Abstract
Introduction
Coronavirus disease 2019 is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which enters host cells through the cell surface proteins ACE2 and TMPRSS2.
Methods
Using a variety of normal and malignant models and tissues from the aerodigestive and respiratory tracts, we investigated the expression and regulation of ACE2 and TMPRSS2.
Results
We find that ACE2 expression is restricted to a select population of epithelial cells. Notably, infection with SARS-CoV-2 in cancer cell lines, bronchial organoids, and patient nasal epithelium induces metabolic and transcriptional changes consistent with epithelial-to-mesenchymal transition (EMT), including up-regulation of ZEB1 and AXL, resulting in an increased EMT score. In addition, a transcriptional loss of genes associated with tight junction function occurs with SARS-CoV-2 infection. The SARS-CoV-2 receptor, ACE2, is repressed by EMT through the transforming growth factor-β, ZEB1 overexpression, and onset of EGFR tyrosine kinase inhibitor resistance. This suggests a novel model of SARS-CoV-2 pathogenesis in which infected cells shift toward an increasingly mesenchymal state, associated with a loss of tight junction components with acute respiratory distress syndrome-protective effects. AXL inhibition and ZEB1 reduction, as with bemcentinib, offer a potential strategy to reverse this effect.
Conclusions
These observations highlight the use of aerodigestive and, especially, lung cancer model systems in exploring the pathogenesis of SARS-CoV-2 and other respiratory viruses and offer important insights into the potential mechanisms underlying the morbidity and mortality of coronavirus disease 2019 in healthy patients and patients with cancer alike.
Keywords: SARS-CoV-2, EMT, ACE2, NSCLC, ZEB1, AXL
Introduction
In December 2019, reports of a viral illness with severe respiratory symptoms emerged from Wuhan, People’s Republic of China.1 , 2 The novel virus was classified as a coronavirus and subsequently named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It exhibited rapid global spread and met the WHO’s criteria of a pandemic within 3 months after the first reported diagnosis (https://covid19.who.int/). As of April 2021, more than 148 million individuals have been infected worldwide with more than 3.1 million deaths (covid19.who.int). SARS-CoV-2 infection is the cause of the respiratory illness coronavirus disease 2019 (COVID-19), which presents most frequently with symptoms including cough and dyspnea, accompanied by a moderate-to-high fever.2 , 3 The severity of patient symptoms after SARS-CoV-2 infection varies widely from asymptomatic carrier status to critical illness. Given the specific complications associated with the infection, it is not initially surprising that patients with cancer and, specifically, thoracic malignancies, seem to experience poorer clinical outcomes.4 , 5 Nevertheless, it is unclear whether these poorer outcomes represent the impact of demographics (e.g., age, smoking status, sex) or cellular and molecular changes associated with the tumor and its microenvironment.
Although there are currently no validated molecular biomarkers for susceptibility to, or severity of, SARS-CoV-2 infection, it has recently been described that, as with SARS-CoV, SARS-CoV-2 cell entry requires interactions with ACE2 and TMPRSS2 on the surface of the host cell.6 Specifically, ACE2 binds to a subunit of the SARS-CoV-2 spike (S) protein, whereas TMPRSS2 is responsible for S-protein cleavage, or priming, to allow fusion of viral and host cellular membranes.6 Differences in these receptors have been described in certain populations harboring higher allelic frequency of mutations and coding variants associated with higher ACE2 expression.7 Notably, ACE2 has previously been found to exert a protective effect toward the development of acute respiratory distress syndrome (ARDS)—one of the most common and lethal complications of SARS-CoV-2 infection.8 , 9 Given the propensity for ARDS in SARS-CoV-2–infected patients, this protective effect initially seems paradoxical, but previous data on SARS-CoV-1 suggest that subsequent to infection, ACE2 expression is down-regulated, thus tipping the balance in favor of acute lung injury.10
As the presence of ACE2 or TMPRSS2 may be rate limiting for SARS-CoV-2 infection, we used bulk and single-cell transcriptional data from a combination of normal and malignant tissue samples and models from the aerodigestive and respiratory tracts to explore mechanisms governing the expression of ACE2 and TMPRSS2. These data were assembled from a variety of sources, including current and previous works from our laboratory, department, and data sets generated as part of our longstanding joint The University of Texas Southwestern/MD Anderson Cancer Center National Cancer Institute (NCI) Lung SPORE program, including public data sets derived worldwide11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 and represent a novel repurposing of data to understand more of ACE2 expression and consequences of infection. Our bulk data suggest that aerodigestive and lung cancer cell lines have a broad range of ACE2 and TMRPSS2 expression and would serve as good models for studying SARS-CoV-2 infection, consistent with the observation that SARS-CoV-2 virus has the ability to infect several lung cancer cell lines (e.g., Calu-3 and A549).6 , 35 Furthermore, although the normal aerodigestive and respiratory tracts represent key points of viral entry and infection, limiting an investigation to normal tissue may unnecessarily limit available samples for analysis and, especially, limit the range of cellular and molecular phenomena represented by those samples.
Meanwhile, single-cell analyses reveal that although TMPRSS2 is widely expressed in normal respiratory epithelium, ACE2 expression is limited to a small collection of cells36 and, therefore, may be a limiting factor for infection or drive dependence on other mechanisms, such as putative AXL-mediated cell entry.37, 38, 39, 40, 41 There have been numerous reports of patients developing loss of chemosensation (taste, smell, etc.) after SARS-CoV-2 infection—an observation that pointed to another small collection of cells.42 , 43 Tuft, or brush, cells are rare chemosensory cells present in the aerodigestive and respiratory tracts, among other sites, that mediate taste and smell, along with innate immune response.44 , 45 These cells, which are epithelial in nature, strongly express POU2F3, a transcription factor that is also highly expressed in certain lung cancers, including a subset of SCLC.46, 47, 48, 49 Our analyses indeed suggest that ACE2 expression is enriched in tuft cells compared with nontuft cells within the respiratory tract. Nevertheless, this was not absolute and, thus, we considered broader regulatory mechanisms that might govern ACE2 expression. Previous literature has proposed, inconsistently, both positive and negative associations between ACE2 expression and epithelial differentiation,50 , 51 although tuft cells, in which ACE2 is enriched in our analysis, are highly epithelial.45 We evaluated the relationship between ACE2 and epithelial differentiation in numerous aerodigestive and respiratory datasets and found striking and consistent positive correlations with transcriptional, microRNA (miRNA), and metabolic signifiers of epithelial differentiation in virtually all data sets analyzed.
Finally, we consider the role that regulators of epithelial-to-mesenchymal transition (EMT) may play in modulation of ACE2 expression. EMT is a well-characterized phenomenon critical to normal developmental processes but co-opted by tumor cells to promote metastasis and resistance to treatments ranging from chemotherapies to radiation.52 We provide evidence that the miR-200 family, ZEB1 pathway, which is an established regulator of EMT,53 , 54 also directly regulates ACE2 expression, likely through putative ZEB1 repressor sites located in the ACE2 promoter. Furthermore, we highlight that SARS-CoV-2 infection both in vitro and in patients yields increased expression of EMT-associated genes, including ZEB1 and AXL and metabolic derangements (e.g., a shift away from glutamine and toward glutamate) characteristic of EMT.55 , 56 Inhibition of AXL, a mesenchymal receptor tyrosine kinase, with bemcentinib both induces ACE2 and represses ZEB1 and may serve as a therapy to shift SARS-CoV-2–infected cells away from a mesenchymal phenotype (i.e., reverse EMT). In addition, viral infection of epithelial cells, which are glutamine dependent with high GLUL expression and presumably more metabolically primed for replication, promotes rapid replication owing to the enhanced dependence on glutamine for nucleotide synthesis and subsequently depletes glutamine stores.56 Other viruses have been found to induce host cell metabolic reprogramming to satisfy the biosynthetic and energy requirements needed for replication.57, 58, 59, 60
The association between EMT and lung injury or ARDS has been described previously, with the central pathogenesis being the breakdown of the alveolar-epithelial barrier, owing in part to disruption of tight junctions.61, 62, 63 Furthermore, EMT is known to down-regulate tight junction components, including several members of the Claudin protein family.64 Claudins are integral membrane proteins localized at tight junctions that are involved in regulating epithelial cell polarity and paracellular permeability. Intriguingly, previous data revealed that overexpression of miR-200 family members (which inhibit ZEB1 expression) or direct exogenous silencing of ZEB1 has a protective effect in a murine, lipopolysaccharide-induced model of ARDS.65 In this context, our findings support a novel model for the pathogenesis of SARS-CoV-2 in which the virus initially infects a small pool of highly epithelial cells of the aerodigestive and respiratory tracts followed by those infected cells undergoing molecular alterations typical of EMT (i.e., ZEB1 or AXL up-regulation). These EMT-like alterations, in turn, result directly in the down-regulation of genes associated with tight junctions and, in doing so, eradicate the proposed ARDS-protective effect of these epithelial, ACE2-positive cells.
