of blebbing in mutant PGCs, but without any particular orientation. Increased blebs can be found associated with apoptosis [62], and consistently, we found that a proportion of mutant PGCs undergo cell death during somitogenesis. Together, the initial loss of germplasm RNAs combined with altered blastoderm and germ cell position, and defects in chemokine guidance likely results in the progressive reduction and eventual loss of PGCs in igf2bp3 mutant embryos. Early loss of PGCs is known to promote testes formation [63] and might underlie the male bias found in igf2bp3 mutant adults.
The mechanism through which Igf2bp3 acts to regulate the germplasm and the germline remains unknown. The mechanisms that determine sex in zebrafish are also not understood [64]. Many proteins associated with m6A modifications, known as ‘m6A readers’, have been shown to be important for fertility and germline development, such as Ythdf2 [31], Ythdc2 [30,65,66] and a closely related Drosophila homolog, Bcgn [67]. Igf2bp proteins were found to be associated with m6A modifications [32]. Our findings, showing an essential role for Igf2bp3 in embryonic versus extraembryonic and germline progenitors in zebrafish, are consistent with these reports, and identifies Igf2bp3 as part of a potentially conserved network of proteins involved in RNA regulation in early embryos and in the germline.
Supporting information
A. RNA-seq expression data for all four zebrafish igf2bp genes during zebrafish development shows that igf2bp3 is expressed throughout zebrafish development. Data from [68]. B. Whole mount in-situ hybridisation (WISH) to detect igf2bp3 in zebrafish embryos at cleavage, blastula, gastrula, somitogenesis and 24 hpf. C. Sex ratios in igf2bp3Δ7/+, igf2bp3Δ7/Δ7, igf2bp3la659Tg/+, and igf2bp3la659Tg/la659Tg (i.e. zygotic igf2bp3la659Tg) adult fish harbouring the Tg(buc:buc-eGFP) transgene show that transgenic zygotic igf2bp3Δ7/Δ7 mutants manifest a strong male bias. Scale bar in B, 200 μm.
(TIF)
A. Schematic representation of the igf2bp3la659Tg mutant locus showing the site of the 6 kb Tg(nlacZ-GT virus) gene trap retroviral insertion, and coloured triangles indicate the position of RT-PCR primers. B,C. MZigf2bp3la659Tg mutant embryos show significantly reduced or undetectable expression of igf2bp3. RT-PCR (gels on left) using exon spanning primers for igf2bp3 and qRT-PCR (bar graphs, right) at performed at six early developmental stages, show reduced igf2bp3 transcripts in mutant embryos relative to gapdh control. C. Igf2bp3 protein is not detectable in MZigf2bp3la659Tg embryos. Western blots on cleavage to early gastrula stage embryos show a 60 kD band in wild type (WT) lysates. No Igf2bp3 band is detected in MZigf2bp3la659Tg lysates. D. WISH shows that shh expression in the midline (top panels) and myoD expression in the myotome (bottom panels) in MZigf2bp3la659Tg mutants is similar to wild type (WT) controls. E. Images of live five-day larvae show that the swim bladder inflates normally in igf2bp3la659Tg/+, zygotic igf2bp3la659Tg and MZigf2bp3la659Tg mutants, and there are no overt morphological defects in mutant larvae. F. Early cell divisions and cytokinesis are similar in WT and MZigf2bp3la659Tg mutant embryos. G. igf2bp family genes are not significantly altered in MZigf2bp3la659Tg mutants. Expression of igf2bp1, igf2bp2a (not expressed prior up to 1K-cell) and igf2bp2b were measured at 1-cell, 1K-cell and 50%-epiboly in WT and MZigf2bp3la659Tg mutants, revealing no changes in expression. Scale bars in D,E, 200 μm.
