BMP-Sox9 signaling drives tumor cells to transdifferentiate into astrocytes, which contributes to the recurrence of SHH-driven medulloblastoma, a common type of pediatric brain tumor.
Abstract
Astrocytes, a major glial cell type in the brain, play a critical role in supporting the progression of medulloblastoma (MB), the most common malignant pediatric brain tumor. Through lineage tracing analyses and single-cell RNA sequencing, we demonstrate that astrocytes are predominantly derived from the transdifferentiation of tumor cells in relapsed MB (but not in primary MB), although MB cells are generally believed to be neuronal-lineage committed. Such transdifferentiation of MB cells relies on Sox9, a transcription factor critical for gliogenesis. Our studies further reveal that bone morphogenetic proteins (BMPs) stimulate the transdifferentiation of MB cells by inducing the phosphorylation of Sox9. Pharmacological inhibition of BMP signaling represses MB cell transdifferentiation into astrocytes and suppresses tumor relapse. Our studies establish the distinct cellular sources of astrocytes in primary and relapsed MB and provide an avenue to prevent and treat MB relapse by targeting tumor cell transdifferentiation.
Graphical Abstract
Introduction
Medulloblastoma (MB) is the most common malignant brain tumor in children. Aggressive treatment including surgical resection, chemotherapy, and radiotherapy has significantly improved MB outcomes; however, patients who survive MB often suffer severe side effects due to the intensive treatment (Louis et al., 2007). An estimated 20–30% of human MB cases finally relapse, and relapsed MB is almost uniformly fatal (Zeltzer et al., 1999). Molecular analyses have revealed four major MB subgroups: Wnt, Sonic Hedgehog (SHH), Group 3, and Group 4 (Taylor et al., 2012). Various subgroups either recur locally (SHH-MB) or relapse at distant sites (Groups 3 and 4; Ramaswamy et al., 2013).
SHH-MB, which exhibits an aberrant activation of the SHH pathway in tumor cells, accounts for ∼30% of human MB. Loss of function in Patched1 (Ptch1, an antagonizing receptor of SHH pathway) represents the most common oncogenic mutations in SHH-MB (Kool et al., 2014; Northcott et al., 2012). Ptch1-deficient mice are important models for the basic research and preclinical studies of SHH-MB. Conditional deletion of Ptch1 in cerebellar granule neuron precursors (GNPs) caused MB formation in Math1-Cre/Ptch1fl/fl mice with 100% penetrance (Schüller et al., 2008; Yang et al., 2008), suggesting that cerebellar GNPs can serve as the cell of origin for SHH-MB. Tumor cells in SHH-MB are generally believed to be neuronal-lineage committed (Hovestadt et al., 2019; Selvadurai et al., 2020).
We previously reported that astrocytes, a major component of tumor microenvironment in SHH-MB, play an important role in supporting MB progression through secretion of SHH ligand. Genetic ablation of astrocytes significantly repressed tumor cell proliferation and promoted the differentiation of MB cells, thereby inhibiting MB growth (Liu et al., 2017). Additionally, a recent study reported that astrocytes in SHH-MB promoted tumor growth by stimulating the production of insulin-like growth factor 1 from microglia (Yao et al., 2020).
Bone morphogenetic proteins (BMPs) are soluble proteins that belong to the TGF-β superfamily. BMPs initiate signal transduction by binding to cell-surface receptors that form a heterotetrameric complex consisting of two type I (BMPRI) and two type II (BMPRII) receptors. Upon ligand binding, BMPRII phosphorylates the BMPRI at a glycine-serine–rich motif known as the GS domain. This consequently activates BMPRI and allows phosphorylation of the immediate substrate protein known as Smads. Phosphorylated Smads translocates into cell nuclei to induce expression of target genes such as Id1/2 and Hey1 (Heldin et al., 1997; Schmierer and Hill, 2007). The transcription factor Sox9 has been found to play a key role in chondrogenesis supported by BMP signaling (Pan et al., 2008; Zehentner et al., 1999). BMPs directly up-regulated the transcription of Sox9 along with the differentiation of chondrocytes, as well as in the semilunar valve cells in hearts (Jo et al., 2014; Zhao et al., 2007). Forced expression of stabilized mutant N-mycT58A in neural stem cells resulted in the formation of SHH-MB that highly expressed Sox9 and Math1. Overexpression of Sox9 with N-mycT58A in cerebellar neural stem cells caused MB with shorter latencies and higher penetrance compared with that driven by N-mycT58A (Swartling et al., 2012). These studies revealed potential cooperative interaction between N-myc and Sox9 in the development of brain tumors. Interestingly, forced expression of Sox9 confers cisplatin resistance in Daoy cells and Group 3 MB (Suryo Rahmanto et al., 2016), suggesting that Sox9 plays an important role in MB progression and drug responses.
We investigated the cellular origin of astrocytes in SHH-MB by lineage tracing analyses and single-cell RNA sequencing (sc-RNAseq). Our studies revealed that astrocytes were derived from the transdifferentiation of tumor cells in relapsed MB, although astrocytes were lineage-independent of tumor cells in primary MB. Expression of BMP2, BMP5, and BMP7 was significantly up-regulated in tumor tissues from relapsed MB compared with primary MB. Treatment with BMPs markedly induced astrocytogenesis and Sox9 phosphorylation in tumor cells. Forced expression of phosphorylated Sox9, but not wild-type Sox9, effectively stimulated tumor cells to transdifferentiate into astrocytes. These data suggest that the transdifferentiation of MB cells is mediated by BMP-Sox9 phosphorylation signaling. Finally, LDN-193189 and LDN-214117, two antagonists of BMPRI, significantly inhibited the transdifferentiation of tumor cells and repressed tumor relapse following radiotherapy. Our studies establish the distinct astrocytogenesis in relapsed MB and demonstrate that blockage of BMP signaling can suppress MB relapse by inhibting tumor cell transdifferentiation.
Results
Tumor cells and astrocytes are lineage-independent in primary MB
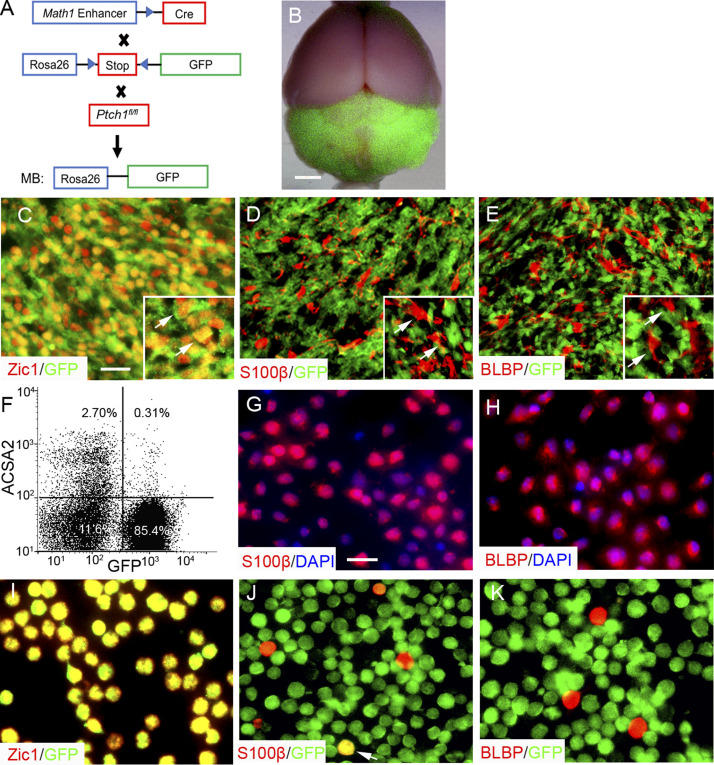
To examine the lineage relationship between tumor cells and tumor-derived astrocytes (TAs) in MB tissue, we lineage-traced tumor cells by using Math1-Cre/Ptch1fl/fl/Rosa-GFP mice (MPG mice), in which tumor cells permanently express GFP in MB tissue (Fig. 1 A; Cheng et al., 2020a; Yang et al., 2008). After tumors were established in MPG mice at ∼8 wk of age, intensified GFP fluorescence was observed in MB located in the cerebellum (Fig. 1 B). As expected, the majority of GFP+ cells were positive for Zic1, a putative marker for tumor cells in MB (Fig. 1 C; Aruga et al., 1994; Yokota et al., 1996), consistent with the notion that Math1+ GNPs give rise to MB after Ptch1 deletion (Schüller et al., 2008; Yang et al., 2008). However, most TAs (S100β+ and BLBP+) were negative for GFP (Fig. 1, D and E), suggesting they were not derived from MB cells or Math1-expressing GNPs.
Figure 1.
Lineage independence of TAs and tumor cells in primary MB.(A) The strategy of generating MPG mice for lineage-tracing experiments. (B) A whole-mount picture of brain from an MPG mouse bearing a GFP-positive tumor in the cerebellum. (C–E) Sagittal frozen sections from MB in MPG mice were immunostained for Zic1/GFP (C), S100β/GFP (D), and BLBP/GFP (E). Insets in C–E show magnified views of designated stainings. Arrows point to tumor cells (C) and astrocytes (D and E). Representative images from three to five experiments are shown. (F–H) Flow cytometry analysis of cells dissociated from MB tissue in MPG mice after immunolabeling with an antibody against ACSA2 (F). ACSA2+ cells were collected to examine S100β (G) and BLBP (H) by immunocytochemistry. DAPI was used to counterstain cell nuclei. (I–K) Cells dissociated from MB tissue in MPG mice were immunostained for Zic1/GFP (I), S100β/GFP (J), and BLBP/GFP (K), right after being plated in vitro. The arrow in J points to a cell double positive for S100β and GFP. Representative flow cytometry data and images from three to five experiments are shown. Scale bars: 4 mm (B); 25 µm (C–E); 20 µm (G–K).
We next dissociated cells from MB tissue in MPG mice and further analyzed GFP expression in TAs by FACS using an antibody against astrocyte cell surface antigen 2 (ACSA-2; Barry and McDermott, 2005; Liu et al., 2017; Sharma et al., 2015). As shown in Fig. 1 F, >85% of cells in MB tissue were GFP+ tumor cells, whereas TAs (ACSA2+) accounted for only 2.7 ± 0.6% of cells (n = 6) in MB from MPG mice. Most TAs (ACSA2+) were negative for GFP, although <10% of TAs were GFP+. Most purified ACSA2+ cells were found to be positive for S100β (Fig. 1 G) or BLBP (Fig. 1 H). We then plated the cells dissociated from MB tissue in MPG mice and found that most GFP+ cells expressed Zic1 (Fig. 1 I), further confirming that GFP+ cells in MB of MPG mice were predominantly tumor cells. Most TAs (S100β+, BLBP+) were found to be negative for GFP by immunocytochemistry (Fig. 1, J and K). These data suggest that TAs and tumor cells were essentially lineage-independent in primary MB, and TAs in primary MB likely migrate from the surrounding brain tissue during tumor initiation.
TAs are derived from tumor cells in transplanted tumors as well as relapsed tumors
In previous studies, subcutaneous tumor models have been used in basic research and preclinical studies of MB (Gordon et al., 2018; Kim et al., 2010). Having observed that TAs in primary MB tissues may migrate from the surrounding brain tissues, we then examined the source of TAs in subcutaneous MB in mouse flanks (Fig. 2 A), where there are no resident astrocytes or other neural cells. For this purpose, we purified tumor cells (GFP+) from MPG mice by FACS and transplanted them into the flanks of CB17/SCID mice as previously described (Gordon et al., 2018). After tumors were established in the flank, we sectioned the tumor tissue for immunohistochemistry. As shown in Fig. 2, B and C, astrocytes (S100β+ and BLBP+) were detected in tissue sections from flank tumors, suggesting that MB developed in mouse flanks also contain astrocytes, as do primary MB developed in the cerebellum. Moreover, most astrocytes were found to be positive for GFP, suggesting that astrocytes in flank tumors originated from the transplanted MB cells. We then intracranially transplanted purified MB cells (GFP+) into mouse cerebella, where there should be abundant astrocytes (Fig. 2 A). In the tumors formed in the cerebella after transplantation, TAs were also found to be positive for GFP (Fig. 2, D and E), indicating that TAs in the intracranial tumors were also derived from tumor cells. These data suggest that TAs in the transplanted tumors originate from MB cells, regardless of the availability of astrocytes in the surrounding tissue.