Materials and Methods
Transcriptional Sequencing Data Sets
RNA sequencing (RNAseq) analysis data sets from normal tissues were obtained from Genotype-Tissue Expression portal (http://www.gtexportal.org), and tumor samples were retrieved from published and unpublished data sets in multiple human tissues, including The Cancer Genome Atlas (TCGA) lung adenocarcinoma (LUAD),11 TCGA head and neck squamous cell (tumors) (HNSC),12 TCGA Lung Squamous Cell Carcinoma, PROSPECT,13 BATTLE 1,14 and BATTLE 2.15 Cell line RNAseq data were obtained from multiple sources, including the Cancer Cell Line Encyclopedia upper aerodigestive tract (n = 32), LCNEC cell lines (n = 15),16 NSCLC (n = 88),17 , 18 head and neck squamous cell carcinoma (cell lines) (HNSCC) (n = 70),19 and SCLC (n = 62).66
Transcriptional data from experimental data sets were publicly available, including EMT induction by 3 weeks of transforming growth factor-β (TGF-β),20 393P murine cells with Zeb1 overexpression,21 erlotinib resistance through EMT in EGFR-mutated HCC4006 and HCC827 NSCLC cell lines,22 gefitinib-resistant PC-9 cells through EMT,23 T790M-mediated gefitinib resistance in PC-9 cells,24 and Calu-3 or A549 transduced with a vector-expressing human ACE2 that were mock infected or infected with SARS-CoV-2 (USA-WA1/2020),25 forced miR-200 expression in 344SQ LUAD cells with high metastatic potential,26 human bronchial organoids that were infected with SARS-CoV-2 for 5 days,27 and nasopharyngeal swabs from patients with positive or negative SARS-CoV-2 polymerase chain reaction (PCR) results.28
Single-cell RNAseq (scRNAseq) data were obtained from publicly available sources, including bronchial epithelial cells obtained by bronchial brushings from six never smokers and six current smokers,29 oral cavity tumors from 18 tumors,30 five normal donor and eight fibrotic lungs,31 freshly resected parenchymal lung tissue from three patients,32 bronchial biopsy, nasal brushing, nasal turbinate specimens (GSE121600), and PC-9 EGFR-mutant NSCLC cell xenograft tumors treated with vehicle or osimertinib for 3 weeks.33
Chromatin immunoprecipitation sequencing analysis of ZEB1 binding in the ACE2 promoter of HepG2, human hepatocyte carcinoma cells, was as previously described.34
Flow Cytometry
One million cells each for HCC2302, H3255, HCC827, H1944, H441, H2023, HCC364, A549, H1355, and HCC827 vectors and HCC827 ZEB1 were surfaced stained for ACE2 (Santa Cruz; sc-390851), fixed in 2 % perfluoroalkoxy alkanes, and permeabilized using 1× Intracellular Staining Perm Wash Buffer (BioLegend; 421002) before intracellular staining for vimentin (BD Pharmingen; 562338). The samples were analyzed on a BD LSRFortessa Flow Cytometer, and data were analyzed using FlowJo 10.7.1. Graphs and statistical analysis were done using GraphPad Prism 8.
Metabolite Assay
One million cells each for 393P ZEB1 (DMSO 16 h), 393P ZEB1 (DOX 16 h), H441, H1944, HCC2302, HCC827, H2023, and H1355 were harvested. Cell lysates were prepared by addition of 0.3 N hydrochloric acid and then 450 nM Tris (pH = 8), and metabolites were analyzed using the Glutamine/Glutamate Glo Assay (Promega; J8021) per manufacturer’s instructions.
Drug Treatment of Cells
393P-Zeb1–inducible murine NSCLC cells were treated with AXL inhibitor, bemcentinib (0.5 μM), or DMSO for a total of 24 hours (8-h pretreatment with bemcentinib or DMSO, followed by induction of Zeb1 with doxycycline [2 μg/mL], and an additional 16-h drug treatment). Five NSCLC cell lines with varying levels of ACE2 were treated with hydroxychloroquine sulfate (Sigma) for 48 hours. Cell lysates were harvested for Western blot or reverse-phase protein array (RPPA) analysis. Antibodies used for Western analysis include ACE2 (MA5-32307, ThermoFisher), GLUL (80636, Cell Signaling Technology), Vimentin (3932, Cell Signaling Technology), ZEB1 (3396, Cell Signaling Technology), and vinculin (Sigma, V9131) as a loading control. The cell line drug-screening data were generated internally. Three measurements were performed for each drug at each concentration to produce dose-response data and determine concentration that inhibits 50 % value.
Reverse-Phase Protein Array
Protein lysates from TCGA LUAD, NSCLC cell lines, and Calu-1 or 393P cells treated with bemcentinib were quantified, and protein arrays were printed and stained.67 Images were quantified with MicroVigene 4.0 (VigeneTech, Carlisle, MA). The spot-level raw data were processed with the R package SuperCurve suite, which returns the estimated protein concentration (raw concentration) and a quality control score for each slide, as described previously.67 Only slides with a quality control score of greater than 0.8 were used for downstream analysis. The raw concentration data were normalized by median centering each sample across all the proteins to correct loading bias.
miRNA Arrays
Total RNAs from 55 NSCLC, 30 SCLC, and 49 HNSCC cell lines were analyzed with Affymetrix miRNA 4.0 arrays. The expression data of a curated list of miR-200 family members (hsa-miR-200b-5p, hsa-miR-200b-3p, hsa-miR-200c-5p, hsa-miR-200c-3p, hsa-miR-200a-5p, hsa-miR-200a-3p, hsa-miR-429, hsa-miR-141-5p, hsa-miR-141-3p) known to be involved with EMT in NSCLC68 were compared with ACE2 expression data.
Metabolite Analyses
A total of 225 metabolites were profiled in the Cancer Cell Line Encyclopedia upper aerodigestive tract cell lines using liquid chromatography–mass spectrometry.69 We compared ACE2 mRNA expression to abundance of metabolites.
Computational Prediction of Binding Sites
To generate the predicted ZEB1 (E-Box) binding sites on the ACE2 promoter, the promoter sequence for human ACE2 was downloaded and used in the matrix profile search on the JASPAR web portal.70 For the search, the vertebrate database selecting ZEB1-binding motifs was selected to search through the promoter sequence of ACE2 with a relative profile threshold score cutoff of 80 % or higher. The resulting sites were ranked, and accordingly, the highest scoring motifs were annotated on the promoter segment using SnapGene.
scRNAseq Analysis
Raw data for single-cell datasets were downloaded from the Gene Expression Omnibus (GEO) database and processed using the Seurat Package version 2.3.1.71 First, the raw read counts were normalized and scaled using “NormalizeData” and “ScaleData” function. The most variable genes were selected using the “FindVariableGenes” function with default parameters, and principle component analysis (PCA) was performed on these variable genes. We selected the first N principle components that account for at least 80 % of the total variances to perform the t-distributed stochastic neighbor embedding (tSNE) transformation. For each gene, the expression status is defined as positive if the cell has nonzero expression value, or negative otherwise. For GSE122960, samples were grouped to donor samples (including GSM3489182, GSM3489187, GSM3489189, GSM3489191, and GSM3489193) and fibrotic samples (including GSM3489183, GSM3489184, GSM3489188, GSM3489190, GSM3489192, GSM3489194, GSM3489196, and GSM3489198). Each group was subsampled to 20,000 total cells. EMT score was calculated on the basis of the EMT signature as described previously.72 , 73 Cell-specific expression of SFTPC (alveolar type II cells), FOXJI, PIFO (ciliated cells), and SCGB1A1 (club cells) was visualized in tSNE feature plots to identify epithelial clusters containing ACE2-positive cells.
Data Quality Assessments
Before performing any analyses, the data sets were screened for data quality. One example of a data set used in these analyses is the NCI TCGA, a landmark cancer genomics program that characterized more than 20,000 primary cancer and adjacent normal tissues from 33 different cancer types. These data are publicly available to the research community for the purpose of investigating novel hypotheses. RNAseq data were inspected for total read counts across all samples as a quality check for library size. Furthermore, datasets were excluded from our analyses owing to highly volatile total read counts. In both microarray and RNAseq data sets, PCA using all features was performed to make sure that no outlier samples were present. When distinct grouping information was included (i.e., control versus infected, positive versus negative), we performed PCA analysis to make sure that there was a good separation in the PCA plot. In addition to data quality assessments, we also considered sample size for each individual data set with a minimum of three samples per group required for further analyses.
The public microarray gene expression data were directly downloaded in the original processed data format from the GEO site. For scRNAseq data, we used the read counting and unique molecular identifier count data matrices in GEO. No additional data-filtering steps were applied. All the TCGA data used were level 3 processed data. Data quality across data sets is not comparable and contains considerable batch effects, especially with the use of different technologies. Therefore, data sets were not combined and all analyses were performed on individual data sets.
Statistical Analyses
All statistic and bioinformatics analyses were performed using R. Paired and unpaired two-sample t tests were used for two-group comparisons on paired and unpaired experimental design experiments. Pearson and Spearman correlations were used for correlating genomic and proteomic measurements, including correlating drug-screening data. In all the analyses, p value less than 0.05 was considered statistically significant.
The drug-target constellation map was generated on the basis of drug-screening data using a suite of R packages, including ggplot2, ggraph, and igraph. A list of drugs was selected on the basis of Spearman coefficient (>0.3) and p value (<0.1), and only targets with more than five associated drugs were revealed in the figure.
Data Set Availability
Publicly available data were obtained from the following GEO data sets: GSE122960, GSE131391, GSE103322, GSE42127, GSE122960, GSE130148, GSE138693, GSE121600, GSE49644, GSE61395, GSE114647, GSE75602, GSE147507, GSE15741, GSE150819, GSE154770, and GSE32465 (GSM1010809).
Results
Expression of ACE2 by Cancer Epithelial Cells
Data sets from multiple sources were used in these analyses, including those publicly available in GEO data sets and those generated by our laboratory, department, and the longstanding joint The University of Texas Southwestern/MD Anderson Cancer Center NCI Lung SPORE program.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 The publicly available data include sets such as TCGA, an NCI resource established for data discovery and hypothesis testing. A rigorous analysis of data quality was performed before inclusion. No comparisons were made across data sets. None of the comparisons or queries were previously performed or published.
ACE2 is widely expressed in normal tissues from Genotype-Tissue Expression Project (http://www.gtexportal.org) and was highest in testis and small intestine and lowest in spleen and blood with moderate expression in lung and salivary glands, which are sites of SARS-CoV-2 transmission or infection (Supplementary Fig. 1 A). Transcriptional data are revealed as normalized relative gene expression.