(TIF)
A. Immunofluorescence to detect membrane β-Catenin (green) and nuclear DAPI (blue) in wild type and MZigf2bp3la659Tg mutant embryos at 3 hpf (left) or 4.5 hpf (right) shows expanded YSL in mutant embryos at 3 hpf but not at 4.5 hpf. B. The distance of the farthest yolk syncytial nuclei (YSN) from the blastoderm margin (dashed yellow lines in embryos shown in A) and the number of YSN in WT and MZigf2bp3la659Tg mutant embryos show an initial increase at 3 hpf in mutant embryos, but becomes similar to WT at 4.5 hpf. N = 20 for both genotypes at 3 hpf and 4.5 hpf; statistical analysis performed with two-tailed unpaired t-test; p * < 0.05, ** < 0.01, *** < 0.001. C-E. qRT-PCR in 1-cell (C), 1000-cell (1K; D) and 50% epiboly stage (E) wild type and MZigf2bp3la659Tg mutant embryos shows significantly reduced igf2bp3 transcript levels in mutant embryos at all stages; mxtx2 is not altered at 1-cell and 1K stages but reduced at 50% epiboly in MZigf2bp3la659Tg mutants, and camsap3 is either unchanged (1-cell and 1K), or slightly increased (50% epiboly) in MZigf2bp3la659Tg; p *** < 0.01, **** <0.001. Scale bar in A, 200 μm.
(TIF)
A. Injection of igf2bp3 mRNA into igf2bp3Δ7 embryos does not rescue YSL expansion or lethality. WT, Migf2bp3Δ7 and MZigf2bp3Δ7 embryos were injected with 200 pg of igf2bp3 mRNA at the 1-cell stage and the YSL phenotype scored at 3 hpf. B. Injection of igf2bp1 or igf2bp3 mRNA into igf2bp3la659Tg does not rescue loss of PGCs. MZigf2bp3la659Tg embryos were injected with 100 pg of igf2bp1 or igf2bp3 mRNA at the 1-cell stage and the number of PGCs quantified at 24 hpf.
(TIF)
A. ΔYBE Sqt-GFP reporter expression (green signals in extracellular space) is premature and elevated in MZigf2bp3la659Tg embryos compared to control embryos at the high, sphere and dome stages. B. Bar graphs show mean GFP signal intensity in the blastoderm of imaged embryos. GFP levels in the blastoderm were normalised to levels of co-injected rhodamine dextran control. Representative examples from three independent experiments are shown. Number of embryos analysed: high (N = 3 WT and 4 MZigf2bp3la569Tg), sphere (N = 6 control and 7 MZigf2bp3la569Tg), dome (N = 4 WT and 5 MZigf2bp3la569Tg). Asterisks indicate level of significance from two-tailed t tests; p *<0.05, **<0.01, ***<0.001. Scale bar, 50 μm.
(TIF)
A,B. Live imaging of PGCs in embryos injected with GFP-nos3’UTR reporter mRNA shows reduced PGC numbers in igf2bp3la659Tg mutants compared to WT embryos at 1-somite, 5-somite, 10-somites and 25-somites. C-E. PGC numbers are reduced and germ cells are ectopically located in MZigf2bp3la659Tg mutants (C, E), and reduced in both Migf2bp3la659Tg and MZigf2bp3la659Tg embryos (D). F. The germline is also mis-regulated in a second transgenic insertion mutant line, igf2bp3la361Tg with significantly reduced PGCs in MZigf2bp3la361Tg embryos (F, G). N = 15 WT, 20 Pigf2bp3la659Tg 20 Migf2bp3la659Tg and 25 MZigf2bp3la659Tg embryos in C-E, and 5 WT and 23 MZigf2bp3la361Tg mutants in F,G. Scale bars in C and F, 200 μm; * p<0.05, **<0.01, ***<0.001.
(TIF)
A,B. WISH to detect the germline marker ddx4 (A) shows that PGCs are reduced (quantitation in B), and ectopically located relative to the midline (dashed line) in Migf2bp3Δ7 embryos by Bud stage. N = 39 WT and 42 MZigf2bp3Δ7 mutants; Scale bar in A, 200 μm; p* < 0.05, ** < 0.01, *** < 0.001.
(TIF)
A-F. qRT-PCR of Migf2bp3Δ7, MZigf2bp3Δ7 or MZigf2bp3Δ7 in a Tg(buc:buc-egfp); bucp106+/- background and MZigf2bp3la659Tg (G-K) mutant embryos compared to wildtype controls, shows variable reduction in nos1, ddx4, dnd1 and dazl expression in one-cell stage mutant embryos. Reduction in ddx4, nos1, dnd1 and gran is observed in MZigf2bp3Δ7 and MZigf2bp3la659Tg embryos at the 1K stage. C, H. At the 1K stage, expression of the chemokine receptors, cxcr4a and cxcr4b, is elevated in igf2bp3 mutant embryos, whereas at 50% epiboly (E, J) ddx4 and cxcl12a levels are reduced in the mutants. At 1K-cell stage, there is an increase in sqt and a variable increase in tbxta in both mutants (D, I). At 50% epiboly (F, K), sqt is significantly reduced or reverts to normal levels in MZigf2bp3la659Tg and MZigf2bp3Δ7 embryos, respectively; gsc is significantly increased in both mutants. Data from 3 biological replicates of 25–50 embryos each; p *<0.09, **<0.05, ***<0.01, ****<0.001.