Figure 2.
TAs are derived from tumor cells in relapsed MB.(A) The strategy of generating MB models by injecting tumor cells into flanks or cerebella of CB17/SCID mice. (B–E) Frozen sections from MB from mouse flanks (B and C) and mouse brains (D and E) were immunostained for S100β/GFP (B and D) and BLBP/GFP (C and E). Representative images from six experiments are shown. (F) The strategy to generate a model for relapsed MB. MPG mice were irradiated at 4 wk of age and collected at 3 wk following the radiation to examine tumor relapse. (G–J) MRI brain images of an MPG mouse bearing MB at 8 wk of age before the irradiation (G), after the irradiation (H), and 3 wk following the irradiation (I); H&E-stained image of relapsed MB 3 wk after the irradiation (J). Representative images from three experiments are shown. (K and L) Frozen sections from relapsed MB in MPG mice were immunostained for S100β/GFP (K) and BLBP/GFP (L). Arrows point to astrocytes. Representative images from four experiments are shown. (M) The numbers of TAs based on immunofluorescence of S100β or BLBP in primary MB (Fig. 1, D and E) and relapsed MB (Fig. 2, K and L) were quantified per section (n = 4). Significance by Student’s t test. (N and O) Flow cytometry analysis of cells dissociated from relapsed MB tissue in MPG mice after immunostaining with an antibody against ACSA2 (N). ACSA2+ cells purified by FACS were plated and examined for S100β and GFP by immunocytochemistry (O). Representative flow cytometry data and images from four experiments are shown. Scale bars: 25 µm (B–E, K, L); 1 mm (G–J); 20 µm (O).
Tumor cells gave rise to astrocytes in regenerated MB after the transplantation, prompting us to further examine whether tumor cells produce astrocytes in relapsed MB. For this purpose, we generated a MB relapse model using MPG mice through irradiation (Fig. 2 F), as previously described (Vanner et al., 2014). Compared with primary MB before irradiation, tumor size in MPG mice was significantly reduced right after the 2-Gy irradiation (Fig. 2, G and H). Cleaved caspase-3 was readily detected in tumor tissue after the irradiation (Fig. S1 A), suggesting that tumor cells and TAs underwent extensive apoptosis following the irradiation. MB was reestablished/relapsed at 3 wk following the irradiation (Fig. 2, I and J). We harvested the relapsed tumor and examined GFP expression in TAs by immunohistochemistry. As shown in Fig. 2, K and L, a majority of TAs in the relapsed MB were GFP+, suggesting that, similar to those in the transplanted MB, astrocytes in relapsed MB were also derived from tumor cells. However, the number of astrocytes remained comparable in primary MB and relapsed MB (Fig. 2 M). As shown in Fig. 2 N, no significant difference in the percentage of TAs (ACSA2+) was found in relapsed MB (3.0 ± 0.7%, n = 6) and primary MB (2.7 ± 0.6%, n = 6, P > 0.05) by FACS, indicating that the abundance of TAs was not altered during MB relapse. However, most TAs in the relapsed MB were GFP+, with only 0.5 ± 0.2% of astrocytes (ACSA2+) appearing to be negative for GFP (Fig. 2 N). After being plated, >90% of purified ACSA2+ cells were found to be positive for GFP (Fig. 2 O). These data suggest that TAs predominantly originate from tumor cells in relapsed MB.
Figure S1.
Sox9 expression was up-regulated in TAs from relapsed MB.(A)Math1-Cre/ptch1fl/fl mice were irradiated at 4 wk of age. 3 d after the radiation, tumor tissues were collected to examine the astrocytes (S100β+) and apoptotic cells (CC3+) by immunohistochemistry. Tumor tissues without radiation were used as a control. Note that apoptotic cells were found among astrocytes (arrows) in tumor tissue after the irradiation but were absent among those in tumor tissue without irradiation. (B and C) Expression levels of Sox8, Sox9, and Sox10 mRNAs in astrocytes purified from primary and relapsed MB by FACS, examined by Q-PCR (B; n = 3). Levels of Sox9 protein in primary and relapsed MB tissue, examined by Western blotting (C). A representative image from three experiments is shown. **, P < 0.01, Student’s t test.
sc-RNaseq defines the distinct origin of TAs in primary and relapsed MB
It is well established that MB cells are neuronal-lineage committed (Schüller et al., 2008; Yang et al., 2008). Here we have described that TAs largely originate from tumor cells in relapsed MB, suggesting that MB cells may undergo transdifferentiation during tumor relapse. To further investigate the possible transdifferentiation of tumor cells, we harvested cells from primary MB tissue and relapsed MB in Math1-Cre/Ptch1fl/fl mice for sc-RNAseq. In total, 17,016 cells (9,164 cells from primary MB and 7,852 cells from relapsed MB) were harvested and used for transcriptomic analysis. We defined subpopulations of cells by principal component analysis (PCA) and t-stochastic neighbor embedding (tSNE). Consistent with our recent studies (Cheng et al., 2020b), a total of six cell clusters were identified based on their distinct transcriptomes: tumor cells that consist of quiescent cells (including cells at G0/G1 phase), dividing cells and differentiated cells; and stromal cells including astrocytes, microglia, and oligodendrocytes (Fig. 3, A and B). The majority (>90%) of dissociated cells were tumor cells that expressed Zic1 and Pax6 (Fig. 3 C). Approximately 22% of dissociated cells were dividing tumor cells with high expression levels of cell cycle–associated genes such as Ccna2, Cdk1, and Ccnb1 (Fig. 3 D). Approximately 20% of dissociated cells were differentiated and expressed NeuroD1, Cntn2, and Tubb3 (Fig. 3 E), as we recently reported (Cheng et al., 2020b). In addition, 47.6% of cells appeared to be quiescent, with declined expression of cell cycle genes as well as SHH pathway genes such as Gli1 and Gli2 (Fig. 3 B). TAs accounted for 3.1% of the total number of cells isolated from tumor tissues (including both primary and relapsed MB), based on the expression of astrocytic genes including Fabp7, Slc1a3, and Gfap (Fig. 3 F).
Figure 3.
Different cellular source of TAs in primary and relapsed MB revealed by sc-RNAseq.(A) tSNE plot showing cell clusters based on transcriptomes from cells isolated from primary and relapsed mouse MB. Cell clusters are color coded. (B) Heatmap of single-cell data based on the tSNE plot. Columns represent individual cells, and rows represent genes. (C–F) tSNE plots showing the expression levels of Zic1 and Pax6 (C); Ccna2, Cdk1, and Ccnb1 (D); NeuroD1, Cntn2, and Tubb3 (E); and Fabp7, Slc1a3, and GFAP (F) in all clusters. (G and H) tSNE plot showing cell clusters from primary MB and relapsed MB based on their transcriptomes (G). Cells from primary MB and relapsed MB are color coded. Percentages of cells from primary and relapsed MB in each cluster are shown in H. (I) Pseudotime analysis of cell clusters in primary MB and relapsed MB. Schematic diagrams show the evolutionary route of astrocytes in primary and relapsed MB. (J) tSNE plot showing cell clusters based on transcriptomes from all astrocytes in primary and relapsed MB. Percentage of astrocytes from primary MB and relapsed MB in cluster 0 and cluster 1. (K) Volcano plot shows the genes differentially expressed in astrocytes from relapsed MB compared with astrocytes in primary MB, according to the statistical P value (y axis; adjusted P value [padj]) and the relative abundance ratio (x axis; log2 fold changes). (L–N) Heatmaps show the expression levels of genes associated with SHH pathway (L), astrocyte activation (M), and SoxE family (N) in astrocytes from primary and relapsed MB.
We then separated primary MB cells and relapsed MB cells in the PCA and tSNE analyses. As shown in Fig. 3 G, tumor cells from primary and relapsed MB significantly overlap in the tSNE plot, suggesting that no major genetic alterations are evident in MB cells after tumor relapse. The ratio of the number of cells from primary and relapse MB was ∼1:1 in the populations of quiescent, dividing, and differentiated cells (Fig. 3 H). These data suggest that the composition of tumor cells was comparable between primary and relapsed MB.
To examine the developmental trajectory of TAs in primary and relapsed MB, we performed unsupervised pseudotime analysis based on the transcriptomes of cells from primary and relapsed tumors. In primary MB, dividing tumor cells progressively became quiescent, and a proportion of tumor cells underwent spontaneous differentiation (Fig. 3 I), as we previously reported (Cheng et al., 2020b). TAs together with microglia and oligodendrocytes were well separated from tumor cell populations, consistent with our findings that TAs and tumor cells were lineage independent in primary MB (Fig. 1). In relapsed MB, dividing tumor cells gradually became differentiated, as observed in primary MB (Fig. 3 I). However, a subset of tumor cells, predominantly those quiescent tumor cells, proceeded through a separate route to give rise to astrocytes (Fig. 3 I). The above trajectory analyses further confirm our findings that tumor cells transdifferentiate into astrocytes in relapsed MB, but not in primary MB.
Finally, we focused on TAs from primary and relapsed MB and classified the cell subpopulations based on their transcriptomes from sc-RNAseq. As shown in Fig. 3 J, two major cell subsets were defined: subset 0 and subset 1. TAs from primary MB were mainly present in subset 0, whereas cells in subset 1 were predominantly TAs from relapsed MB (Fig. 3 J). These data indicate that TAs from primary MB have distinct expression profiles compared with their counterparts in relapsed MB.
Sox9 expression is up-regulated in TAs from relapsed MB
To further examine the transcriptional difference between TAs in primary MB and those in relapsed MB, we purified TAs (ACSA2+) from primary MB (n = 3) and relapsed MB (n = 3) in Ptch1+/− mice by FACS, as we previously described (Liu et al., 2017). Purified TAs were then analyzed by RNAseq. Based on the transcriptome, genes differentially expressed between TAs in primary MB and those from relapsed MB were defined (P < 0.05 and log2 fold changes ≥1). In all 1,524 transcripts that were differentially expressed, 894 genes were up-regulated, and 630 genes were down-regulated in TAs from relapsed MB compared with astrocytes from primary MB (Fig. 3 K). Among them, the expression levels of SHH pathway target genes including Gli1, Gli2, and Ptch2 were significantly elevated in TAs from relapsed MB, compared with those from primary MB (Fig. 3 L). These findings further support that TAs from relapsed MB are derived from tumor cells with Ptch1 deletion. Moreover, the expression of genes associated with astrocyte activation including mKi67, Nestin, and GFAP (Liddelow and Barres, 2017; Zamanian et al., 2012) was overall up-regulated in TAs from relapsed MB, compared with those in primary MB (Fig. 3 M). These data suggest that TAs from relapsed MB were more activated than TAs from primary MB.
SoxE transcription factors (Sox8, Sox9, and Sox10) are associated with gliogenesis in developing brains and spinal cords (Scott et al., 2010; Stolt et al., 2003). As shown in Fig. 3 N, expression levels of Sox9 were significantly elevated, whereas expression of Sox8 and Sox10 was down-regulated in TAs from relapsed MB, compared with TAs in primary MB. Differential expressions of Sox8, Sox9, and Sox10 in TAs from relapsed MB and primary MB were further confirmed by quantitative PCR (Q-PCR; Fig. S1 B). Elevated levels of Sox9 protein were detected in relapsed MB tissue compared with primary MB (Fig. S1 C). These data imply that up-regulation of Sox9 expression may be involved in the astrocytogenesis in relapsed MB.
Sox9 is required but not sufficient for MB cell transdifferentiation into astrocytes
Sox9 has been previously found to regulate astrocyte specification and expansion (Kang et al., 2012; Sun et al., 2017). In MB tissue from ptch1+/− mice, Sox9 protein was detected in both tumor cells (Zic1+; Fig. 4 A) and TAs (S100β+; Fig. 4 B) in MB tissues, although the expression level of Sox9 mRNA in TAs was significantly greater than in tumor cells (Fig. 4 C). Based on our sc-RNAseq data, Sox9 expression was found only in tumor cells and astrocytes, but not observed in microglia or oligodendrocytes (Fig. 4 D).