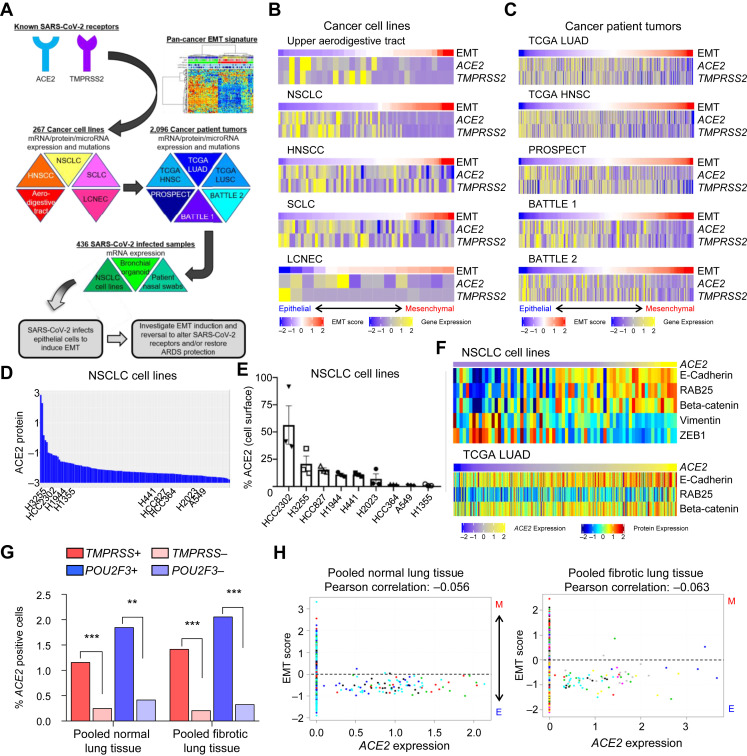
To leverage cancer cell lines and patient tumors to SARS-CoV-2 research, we explored expression of the receptors, ACE2 and TMPRSS2, and discovered a negative correlation with EMT. This was validated in SARS-CoV-2–infected samples and led to investigation of ways to manipulate EMT in an effort to minimize infection or ARDS in patients (Fig. 1 A). ACE2 expression in aerodigestive and respiratory cancer cell lines (Fig. 1 B) and both normal and tumor specimens (Fig. 1 C) is consistently higher in specimens with a low EMT score, suggesting ACE2 is primarily expressed by epithelial cells in these cancers.11, 12, 13, 14, 15, 16, 17, 18, 19 , 66 , 74 , 75 The EMT score uses 76 genes to define the degree to which cells or tissues have undergone EMT and has been applied in TCGA projects.72 , 76 , 77 This observation is confirmed by a consistent negative correlation of ACE2 and EMT score in all nine cell line and tumor data sets (Table 1 ). TMPRSS2, the protease also required for SARS-CoV-2 infection, is similarly expressed in models with epithelial gene signatures (Fig. 1 B and C) and highly correlated with ACE2 expression in cancer models (Table 1). To determine whether expression levels of ACE2 were different in normal tissues and tumors, we compared expression in TCGA tumor data sets with paired normal, adjacent samples. Both normal and LUAD tissues have similar ACE2 expression (TCGA LUAD p = 0.21) and EMT score (TCGA LUAD p = 0.87) (Supplementary Fig. 1 B), although a greater range of expression (both higher and lower) could be observed in tumors.78, 79, 80 Interestingly, and in contrast to some previous reports, no difference in ACE2 expression was detected in normal lung tissue (adjacent to TCGA LUAD) with regard to smoking status, gender, or age (Supplemental Fig. 1 C).
Figure 1.
The SARS-CoV-2 receptor, ACE2, is expressed by epithelial cells in cancer models. (A) Schematic depicting an overview of the strategy to investigate ACE2 biology in the context of EMT using cancer cell lines and tumor samples before applying to SARS-CoV-2–infected samples to develop our hypotheses. (B) ACE2 and TMPRSS2 expression and EMT score in aerodigestive and lung cancer cell lines (ACE2 versus EMT score, p < 0.0001 for all) and (C) in patient tumor biopsies (ACE2 versus, EMT score, p < 0.0001 for all). TCGA sets (LUAD and HNSC) refer to LUAD and HNSCC, respectively. The BATTLE-1, BATTLE-2, and PROSPECT trials include only NSCLC. Rho values are listed in Table 1. (D) ACE2 protein levels in NSCLC cell lines. (E) Cell surface ACE2 in a subset of NSCLC cell lines. (F) NSCLC cell lines and TCGA LUAD tumor biopsies were ranked by ACE2 mRNA expression and reveal protein expression of common epithelial (E-cadherin, RAB25, beta-catenin) or mesenchymal (vimentin, ZEB1, fibronectin) markers. (G) Frequency of ACE2-positive cells in TMPRSS2-positive or -negative cells and POU2F3-positive or negative cells as determined by single-cell RNAseq of donor or fibrotic lungs. (H) EMT score values for each ACE2-positive cell present in five donor and eight fibrotic lungs. Each colors represent cells from an individual lung sample. ∗∗p < 0.01, ∗∗∗p < 0.001. ARDS, acute respiratory distress syndrome; EMT, epithelial-to-mesenchymal transition; HNSC, head and neck squamous cell (tumors); HNSCC, head and neck squamous cell carcinoma (cell lines); LCNEC, large cell neuroendocrine carcinoma; LUAD, lung adenocarcinoma; RNAseq, RNA sequencing; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Table 1.
ACE2 Rho Correlation Values in Aerodigestive and Respiratory Tract Cell Line and Tumor Biopsy Specimens
| Dataset | TMPRSS2 | POU2F3 | EMT Score | ZEB1 | AXL | GLUL | GLS |
|---|---|---|---|---|---|---|---|
| NSCLC cell lines | 0.565a | 0.668a | −0.694a | −0.801a | −0.410a | 0.394a | −0.202b |
| HNSCC cell lines | 0.230b | 0.188 | −0.408a | −0.388a | −0.078 | 0.306c | −0.042 |
| SCLC cell lines | 0.069 | 0.394c | −0.401a | −0.429a | −0.351b | 0.284b | −0.305b |
| TCGA LUAD | 0.228a | 0.328a | −0.286a | −0.28a | −0.083 | 0.296a | −0.197a |
| TCGA HNSC | 0.082 | 0.159a | −0.402a | 0.238a | −0.239a | 0.037 | −0.071 |
| TCGA LUSC | 0.242a | 0.033 | −0.239a | −0.070 | −0.121c | 0.404a | −0.179a |
| PROSPECT | 0.195a | 0.065 | −0.279a | −0.117b | −0.090c | −0.091 | 0.126b |
| BATTLE 1 | 0.437a | 0.134 | −0.367a | −0.166b | −0.222c | 0.339a | −0.082 |
| BATTLE 2 | 0.373a | 0.249∗∗ | −0.337a | −0.196b | −0.235c | 0.208c | −0.064 |
HNSC, head and neck squamous cell (tumors); HNSCC, head and neck squamous cell carcinoma (cell lines); LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
p value less than 0.001.
p value less than 0.05.
p value less than 0.01.
In addition to ACE2 mRNA expression, we screened a cohort of NSCLC and SCLC cell lines for ACE2 protein levels by RPPA (Fig. 1 D), and within a subset of NSCLC cell lines, we analyzed cell surface expression of NSCLC cell lines by flow cytometry (Fig. 1 E). Similar to gene expression, ACE2 levels negatively correlate with mesenchymal proteins, including vimentin (Supplemental Fig. 1 D). To go beyond the quantitative, gene-based EMT score, we investigated expression of ACE2 mRNA compared with specific epithelial (i.e., E-cadherin, RAB25, beta-catenin) or mesenchymal (i.e., vimentin, ZEB1) proteins by RPPA analysis. ACE2 was higher in NSCLC cell lines (p < 0.008) and TCGA tumors (p < 0.001; Fig. 1 F) with higher levels of epithelial proteins, consistent with the mRNA associations and EMT score.
Our analyses reveal that ACE2 is only expressed by a small subset of cells (0%–4.7 %) at the single-cell transcriptional level in normal respiratory tissues, including in oral cavity tumors and lung cancer xenograft models (Table 2 ).30, 31, 32, 33 , 81 Nevertheless, in all data sets, the rare, ACE2-positive cells consistently have a low, or more epithelial, EMT score. In both fibrotic and donor lung samples, TMPRSS2 is more widely expressed than ACE2, whereas ACE2 expression is found more frequently in TMPRSS2-positive cells (Fig. 1 G and Supplementary Table 1) relative to TMPRSS2-negative cells. As studies suggest that both are required for infection, these data indicate that ACE2 expression is more limiting than TMPRSS2 expression for SARS-CoV-2 infection within respiratory epithelium.
Table 2.
Total Number of ACE2-Positive Cells and EMT Score
| Data Sets | Total No. of Cells/Samples | ACE2+ Cells (%) | EMT Score in ACE2+ Cells |
|---|---|---|---|
| Bronchial brushings (GSE131391) | 1146 cells (12 patients) | 54 (4.71) | −0.13 |
| Oral cavity tumors (GSE103322) | 5902 cells (18 tumors) | 87 (1.47) | −0.54 |
| Donor lungs (GSE122960) | 20,000 cells (5 lungs) | 94 (0.47) | −0.52 |
| Fibrotic lungs (GSE122960) | 19,587 cells (8 lungs) | 72 (0.37) | −0.75 |
| Parenchymal lung (GSE130148) | 10,360 cells (3 lungs) | 14 (0.14) | −0.26 |
| PC9 xenograft DMSO treated (GSE138693) | 3766 cells (1 xenograft) | 29 (0.77) | −0.40 |
| PC9 xenograft osimertinib treated (GSE138693) | 2332 cells (1 xenograft) | 0 (0) | N/A |
| Bronchial biopsy/nasal brushing/turbinate (GSE121600) | 12,000 cells (3 specimens) | 4 (0.03) | −0.49 |
EMT, epithelial-to-mesenchymal transition; N/A, not applicable.