(TIF)
GFP-labelled PGCs in a late gastrula (80% epiboly) MZigf2bp3la659Tg embryo show convoluted migration paths and considerable spread along the animal-vegetal axis. In the clip shown, dorsal is to the right, animal pole towards the top and the gastrula margin to the bottom.
(AVI)
PGCs expressing GFP in 10 hpf WT embryos tracked with spinning disk confocal microscopy. Cells cluster together and migrating rapidly towards the midline (red dashed line).
(AVI)
PGCs expressing GFP in 10 hpf mutant embryos, imaged with spinning disk confocal microscopy. Reduced number of cells cluster and migrate towards the midline in the mutant.
(AVI)
Ectopic and dispersed PGCs in an MZigf2bp3la659Tg mutant embryo at bud stage. The PGCs move in a non-linear fashion.
(AVI)
PGCs in a 10 hpf WT embryo, expressing Farnesylated-eGFP, tracked with spinning disk confocal microscopy, showing cellular projections towards the midline (right, indicated by a white arrow).
(AVI)
PGCs expressing Farnesylated-eGFP in a MZigf2bp3la659Tg embryo tracked with spinning disk confocal microscopy. Extensive blebbing is observed and projections are not directed towards the midline (right) in the mutant embryo.
(AVI)
PGCs expressing GFP in MZigf2bp3la659Tg embryos tracked with spinning disk confocal microscopy. One PGC fragments (red arrow) and scatters during migration towards the midline (dashed red line, left) in the mutant embryo.
(AVI)
PGCs expressing GFP in MZigf2bp3la659Tg embryos tracked with spinning disk confocal microscopy. One PGC fragments (red arrow) and scatters during its migration towards the midline (dashed red line, right) in the mutant.
(AVI)
Primers for qRT-PCR, genotyping, and sequence of aptamers for tobramycin affinity pull-downs.
(DOCX)
Acknowledgments
We thank members of the Sampath laboratory for suggestions, Shawn Burgess for the igf2bp3 retroviral insertion lines, Bernardo Chapa y Lazo and Mohan Balasubramanian for spinning disk microscopy facilities, the University of Warwick Research Technology Platform for Proteomics and the Aquatics facility for zebrafish care.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
YHV was supported by a doctoral scholarship from the BBSRC Midlands Integrative Biosciences Training Programme. LS was supported by BBSRC project grant (BB/L007525/1) to KS. RL is supported by a doctoral scholarship from the MRC DTP. AZ is supported by a Leverhulme Trust project grant (RPG-2017-407) to KS. AJ is supported by the School of Life sciences, University of Warwick. KS is supported by the BBSRC and Warwick Medical School. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gall JG. Small granules in the amphibian oocyte nucleus and their relationship to RNA. J Biophys Biochem Cytol. 1956;2(4 Suppl):393–6. doi: 10.1083/jcb.2.4.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guil S, Gattoni R, Carrascal M, Abián J, Stévenin J, Bach-Elias M. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol Cell Biol. 2003;23(8):2927–41. doi: 10.1128/MCB.23.8.2927-2941.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talukdar I, Sen S, Urbano R, Thompson J, Yates JR, Webster NJ. hnRNP A1 and hnRNP F modulate the alternative splicing of exon 11 of the insulin receptor gene. PLoS One. 2011;6(11):e27869. doi: 10.1371/journal.pone.0027869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamberi C, Izaurralde E, Beisel C, Mattaj IW. Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol Cell Biol. 1997;17(5):2587–97. doi: 10.1128/MCB.17.5.2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazim M, Masuda A, Rahman MA, Nasrin F, Takeda JI, Ohe K, et al. Competitive regulation of alternative splicing and alternative polyadenylation by hnRNP H and CstF64 determines acetylcholinesterase isoforms. Nucleic Acids Res. 2017;45(3):1455–68. doi: 10.1093/nar/gkw823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134(6):1365–73. doi: 10.1083/jcb.134.6.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weidensdorfer D, Stöhr N, Baude A, Lederer M, Köhn M, Schierhorn A, et al. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15(1):104–15. doi: 10.1261/rna.1175909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6):1188–203. doi: 10.1016/j.cell.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samuels TJ, Järvelin AI, Ish-Horowicz D, Davis I. Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. Elife. 2020;9: e51529. doi: 10.7554/eLife.51529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, et al. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24(10):4448–64. doi: 10.1128/MCB.24.10.4448-4464.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao L, Yuan Y, Cheng F, Fang J, Zhou F, Ma W, et al. Translation repression by maternal RNA binding protein Zar1 is essential for early oogenesis in zebrafish. Development. 2017;144(1):128–38. doi: 10.1242/dev.144642 [DOI] [PubMed] [Google Scholar]
- 12.Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382(6593):713–6. doi: 10.1038/382713a0 [DOI] [PubMed] [Google Scholar]
- 13.Knaut H, Pelegri F, Bohmann K, Schwarz H, Nüsslein-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149(4):875–88. doi: 10.1083/jcb.149.4.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13(4):424–36. doi: 10.1101/gad.13.4.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braat AK, Zandbergen T, van de Water S, Goos HJ, Zivkovic D. Characterization of zebrafish primordial germ cells: morphology and early distribution of vasa RNA. Dev Dyn. 1999;216(2):153–67. doi: [DOI] [PubMed] [Google Scholar]
- 16.Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, et al. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol. 2003;13(16):1429–34. doi: 10.1016/s0960-9822(03)00537-2 [DOI] [PubMed] [Google Scholar]
- 17.Hartung O, Forbes MM, Marlow FL. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol Reprod Dev. 2014;81(10):946–61. doi: 10.1002/mrd.22414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roovers EF, Kaaij LJT, Redl S, Bronkhorst AW, Wiebrands K, de Jesus Domingues AM, et al. Tdrd6a Regulates the Aggregation of Buc into Functional Subcellular Compartments that Drive Germ Cell Specification. Dev Cell. 2018;46(3):285–301.e9. doi: 10.1016/j.devcel.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J, Yan L, Shen W, Meng A. Maternal Ybx1 safeguards zebrafish oocyte maturation and maternal-to-zygotic transition by repressing global translation. Development. 2018;145(19):dev166587. doi: 10.1242/dev.166587 [DOI] [PubMed] [Google Scholar]
- 20.Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, et al. Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol. 2009;19(5):414–22. doi: 10.1016/j.cub.2009.01.038 [DOI] [PubMed] [Google Scholar]
- 21.Kumari P, Gilligan PC, Lim S, Tran LD, Winkler S, Philp R, et al. An essential role for maternal control of Nodal signaling. Elife. 2013;2:e00683. doi: 10.7554/eLife.00683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gore AV, Maegawa S, Cheong A, Gilligan PC, Weinberg ES, Sampath K. The zebrafish dorsal axis is apparent at the four-cell stage. Nature. 2005;438(7070):1030–5. doi: 10.1038/nature04184 [DOI] [PubMed] [Google Scholar]
- 23.Gilligan PC, Kumari P, Lim S, Cheong A, Chang A, Sampath K. Conservation defines functional motifs in the squint/nodal-related 1 RNA dorsal localization element. Nucleic Acids Res. 2011;39(8):3340–9. doi: 10.1093/nar/gkq1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaucker A, Nagorska A, Kumari P, Hecker N, Wang Y, Huang S, et al. Translational co-regulation of a ligand and inhibitor by a conserved RNA element. Nucleic Acids Res. 2018;46(1):104–19. doi: 10.1093/nar/gkx938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haston KM, Tung JY, Reijo Pera RA. Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS One. 2009;4(5):e5654. doi: 10.1371/journal.pone.0005654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maegawa S, Yamashita M, Yasuda K, Inoue K. Zebrafish DAZ-like protein controls translation via the sequence ’GUUC’. Genes Cells. 2002;7(9):971–84. doi: 10.1046/j.1365-2443.2002.00576.x [DOI] [PubMed] [Google Scholar]