Figure 4.
Sox9 is required for MB cell transdifferentiation.(A and B) Sagittal sections of MB from Ptch1+/− mice were immunostained for Zic1/Sox9 (A) and S100β/Sox9 (B). Arrows point to tumor cells expressing Sox9, whereas arrowheads point to Sox9+ astrocytes. Representative images from four experiments are shown. (C) Tumor cells and astrocytes were purified from MB in Ptch1+/− mice (n = 4) by FACS and examined for expression of Sox9 mRNA by Q-PCR. ***, P < 0.001, Student’s t test. (D) Violin plot of Sox9 expression in tumor cells, astrocytes, microglia, and oligodendrocytes (oligodendro), based on sc-RNAseq data of cells in tumor tissues from Math1-Cre/ptch1fl/fl mice. (E) Sox9 protein levels in MB tissue from MPS mice after treatment with tamoxifen or corn oil as a control. A representative image from four Western blotting experiments is shown. (F) The strategy for deleting Sox9 in tumor cells in relapsed MB. MPS mice bearing MB were irradiated once at 2 Gy and treated with tamoxifen or corn oil by oral gavage, once a day for 7 d. Mice were monitored for survival or sacrificed 3 wk following the irradiation to examine tumor cell transdifferentiation by immunohistochemistry (IHC). (G) Survival curves of MPS mice after irradiation and treatment with tamoxifen or corn oil. All MPS mice (n = 6) with corn oil treatment developed MB (median survival, 38 d), whereas only one mouse (n = 6) with tamoxifen treatment formed tumor. P < 0.0001, log-rank test. (H and I) Sagittal sections of MB from MPS mice after irradiation and treatment with tamoxifen or corn oil underwent H&E staining (H), as well as immunostaining for Sox9, BLBP, Ki67, NeuN, and CC3 (I). The percentage of cells positive for Sox9, Ki67, NeuN, or CC3 in MB tissue, or the number of astrocytes (BLBP+) per tumor section, was quantified based on the immunostaining results (n = 4), as shown at the right. ***, P < 0.001, Student’s t test. DAPI was used to counterstain cell nuclei in tumor sections. Representative images from four experiments are shown. Scale bars: 25 µm (A and B); 1 mm (H); 30 µm (I).
To investigate the possible functions of Sox9 in tumor cell transdifferentiation, we first examined possible alterations in the growth of primary MB after Sox9 deletion in tumor cells. For this purpose, conditional Sox9 mice (Sox9fl/fl mice; Akiyama et al., 2002) were crossed with Ptch1+/− mice and Math1-CreERT2 mice that carry an inducible Cre recombinase in MB cells (Du et al., 2019; Machold and Fishell, 2005). After MB was established in Math1-CreER/Ptch1+/-/Sox9fl/fl (MPS) mice, they were treated with tamoxifen or corn oil by oral gavage. After treatment, tumor tissues were harvested to examine Sox9 protein by Western blotting. As shown in Fig. 4 E, the protein levels of Sox9 were markedly declined in tumor tissue after tamoxifen treatment, indicating the effective deletion of Sox9 in MB tissue from MPS mice. However, no obvious difference in the proliferation (Ki67+) or differentiation (NeuN+) of tumor cells was observed after tamoxifen treatment, compared with corn oil treatment (Fig. S2, A–F). No alterations were found in the number of TAs in tumor tissue after tamoxifen treatment (Fig. S2, G–I). More importantly, no difference was observed in the latency of primary MB in MPS mice after Sox9 deletion in tumor cells (Fig. S2 J). These data suggest that Sox9 in tumor cells is dispensable for tumor cell proliferation as well as astrocytogenesis in primary MB.
Figure S2.
No alterations in the proliferation of MB cells after Sox9 deletion.(A–J) Tumor-bearing MPS mice (confirmed by MRI) were treated with corn oil or tamoxifen by oral gavage once a day for 7 consecutive days. After the treatment, tumor tissues were harvested to examine tumor cell proliferation (Ki67, A and B) and differentiation (NeuN, D and E), as well as astrocytes (BLBP, G and H), by immunohistochemistry. Representative images from three experiments are shown. The percentage of Ki67+ cells (C), NeuN+ cells (F), and astrocytes (BLBP+, I) in tumor tissue was quantified (n = 3). Survival curves of MPS mice after treatment with tamoxifen or corn oil (J). DAPI was used to counterstain cell nuclei. (K) Increased levels of NeuN protein in tumor tissue from irradiated MPS mice after tamoxifen treatment, compared with vehicle treatment, was confirmed by Western blotting. Representative images from three experiments are shown. Scale bars: 40 µm.
We next tested whether Sox9 in MB cells is necessary for tumor cell transdifferentiation during tumor relapse. After tumors were established in MPS mice, tumor-bearing mice were irradiated once at 2 Gy and treated with tamoxifen or corn oil as a control (Fig. 4 F) by oral gavage (once a day for 2 wk). All control MPS mice succumbed to MB within 2 mo following the irradiation (n = 8; median survival, 38 d); however, the majority (8/9) of MPS mice treated with tamoxifen survived (Fig. 4 G). We harvested cerebella in MPS mice 3 wk after irradiation to examine the transdifferentiation of tumor cells by immunohistochemistry. Compared with the control treated with corn oil, tamoxifen treatment significantly reduced tumor size (Fig. 4 H). The number of Sox9+ cells in MB tissue was decreased after tamoxifen treatment (78 ± 8% vs. 20 ± 4%; Fig. 4 I), suggesting that Sox9 gene was effectively deleted in most tumor cells. The abundance of astrocytes (BLBP+) in MB tissue in MPS mice was markedly reduced (344 ± 51 vs. 27 ± 8 per section) after tamoxifen treatment, compared with corn oil treatment. As a comparison, the number of astrocytes was not altered in primary MB after Sox9 deletion in tumor cells (Fig. S2, G–I). These data suggest that Sox9 deletion in tumor cells suppressed their transdifferentiation during MB relapse. As expected, tumor cell proliferation (Ki67+) in relapsed MB in MPS mice was significantly inhibited, whereas the differentiation and apoptosis of tumor cells was increased in relapsed tumors in MPS mice after tamoxifen treatment (Fig. 4 I), consistent with the critical role of TAs in supporting the proliferation and survival of MB cells (Liu et al., 2017). Enhanced differentiation of MB cells in tumor tissues that lack astrocytes was confirmed by an increased number of NeuN+ cells (Fig. 4 I) and increased levels of NeuN protein (Fig. S2 K) in tumor tissue after tamoxifen treatment. The above data suggest that Sox9 is necessary for the transdifferentiation of MB cells during tumor relapse.
To further examine whether up-regulation of Sox9 in MB cells is sufficient to induce transdifferentiation of tumor cells, we purified MB cells from Ptch1+/− mice by FACS and infected them with a lentivirus carrying Sox9-expressing vector or an empty vector as a control. 48 h following the infection, MB cells were harvested to examine the cell lineage of infected cells by immunocytochemistry. Cells with forced expression of Sox9 or empty vector were still positive for Zic1 but negative for S100β or BLBP (not depicted), indicating that overexpression of Sox9 alone failed to induce the transdifferentiation of MB cells. Altogether, these data suggest that Sox9 is necessary but not sufficient to induce tumor cell transdifferentiation in relapsed MB.
BMPs stimulate MB cells to transdifferentiate into astrocytes
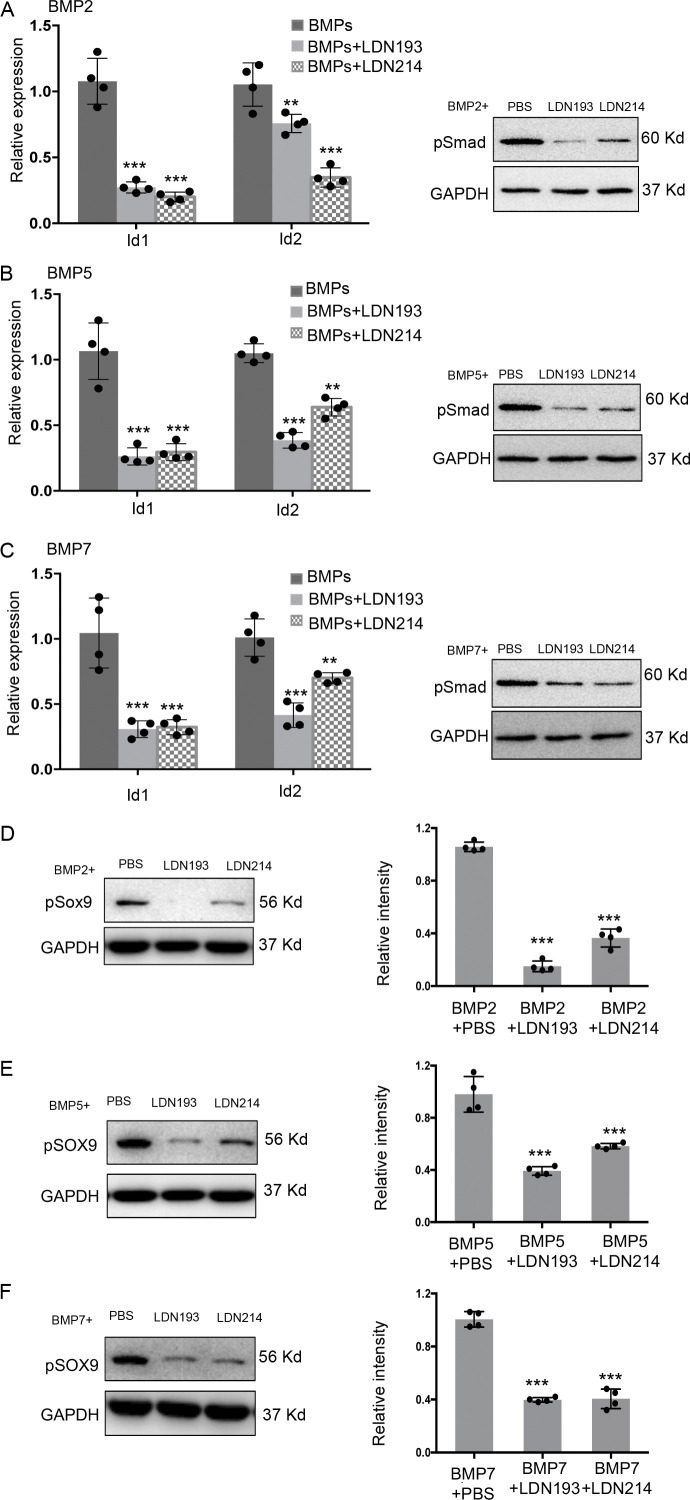
Previous studies have demonstrated that BMP signaling can induce cerebellar GNPs to transdifferentiate into astrocytes (Okano-Uchida et al., 2004). This prompted us to investigate whether BMP signaling drives the transdifferentiation of tumor cells in relapsed MB. We compared the expression of all brain-related BMPs, BMP1-7 and BMP8b (Ebendal et al., 1998; Mehler et al., 1997), in primary mouse MB tissues and relapsed MB by Q-PCR. Among eight BMP genes that we examined, expression of BMP2, BMP5, and BMP7 mRNA was significantly up-regulated in tumor tissues from relapsed MB compared with primary MB in MPG mice (Fig. 5 A). Consistent with this, phosphorylation of Smad (pSmad) was markedly increased in relapsed MB compared with primary MB (Fig. 5 B). In addition, enhanced expression of BMP2, BMP5, and BMP7 mRNA was also observed in human relapsed SHH-MB tissue compared with the paired primary MB tissue (n = 2; Fig. 5 C). We also analyzed a transcriptomic dataset of a human MB cohort (including three paired primary and relapsed SHH-MB: #11, #12, and #31; Morrissy et al., 2016). We found that 493 genes were differentially expressed between primary and relapsed MB, and the expression of 478 genes was significantly up-regulated in relapsed MB (log2 fold-change >1; at least two patients showed the same regulatory trend; Table S1). Gene ontology analysis of those 478 genes revealed that the expression of BMP pathway genes including Id3 and Id4 was overall elevated in relapsed MB compared with primary MB (Fig. 5 D). Expression levels of BMP2, BMP5, and BMP7 were overall elevated in relapsed MB compared with matched primary MB, although expression patterns of these BMPs were not very consistent in these pairs of tumor tissues (Fig. 5 D). These data suggest that the BMP pathway is activated in tumor tissue during MB relapse.