POU2F3 is a marker of tuft cell expression and is correlated with ACE2 expression in many aerodigestive and lung cancer data sets (Table 1). ACE2 protein levels also correlate with POU2F3 in both NSCLC (Rho = 0.221, p = 0.01) and SCLC (Rho = 0.673, p < 0.0001) cell lines. Similar to the observation for co-expression with TMPRSS2, ACE2 expression is more often detected in POU2F3-positive than -negative cells (Fig. 1 G and Supplementary Table 1). This suggests that tuft cells may be preferentially targeted by SARS-CoV-2 infection in aerodigestive tissues. These rare, ACE2-positive cells can be visualized in tSNE space (Supplemental Fig. 2 A) and had a low EMT score on a cell-by-cell basis in both donor and fibrotic lung samples (Fig. 1 H) and oral cavity tumors (Supplemental Fig. 2 B). These ACE2-positive cells are localized within epithelial cell clusters with low EMT score that primarily consist of alveolar type II ciliated and club cells (Supplemental Fig. 2 C) on the basis of expression of cell-type specific markers including SFTPC (alveolar type II cells), FOXJI (ciliated cells), and SCGB1A1 (club cells).
SARS-CoV-2 Infection Induces EMT
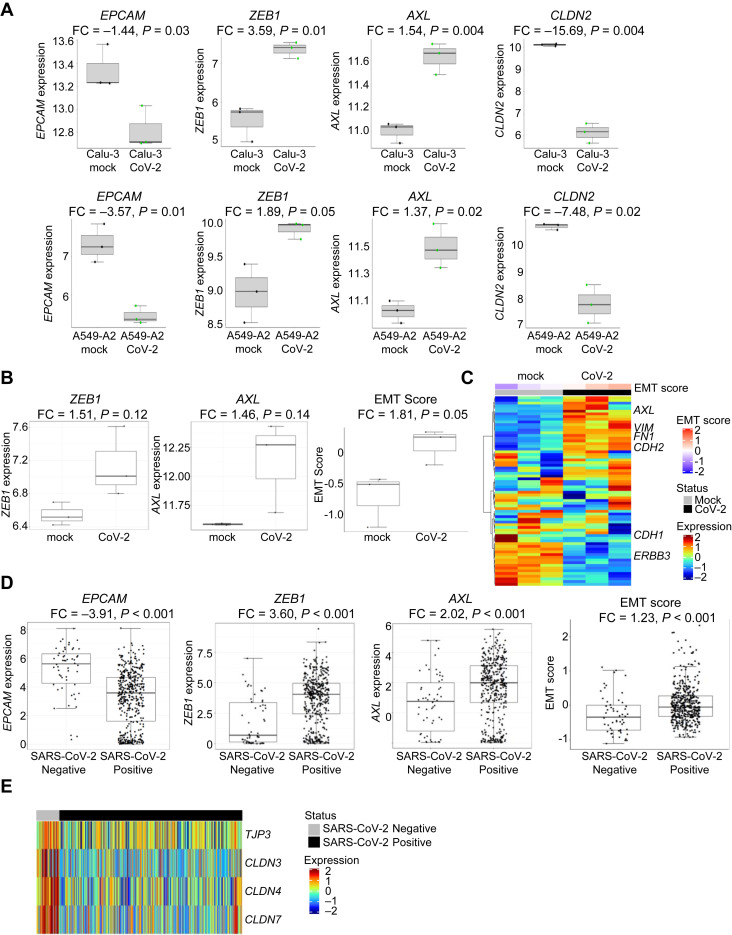
We next sought to reveal the impact of SARS-CoV-2 infection on the EMT status of the infected cell. We analyzed RNAseq data from NSCLC cell lines (A549 and Calu-3) infected for 24 h with SARS-CoV-2 (GSE147507).25 These are cell lines that are frequently used for lung cancer research. Because A549 cells have low endogenous levels of ACE2 expression, likely owing to their mesenchymality, the authors also transduced these cells with a vector overexpressing human ACE2 to improve infection. Expression of the epithelial gene EPCAM was down-regulated after viral infection (Calu-3, p = 0.03, A549+ACE2, p = 0.01, Fig. 2 A; A549, p = 0.04, Supplemental Fig. 3 A), suggesting infection is inducing a shift away from an epithelial phenotype. This was confirmed by ZEB1 up-regulation in all three cell lines (Calu-3, p = 0.01, A549+ACE2, p = 0.05, Fig. 2 A; A549, p = 0.003, Supplementary Fig. 3 A) after infection.
Figure 2.
SARS-CoV-2 induces EMT. (A) Effects of SARS-CoV-2 infection of Calu-3 or A549+ACE2 for 24 hours on EPCAM, ZEB1, AXL, and CLDN2 expression. (B) Effects of SARS-CoV-2 infection on human bronchial organoids for 5 days on ZEB1 and AXL expression and EMT score. (C) Heatmap revealing differences in gene expression between mock and SARS-CoV-2–infected bronchial organoids in EMT-associated genes. (D) Effects of SARS-CoV-2 infection in nasal swab samples from 430 individuals with PCR-confirmed SARS-CoV-2 and 54 negative controls on EPCAM, ZEB1, AXL expression and EMT score. (E) Effects of SARS-CoV-2 infection in nasal swabs on genes associated with tight junctions. RNAseq data are revealed as normalized relative gene expression. CoV-2, coronavirus 2; EMT, epithelial-to-mesenchymal transition; FC, fold change; PCR, polymerase chain reaction; RNAseq, RNA sequencing; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
An alternative EMT regulator, AXL, which is a TAM (Tyro3, AXL, Mer) family receptor tyrosine kinase strongly associated with a mesenchymal phenotype, has emerged as a key determinant of therapeutic resistance in NSCLC and other cancer types.72 , 82 Similar to ZEB1, AXL is inversely correlated with ACE2 in cell line and tumor samples (Table 1). Furthermore, infection of cells with SARS-CoV-2 up-regulated AXL expression in all three cell lines (Calu-3, p = 0.004, A549+ACE2 p = 0.02, Fig. 2 A; A549, p < 0.001, Supplementary Fig. 3 A). A similar induction of mesenchymal genes with viral infection was observed for SNAI1 (Calu-3: fold change [FC] = 1.84, p = 0.009; A549+ACE2: FC = 7.43, p = 0.01) and ZEB2 (Calu-3: FC = 6.74, p = 0.002; A549+ACE2: FC = 5.17, p = 0.0001). Nevertheless, EMT score was not different between mock- or SARS-CoV-2–infected Calu-3 (FC = −1.09, p = 0.14) or A549+ACE2 cells (FC = −1.03, p = 0.48), and this may be due to the relatively short period of viral infection (24 h).
We next investigated whether SARS-CoV-2 infection directly altered Claudin expression. CLDN2 is expressed in respiratory epithelium, but a role in mediating alveolar edema has not been described.83 Nevertheless, of all Claudin family members, CLDN2 was down-regulated in all three cell lines (Calu-3, p = 0.004, A549+ACE2, p = 0.02 Fig. 2 A; A549, p = 0.008, Supplemental Fig. 3 A), suggesting a change in epithelial and endothelial cell permeability that may contribute to ARDS in patients with COVID-19. Similarly, there was a reduction in TJP3, the gene encoding a scaffolding protein that links tight junction transmembrane proteins, such as claudins, to the actin cytoskeleton (Calu3: FC = −1.48, p = 0.04; A549+ACE2: FC = −2.89, p = 0.10) after viral infection. Evidence of a metabolic shift away from glutamine was observed postinfection, as GLUL expression was reduced in ACE2-high cell lines, but not in nontransfected A549 (Calu-3 p = 0.01, A549+ACE2 p = 0.01, Supplemental Fig. 3 B, A549, p = 0.56). The ZEB1 and AXL increase and EPCAM and GLUL decrease point toward a SARS-CoV-2–induced shift from an epithelial to a mesenchymal phenotype.
To reveal whether longer exposure to virus may induce additional EMT changes, we analyzed RNAseq data from human bronchial organoids developed from commercially available, cryopreserved adult bronchial epithelial cells infected with SARS-CoV-2 for 5 days (GSE150819).27 There was a subtle increase in both ZEB1 (p = 0.12) and AXL (p = 0.14) and other mesenchymal genes, including VIM (FC = 1.56, p = 0.01), ZEB2 (FC = 3.23, p = 0.22), SNAI1 (FC = 4.75, p = 0.05), and CDH2 (FC = 2.06, p = 0.02). Importantly, longer SARS-CoV-2 infection increased the EMT score (p = 0.05; Fig. 2 B). Although these shifts are not as strong as those found in the cell lines, there is a clear pattern for expression of the epithelial and mesenchymal genes making up the EMT score by heatmap (Fig. 2 C). Similar to the NSCLC cell lines, SARS-CoV-2 infection down-regulated expression of tight junction-related genes, including CLDN8 (FC = −2.66, p = 0.009) and CLDN17 (FC = −1.62, p = 0.05).
Finally, to evaluate whether viral-induced EMT also occurs in patients, we analyzed metagenomic next-generation sequencing data of human nasopharyngeal swabs from 430 PCR-confirmed SARS-CoV-2 and 54 negative control individuals (GSE154770).28 SARS-CoV-2 infection down-regulated expression of EPCAM (p < 0.0001) and up-regulated ZEB1 (p < 0.0001) and AXL (p < 0.0005; Fig. 2 D). Similar patterns were found with other epithelial genes, including RAB25 (FC = −2.35, p < 0.0001) and CTNNB1 (FC = −3.38, p < 0.0001), or mesenchymal genes, including ZEB2 (FC = 8.14, p < 0.0001). As expected, viral infection was associated with an increased EMT score (p < 0.0001; Fig. 2 D) and TGFB1 expression (FC = 2.75, p < 0.0001). Interestingly, EMT gene expression was not affected by viral load (Supplemental Fig. 3 C), as defined by the cycle threshold of the SARS-CoV-2 nucleocapsid gene region 1 (N1) target during diagnostic PCR.28 This suggests that EMT is induced in cells, regardless of the viral burden in the patient. Consistent with the previous metabolic data, SARS-CoV-2 infection induces expression of GLS, indicating a shift away from glutamine production (p = 0.009; Supplemental Fig. 3 D). Similar to the cell lines and bronchial organoids, SARS-CoV-2 infection reduces expression of genes associated with tight junctions (Fig. 2 E), including CLDN4 (FC = −6.63, p < 0.0001), CLDN7 (FC = −6.59, p < 0.0001), CLDN3 (FC = −5.14, p < 0.0001), CLDN9 (FC = −1.72, p < 0.0001), CLDN23 (FC = −1.95, p = 0.0002), TJP2 (FC = −1.58, p = 0.05), and TJP3 (FC = −2.22, p < 0.0001).