- 27.Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24(14):2656–66. doi: 10.1038/sj.emboj.7600738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife. 2017;6:e26116. doi: 10.7554/eLife.26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115–27. doi: 10.1038/cr.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542(7642):475–8. doi: 10.1038/nature21355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–95. doi: 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wächter K, Köhn M, Stöhr N, Hüttelmaier S. Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Biol Chem. 2013;394(8):1077–90. doi: 10.1515/hsz-2013-0111 [DOI] [PubMed] [Google Scholar]
- 34.Ren F, Lin Q, Gong G, Du X, Dan H, Qin W, et al. Igf2bp3 maintains maternal RNA stability and ensures early embryo development in zebrafish. Commun Biol. 2020;3(1):94. doi: 10.1038/s42003-020-0827-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz SP, Aisenthal L, Elisha Z, Oberman F, Yisraeli JK. A 69-kDa RNA-binding protein from Xenopus oocytes recognizes a common motif in two vegetally localized maternal mRNAs. Proc Natl Acad Sci U S A. 1992;89(24):11895–9. doi: 10.1073/pnas.89.24.11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elisha Z, Havin L, Ringel I, Yisraeli JK. Vg1 RNA binding protein mediates the association of Vg1 RNA with microtubules in Xenopus oocytes. EMBO J. 1995;14(20):5109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon S, Abramson T, Munro TP, John CM, Köhrmann M, Schnapp BJ. UUCAC- and vera-dependent localization of VegT RNA in Xenopus oocytes. Curr Biol. 2002;12(7):558–64. doi: 10.1016/s0960-9822(02)00740-6 [DOI] [PubMed] [Google Scholar]
- 38.Yaniv K, Fainsod A, Kalcheim C, Yisraeli JK. The RNA-binding protein Vg1 RBP is required for cell migration during early neural development. Development. 2003;130(23):5649–61. doi: 10.1242/dev.00810 [DOI] [PubMed] [Google Scholar]
- 39.Toledano H D ’Alterio C, Czech B, Levine E, Jones DL. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485(7400):605–10. doi: 10.1038/nature11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 5th ed. Eugene: Univ. of Oregon Press; 2007. [Google Scholar]
- 41.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, et al. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci U S A. 2002;99(26):16719–24. doi: 10.1073/pnas.262483899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, et al. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3’ UTR structures. RNA Biol. 2011;8(6):1173–86. doi: 10.4161/rna.8.6.17836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatano T, Alioto S, Roscioli E, Palani S, Clarke ST, Kamnev A, et al. Rapid production of pure recombinant actin isoforms in Pichia pastoris. J Cell Sci. 2018;131(8):jcs213827. doi: 10.1242/jcs.213827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–40. doi: 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 45.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11(12):2301–19. doi: 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- 46.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44(W1):W272–6. doi: 10.1093/nar/gkw398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015;25(7):1030–42. doi: 10.1101/gr.186379.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Köprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15(21):2877–85. doi: 10.1101/gad.212401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weidinger G, Wolke U, Köprunner M, Thisse C, Thisse B, Raz E. Regulation of zebrafish primordial germ cell migration by attraction towards an intermediate target. Development. 2002;129(1):25–36. [DOI] [PubMed] [Google Scholar]
- 50.Lim S, Kumari P, Gilligan P, Quach HN, Mathavan S, Sampath K. Dorsal activity of maternal squint is mediated by a non-coding function of the RNA. Development. 2012;139(16):2903–15. doi: 10.1242/dev.077081 [DOI] [PubMed] [Google Scholar]
- 51.Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4 [DOI] [PubMed] [Google Scholar]
- 52.Safura S, Roba G and Freeman E. Evaluating the Effects of Bisphenols F and S with Respect to Bisphenol A on Primordial Germ Cell Migration in Zebrafish (Danio rerio) Embryos Using Immunofluorescence Microscopy. American Journal of Undergraduate Research. 2019;16(3): 69–77. doi: 10.33697/ajur.2019.031 [DOI] [Google Scholar]