Figure 5.
BMPs induce the transdifferentiation of MB cells into astrocytes.(A and B) Primary tumor tissue (n = 3) and relapsed tumor tissue (n = 3) in MPG mice were collected to examine expression of BMP mRNAs by Q-PCR (A; ***, P < 0.001, Student’s t test) and phosphorylation of Smad (pSmad) by Western blotting (B). GAPDH was used as a loading control. (C) Tumor tissues from matched primary and relapsed human SHH-MB were collected to examine expression of BMP mRNAs by Q-PCR (n = 3). ***, P < 0.001, Student’s t test. (D) Heatmap shows the expression levels of genes associated with BMP signaling in human primary MB and relapsed MB (including three paired SHH-MB: #11, #12, and #31). P < 0.05 by one-way ANOVA. (E–H) Tumor cells from MPG mice were treated with DPBS (control) or BMPs (BMP2, BMP5, or BMP7, all at 80 ng/ml) for 72 h, and then collected to examine S100β and GFP by immunocytochemistry (E). Representative images from four independent experiments are shown. The percentage of cells positive for S100β in GFP+ tumor cells was quantified (F; n = 4). ***, P < 0.001 by Student’s t test. After the treatment, tumor cells were harvested to examine mRNA expression of S100β and BLBP by Q-PCR (G; ***, P < 0.001, Student’s t test) or levels of pSmad by Western blotting (H). Scale bars: 20 µm (E).
To examine whether BMP signaling stimulates the transdifferentiation of MB cells into astrocytes, we isolated tumor cells (GFP+) from MPG mice and treated them in vitro with 80 ng/ml BMP2, BMP5, or BMP7. MB cells were also treated with Dulbecco’s PBS (DPBS) as a control. 72 h following the treatment, no astrocytes (S100β+) were detected in MB cells (GFP+) after DPBS treatment; however, a significant proportion of tumor cells (GFP+) expressed S100β (Fig. 5, E and F) after treatment with BMPs, suggesting that BMP treatment promoted the transdifferentiation of tumor cells into astrocytes. In addition, mRNA expressions of S100β and BLBP in tumor cells were significantly up-regulated following BMP treatment compared with DPBS control (Fig. 5 G). The activation of BMP pathway was confirmed by increased levels of phosphorylated Smad in tumor cells after BMP treatment (Fig. 5 H). These data suggest that BMP signaling can induce the transdifferentiation of MB cells into astrocytes.
Sox9 phosphorylation mediates BMP-stimulated transdifferentiation of MB cells
We next examined whether Sox9 is required for BMP-induced transdifferentiation of MB cells. To this end, we reduced Sox9 expression in tumor cells by viral infection of RFP-tagged shRNAs. Tumor cells were also virally infected with scrambled shRNA as a control. 48 h following infection, expression levels of Sox9 were markedly decreased in MB cells infected with shRNA specific for Sox9 (Sh-sox9), compared with scrambled shRNA (Sh-scr; Fig. 6 A). We then treated those infected MB cells with BMP2 for an additional 72 h. Approximately 50% of MB cells infected with scrambled shRNA expressed S100β after BMP2 treatment, as expected. However, a significantly reduced number of astrocytes were detected in Sox9-deficient MB cells (RFP+) in the presence of BMP2 (Fig. 6, B and C). Sox9 knockdown also prohibited tumor cells from generating astrocytes following treatment with BMP5 and BMP7 (Fig. 6 C). These data suggest that Sox9 deficiency suppressed BMP-induced transdifferentiation of tumor cells (Fig. 6 C), further confirming that Sox9 is required for MB cell transdifferentiation into astrocytes.
Figure 6.
Sox9 phosphorylation mediates BMP-induced transdifferentiation of MB cells.(A–C) MB cells were virally infected with shRNA specific for Sox9 (Sh-Sox9) or scrambled shRNA (Sh-scr) for 48 h, before being harvested to examine Sox9 protein by Western blotting (A; n = 5). In addition, 24 h following the above infection, tumor cells were treated with 80 ng/ml BMP2 or DPBS for 48 h and collected to examine S100β and RFP expression by immunocytochemistry (B). Representative images from five experiments are shown. The percentage of astrocytes (S100β+, RFP+) in infected cells (RFP+) was quantified (C; n = 5), ***, P < 0.001 by Student’s t test. (D) MB cells were treated with 80 ng/ml BMP2, BMP5, BMP7, or DPBS in vitro for 4 h before being collected to examine Sox9 and phosphorylated Sox9 (p-Sox9) by Western blotting. Representative images from three experiments are shown. (E–G) MB cells were virally infected with GFP-tagged Sox9S181D, wild-type Sox9 (Sox9WT), or an empty vector as a control. 48 h following the infection, tumor cells were harvested to examine Sox9 and p-Sox9 by Western blotting (E). In addition, tumor cells were collected to examine S100β and GFP by immunocytochemistry (F). Representative images from four experiments are shown. The percentage of astrocytes (S100β+) among infected cells (GFP+) was quantified (G; n = 4). ***, P < 0.001; by Student’s t test. (H) MPS mice were treated with tamoxifen (TM) or corn oil by oral gavage once a day for 7 d. MB cells were harvested to examine Sox9 protein by Western blotting. GAPDH was used a loading control. (I–K) Sox9-null MB cells were virally infected with GFP-tagged Sox9WT, Sox9S181A, or empty vector and treated with 80 ng/ml BMP2 or PBS for 72 h. Sox9 protein and phosphorylated Sox9 in infected MB cells were examined by Western blotting (I). Astrocytes (S100β+) among infected cells (GFP+) after BMP2 treatment were examined by immunocytochemistry (J). Representative images from three experiments are shown. The percentage of astrocytes (S100β+) among infected cells (GFP+) was quantified (K; n = 3). ***, P < 0.001 by Student’s t test. Scale bars: 20 µm (B, F, and J).
The transcriptional activities of Sox9 are regulated by its phosphorylation, especially the phosphorylation of S181 (Liu et al., 2013; Zuo et al., 2018). Having observed that Sox9 is required but not sufficient to induce MB cell transdifferentiation, we next investigated whether Sox9 is phosphorylated in MB cells after BMP treatment. MB cells isolated from Math1-Cre/Ptch1fl/fl mice were treated with 80 ng/ml BMPs or DPBS for 4 h before being harvested for examination of Sox9 phosphorylation by Western blotting, using an antibody recognizing phosphorylated Sox9 at the S181 residue. As shown in Fig. 6 D, the levels of phosphorylated Sox9 were significantly elevated in BMP-treated cells, compared with those in cells treated with DPBS. The levels of total Sox9 proteins in tumor cells were not altered in MB cells within 4 h following the BMP treatment. These data suggest that BMP treatment induces the phosphorylation of Sox9 in MB cells.
To determine whether phosphorylated Sox9 can direct the transdifferentiation of tumor cells, we infected MB cells with a lentiviral vector encoding GFP-tagged phosphomimetic Sox9 by replacing serine with aspartate at the 181 residue (Sox9S181D) that mimics phosphorylated Sox9 (Liu et al., 2013). MB cells were also virally infected with wild type Sox9 (Sox9WT) or an empty vector (GFP) as controls. 48 h after infection, increased levels of Sox9 proteins were detected in MB cells infected with Sox9WT, compared with the GFP control (Fig. 6 E). Sox9 phosphorylation was readily observed in MB cells infected with Sox9S181D, but not in tumor cells with forced expression of Sox9WT or GFP alone (Fig. 6 E). A significant proportion of MB cells with Sox9S181D infection started to express S100β (52 ± 6.7%; Fig. 6 F), whereas <5% of tumor cells expressed S100β after infection with Sox9WT or GFP alone (Fig. 6, F and G). These data demonstrate that phosphorylated Sox9 can drive the transdifferentiation of MB cells. We next examined whether MB cell transdifferentiation can be blocked by a phosphorylation-resistant form of Sox9 with a mutation at the phosphorylation site (Sox9S181A; Kumar and Lassar, 2009; Liu et al., 2013). To remove the endogenous Sox9 in MB cells, we orally treated tumor-bearing MPS mice with tamoxifen or corn oil for 1 wk (once a day). Deletion of Sox9 in tumor cells was confirmed by Western blotting (Fig. 6 H). Sox9-null MB cells were then virally infected with GFP-tagged Sox9S181A, Sox9WT, or an empty GFP vector. Infected MB cells were treated with 80 ng/ml BMP2 or PBS for 72 h. As shown in Fig. 6 I, high levels of Sox9 protein were detected in MB cells infected with Sox9WT, in which Sox9 phosphorylation was induced by BMP2 treatment. However, Sox9 or phosphorylated Sox9 was barely found in Sox9-null MB cells infected with Sox9S181A or the GFP control, regardless of BMP2 treatment (Fig. 6 I). As expected, a significant number of MB cells infected with Sox9WT (GFP+) were transdifferentiated into astrocytes (S100β+) after BMP2 treatment (Fig. 6 J). However, almost no astrocytes were found in MB cells infected with GFP or Sox9S181A (Fig. 6, J and K). These data further confirm that Sox9 phosphorylation is critical for BMP-induced transdifferentiation of MB cells.
Inhibition of BMP signaling suppresses the transdifferentiation of tumor cells
Having observed that BMP signaling induced the transdifferentiation of tumor cells into astrocytes during MB relapse, we then examined whether inhibition of BMP signaling could repress the astrocytogenesis in relapsed tumors. LDN193189 (LDN193) and LDN214117 (LDN214) are two potent inhibitors of the BMP pathway by antagonizing BMPRI (Mohedas et al., 2014; Vogt et al., 2011). To test the efficacies of LDN193 and LDN214 in repressing BMP signaling in MB cells, we treated tumor cells with BMPs, together with LDN193, LDN214, or DMSO for 48 h. As shown in Fig. S3, A–C, LDN193 and LDN214 significantly repressed levels of Id1 and Id2 mRNAs (BMP pathway target genes) and Smad phosphorylation in BMP-treated tumor cells. These data indicated that LDN193 and LDN214 effectively inhibited BMP signaling in MB cells. As expected, a significant proportion of MB cells expressed S100β after treatment with BMP2, BMP5, or BMP7 (Fig. 7 A). However, the number of S100β+ cells in the BMP-treated cultures was markedly reduced after treatment with LDN193 or LDN214, compared with DMSO treatment (Fig. 7, A and B), suggesting that LDN193 and LDN214 effectively inhibited BMP-induced transdifferentiation of tumor cells. The levels of Sox9 phosphorylation in BMP-treated tumor cells were significantly declined following the treatment with LDN193 or LDN214 (Fig. S3, D–F). In addition, comparable levels of 5-ethynyl-2′-deoxyuridine (EdU) incorporation were observed in MB cells treated with LDN193 or LDN214 (Fig. S4, A and B). No alterations in the number of astrocytes were found in primary MB after treatment with LDN193 or LDN24 (Fig. S4, C and D). The survival of MPG mice was not changed by treatment of LDN193 or LDN214 (Fig. S4 E). These data suggest that the progression of primary MB is not affected by inhibition of the BMP pathway.
Figure S3.