Induction of EMT Represses ACE2 Expression
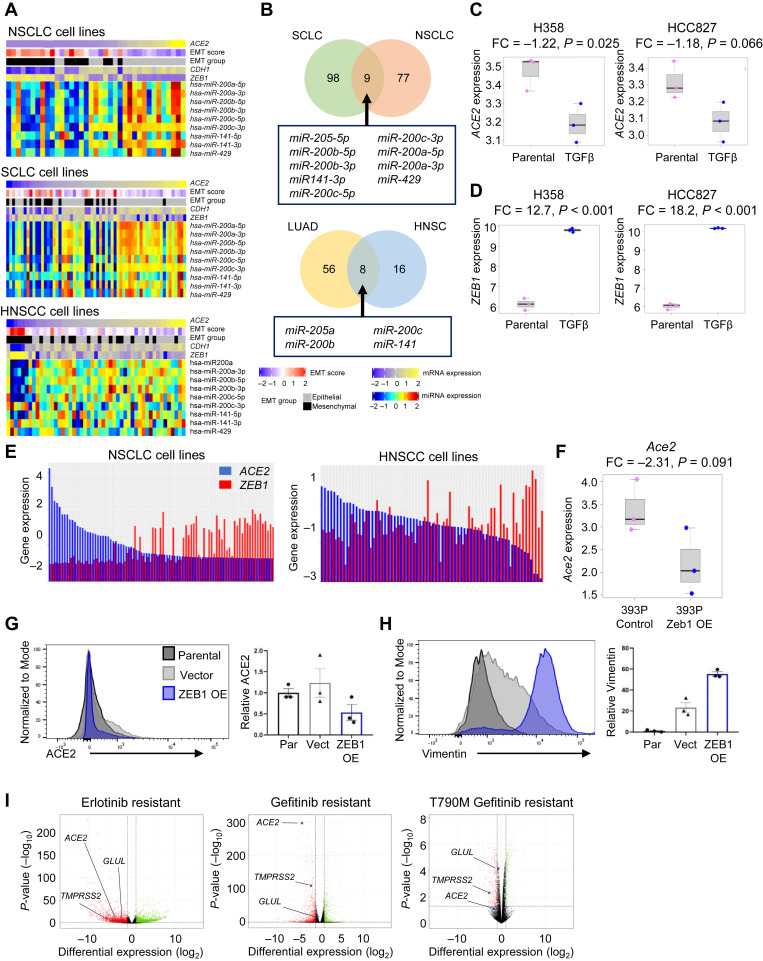
To connect differences in ACE2 expression to a more specific EMT regulatory program, we investigated miRNAs in both lung cancer and head and neck cell lines and tumor samples. Interestingly, miRNAs from the miR-200 family were consistently positively correlated with ACE2.68 As expected, given their known role as inhibitors of EMT, high miR-200 family expression was found in cells and tumors with high CDH1 (an epithelial-specific marker) and low EMT score and ZEB1, the latter a transcriptional promoter of EMT and target of miR-200 family (Fig. 3 A and Supplementary Fig. 3 E). The miR-200 family was correlated with ACE2 expression in NSCLC,21 SCLC, and HNSCC cell lines (p < 0.01; Fig. 3 B and Supplementary Fig. 3 E)84 and both TCGA LUAD and HNSC tumors (Fig. 3 B). Nevertheless, forced miR-200 expression in 344SQ LUAD cells with high metastatic potential was not sufficient to alter ACE2 expression (Supplementary Fig. 3 F).26 miR-200 family members repress ZEB1 expression, so unsurprisingly ACE2 is negatively correlated with ZEB1 in cancer cell lines and tumors (Table 1). ACE2 similarly is negatively correlated with a number of other EMT-associated genes, including ZEB2, SNAI1, SNAI2, TWIST1, and CDH2 (Supplementary Table 2). Although these correlations are almost invariably negative, as expected, none of these genes reveal a negative correlation as robust and consistent across data sets between ACE2 as EMT score or ZEB1.
Figure 3.
ACE2 repression by EMT in lung cancer cells. (A) ACE2, EMT score, EMT group (epithelial or mesenchymal), CDH1, and ZEB1 expression with miR200 family miRNAs in NSCLC, SCLC, and HNSCC cell lines. (B) Venn diagrams revealing miRNAs correlated with ACE2 expression in both NSCLC and SCLC cell lines (upper) and TCGA LUAD and HNSC tumor biopsies (lower). RNAseq data are revealed as normalized relative gene expression. (C, D) Expression of (C) ACE2 and (D) ZEB1 after induction of EMT by TGF-β treatment in H358 and HCC827 NSCLC cell lines. (E) Expression of ACE2 and ZEB1 in NSCLC and HNSCC cell lines. (F) Expression of Ace2 in 393P murine lung adenocarcinoma cells with Zeb1 overexpression. Cell surface expression of (G) ACE2 and (H) Vimentin in HCC827 cells with constitutive overexpression of ZEB1. (I) ACE2, TMPRSS2, and GLUL expression in NSCLC cell lines with acquired resistance to EGFR TKIs that occurred through EMT but not when resistance occurred through EGFR T790M resistance mutations. RNAseq data are revealed as normalized relative gene expression. EMT, epithelial-to-mesenchymal transition; FC, fold change; HNSC, head and neck squamous cell (tumors); HNSCC, head and neck squamous cell carcinoma (cell lines); LUAD, lung adenocarcinoma; miRNA, microRNA; OE, overexpressing; Par, parental; RNAseq, RNA sequencing; TCGA, The Cancer Genome Atlas; TGF-β, transforming growth factor-β; TKI, tyrosine kinase inhibitor; Vect, vector.
To reveal whether transformation from an EMT directly alters ACE2 expression, we used various methods to induce EMT, including treatment with TGFβ, overexpression of ZEB1, and EGFR tyrosine kinase inhibitor (TKI) resistance. Two highly epithelial NSCLC cell lines cultured with TGFβ for 3 to 5 weeks20 increased the EMT score (Supplemental Fig. 4 A). Induction of EMT resulted in a loss of ACE2 in H358 and HCC827 (p = 0.03 and p = 0.06, respectively; Fig. 3 C). In addition, both cell lines exhibited increased ZEB1 after EMT induction (p < 0.0001; Fig. 3 D). The experiment also included EMT induction in A549 cells, but this line is more mesenchymal with low endogenous ACE2 expression at baseline and TGFβ was unable to reduce already minimal ACE2 (p = 0.23, data not found).
Accordingly, NSCLC and HNSCC cell lines reveal an inverse correlation between ACE2 and ZEB1 (Fig. 3 E). Computational investigation of the ACE2 promoter using the JASPAR database revealed five putative ZEB1 binding sites, suggesting ZEB1 may directly repress ACE2 expression.70 Two of these predicted binding sites (ZEB1-EBOX3, ZEB1-EBOX4) were confirmed by chromatin immunoprecipitation sequencing to be functional binding sites for ZEB1 in the ACE2 promoter in HepG2, a hepatocellular cell line (Supplemental Fig. 4 B). Consistent with this, induction of EMT by means of overexpression of Zeb1 in 393P murine LUAD cells results in a more than twofold reduction in Ace2 expression (p = 0.091; Fig. 3 F).21 Furthermore, constitutive overexpression of ZEB1 in HCC827 cells21 , 85 , 86 reduced cell surface expression of ACE2 compared with either the parental cell line (p = 0.05) or the vector control (p = 0.07; Fig. 3 G). As expected, overexpression of ZEB1 increased vimentin levels (compared with parental, p < 0.0001, or vector control, p = 0.002; Fig. 3 H). ACE2 is repressed by EMT, and this is mediated, at least in part, by ZEB1.
EGFR-mutated NSCLCs are routinely and effectively treated with EGFR TKIs, but resistance inevitably develops by means of characteristic mechanisms, including secondary EGFR resistance mutations (i.e., T790M) and, notably, EMT.87 Thus, we evaluated expression of ACE2 in EGFR-mutated NSCLC cells (HCC4006 and HCC827) treated with the EGFR TKI erlotinib until resistance occurred.22 ACE2 and TMPRSS2 were down-regulated in EGFR TKI-resistant lines that had undergone EMT in comparison to parental (EGFR TKI-sensitive cells) (Fig. 3 I). Separately, PC-9 cells, which also harbor EGFR-activating mutations and display high baseline levels of ACE2, were treated with the EGFR TKI gefitinib for 3 weeks until resistance emerged through EMT23 or acquired T790M resistance mutations.24 ACE2 was down-regulated in gefitinib-resistant cells that had undergone an EMT but not in T790M+-resistant cells that maintained an epithelial phenotype (Fig. 3 I). These findings are consistent with the notion of ACE2 repression by the EMT regulator, ZEB1. In xenograft models, mice bearing PC-9 xenograft tumors treated with vehicle or the EGFR TKI osimertinib for 3 weeks to induce resistance through the EMT were transcriptionally profiled by scRNAseq.33 ACE2 was only detectable in vehicle-treated tumor cells, suggesting ACE2 expression was lost in tumors that had undergone EMT.