- 53.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2003;60(5):523–33. doi: 10.1007/s00253-002-1158-6 [DOI] [PubMed] [Google Scholar]
- 54.Tian J, Andrée B, Jones CM, Sampath K. The pro-domain of the zebrafish Nodal-related protein Cyclops regulates its signaling activities. Development. 2008;135(15):2649–58. doi: 10.1242/dev.019794 [DOI] [PubMed] [Google Scholar]
- 55.Wilson K and Hardy ICW. Statistical analysis of sex ratios: an introduction. In: Sex Ratios. Cambridge University Press; 2002. pp. 48–92. doi: 10.1017/CBO9780511542053.004 [DOI] [Google Scholar]
- 56.Varshney GK, Lu J, Gildea DE, Huang H, Pei W, Yang Z, et al. A large-scale zebrafish gene knockout resource for the genome-wide study of gene function. Genome Res. 2013;23(4):727–35. doi: 10.1101/gr.151464.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu LT, Fong SH, Kondrychyn I, Loh SL, Ye Z, Korzh V. Yolk syncytial layer formation is a failure of cytokinesis mediated by Rock1 function in the early zebrafish embryo. Biol Open. 2012;1(8):747–53. doi: 10.1242/bio.20121636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell. 2004;6(6):771–80. doi: 10.1016/j.devcel.2004.05.002 [DOI] [PubMed] [Google Scholar]
- 59.Yabe T, Ge X, Lindeman R, Nair S, Runke G, Mullins MC, et al. The maternal-effect gene cellular island encodes aurora B kinase and is essential for furrow formation in the early zebrafish embryo. PLoS Genet. 2009;5(6):e1000518. doi: 10.1371/journal.pgen.1000518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bussmann J, Raz E. Chemokine-guided cell migration and motility in zebrafish development. EMBO J. 2015;34(10):1309–18. doi: 10.15252/embj.201490105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren F, Miao R, Xiao R and Mei J. m6A reader Igf2bp3 enables germ plasm assembly by m6A-dependent regulation of gene expression in zebrafish. Sci Bull. 2021;66(11): 1119–1128. doi: 10.1016/J.SCIB.2021.02.003 [DOI] [PubMed] [Google Scholar]
- 62.Paluch EK, Raz E. The role and regulation of blebs in cell migration. Curr Opin Cell Biol. 2013;25(5):582–90. doi: 10.1016/j.ceb.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tzung KW, Goto R, Saju JM, Sreenivasan R, Saito T, Arai K, et al. Early depletion of primordial germ cells in zebrafish promotes testis formation. Stem Cell Reports. 2015;4(1):61–73. doi: 10.1016/j.stemcr.2014.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kossack ME, Draper BW. Genetic regulation of sex determination and maintenance in zebrafish (Danio rerio). Curr Top Dev Biol. 2019;134:119–49. doi: 10.1016/bs.ctdb.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soh YQS, Mikedis MM, Kojima M, Godfrey AK, de Rooij DG, Page DC. Meioc maintains an extended meiotic prophase I in mice. PLoS Genet. 2017;13(4):e1006704. doi: 10.1371/journal.pgen.1006704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jain D, Puno MR, Meydan C, Lailler N, Mason CE, Lima CD, et al. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. Elife. 2018;7:e30919. doi: 10.7554/eLife.30919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gateff E. Cancer, genes, and development: the Drosophila case. Adv Cancer Res. 1982;37:33–74. doi: 10.1016/s0065-230x(08)60881-7 [DOI] [PubMed] [Google Scholar]
- 68.White RJ, Collins JE, Sealy IM, Wali N, Dooley CM, Digby Z, et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife. 2017;6:e30860. doi: 10.7554/eLife.30860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. RNA-seq expression data for all four zebrafish igf2bp genes during zebrafish development shows that igf2bp3 is expressed throughout zebrafish development. Data from [68]. B. Whole mount in-situ hybridisation (WISH) to detect igf2bp3 in zebrafish embryos at cleavage, blastula, gastrula, somitogenesis and 24 hpf. C. Sex ratios in igf2bp3Δ7/+, igf2bp3Δ7/Δ7, igf2bp3la659Tg/+, and igf2bp3la659Tg/la659Tg (i.e. zygotic igf2bp3la659Tg) adult fish harbouring the Tg(buc:buc-eGFP) transgene show that transgenic zygotic igf2bp3Δ7/Δ7 mutants manifest a strong male bias. Scale bar in B, 200 μm.