LDN193 and LDN214 suppressed BMP signaling and Sox9 phosphorylation in MB cells.(A–C) MB cells isolated from MPG mice were treated with BMPs combined with DMSO, LDN193, or LDN214 for 48 h, before being collected to examine the expression of Id1 and Id2 by Q-PCR (n = 4) and the levels of phosphorylation of Smad by Western blotting. Representative images from four experiments are shown. The expression of Id1 or Id2 in tumor cells after combined treatment with BMPs and LDN193 or LDN214, relative to that with the treatment of BMPs alone: BMP2 (A), BMP5 (B), and BMP7 (C). GAPDH was used as a loading control. (D–F) MB cells from MPG mice were treated with 80 ng/ml BMPs together with DMSO, LDN193, or LDN214 (1 µM) for 4 h, before being harvested to examine p-Sox9 by Western blotting. GAPDH was used for a loading control. Representative images from four experiments are shown. Levels of p-Sox9 in tumor cells treated with LDN193 or LDN214, relative to that in DMSO-treated tumor cells, were quantified (n = 4). BMP2 (D), BMP5 (E), and BMP7 (F). **, P<0.01; ***, P < 0.001, Student’s t test.
Figure 7.
BMP inhibitors repress MB cell transdifferentiation during MB relapse.(A and B) MB cells isolated from MPG mice were treated with BMP combined with DMSO, LDN193, or LDN214 for 72 h, before being collected to detect S100β and GFP by immunocytochemistry (A). Representative images from five experiments are shown. The percentage of astrocytes (S100β+) in the culture was quantified (B; n = 5). ***, P < 0.001 by Student’s t test. (C) MPG mice at 4 wk of age were irradiated once at 2 Gy and treated with LDN193 (25 mg/kg), LDN214 (25 mg/kg), or MCT by oral gavage, once a day for 3 wk. Mice were then monitored for survival or sacrificed to examine tumor formation by immunohistochemistry (IHC). (D) Survival curves of MPG mice after irradiation and treatment with LDN193 (n = 8), LDN214 (n = 8), or MCT (n = 8). P < 0.0001, log-rank test. (E and F) Cerebella in MPG mice were harvested after irradiation and treatment with LDN193, LDN214, or MCT to examine tumor relapse by H&E staining (upper panels) and to detect astrocytes (BLBP+, lower panels) by immunohistochemistry (E). Arrows point to ectopic lesions in mutant cerebella (upper panels). Images in lower panels correspond to boxed regions in H&E stains of upper panels. DAPI was used to counterstain cell nuclei. Representative images from five experiments are shown. The number of astrocytes per section was quantified (F; n = 5). ***, P < 0.001 by Student’s t test. (G–I) PDX models of SHH-MB were irradiated and treated with LDN193, LDN214, or MCT as a control. The survival curves of CB17/SCID mice after the drug treatment (G; P < 0.0001, log-rank test). 3 wk after irradiation, mouse cerebella were harvested for H&E staining (upper panels) and immunohistochemistry (lower panels) to detect astrocytes (BLBP+; H). Arrows point to the tumor mass (in the control cerebellum) or injection sites (in the cerebella after treatment with LDN193 or LDN214). The lower panels correspond to boxed regions in upper panels. Representative images from three experiments are shown. The number of astrocytes per cerebellar section was quantified (I; ***, P < 0.001 by Student’s t test; n = 3). Scale bars: 30 µm (A); 1 mm (E and H, upper panels) and 25 µm (E and H, lower panels).
Figure S4.
Tumor cell proliferation and TA number were not altered in primary MB after treatment with LDN193 or LDN214.(A and B) MB cells from Math1-Cre/Ptch1fl/fl mice were treated with DMSO, LDN193, or LDN214 (both at 1 µM) for 48 h and pulse-labeled with EdU for 2 h before being harvested to examine EdU incorporation by immunocytochemistry (A). DAPI was used to counterstain cell nuclei. Representative images from four experiments are shown. The percentage of EdU+ cells in tumor cells treated with DMSO, LDN193, or LDN214 was quantified (n = 4; B). (C–E) Math1-Cre/Ptch1fl/fl mice, 4 wk of age, were treated with 25 mg/kg LDN193 (n = 5), LDN214 (n = 4), or MCT as a control (n = 6) by oral gavage once a day for 3 wk. After the treatment, tumor tissues were collected to examine astrocytes (S100β+) by immunohistochemistry (C). DAPI was used to counterstain cell nuclei. Representative images from four experiments are shown. The percentage of astrocytes (S100β+) in tumor tissue was quantified (n = 4; D). Survival curves of Math1-Cre/Ptch1fl/fl mice after treatment with LDN193, LDN214, or MCT (E). Scale bars: 20 µm (A); 15 µm (C).
We then examined whether MB relapse is suppressed after inhibition of BMP signaling with LDN193 or LDN214. For this purpose, we irradiated MPG mice at 4 wk of age, followed by treatment with 25 mg/kg LDN193, LDN214, or vehicle (0.5% methylcellulose plus 0.2% Tween-80 [MCT], n = 8 in each group) once a day for 3 wk (Fig. 7 C). The effective brain penetration of these two compounds was recently established (Carvalho et al., 2019). MB was relapsed in all vehicle-treated mice within 2 mo after the irradiation (median survival, 35 d), whereas the majority of mice treated with LDN193 or LDN214 survived >60 d after irradiation (Fig. 7 D). These data suggest that LDN193 or LDN214 effectively prevents MB relapse in mice. After the treatment with LDN193, LDN214, or vehicle control for 3 wk, we harvested the cerebella to examine tumor relapse. As shown in Fig. 7 E, tumor cells were readily detected in the cerebella treated with MCT, indicative of the recurrence of MB following the irradiation. As a comparison, tumors were not fully established in Ptch1-deficient mice after the treatment with LDN193 or LDN214, except a few ectopic lesions detected in the treated cerebella (Fig. 7 E). The number of TAs (BLBP+) was significantly reduced in ectopic lesions in the cerebella after LDN193 or LDN214 treatment, compared with the control (MCT treatment; Fig. 7, E and F). The expression levels of Id1 and Id2 mRNA (Fig. S5 A) as well as phosphorylation of Smad (Fig. S5 B), in mutant cerebella were significantly repressed after treatment with LDN193 or LDN214, compared with MCT treatment. These data confirm that BMP signaling in the mutant cerebella was effectively repressed by the treatment with LDN193 or LDN214. The levels of both total Sox9 and phosphorylated Sox9 also declined in the cerebella treated with LDN193 or LDN214 (Fig. S5 C). As a comparison, Math1-Cre/Ptch1fl/fl mice were treated with 25 mg/kg LDN193, LDN214, or MCT by oral gavage for a week. The number of TAs was not altered in primary tumors after treatment with LDN193 or LDN214, compared with the control (Fig. S4, C and D). These data suggest that astrocytogenesis in primary MB is not affected by BMP pathway inhibition.
Figure S5.
BMP pathway was inhibited in tumor tissue after treatment with LDN193 or LDN214.(A–C) Cerebella in MPG mice after irradiation and drug treatment were harvested to examine the expression of Id1 and Id2 mRNAs by Q-PCR (A) and levels of phosphorylated Smad (B) and Sox9 (total Sox9 and phosphorylated Sox9; C) by Western blotting. Representative images from four experiments are shown. Levels of pSmad and pSox9 were quantified and normalized to the GAPDH loading control (right panels in B and C; n = 4). *, P < 0.05; ***, P < 0.001, Student’s t test.
We next generated patient-derived xenograft (PDX) MB models by transplanting human SHH-MB cells (ICb-5610MB) into the cerebella of CB17/SCID mice. 3 wk following the transplantation, we irradiated the recipient cerebella and treated the mice with LDN193, LDN214, or vehicle control (MCT) by oral gavage (once a day for 3 wk). Finally, tumors relapsed in all mice after control treatment (5/5; median survival, 45 d; Fig. 7 G). However, tumors were detected in only two of five mice treated with LDN193 or LDN214 (Fig. 7 G), suggesting that tumor relapse was repressed after inhibition of the BMP pathway. We then collected the cerebella at 3 wk following the drug treatment. As shown in Fig. 7 H, a relapsed tumor was found in the cerebellum after control treatment. Moreover, astrocytes were readily detected in the tumor. However, only microscopic tumor foci were observed around the injection sites in the cerebella after treatment with LDN193 or LDN214 (Fig. 7, H and I). Very few astrocytes were detected in those ectopic foci in the cerebella. These data further confirm that BMP pathway inhibitors block the transdifferentiation from MB cells and suppress tumor relapse (Fig. 7 H).
Discussion
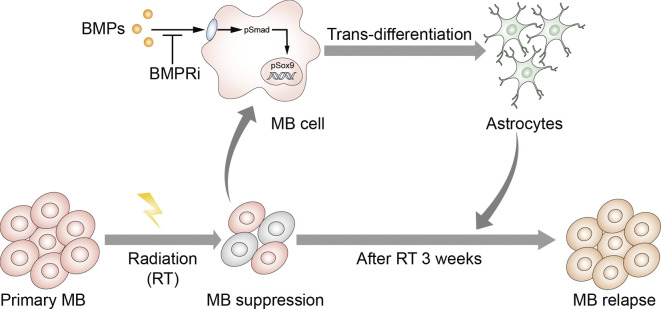
Astrocytes are important for normal brain development as well as pathological processes in the central nervous system such as neurodegenerative diseases and brain tumors. Previous studies established that TAs play a critical role in supporting the progression of MB (Gronseth et al., 2020; Liu et al., 2017). Our lineage-tracing analyses in this study suggest that TAs in primary MB may migrate from the surrounding brain tissues. However, TAs were found to originate from tumor cells in MB reestablished subcutaneously or intracranially after the transplantation. More importantly, TAs were also derived from tumor cells in relapsed MB, as further confirmed by sc-RNAseq of tumor cells from primary and relapsed MB. These studies, for the first time, reveal the distinct origins of TAs and underlying mechanisms of astrocytogenesis in relapsed MB (Fig. 8). Our studies here suggest that the tumor microenvironment, especially astrocytes with different cellular sources, may also contribute to tumor relapse.
Figure 8.
Schematic diagram showing the transdifferentiation of tumor cells during MB relapse. During MB recurrence after irradiation, the BMP pathway is activated in tumor cells. BMP signaling induces the phosphorylation of Sox9, which drives the transdifferentiation of tumor cells into astrocytes. Astrocytes promote the proliferation of MB cells, supporting tumor relapse. Inhibitors of BMP receptors (BMPRi) block tumor cell transdifferentiation, thereby suppressing the recurrence of MB.
Relapsed MB (including local and metastatic recurrence) is almost always fatal, despite a multitude of therapies including re-resection or high-dosage irradiation or chemotherapy (Pizer et al., 2011). Although MB does not change subgroups at the time of recurrence, therapeutic responses of relapsed MB are very different from those of primary MB (Ramaswamy et al., 2013). Our studies suggest that TAs with different cells of origin may be involved in the divergent susceptibility to therapy in primary and relapsed MB. Indeed, in our studies, TAs have Ptch1 deficiency and become reactivated in relapsed MB. Future studies are needed to further investigate whether this can result in distinct therapeutic responses of relapsed tumors. In addition, the Taylor group demonstrated that MB can metastasize remotely through circulating tumor cells, in addition to dissemination through the cerebrospinal fluid (Garzia et al., 2018). Extracranial metastasis, including bone, lymph nodes, and lung, of human MB has been previously reported (Rochkind et al., 1991; Rudin et al., 2009). Due to the lack of enough metastasis samples and relevant mouse models, little is known about how the MB microenvironment is built in extracranial locations. It is intriguing to think that tumor cells may generate those stromal cells to facilitate tumor establishment outside of brain, as evidenced by our findings in the subcutaneous MB model.
Granule neuron–lineage commitment appears to be critical for the tumorigenesis of SHH-MB (Schüller et al., 2008; Yang et al., 2008). Tumor cells in SHH-MB are generally believed to be neuronal-lineage restricted. However, several groups have identified a population of cancer stem cells responsible for tumor progression as well as tumor relapse (Arnold et al., 2011; Ward et al., 2009; Zhang et al., 2019). Those studies indicated the plasticity of tumor cells in SHH-MB and the essential functions of tumor cell multipotency in MB development. Here, our studies demonstrated that MB cells can transdifferentiate into astrocytes, further confirming the multipotency of tumor cells in MB.