Metabolic Signifiers of ACE2-Positive Cells
To identify metabolic features of cells able to be infected by SARS-CoV-2, metabolites associated with ACE2 expression were investigated. ACE2 expression correlates with glutamine in upper aerodigestive tract cell lines (p < 0.01; Supplemental Fig. 4 D),69 and ACE2 levels correlate with glutamine in a subset of NSCLC cell lines (rho = 0.89, p = 0.02; Supplementary Fig. 4 E). Consistent with this finding, ACE2 expression correlation with a list of 253 metabolism-associated genes and GLUL, which encodes an enzyme (glutamine synthetase) responsible for conversion of glutamate to glutamine, was identified in NSCLC, HNSCC, and SCLC cell lines (Supplementary Fig. 4 F) and was confirmed in the tumor data sets (Table 1). In addition to GLUL, DERA and SLC6A14 were identified and are implicated in nucleotide metabolism and amino acid (including glutamine) transport, respectively (Supplemental Fig. 4 F). Interestingly, GLS, which encodes the enzyme (glutaminase) that catalyzes the opposing reaction, in which glutamine is converted to glutamate, is negatively correlated with ACE2 expression (Table 1). Next, we evaluated whether ZEB1 is a regulator of metabolism. Unlike Ace2, Zeb1 is unable to directly regulate Glul expression in murine 393P cells overexpressing Zeb1 (p = 0.23, Supplementary Fig. 4 G). This was confirmed by ZEB1 gene silencing and overexpression analyses (data not found), suggesting the metabolic changes are not directly controlled by ZEB1. Interestingly, GLUL was down-regulated in EGFR TKI gefitinib-resistant cells that had undergone an EMT but not in T790M+-resistant cells that maintained an epithelial phenotype. (Fig. 3 I). Similarly, GLUL expression was reduced after osimertinib resistance that occurred through the EMT (Supplementary Fig. 4 C). Taken together, glutamine synthetase-catalyzed conversion of glutamate to glutamine in epithelial cells aims to increase production of amino acids and nucleotides, whereas in mesenchymal cells, glutaminase converts glutamine to glutamate to preferentially shift toward the tricarboxylic acid cycle, thus priming epithelial cells for viral infection.
EGFR/HER Family Link to ACE2
NSCLC encompasses a heterogeneous group of lung cancers with unique molecular features. To determine whether any previously characterized NSCLC subsets were linked to differential ACE2 expression, we compared common driver mutation subsets of the TCGA LUAD tumors and found that EGFR-mutant LUAD tumors had significantly higher ACE2 expression (p = 0.001; Supplemental Fig. 4 H). EGFR-mutated tumors have constitutive activation of the EGFR and downstream signal transduction pathway. Accordingly, higher ACE2 expression was associated with increased sensitivity to EGFR TKIs, as visualized by drug-target constellation map (Supplemental Fig. 3 I). Moreover, ACE2 expression was broadly correlated with increased expression of several EGFR/HER family members, including EGFR, ERBB2, and ERBB3 in cell lines and tumors (Supplemental Fig. 3 J), consistent with its preferential expression in epithelial cells. A similar link between EGFR inhibition and patients with COVID-19 has been described previously.88
Reversal of EMT
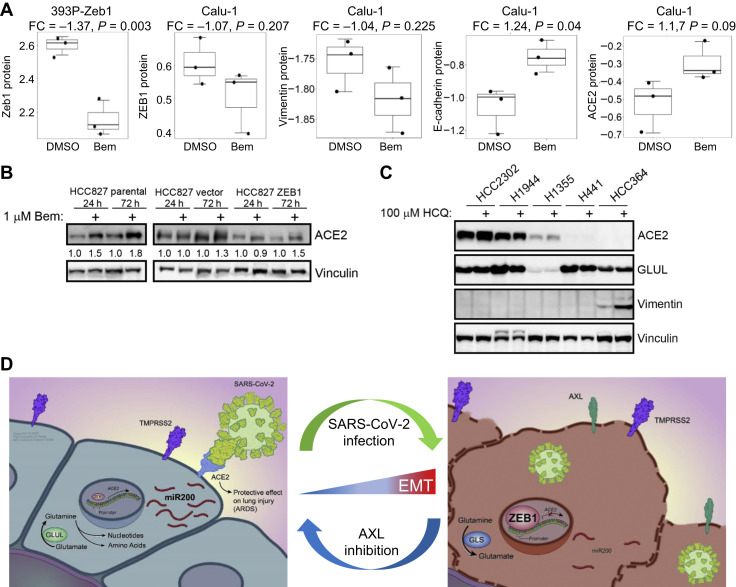
We hypothesized that bemcentinib (BGB324, or R428), an inhibitor of the mesenchymal receptor tyrosine kinase AXL with a known antiviral effect, may offer therapeutic benefit through a reversal of EMT.89 Bemcentinib down-regulated ZEB1 expression in mesenchymal cell lines, 393P overexpressing Zeb1 (p = 0.003) and Calu-1 (p = 0.21) (Fig. 4 A). In Calu-1 cells, bemcentinib also down-regulated vimentin and up-regulated both E-cadherin and ACE2. Similarly, 48 hours of bemcentinib treatment in HCC827 parental, vector control, and ZEB1-constitutive overexpressing lines21 , 85 , 86 induced ACE2, as confirmed by densitometry normalized to vinculin controls (Fig. 4 B). In contrast, hydroxychloroquine treatment was unable to consistently alter ACE2 or GLUL levels in a range of NSCLC cell lines (Fig. 4 C), indicating this treatment is unable to alter EMT.
Figure 4.
EMT modulation for treatment of SARS-CoV-2. (A) ZEB1 expression after 24 h of 0.5 μM Bem treatment in mesenchymal Calu-1 cells or after overexpression of Zeb1 in 393P murine cells, including vimentin, E-cadherin, and ACE2 in Calu-1 cells. (B) ACE2 levels after 24 h and 72 of 1 μM Bem treatment in HCC827 cells with constitutive overexpression of ZEB1. (C) ACE2, GLUL, and Vimentin levels in a subset of NSCLC cell lines after 48 h treatment with 100 μM hydroxychloroquine. (D) Working model revealing aerodigestive/respiratory cells with ACE2 expression are epithelial and metabolically primed for replication (left), whereas infected cells reveal a shift to a mesenchymal phenotype with reversal of glutamine production, a leaky cell membrane, and increased ZEB1 to induce EMT (right). ARDS, acute respiratory distress syndrome; Bem, bemcentinib; EMT, epithelial-to-mesenchymal transition; FC, fold change; h, hour; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Together, these data suggest a potential model in which SARS-CoV-2 infects rare, epithelial, ACE2- and TMPRSS2-positive cells in the aerodigestive and respiratory tracts that are metabolically primed for glutamine synthesis and rapid replication (Fig. 4 D). Once a cell is infected with SARS-CoV-2, there is a shift to a more mesenchymal phenotype characterized by high ZEB1 and AXL, putatively lower levels of miR-200, and a decreased dependence on glutamine synthesis. Viral infection and the resultant mesenchymal shift and ZEB1 increase may have devastating implications on the risk and pathogenesis of ARDS in patients with COVID-19 (Fig. 4 D).
Discussion
The ultimate public health impact of the SARS-CoV-2 pandemic has not yet been fully realized. Although increased molecular understanding of the virus-host interactions is sorely needed, the impact of the pandemic itself has created practical limitations on laboratory-based research owing to wide-ranging institutional restrictions. Despite these restrictions, an unprecedented collaborative research effort has ensued, with little regard for geographic and public-private boundaries that so often impose their own limitations. In the above-mentioned study, we present an effort that not only incorporates our own unique cancer data sets but also uses a plethora of publicly available data from basic scientists, clinicians, and bioinformaticians around the world, including both normal and malignant tissues. The linkage of regulation of ACE2 expression and EMT, the latter an uncommon phenomenon in normal tissue but well defined in cancer cells, highlights the value of considering malignant tissue and models for the investigation of SARS-CoV-2. Although tumor tissues and models undoubtedly have unique genetic and epigenetic features not routinely observed in nonmalignant SARS-CoV-2–infected cells in patients, many regulatory pathways and cellular behaviors, including those involved in EMT, are conserved between normal and malignant settings. Malignant tissues, however, offer a marked advantage than normal tissues in that they have been extensively profiled genomically, transcriptomically, and proteomically, including in circumstances in which processes such as EMT have been manipulated. The extent of available malignant tissue and model data, thus, allowed us to develop and preliminarily pursue novel hypotheses related to SARS-CoV-2 in data sets that were largely published before the emergence of the virus itself—another vital advantage when investigating a novel pathogen.
In this study, we describe a novel model for the regulation of ACE2, which others have found is essential for the host cell entry of SARS-CoV-2.6 In contrast to reports of patient susceptibility to SARS-CoV-2 owing to age, gender, smoking history, or thoracic malignancies,4 , 5 , 90 , 91 we were unable to identify consistent differences in ACE2 expression on the basis of these variables. Our data suggest that ACE2 expression is restricted in both normal and malignant aerodigestive and respiratory tissues almost exclusively to epithelial cells, including highly specialized POU2F3-positive tuft cells, and that this restriction reflects regulatory mechanisms shared with EMT. Although others have observed a putative relationship in other contexts between ACE2 and EMT, these observations were inconsistent, supporting roles for ACE2 as an inducer and an inhibitor of EMT.50 , 51 In virtually all explored datasets, ranging from cancer cell lines to normal respiratory tissue, we observed a strong and consistent negative correlation between ACE2 expression and EMT. This relationship is observed even on a cell-by-cell basis, as scRNAseq illustrates that virtually every cell with detectable ACE2 expression is epithelial on the basis of our published EMT score.76
The negative correlation between EMT and ACE2 was not limited to expression analyses, as metabolomic analysis revealed that ACE2 expression is strongly correlated with high levels of glutamine and the expression of GLUL, the enzyme responsible for the conversion of glutamate to glutamine. High levels of glutamine are characteristic of epithelial differentiation, whereas a shift from glutamine to glutamate is associated with EMT. We propose that SARS-CoV-2 targets these ACE2-expressing, metabolically primed epithelial cells to exploit the abundant nucleotides for rapid replication and viral spread. The consequence of SARS-CoV-2 infection is induction of EMT and subsequent up-regulation of GLS, resulting in a depletion of glutamine after SARS-CoV-2 infection in patients.56
Our subsequent analyses reveal that, beyond mere correlations, various strategies to manipulate EMT result in predictable alterations in the expression of ACE2. For example, treatment of cells with TGFβ or overexpression of ZEB1 induces EMT and decreased expression of ACE2. Similarly, in EGFR-mutant LUAD, in which there is relatively higher ACE2 expression, prolonged treatment with an EGFR-targeting TKI may result in multiple mechanisms of therapeutic resistance, of which EMT is a common one. Our results reveal that ACE2 is down-regulated when EMT is the underlying resistance mechanism, but not when resistance emerges owing to development of secondary resistance mutations in EGFR. Coincident with the loss of ACE2 expression is the down-regulation of GLUL to promote glutamate and enable production of tricarboxylic acid intermediates and energy.