(TIF)
A. Schematic representation of the igf2bp3la659Tg mutant locus showing the site of the 6 kb Tg(nlacZ-GT virus) gene trap retroviral insertion, and coloured triangles indicate the position of RT-PCR primers. B,C. MZigf2bp3la659Tg mutant embryos show significantly reduced or undetectable expression of igf2bp3. RT-PCR (gels on left) using exon spanning primers for igf2bp3 and qRT-PCR (bar graphs, right) at performed at six early developmental stages, show reduced igf2bp3 transcripts in mutant embryos relative to gapdh control. C. Igf2bp3 protein is not detectable in MZigf2bp3la659Tg embryos. Western blots on cleavage to early gastrula stage embryos show a 60 kD band in wild type (WT) lysates. No Igf2bp3 band is detected in MZigf2bp3la659Tg lysates. D. WISH shows that shh expression in the midline (top panels) and myoD expression in the myotome (bottom panels) in MZigf2bp3la659Tg mutants is similar to wild type (WT) controls. E. Images of live five-day larvae show that the swim bladder inflates normally in igf2bp3la659Tg/+, zygotic igf2bp3la659Tg and MZigf2bp3la659Tg mutants, and there are no overt morphological defects in mutant larvae. F. Early cell divisions and cytokinesis are similar in WT and MZigf2bp3la659Tg mutant embryos. G. igf2bp family genes are not significantly altered in MZigf2bp3la659Tg mutants. Expression of igf2bp1, igf2bp2a (not expressed prior up to 1K-cell) and igf2bp2b were measured at 1-cell, 1K-cell and 50%-epiboly in WT and MZigf2bp3la659Tg mutants, revealing no changes in expression. Scale bars in D,E, 200 μm.
(TIF)
A. Immunofluorescence to detect membrane β-Catenin (green) and nuclear DAPI (blue) in wild type and MZigf2bp3la659Tg mutant embryos at 3 hpf (left) or 4.5 hpf (right) shows expanded YSL in mutant embryos at 3 hpf but not at 4.5 hpf. B. The distance of the farthest yolk syncytial nuclei (YSN) from the blastoderm margin (dashed yellow lines in embryos shown in A) and the number of YSN in WT and MZigf2bp3la659Tg mutant embryos show an initial increase at 3 hpf in mutant embryos, but becomes similar to WT at 4.5 hpf. N = 20 for both genotypes at 3 hpf and 4.5 hpf; statistical analysis performed with two-tailed unpaired t-test; p * < 0.05, ** < 0.01, *** < 0.001. C-E. qRT-PCR in 1-cell (C), 1000-cell (1K; D) and 50% epiboly stage (E) wild type and MZigf2bp3la659Tg mutant embryos shows significantly reduced igf2bp3 transcript levels in mutant embryos at all stages; mxtx2 is not altered at 1-cell and 1K stages but reduced at 50% epiboly in MZigf2bp3la659Tg mutants, and camsap3 is either unchanged (1-cell and 1K), or slightly increased (50% epiboly) in MZigf2bp3la659Tg; p *** < 0.01, **** <0.001. Scale bar in A, 200 μm.
(TIF)
A. Injection of igf2bp3 mRNA into igf2bp3Δ7 embryos does not rescue YSL expansion or lethality. WT, Migf2bp3Δ7 and MZigf2bp3Δ7 embryos were injected with 200 pg of igf2bp3 mRNA at the 1-cell stage and the YSL phenotype scored at 3 hpf. B. Injection of igf2bp1 or igf2bp3 mRNA into igf2bp3la659Tg does not rescue loss of PGCs. MZigf2bp3la659Tg embryos were injected with 100 pg of igf2bp1 or igf2bp3 mRNA at the 1-cell stage and the number of PGCs quantified at 24 hpf.
(TIF)
A. ΔYBE Sqt-GFP reporter expression (green signals in extracellular space) is premature and elevated in MZigf2bp3la659Tg embryos compared to control embryos at the high, sphere and dome stages. B. Bar graphs show mean GFP signal intensity in the blastoderm of imaged embryos. GFP levels in the blastoderm were normalised to levels of co-injected rhodamine dextran control. Representative examples from three independent experiments are shown. Number of embryos analysed: high (N = 3 WT and 4 MZigf2bp3la569Tg), sphere (N = 6 control and 7 MZigf2bp3la569Tg), dome (N = 4 WT and 5 MZigf2bp3la569Tg). Asterisks indicate level of significance from two-tailed t tests; p *<0.05, **<0.01, ***<0.001. Scale bar, 50 μm.