BMP signaling has complex functions in cerebellar development and MB progression. BMP2 represses the hedgehog pathway–induced proliferation of cerebellar GNPs, through regulation of N-Myc or phosphorylation of Smad5 (Alvarez-Rodríguez et al., 2007; Rios et al., 2004). In addition, BMP2 and BMP4 inhibit the proliferation of cerebellar GNPs and MB cells through rapid degradation of Math1 (Zhao et al., 2008). Further, BMP2 has also been found to induce apoptosis in MB cell lines (Hallahan et al., 2003). In our studies, we found that BMPs including BMP2, BMP5, and BMP7 can induce the transdifferentiation of MB cells into astrocytes in vitro. Pharmaceutical inhibition of BMP signaling significantly repressed tumor cell transdifferentiation during MB relapse. These findings reveal a novel function of BMP signaling in driving the transdifferentiation of tumor cells in relapsed MB.
Previous studies found that Sox9 is a master regulator in triggering the switch from neurogenic to gliogenic program at the germinal zone of different neural tissues (Deneen et al., 2006; Kang et al., 2012; Stolt et al., 2003). Forced expression of NF1 and Sox9 in neural progenitors, or even in cultured mouse fibroblasts, promoted astrocyte generation (Caiazzo et al., 2015). Ectopic expression of NF1A and Sox9 repressed neuronal differentiation and facilitated astrocytogenesis from human pluripotent stem cells (Li et al., 2018). Sox9 was found to be predominantly expressed by astrocytes (Sun et al., 2017); however, we found basal levels of Sox9 expression in MB cells, despite more intense expression of Sox9 in astrocytes. Sox9 mRNA expression was significantly enhanced in astrocytes from relapsed MB, compared with their counterparts in primary MB. By using conditional knockout mice, we demonstrated that Sox9 deletion in MB cells did not affect their proliferation. However, Sox9 deletion clearly blocked the transdifferentiation of MB cells into astrocytes in vivo and in vitro, highlighting the essential role of Sox9 in the transdifferentiation of tumor cells. In our studies, BMP treatment markedly increased the phosphorylation of Sox9 in tumor cells. Forced expression of a phosphomimetic form of Sox9 (but not a wild-type form of Sox9) effectively stimulated the transdifferentiation of MB cells into astrocytes. Moreover, expression of Sox9 with a mutation at the phosphorylation site abolished BMP-induced transdifferentiation in MB cells. These results demonstrate the important role of Sox9 phosphorylation in BMP-induced transdifferentiation of MB cells, consistent with recent studies that Sox9 phosphorylation is required for the transcriptional functions of Sox9 in neural crest delamination (Liu et al., 2013; Zuo et al., 2018).
The Zong group recently demonstrated the transdifferentiation of tumor cells in SHH-MB using a mouse genetic system called mosaic analysis with double markers (Yao et al., 2020). We found only a small population of TAs in mouse primary MB in our studies; however, almost all TAs originated from tumor cells in the primary MB model in the studies of Zong and colleagues. This could be due to the different MB mouse models used in our laboratory and the Zong group (Yao et al., 2020). We used Ptch1-deficient mice (including Ptch1+/− and Math1-Cre/Ptch1fl/fl) that develop MB with no P53 mutations, whereas Ptch1+/−, P53−/− mice were used in the study of the Zong group. P53 deficiency could promote the dedifferentiation or transdifferentiation of tumor cells as previously reported (Piechowski, 2017; Rasmussen et al., 2014; Tapia and Schöler, 2010). Future studies are needed to investigate whether P53 mutations affect the cellular origins of TAs in human MB. Nevertheless, the capacity of MB cell transdifferentiation into astrocytes is established in both studies. In our studies, almost all tumor cells were found to transdifferentiate into astrocytes following BMP treatment in vitro, suggesting that the transdifferentiation capacity is not limited to particular subsets of tumor cells. However, the number of TAs was not overall increased in relapsed MB, suggesting that the transdifferentiation of tumor cells is tightly controlled during MB recurrence. Based on our sc-RNAseq data, the expression of BMPs was up-regulated in microglia from relapsed MB, compared with their counterparts in primary MB (not depicted). It is possible that microglia secrete BMPs and induce the transdifferentiation of tumor cells during tumor relapse. We are currently investigating the role of microglia in the transdifferentiating and recurrence of MB cells.
In our studies, in vitro treatment with LDN-193 or LDN-214 caused no alterations in the survival and proliferation of tumor cells and TAs from either primary or relapsed MB (Fig. S4 and not depicted). However, treatment with LDN-193 or LDN-214 following the irradiation effectively blocked tumor cell transdifferentiation and suppressed tumor relapse. These findings further support the essential role of BMP signaling underlying the transdifferentiation of tumor cells into astrocytes. In addition, our findings suggest that inhibition of the BMP pathway may prevent the recurrence of SHH-MB. Future studies are necessary to test the efficacies of BMP pathway inhibitors in the treatment of relapsed MB after tumor is reestablished (with preexisting transdifferentiation of tumor cells). Nevertheless, our studies demonstrate the good potency of LDN-193 and LDN-214 in repressing BMP signaling as well as in inhibiting MB recurrence. With attractive physicochemical properties consistent with their blood–brain barrier penetration and good pharmacokinetics (Carvalho et al., 2019), LDN-193 and LDN-214 represent promising lead compounds in developing agents to prevent MB recurrence.
Materials and methods
Data availability
Transcriptional data generated in this study, including RNAseq data and sc-RNAseq data, were deposited in NCBI Gene Expression Omnibus database under accession no. GSE175416.
Mice and relapsed tumor models
Math1-CreERT2, Math1-Cre, Rosa-GFP, Sox9fl/fl, ptch1+/−, ptch1fl/fl, and Rosa-GFP mice were purchased from The Jackson Laboratory. All animals were maintained in the Laboratory Animal Facility at Fox Chase Cancer Center, and all experiments were performed in accordance with procedures approved by the Fox Chase Cancer Center Animal Care and Use Committee. All mice were bred and genotyped as recommended by The Jackson Laboratory. For all mouse studies, mice of either sex were used. Ages of all mice used in experiments are indicated in the figure legends.
Two types of ptch1-deficient mice were used in this study, including Math1-Cre/ptch1fl/fl mice that develop tumors at 6–8 wk of age with 100% penetrance (Yang et al., 2008) and ptch1+/− mice that develop MB at 20–27 wk with tumor incidence of 15–20% (Goodrich et al., 1997). There is no significant difference in the percentage of astrocytes and Sox9+ cells in tumor tissues from these mice. To generate relapsed MB models, ptch1+/− mice or mice bearing PDX were irradiated once with 2 Gy γ rays from a cesium-137 source, when the mice exhibited signs for cerebellar tumors including ataxia and domed head (∼18-22 wk of age). Math1-Cre/ptch1fl/fl mice, 4 wk of age, were irradiated once with 2 Gy γ rays from a cesium-137 source.
Magnetic resonance imaging (MRI)
Tumor formation in ptch1-deficient mice was examined by MRI in some experiments. For MRI of mouse brains, anesthesia was performed using ketamine (100 mg/kg). All mice were kept in a supine position. ptch1-deficient mice were sagittal scanned in a GE MRI scanner. MRI analyses were performed using a T2-fast spin echo sequence. Some parameters were repetition time, 3,450 ms; echo time, 159 ms; and 12 slices at 0.8 mm per slice. Generated images were analyzed using a media viewer (GE Healthcare).
Cell culture and drug treatment
For examining in vivo efficacies, stock solutions of LDN193 (in DPBS, 100 mg/ml) and LDN214 (in DMSO, 100 mg/ml) were diluted using MCT and administrated to mice by oral gavage. Tamoxifen (T5648; Sigma-Aldrich) was dissolved at a concentration of 20 mg/ml in corn oil and administrated by oral gavage at 200 mg/kg. For survival analyses, mice after treatment with tamoxifen or BMP inhibitors were monitored daily and sacrificed when they exhibited brain tumor symptoms (ataxia and tilted heads). Log-rank survival analyses were performed in GraphPad Prism 7.
To isolate tumor cells, tumor tissue from Ptch1-deficient mice were digested in a papain solution to obtain a single-cell suspension and then centrifuged through a 35 and 65% Percoll gradient. Cells from the 35–65% interface were suspended in DPBS plus 0.5% BSA. Cells were then suspended in NB-B27 (neurobasal with 1 mM sodium pyruvate, 2 mM L-glutamine, B27 supplement, and 1% penicillin/streptomycin, all from Invitrogen) and plated on poly-D-lysine–coated coverslips (BD Biosciences). Human recombinant BMP2, BMP5, and BMP7 (Peprotech) were used at 80 ng/ml for in vitro treatment of tumor cells.
Human tumor samples
All human patient samples (including two paired and relapsed SHH-MB tissues) were obtained from the Children’s Brain Tumor Tissue Consortium at the Children’s Hospital of Philadelphia, with consent under approval and oversight by the institutional review board committees of the Children’s Hospital of Philadelphia. The SHH type of human MB samples was identified by immunochemistry GAB1 and YAP. Human SHH-MB cells (ICb-5610MB), kindly provided by Dr. Xiaonan Li from Northwestern University, Evanston, IL, were used to generate PDX MB models by intracranial transplantation.
Immunostaining, flow cytometry, and Western blotting
Immunofluorescent staining of sections and cells was performed according to standard methods. Briefly, sections or cells were blocked and permeabilized for 1 h with PBS containing 0.1% Triton X-100 and 10% normal goat serum, stained with primary antibodies overnight at 4°C, and incubated with secondary antibodies for 2 h at room temperature. Sections or cells were counterstained with DAPI and mounted with Fluoromount-G (Southern Biotech) before being visualized using a Nikon Eclipse Ti microscope. For purifying astrocytes, tumor cell suspension was incubated with the primary antibody against ACSA2 (1:200; Cell Signaling) on ice for 30 min. After washing with PBS three times, tumor cells were incubated with Texas Red–conjugated secondary antibody for 30 min. Astrocytes (ACSA2+) were then fractionated by FACS. Details of all other antibodies are available upon request.
For Western blot analysis, cells were lysed in radioimmunoprecipitation assay buffer (Thermo Fisher Scientific) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific). Total lysate containing equal amounts of protein were separated by SDS-PAGE gel and subsequently transferred onto polyvinylidene fluoride membrane. Membranes were then subjected to probing with antibodies. Western blot signals were detected by using SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher Scientific) and exposed on films.
Q-PCR
RNA was isolated using the RNAqueous kit (Ambion) and treated with DNA-free DNase (Ambion). cDNA was synthesized using oligo(dT) and Superscript II reverse transcription (Invitrogen). Q-PCR reactions were performed in triplicate using iQ SYBR Green Supermix (Bio-Rad) and the Bio-Rad iQ5 Multicolor Real-Time PCR Detection System. Primer sequences are available upon request.
Three paired primary and relapsed human MB (SHH-group) tissues were provided by the Children’s Brain Tumor Tissue Consortium at the Children’s Hospital of Philadelphia. Total RNAs were extracted from tumor tissues using the RNAqueous kit to examine the expression of BMPs by Q-PCR.
sc-RNAseq and data analyses
Viable single cells were loaded on to a 10x Genomics Chromium Controller instrument to generate single-cell barcoded droplets (gel beads in emulsion [GEMs]) using the 10x Single Cell 3′ Gel Bead Kit and 10x Chromium system at GENEWIZ following the manufacturer’s protocol. Briefly, single cells were partitioned into GEMs in the Chromium Controller instrument with cell lysis and barcoded reverse transcription of RNA, followed by cDNA amplification, shearing, library construction, and indexing by PCR amplification. Subsequently, libraries were sequenced by Illumina 2× 150-bp sequencing.