EMT is a complex process regulated by an intricate molecular network, and we hoped to discern whether any of the specific pathway(s) that regulate EMT also mediate ACE2 expression. An investigation into one of the transcriptional mechanisms underlying EMT and its association with ACE2 revealed consistent positive correlations across tissue and sample types with the miR-200 family of miRNAs. The miR-200 family has a well-established role as an inhibitor of EMT by means of the down-regulation of the transcriptional repressor ZEB1. Predictably, a strong negative correlation was observed between ACE2 and ZEB1. The repression of ACE2 by ZEB1 is mediated by means of two repressor binding sites for within the ACE2 promoter, and forced overexpression of ZEB1 results in a more than twofold decrease in ACE2 expression, but not GLUL. ZEB1 regulation occurs by means of SNAI1 or TWIST1, whereas SNAI1 induces nuclear translocation of ETS1, which is required for ZEB1 expression.92 It is likely that EMT induction of these genes by SARS-CoV-2 results in increased ZEB1. Finally, we observed that SARS-CoV-2–infected lung cancer cell lines, bronchial organoids, and patient nasopharyngeal swabs had substantial shifts toward several EMT-like features, including up-regulation of ZEB1 and down-regulation of EPCAM.
Together, our data, in the context of the ever-growing literature on SARS-CoV-2, suggest that ACE2 expression is necessary for initial viral entry and, therefore, is relatively high in those cells infected by the virus. These cells are epithelial and, in our data sets, unexpectedly rare, considering the devastating impact of this infection. After viral entry, however, SARS-CoV-2 infection induces molecular changes within the cells that are reminiscent of EMT—especially the increased expression of ZEB1. These SARS-CoV-2–induced changes, compounded by the down-regulation of genes, including tight junction components, which play a critical role in restricting epithelial and endothelial permeability, exposes respiratory cells to increased risk of edema and ARDS. Interestingly, previous reports with SARS-CoV(-1) reveal that lung cells, including alveolar-epithelial cells, express higher levels of TGFβ, a key regulator of EMT.93 It is not surprising that a similar mechanism would occur with a similar coronavirus, such as SARS-CoV-2.
Although these data reveal an intriguing mechanism for the molecular virus-host interaction, it is critical to consider how these observations may be harnessed to develop strategies to improve patient outcomes. This, as always, is complex. Would a therapeutic effort focused on prevention of the initial infection by decreasing ACE2 levels, for example with an inhibitor of the miR-200 family, have unforeseen consequences, such as increased risk of ARDS? Similarly, would therapeutic efforts postinfection aimed at preventing SARS-CoV-2–induced ACE2 down-regulation, for example with a miR-200 mimetic, serve to place adjacent, newly ACE2-positive cells at risk for infection? Several large-cohort observational studies on ACE inhibitors and angiotensin II receptor blockers, which up-regulate ACE2 expression, did not reveal association with either SARS-Cov-2 infection or risk of severe COVID-19.94 Interestingly, a human recombinant soluble ACE2, which has already been tested in phase 1 and phase 2 clinical trials for ARDS, revealed inhibition of SARS-CoV-2 infection in human capillary organoids and human kidney organoids.95
On the basis of the findings in our study, an attractive alternative therapeutic strategy would be to reverse EMT with an AXL inhibitor. AXL is overexpressed in several solid and hematologic malignancies, including NSCLC, acute myeloid leukemia, breast, and prostate cancer, among others. In the context of malignancy, AXL overexpression can drive EMT, tumor angiogenesis, and decreased antitumor immune response.82 Furthermore, AXL inhibitor, gilteritinib, was identified to have antiviral activity against SARS-CoV-2.96 Bemcentinib, a highly selective and potent inhibitor of AXL, is currently being tested in phase 2 trials in various malignancies, including NSCLC, acute myeloid leukemia, and breast cancer. In preclinical models, bemcentinib has already revealed potent antiviral activity in Ebola and Zika viruses.38 , 97 Recent data suggest that bemcentinib inhibits SARS-CoV-2 viral entry into cells and prevents inhibition of type I interferon, which is the host cells’ innate antiviral immune response. In contrast to AXL inhibition, hydroxychloroquine, an alternative treatment for patients with COVID-19, does not reveal any ability to reverse EMT. Accordingly, a phase II clinical trial (BGBC020) recently completed accrual of 115 patients, evaluating the efficacy and safety of bemcentinib for the treatment of hospitalized patients with COVID-19 alone or in combination with standard of care.98 This trial reported a numerically lower number of deaths in the bemcentinib arm (one death in 28 patients) compared with the standard of care (five deaths in 32 patients), supporting our hypothesis that reversal of EMT in combination with antiviral activity warrants further investigation as a treatment for patients with clinically significant SARS-CoV-2 infection.99 Separately, a recent report independently identified both bemcentinib (AXL inhibitor) and dacomitinib (EGFR inhibitor) as possessing SARS-CoV-2 antiviral properties from 3000 drugs screened.100 These data support our findings that EMT reversal by bemcentinib may augment the antiviral activity of AXL inhibitors against SARS-CoV-2, resulting in additional clinical benefit for patients with COVID-19.
CRediT Authorship Contribution Statement
C. Allison Stewart, Carl M. Gay, Lauren Averett Byers: Conceived the project, Analyzed and interpreted the data, Wrote the manuscript.
Kavya Ramkumar, Kasey R. Cargill: Performed the experiments, Interpreted the results.
Robert J. Cardnell, Monique B. Nilsson, Elizabeth M. Park, Carminia Maria Della Corte, Kiran Kundu: Interpreted the results.
Simon Heeke, Samrat T. Kundu, Lixia Diao, Qi Wang, Li Shen, Yuanxin Xi, Jing Wang: Contributed to the analysis and interpretation of data.
Curtis R. Pickering, Faye M. Johnson, Jianjun Zhang, Humam Kadara, John D. Minna, Don L. Gibbons, and John V. Heymach: Contributed to the acquisition of data, administrative, and/or material support.
C. Allison Stewart, Carl M. Gay, Kavya Ramkumar, Kasey R. Cargill, Robert J. Cardnell, Monique B. Nilsson, Simon Heeke, Elizabeth M. Park, Samrat T. Kundu, Lixia Diao, Qi Wang, Li Shen, Yuanxin Xi, Bingnan Zhang, Carminia Maria Della Corte, Youhong Fan, Kiran Kundu, Boning Gao, Kimberley Avila, Curtis R. Pickering, Faye M. Johnson, Jianjun Zhang, Humam Kadara, John D. Minna, Don L. Gibbons, Jing Wang, John V. Heymach, Lauren Averett Byers: Contributed to the writing, review, and/or revision of the manuscript.
Acknowledgments
We wish to acknowledge the patients affected by the coronavirus disease 2019 pandemic, along with their loved ones, the first responders, health care staff, and essential workers who worked tirelessly and selflessly during this devastating period. We thank C.E.S. for graphical elements and David M. Aten for assistance with medical illustrations. We thank P.M.G., M.P.B., W.A.B., G.D.S., P.W.S., R.A.S., R.R.S., R.A.D., B.A.D., V.J.M.C., S.C., S.C., J.P., S.I.1., S.I.2., and C.J.C. for intellectual support provided during quarantine. The work was supported by the National Institutes of Health (NIH)/National Cancer of Institute (NCI) R50-CA243698 (Dr. Stewart), NIH/NCI R01-CA207295 (Dr. Byers), NIH/NCI U01-CA213273 (Drs. Byers, Heymach), CCSG P30-CA01667 (Dr. Byers), University of Texas SPORE in Lung Cancer P5-CA070907 (Drs. Byers, Gibbons, Heymach, Gay, Wang, Ramkumar, Cargill), NIH/NCI R37CA214609 (Dr. Gibbons), the Department of Defense (LC170171; Byers), Khalifa Bin Zayed Al Nahyan Foundation (Dr. Gay), CPRIT Research Training Program RP170067 (Dr. Park), The International Association for the Study of Lung Cancer (Dr. Cargill), The University of Texas MD Anderson Cancer Center-Oropharynx Cancer Program generously supported by Mr. and Mrs. Charles W. Stiefel (Dr. Johnson), through generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shot Program, The Jane Ford Petrin Fund, The Andrew Sabin Family Fellowship, and The Rexanna Foundation for Fighting Lung Cancer.
Footnotes
Drs. Stewart and Gay contributed equally to this work.