(TIF)
A,B. Live imaging of PGCs in embryos injected with GFP-nos3’UTR reporter mRNA shows reduced PGC numbers in igf2bp3la659Tg mutants compared to WT embryos at 1-somite, 5-somite, 10-somites and 25-somites. C-E. PGC numbers are reduced and germ cells are ectopically located in MZigf2bp3la659Tg mutants (C, E), and reduced in both Migf2bp3la659Tg and MZigf2bp3la659Tg embryos (D). F. The germline is also mis-regulated in a second transgenic insertion mutant line, igf2bp3la361Tg with significantly reduced PGCs in MZigf2bp3la361Tg embryos (F, G). N = 15 WT, 20 Pigf2bp3la659Tg 20 Migf2bp3la659Tg and 25 MZigf2bp3la659Tg embryos in C-E, and 5 WT and 23 MZigf2bp3la361Tg mutants in F,G. Scale bars in C and F, 200 μm; * p<0.05, **<0.01, ***<0.001.
(TIF)
A,B. WISH to detect the germline marker ddx4 (A) shows that PGCs are reduced (quantitation in B), and ectopically located relative to the midline (dashed line) in Migf2bp3Δ7 embryos by Bud stage. N = 39 WT and 42 MZigf2bp3Δ7 mutants; Scale bar in A, 200 μm; p* < 0.05, ** < 0.01, *** < 0.001.
(TIF)
A-F. qRT-PCR of Migf2bp3Δ7, MZigf2bp3Δ7 or MZigf2bp3Δ7 in a Tg(buc:buc-egfp); bucp106+/- background and MZigf2bp3la659Tg (G-K) mutant embryos compared to wildtype controls, shows variable reduction in nos1, ddx4, dnd1 and dazl expression in one-cell stage mutant embryos. Reduction in ddx4, nos1, dnd1 and gran is observed in MZigf2bp3Δ7 and MZigf2bp3la659Tg embryos at the 1K stage. C, H. At the 1K stage, expression of the chemokine receptors, cxcr4a and cxcr4b, is elevated in igf2bp3 mutant embryos, whereas at 50% epiboly (E, J) ddx4 and cxcl12a levels are reduced in the mutants. At 1K-cell stage, there is an increase in sqt and a variable increase in tbxta in both mutants (D, I). At 50% epiboly (F, K), sqt is significantly reduced or reverts to normal levels in MZigf2bp3la659Tg and MZigf2bp3Δ7 embryos, respectively; gsc is significantly increased in both mutants. Data from 3 biological replicates of 25–50 embryos each; p *<0.09, **<0.05, ***<0.01, ****<0.001.
(TIF)
GFP-labelled PGCs in a late gastrula (80% epiboly) MZigf2bp3la659Tg embryo show convoluted migration paths and considerable spread along the animal-vegetal axis. In the clip shown, dorsal is to the right, animal pole towards the top and the gastrula margin to the bottom.
(AVI)
PGCs expressing GFP in 10 hpf WT embryos tracked with spinning disk confocal microscopy. Cells cluster together and migrating rapidly towards the midline (red dashed line).
(AVI)
PGCs expressing GFP in 10 hpf mutant embryos, imaged with spinning disk confocal microscopy. Reduced number of cells cluster and migrate towards the midline in the mutant.
(AVI)
Ectopic and dispersed PGCs in an MZigf2bp3la659Tg mutant embryo at bud stage. The PGCs move in a non-linear fashion.
(AVI)
PGCs in a 10 hpf WT embryo, expressing Farnesylated-eGFP, tracked with spinning disk confocal microscopy, showing cellular projections towards the midline (right, indicated by a white arrow).
(AVI)
PGCs expressing Farnesylated-eGFP in a MZigf2bp3la659Tg embryo tracked with spinning disk confocal microscopy. Extensive blebbing is observed and projections are not directed towards the midline (right) in the mutant embryo.
(AVI)
PGCs expressing GFP in MZigf2bp3la659Tg embryos tracked with spinning disk confocal microscopy. One PGC fragments (red arrow) and scatters during migration towards the midline (dashed red line, left) in the mutant embryo.
(AVI)
PGCs expressing GFP in MZigf2bp3la659Tg embryos tracked with spinning disk confocal microscopy. One PGC fragments (red arrow) and scatters during its migration towards the midline (dashed red line, right) in the mutant.
(AVI)
Primers for qRT-PCR, genotyping, and sequence of aptamers for tobramycin affinity pull-downs.
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.