The Cell Ranger Single Cell Software Suite v2.0.1 (https://support.10xgenomics.com/) by 10x Genomics was used to perform sample demultiplexing, barcode processing, and single-cell 3′ gene counting. In brief, base call files in FASTQ format were aligned to the mm10 reference genome that used an aligner called STAR with default settings, and aligned reads were filtered for valid cell barcodes and a unique molecular identifier (UMI) to generate filtered gene–barcode matrices. The preprocessed 10x and DGE matrices were then imported using the Seurat package for the R programming language (v3.5.4), and the data were filtered to include genes detected in >5 cells, cells with ∼500–6,000 detected genes, and UMIs with ∼1,000–30,000 and <15% mitochondrial genes. For single-dataset analyses, log-normalized expression values were obtained by the NormalizeData function, and variable genes were identified using the FindVariableGenes Seurat function. Transcripts per million values were obtained from raw UMI counts by normalizing with total UMI counts per cell. Next, PCA was performed, and significant principal components were used as input for graph-based clustering. 2D visualization of the multidimensional dataset was done with tSNE. For clustering, we used the function FindClusters that implements a shared-nearest-neighbor modularity optimization–based clustering algorithm on 15 PCA components with resolutions of 0.1–0.5, leading to 5–12 clusters. A resolution of 0.25 was chosen for the analysis.
Profiling differentially expressed genes within each cell cluster
To identify differentially expressed genes in each cell cluster, we applied the FindAllMarkers function from Seurat to the normalized gene expression data. We selected a power value >0.25 as the cutoff for gene selection. The gene expression levels plotted by packages in R. For cell type analysis, we performed differentially expressed gene analysis by comparing each cluster with the other clusters using the Wilcoxon rank sum test. Genes with scaled dispersion >0.25 and log-normalized average expression >0.01 were included in the analyses. For each cluster, differentially expressed genes with top 1.5-fold changes were designated as a cell type signature, Thresholds were set as P < 0.05. If a cell type signature gene overlapped among different clusters, the gene was assigned to the cluster having the highest expression value. To characterize clusters, we used known marker genes that were reported to compare the differentially expressed gene using GeneQuery and Enrichment analysis. For heatmap representation, normalized expression of markers inside each cluster was used.
RNAseq
Total RNA was extracted from FACS-purified astrocytes in mouse primary and relapsed MB. Strand-specific mRNAseq libraries for the Illumina platform were generated and sequenced at GENEWIZ following the manufacturer’s protocol. High-quality total RNA was used as input for the so-called dUTP (2'-deoxyuridine 5'-triphosphate) library preparation method. Briefly, the mRNA fraction was purified from total RNA by polyA capture, fragmented, and subjected to first-strand cDNA synthesis with random hexamers in the presence of actinomycin D. Second-strand synthesis was performed incorporating dUTP instead of 2'-deoxythymidine 5'-triphosphate. Barcoded DNA adapters were ligated to both ends of the double-stranded cDNA and subjected to PCR amplification. The resultant library was checked on a Bioanalyzer (Agilent) and quantified. The libraries were multiplexed, clustered, and sequenced on an Illumina HiSeq 2000. The sequencing run was analyzed with the Illumina CASAVA pipeline (v1.8.2), with demultiplexing based on sample-specific barcodes. The raw sequencing data produced was processed by removing the sequence reads that were of too-low quality (only “passing filter” reads were selected) and discarding reads containing adaptor sequences or PhiX control with an in-house filtering protocol.
Quantification and statistical analysis
Student’s t test was performed to determine the statistical significance of the difference, unless stated otherwise. P < 0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). Error bars represent SEM. Data handling and statistical processing were performed using Microsoft Excel and GraphPad Prism software. Details of specific tests and sample numbers are reported in figure legends or in the main text.
Online supplemental material
Fig. S1 shows the apoptosis of astrocytes in primary MB after irradiation; expression of SoxE genes (Sox8, Sox9, and Sox10); and expression of levels of Sox9 protein in astrocytes from primary and relapsed MB. Fig. S2 shows the effects of Sox9 deletion on tumor cell proliferation and differentiation and the abundance of astrocytes and animal survival in MPS mice bearing primary MB. Fig. S3 shows the repression of BMP signaling in tumor cells after treatment with LDN193 or LDN214 and the inhibition of Sox9 phosphorylation in tumor cells following treatment with LDN193 or LDN214. Fig. S4 shows the unaltered proliferation of tumor cells and unchanged number of TAs in primary MB after treatment with LDN193 or LDN214. Fig. S5 shows the inhibition of BMP signaling in MB tissue after treatment with LDN193 or LDN214. Table S1 lists the genes that are up-regulated in relapsed human MB tissue compared with paired primary MB tissue.
Supplementary Material
lists the genes that are up-regulated in relapsed human MB tissue compared with paired primary MB tissue.
Acknowledgments
We would like to thank Drs. Andrey Efimov and James Oesterling for technical assistance, the Children’s Brain Tumor Tissue Consortium at the Children’s Hospital of Philadelphia for providing human MB tissues, and GENEWIZ for sc-RNAseq.
This research was supported by grants from the American Cancer Society (RSG1605301NEC), American Brain Tumor Association (DG1900025), and Pennsylvania CURE Health Research Fund (CURE 4100068716).
Author contributions: Z.-j. Yang and Y. Zhao conceived the project and designed the experiments. D. Guo, Y. Wang, Y. Cheng, S. Liao, J. Hu, F. Du, G. Xu, Y. Liu, and K.Q. Cai performed the experiments and analyzed the data. S. Liao and Y. Zhao performed biostatistical analyses of sequencing data. M. Cheung, B.J. Wainwright, and Q.R. Lu provided essential reagents and critical consultation. All authors contributed to writing the manuscript.
References
- Akiyama, H., Chaboissier M.C., Martin J.F., Schedl A., and de Crombrugghe B.. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16:2813–2828. 10.1101/gad.1017802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Rodríguez, R., Barzi M., Berenguer J., and Pons S.. 2007. Bone morphogenetic protein 2 opposes Shh-mediated proliferation in cerebellar granule cells through a TIEG-1-based regulation of Nmyc. J. Biol. Chem. 282:37170–37180. 10.1074/jbc.M705414200 [DOI] [PubMed] [Google Scholar]
- Arnold, K., Sarkar A., Yram M.A., Polo J.M., Bronson R., Sengupta S., Seandel M., Geijsen N., and Hochedlinger K.. 2011. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 9:317–329. 10.1016/j.stem.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruga, J., Yokota N., Hashimoto M., Furuichi T., Fukuda M., and Mikoshiba K.. 1994. A novel zinc finger protein, zic, is involved in neurogenesis, especially in the cell lineage of cerebellar granule cells. J. Neurochem. 63:1880–1890. 10.1046/j.1471-4159.1994.63051880.x [DOI] [PubMed] [Google Scholar]
- Barry, D., and McDermott K.. 2005. Differentiation of radial glia from radial precursor cells and transformation into astrocytes in the developing rat spinal cord. Glia. 50:187–197. 10.1002/glia.20166 [DOI] [PubMed] [Google Scholar]
- Caiazzo, M., Giannelli S., Valente P., Lignani G., Carissimo A., Sessa A., Colasante G., Bartolomeo R., Massimino L., Ferroni S., et al. 2015. Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Reports. 4:25–36. 10.1016/j.stemcr.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, D., Taylor K.R., Olaciregui N.G., Molinari V., Clarke M., Mackay A., Ruddle R., Henley A., Valenti M., Hayes A., et al. 2019. ALK2 inhibitors display beneficial effects in preclinical models of ACVR1 mutant diffuse intrinsic pontine glioma. Commun. Biol. 2:156. 10.1038/s42003-019-0420-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Franco-Barraza J., Wang Y., Zheng C., Zhang L., Qu Y., Long Y., Cukierman E., and Yang Z.J.. 2020a. Sustained hedgehog signaling in medulloblastoma tumoroids is attributed to stromal astrocytes and astrocyte-derived extracellular matrix. Lab. Invest. 100:1208–1222. 10.1038/s41374-020-0443-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Liao S., Xu G., Hu J., Guo D., Du F., Contreras A., Cai K.Q., Peri S., Wang Y., et al. 2020b. NeuroD1 dictates tumor cell differentiation in medulloblastoma. Cell Rep. 31:107782. 10.1016/j.celrep.2020.107782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen, B., Ho R., Lukaszewicz A., Hochstim C.J., Gronostajski R.M., and Anderson D.J.. 2006. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 52:953–968. 10.1016/j.neuron.2006.11.019 [DOI] [PubMed] [Google Scholar]
- Du, F., Yuelling L., Lee E.H., Wang Y., Liao S., Cheng Y., Zhang L., Zheng C., Peri S., Cai K.Q., et al. 2019. Leukotriene Synthesis Is Critical for Medulloblastoma Progression. Clin. Cancer Res. 25:6475–6486. 10.1158/1078-0432.CCR-18-3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebendal, T., Bengtsson H., and Söderström S.. 1998. Bone morphogenetic proteins and their receptors: potential functions in the brain. J. Neurosci. Res. 51:139–146. [DOI] [PubMed] [Google Scholar]
- Garzia, L., Kijima N., Morrissy A.S., De Antonellis P., Guerreiro-Stucklin A., Holgado B.L., Wu X., Wang X., Parsons M., Zayne K., et al. 2018. A Hematogenous Route for Medulloblastoma Leptomeningeal Metastases. Cell. 173:1549. 10.1016/j.cell.2018.05.033 [DOI] [PubMed] [Google Scholar]
- Goodrich, L.V., Milenković L., Higgins K.M., and Scott M.P.. 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 277:1109–1113. 10.1126/science.277.5329.1109 [DOI] [PubMed] [Google Scholar]
- Gordon, R.E., Zhang L., Peri S., Kuo Y.M., Du F., Egleston B.L., Ng J.M.Y., Andrews A.J., Astsaturov I., Curran T., and Yang Z.J.. 2018. Statins Synergize with Hedgehog Pathway Inhibitors for Treatment of Medulloblastoma. Clin. Cancer Res. 24:1375–1388. 10.1158/1078-0432.CCR-17-2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronseth, E., Gupta A., Koceja C., Kumar S., Kutty R.G., Rarick K., Wang L., and Ramchandran R.. 2020. Astrocytes influence medulloblastoma phenotypes and CD133 surface expression. PLoS One. 15:e0235852. 10.1371/journal.pone.0235852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallahan, A.R., Pritchard J.I., Chandraratna R.A., Ellenbogen R.G., Geyer J.R., Overland R.P., Strand A.D., Tapscott S.J., and Olson J.M.. 2003. BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat. Med. 9:1033–1038. 10.1038/nm904 [DOI] [PubMed] [Google Scholar]
- Heldin, C.H., Miyazono K., and ten Dijke P.. 1997. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 390:465–471. 10.1038/37284 [DOI] [PubMed] [Google Scholar]
- Hovestadt, V., Smith K.S., Bihannic L., Filbin M.G., Shaw M.L., Baumgartner A., DeWitt J.C., Groves A., Mayr L., Weisman H.R., et al. 2019. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature. 572:74–79. 10.1038/s41586-019-1434-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, A., Denduluri S., Zhang B., Wang Z., Yin L., Yan Z., Kang R., Shi L.L., Mok J., Lee M.J., and Haydon R.C.. 2014. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 1:149–161. 10.1016/j.gendis.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, P., Lee H.K., Glasgow S.M., Finley M., Donti T., Gaber Z.B., Graham B.H., Foster A.E., Novitch B.G., Gronostajski R.M., and Deneen B.. 2012. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 74:79–94. 10.1016/j.neuron.2012.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Tang J.Y., Gong R., Kim J., Lee J.J., Clemons K.V., Chong C.R., Chang K.S., Fereshteh M., Gardner D., et al. 2010. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell. 17:388–399. 10.1016/j.ccr.2010.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool, M., Jones D.T., Jäger N., Northcott P.A., Pugh T.J., Hovestadt V., Piro R.M., Esparza L.A., Markant S.L., Remke M., et al. ICGC PedBrain Tumor Project . 2014. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 25:393–405. 10.1016/j.ccr.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D., and Lassar A.B.. 2009. The transcriptional activity of Sox9 in chondrocytes is regulated by RhoA signaling and actin polymerization. Mol. Cell. Biol. 29:4262–4273. 10.1128/MCB.01779-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Tao Y., Bradley R., Du Z., Tao Y., Kong L., Dong Y., Jones J., Yan Y., Harder C.R.K., et al. 2018. Fast Generation of Functional Subtype Astrocytes from Human Pluripotent Stem Cells. Stem Cell Reports. 11:998–1008. 10.1016/j.stemcr.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow, S.A., and Barres B.A.. 2017. Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity. 46:957–967. 10.1016/j.immuni.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Liu, J.A., Wu M.H., Yan C.H., Chau B.K., So H., Ng A., Chan A., Cheah K.S., Briscoe J., and Cheung M.. 2013. Phosphorylation of Sox9 is required for neural crest delamination and is regulated downstream of BMP and canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 110:2882–2887. 10.1073/pnas.1211747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Yuelling L.W., Wang Y., Du F., Gordon R.E., O’Brien J.A., Ng J.M.Y., Robins S., Lee E.H., Liu H., et al. 2017. Astrocytes Promote Medulloblastoma Progression through Hedgehog Secretion. Cancer Res. 77:6692–6703. 10.1158/0008-5472.CAN-17-1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, D.N., Ohgaki H., Wiestler O.D., Cavenee W.K., Burger P.C., Jouvet A., Scheithauer B.W., and Kleihues P.. 2007. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114:97–109. 10.1007/s00401-007-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold, R., and Fishell G.. 2005. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 48:17–24. 10.1016/j.neuron.2005.08.028 [DOI] [PubMed] [Google Scholar]
- Mehler, M.F., Mabie P.C., Zhang D., and Kessler J.A.. 1997. Bone morphogenetic proteins in the nervous system. Trends Neurosci. 20:309–317. 10.1016/S0166-2236(96)01046-6 [DOI] [PubMed] [Google Scholar]
- Mohedas, A.H., Wang Y., Sanvitale C.E., Canning P., Choi S., Xing X., Bullock A.N., Cuny G.D., and Yu P.B.. 2014. Structure-activity relationship of 3,5-diaryl-2-aminopyridine ALK2 inhibitors reveals unaltered binding affinity for fibrodysplasia ossificans progressiva causing mutants. J. Med. Chem. 57:7900–7915. 10.1021/jm501177w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy, A.S., Garzia L., Shih D.J., Zuyderduyn S., Huang X., Skowron P., Remke M., Cavalli F.M., Ramaswamy V., Lindsay P.E., et al. 2016. Divergent clonal selection dominates medulloblastoma at recurrence. Nature. 529:351–357. 10.1038/nature16478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott, P.A., Jones D.T., Kool M., Robinson G.W., Gilbertson R.J., Cho Y.J., Pomeroy S.L., Korshunov A., Lichter P., Taylor M.D., and Pfister S.M.. 2012. Medulloblastomics: the end of the beginning. Nat. Rev. Cancer. 12:818–834. 10.1038/nrc3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano-Uchida, T., Himi T., Komiya Y., and Ishizaki Y.. 2004. Cerebellar granule cell precursors can differentiate into astroglial cells. Proc. Natl. Acad. Sci. USA. 101:1211–1216. 10.1073/pnas.0307972100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q., Yu Y., Chen Q., Li C., Wu H., Wan Y., Ma J., and Sun F.. 2008. Sox9, a key transcription factor of bone morphogenetic protein-2-induced chondrogenesis, is activated through BMP pathway and a CCAAT box in the proximal promoter. J. Cell. Physiol. 217:228–241. 10.1002/jcp.21496 [DOI] [PubMed] [Google Scholar]
- Piechowski, J.2017. Hypothesis about Transdifferentiation As Backbone of Malignancy. Front. Oncol. 7:126. 10.3389/fonc.2017.00126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizer, B., Donachie P.H., Robinson K., Taylor R.E., Michalski A., Punt J., Ellison D.W., and Picton S.. 2011. Treatment of recurrent central nervous system primitive neuroectodermal tumours in children and adolescents: results of a Children’s Cancer and Leukaemia Group study. Eur. J. Cancer. 47:1389–1397. 10.1016/j.ejca.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Ramaswamy, V., Remke M., Bouffet E., Faria C.C., Perreault S., Cho Y.J., Shih D.J., Luu B., Dubuc A.M., Northcott P.A., et al. 2013. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 14:1200–1207. 10.1016/S1470-2045(13)70449-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, M.A., Holst B., Tümer Z., Johnsen M.G., Zhou S., Stummann T.C., Hyttel P., and Clausen C.. 2014. Transient p53 suppression increases reprogramming of human fibroblasts without affecting apoptosis and DNA damage. Stem Cell Reports. 3:404–413. 10.1016/j.stemcr.2014.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios, I., Alvarez-Rodríguez R., Martí E., and Pons S.. 2004. Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling. Development. 131:3159–3168. 10.1242/dev.01188 [DOI] [PubMed] [Google Scholar]
- Rochkind, S., Blatt I., Sadeh M., and Goldhammer Y.. 1991. Extracranial metastases of medulloblastoma in adults: literature review. J. Neurol. Neurosurg. Psychiatry. 54:80–86. 10.1136/jnnp.54.1.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin, C.M., Hann C.L., Laterra J., Yauch R.L., Callahan C.A., Fu L., Holcomb T., Stinson J., Gould S.E., Coleman B., et al. 2009. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 361:1173–1178. 10.1056/NEJMoa0902903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer, B., and Hill C.S.. 2007. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8:970–982. 10.1038/nrm2297 [DOI] [PubMed] [Google Scholar]
- Schüller, U., Heine V.M., Mao J., Kho A.T., Dillon A.K., Han Y.G., Huillard E., Sun T., Ligon A.H., Qian Y., et al. 2008. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 14:123–134. 10.1016/j.ccr.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, C.E., Wynn S.L., Sesay A., Cruz C., Cheung M., Gomez Gaviro M.V., Booth S., Gao B., Cheah K.S., Lovell-Badge R., and Briscoe J.. 2010. SOX9 induces and maintains neural stem cells. Nat. Neurosci. 13:1181–1189. 10.1038/nn.2646 [DOI] [PubMed] [Google Scholar]
- Selvadurai, H.J., Luis E., Desai K., Lan X., Vladoiu M.C., Whitley O., Galvin C., Vanner R.J., Lee L., Whetstone H., et al. 2020. Medulloblastoma Arises from the Persistence of a Rare and Transient Sox2+ Granule Neuron Precursor. Cell Rep. 31:107511. 10.1016/j.celrep.2020.03.075 [DOI] [PubMed] [Google Scholar]
- Sharma, K., Schmitt S., Bergner C.G., Tyanova S., Kannaiyan N., Manrique-Hoyos N., Kongi K., Cantuti L., Hanisch U.K., Philips M.A., et al. 2015. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 18:1819–1831. 10.1038/nn.4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt, C.C., Lommes P., Sock E., Chaboissier M.C., Schedl A., and Wegner M.. 2003. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17:1677–1689. 10.1101/gad.259003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W., Cornwell A., Li J., Peng S., Osorio M.J., Aalling N., Wang S., Benraiss A., Lou N., Goldman S.A., and Nedergaard M.. 2017. SOX9 Is an Astrocyte-Specific Nuclear Marker in the Adult Brain Outside the Neurogenic Regions. J. Neurosci. 37:4493–4507. 10.1523/JNEUROSCI.3199-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryo Rahmanto, A., Savov V., Brunner A., Bolin S., Weishaupt H., Malyukova A., Rosén G., Čančer M., Hutter S., Sundström A., et al. 2016. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J. 35:2192–2212. 10.15252/embj.201693889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling, F.J., Savov V., Persson A.I., Chen J., Hackett C.S., Northcott P.A., Grimmer M.R., Lau J., Chesler L., Perry A., et al. 2012. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 21:601–613. 10.1016/j.ccr.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia, N., and Schöler H.R.. 2010. p53 connects tumorigenesis and reprogramming to pluripotency. J. Exp. Med. 207:2045–2048. 10.1084/jem.20101866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, M.D., Northcott P.A., Korshunov A., Remke M., Cho Y.J., Clifford S.C., Eberhart C.G., Parsons D.W., Rutkowski S., Gajjar A., et al. 2012. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 123:465–472. 10.1007/s00401-011-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanner, R.J., Remke M., Gallo M., Selvadurai H.J., Coutinho F., Lee L., Kushida M., Head R., Morrissy S., Zhu X., et al. 2014. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell. 26:33–47. 10.1016/j.ccr.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, J., Traynor R., and Sapkota G.P.. 2011. The specificities of small molecule inhibitors of the TGFß and BMP pathways. Cell. Signal. 23:1831–1842. 10.1016/j.cellsig.2011.06.019 [DOI] [PubMed] [Google Scholar]
- Ward, R.J., Lee L., Graham K., Satkunendran T., Yoshikawa K., Ling E., Harper L., Austin R., Nieuwenhuis E., Clarke I.D., et al. 2009. Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res. 69:4682–4690. 10.1158/0008-5472.CAN-09-0342 [DOI] [PubMed] [Google Scholar]
- Yang, Z.J., Ellis T., Markant S.L., Read T.A., Kessler J.D., Bourboulas M., Schüller U., Machold R., Fishell G., Rowitch D.H., et al. 2008. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 14:135–145. 10.1016/j.ccr.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, M., Ventura P.B., Jiang Y., Rodriguez F.J., Wang L., Perry J.S.A., Yang Y., Wahl K., Crittenden R.B., Bennett M.L., et al. 2020. Astrocytic trans-Differentiation Completes a Multicellular Paracrine Feedback Loop Required for Medulloblastoma Tumor Growth. Cell. 180:502–520.e19. 10.1016/j.cell.2019.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, N., Aruga J., Takai S., Yamada K., Hamazaki M., Iwase T., Sugimura H., and Mikoshiba K.. 1996. Predominant expression of human zic in cerebellar granule cell lineage and medulloblastoma. Cancer Res. 56:377–383. [PubMed] [Google Scholar]
- Zamanian, J.L., Xu L., Foo L.C., Nouri N., Zhou L., Giffard R.G., and Barres B.A.. 2012. Genomic analysis of reactive astrogliosis. J. Neurosci. 32:6391–6410. 10.1523/JNEUROSCI.6221-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehentner, B.K., Dony C., and Burtscher H.. 1999. The transcription factor Sox9 is involved in BMP-2 signaling. J. Bone Miner. Res. 14:1734–1741. 10.1359/jbmr.1999.14.10.1734 [DOI] [PubMed] [Google Scholar]
- Zeltzer, P.M., Boyett J.M., Finlay J.L., Albright A.L., Rorke L.B., Milstein J.M., Allen J.C., Stevens K.R., Stanley P., Li H., et al. 1999. Metastasis stage, adjuvant treatment, and residual tumor are prognostic factors for medulloblastoma in children: conclusions from the Children’s Cancer Group 921 randomized phase III study. J. Clin. Oncol. 17:832–845. 10.1200/JCO.1999.17.3.832 [DOI] [PubMed] [Google Scholar]
- Zhang, L., He X., Liu X., Zhang F., Huang L.F., Potter A.S., Xu L., Zhou W., Zheng T., Luo Z., et al. 2019. Single-Cell Transcriptomics in Medulloblastoma Reveals Tumor-Initiating Progenitors and Oncogenic Cascades during Tumorigenesis and Relapse. Cancer Cell. 36:302–318.e7. 10.1016/j.ccell.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., Etter L., Hinton R.B. Jr., and Benson D.W.. 2007. BMP and FGF regulatory pathways in semilunar valve precursor cells. Dev. Dyn. 236:971–980. 10.1002/dvdy.21097 [DOI] [PubMed] [Google Scholar]
- Zhao, H., Ayrault O., Zindy F., Kim J.H., and Roussel M.F.. 2008. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 22:722–727. 10.1101/gad.1636408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, C., Wang L., Kamalesh R.M., Bowen M.E., Moore D.C., Dooner M.S., Reginato A.M., Wu Q., Schorl C., Song Y., et al. 2018. SHP2 regulates skeletal cell fate by modifying SOX9 expression and transcriptional activity. Bone Res. 6:12. 10.1038/s41413-018-0013-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the genes that are up-regulated in relapsed human MB tissue compared with paired primary MB tissue.
Data Availability Statement
Transcriptional data generated in this study, including RNAseq data and sc-RNAseq data, were deposited in NCBI Gene Expression Omnibus database under accession no. GSE175416.