Disclosure: Dr. Byers reports serving on advisory committees for AstraZeneca, AbbVie, GenMab, BergenBio, Pharma Mar SA, Sierra Oncology, Merck, Bristol-Myers Squibb, Genentech, and Pfizer and receiving research support from AbbVie, AstraZeneca, GenMab, Sierra Oncology, and Tolero Pharmaceuticals. Dr. Heymach reports serving on advisory committees for AstraZeneca, Boehringer Ingelheim, Catalyst, Genentech, GlaxoSmithKline, Guardant Health, Foundation Medicine, Hengrui Therapeutics, Eli Lilly, Novartis, Spectrum, Sanofi, Takeda, Mirati Therapeutics, Bristol-Myers Squibb, BrightPath Biotherapeutics, Janssen Global Services, Nexus Health Systems, EMD Serono, Pneuma Respiratory, Kairos Venture Investments, Roche, and Leads Biolabs; receiving research support from AstraZeneca, Bayer, GlaxoSmithKline, and Spectrum; having patents PCT/US2019/024353-UTFC.P1354AE, 62/793,565/PCT/US2020/013889, 62/967,282, 62/895,322-17/011,519, 62/929,840/PCT/US2020/058548, and 62/967,275/UTSC.P1499US.P1; and receiving royalties and licensing fees from Spectrum. Dr. Gibbons reports serving on scientific advisory committees for AstraZeneca, GlaxoSmithKline, Sanofi, and Janssen and receiving research support from Janssen, Takeda, Ribon Therapeutics, Astellas, and AstraZeneca. Dr. Johnson reports receiving donor research funds. Dr. Gay reports receiving research funding from AstraZeneca. Dr. Minna reports receiving royalties and licensing from distribution of cell lines from the National Institutes of Health and The University of Texas Southwestern. Dr. Nilsson reports receiving royalties and licensing fees from Spectrum. Zhang reports receiving grants from Merck and Johnson and Johnson and personal fees from Bristol-Myers Squibb, AZ, GenePlus, and Innovent outside of the submitted work. Dr. Kadara reports receiving grant from Johnson and Johnson. The remaining authors declare no conflict of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology at www.jto.org and at https://doi.org/10.1016/j.jtho.2021.07.002.
Supplementary Data
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai M., Liu D., Liu M., et al. Patients with cancer appear more vulnerable to SARS-COV-2: a multicenter study during the COVID-19 outbreak. Cancer Discov. 2020;10:783–791. doi: 10.1158/2159-8290.CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J., Ouyang W., Chua M.L.K., Xie C. SARS-CoV-2 transmission in patients with cancer at a tertiary care hospital in Wuhan, China. JAMA Oncol. 2020;6:1108–1110. doi: 10.1001/jamaoncol.2020.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y., Li L., Feng Z., et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai Y., Kuba K., Rao S., et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X., Yu Y., Xu J., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuba K., Imai Y., Rao S., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang H., Xiao G., Behrens C., et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. 2013;19:1577–1586. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim E.S., Herbst R.S., Wistuba, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papadimitrakopoulou V., Lee J.J., Wistuba, et al. The BATTLE-2 study: a biomarker-integrated targeted therapy study in previously treated patients with advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:3638–3647. doi: 10.1200/JCO.2015.66.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barretina J., Caponigro G., Stransky N., et al. Addendum: the Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2019;565:E5–E6. doi: 10.1038/s41586-018-0722-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou B.B., Peyton M., He B., et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockwood W.W., Chari R., Coe B.P., et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27:4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalu N.N., Mazumdar T., Peng S., et al. Comprehensive pharmacogenomic profiling of human papillomavirus-positive and -negative squamous cell carcinoma identifies sensitivity to aurora kinase inhibition in KMT2D mutants. Cancer Lett. 2018;431:64–72. doi: 10.1016/j.canlet.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y., Daemen A., Hatzivassiliou G., et al. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelial-mesenchymal transition and drug resistance in tumor cells. Cancer Metab. 2014;2:20. doi: 10.1186/2049-3002-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kundu S.T., Byers L.A., Peng D.H., et al. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2016;35:173–186. doi: 10.1038/onc.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson MB Sun H., Robichaux J., et al. The YAP/FOXM1 axis regulates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci Transl Med. 2020;12:eaaz4589. doi: 10.1126/scitranslmed.aaz4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raoof S., Mulford I.J., Frisco-Cabanos H., et al. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene. 2019;38:6399–6413. doi: 10.1038/s41388-019-0887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hata A.N., Niederst M.J., Archibald H.L., et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbons D.L., Lin W., Creighton C.J., et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, Itoh Y, Sakai Y, et al. Generation of human bronchial organoids for SARS-CoV-2 research. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.05.25.115600v2.full.pdf. Accessed June 20, 2020.
- 28.Lieberman N.A.P., Peddu V., Xie H., et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duclos G.E., Teixeira V.H., Autissier P., et al. Characterizing smoking-induced transcriptional heterogeneity in the human bronchial epithelium at single-cell resolution. Sci Adv. 2019;5:eaaw3413. doi: 10.1126/sciadv.aaw3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puram S.V., Tirosh I., Parikh A.S., et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck. Cancer Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyfman P.A., Walter J.M., Joshi N., et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieira Braga F.A., Kar G., Berg M., et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 33.Kurppa K.J., Liu Y., To C., et al. Treatment-induced tumor dormancy through YAP-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. 2020;37:104–122:e12. doi: 10.1016/j.ccell.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gertz J., Savic D., Varley K.E., et al. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52:25–36. doi: 10.1016/j.molcel.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuyama S., Nao N., Shirato K., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziegler C.G.K., Allon S.J., Nyquist S.K., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e1019. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brindley M.A., Hunt C.L., Kondratowicz A.S., et al. Tyrosine kinase receptor Axl enhances entry of Zaire ebolavirus without direct interactions with the viral glycoprotein. Virology. 2011;415:83–94. doi: 10.1016/j.virol.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meertens L., Labeau A., Dejarnac O., et al. Axl mediates Zika virus entry in human glial cells and modulates innate immune responses. Cell Rep. 2017;18:324–333. doi: 10.1016/j.celrep.2016.12.045. [DOI] [PubMed] [Google Scholar]
- 39.Chen J., Yang Y.F., Yang Y., et al. AXL promotes Zika virus infection in astrocytes by antagonizing type I interferon signalling. Nat Microbiol. 2018;3:302–309. doi: 10.1038/s41564-017-0092-4. [DOI] [PubMed] [Google Scholar]
- 40.Brown J. University of Iowa virology research helps facilitate new clinical trial for COVID-19. University of Iowa Hospitals & Clinics. https://uihc.org/news/university-iowa-virology-research-helps-facilitate-new-clinical-trial-covid-19 Accessed June 26, 2020.
- 41.BerGenBio BERGENBIO’S bemcentinib selected to be fast-tracked as potential treatment for COVID-19 through new national UK government clinical trial INITIATIVE. 2020. https://www.bergenbio.com/bergenbios-bemcentinib-selected-to-be-fast-tracked-as-potential-treatment-for-covid-19-through-new-national-uk-government-clinical-trial-initiative/
- 42.Spinato G., Fabbris C., Polesel J., et al. Alterations in smell or taste in mildly symptomatic outpatients with SARS-CoV-2 infection. JAMA. 2020;323:2089–2090. doi: 10.1001/jama.2020.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xydakis M.S., Dehgani-Mobaraki P., Holbrook E.H., et al. Smell and taste dysfunction in patients with COVID-19. Lancet Infect Dis. 2020;20:1015–1016. doi: 10.1016/S1473-3099(20)30293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howitt M.R., Lavoie S., Michaud M., et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 2016;351:1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Leary C.E., Schneider C., Locksley R.M. Tuft cells-systemically dispersed sensory epithelia integrating immune and neural circuitry. Annu Rev Immunol. 2019;37:47–72. doi: 10.1146/annurev-immunol-042718-041505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsumoto I., Ohmoto M., Narukawa M., Yoshihara Y., Abe K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat Neurosci. 2011;14:685–687. doi: 10.1038/nn.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohmoto M., Yamaguchi T., Yamashita J., Bachmanov A.A., Hirota J., Matsumoto I. Pou2f3/Skn-1a is necessary for the generation or differentiation of solitary chemosensory cells in the anterior nasal cavity. Biosci Biotechnol Biochem. 2013;77:2154–2156. doi: 10.1271/bbb.130454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita J., Ohmoto M., Yamaguchi T., Matsumoto I., Hirota J. Skn-1a/Pou2f3 functions as a master regulator to generate Trpm5-expressing chemosensory cells in mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y.H., Klingbeil O., He X.Y., et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev. 2018;32:915–928. doi: 10.1101/gad.314815.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burns W.C., Velkoska E., Dean R., Burrell L.M., Thomas M.C. Angiotensin II mediates epithelial-to-mesenchymal transformation in tubular cells by ANG 1–7/MAS-1-dependent pathways. Am J Physiol Renal Physiol. 2010;299:F585–F593. doi: 10.1152/ajprenal.00538.2009. [DOI] [PubMed] [Google Scholar]
- 51.Qian Y.R., Guo Y., Wan H.Y., et al. Angiotensin-converting enzyme 2 attenuates the metastasis of non-small cell lung cancer through inhibition of epithelial-mesenchymal transition. Oncol Rep. 2013;29:2408–2414. doi: 10.3892/or.2013.2370. [DOI] [PubMed] [Google Scholar]
- 52.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 53.Park S.M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Title A.C., Hong S.J., Pires N.D., et al. Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat Commun. 2018;9:4671. doi: 10.1038/s41467-018-07130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramirez- Peña E., Arnold J., Shivakumar V., et al. The epithelial to mesenchymal transition promotes glutamine independence by suppressing GLS2 expression. Cancers (Basel) 2019;11:1610. doi: 10.3390/cancers11101610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen B., Yi X., Sun Y., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng M.L., Chien K.Y., Lai C.H., Li G.J., Lin J.F., Ho H.Y. Metabolic reprogramming of host cells in response to enteroviral infection. Cells. 2020;9:473. doi: 10.3390/cells9020473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu X., Hu X., Li N., et al. Glutamine and glutaminolysis are required for efficient replication of infectious spleen and kidney necrosis virus in Chinese perch brain cells. Oncotarget. 2017;8:2400–2412. doi: 10.18632/oncotarget.13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chambers J.W., Maguire T.G., Alwine J.C. Glutamine metabolism is essential for human cytomegalovirus infection. J Virol. 2010;84:1867–1873. doi: 10.1128/JVI.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fontaine K.A., Camarda R., Lagunoff M. Vaccinia virus requires glutamine but not glucose for efficient replication. J Virol. 2014;88:4366–4374. doi: 10.1128/JVI.03134-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim K.K., Kugler M.C., Wolters P.J., et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A. 2006;103:13180–13185. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gouda M.M., Shaikh S.B., Bhandary Y.P. Inflammatory and fibrinolytic system in acute respiratory distress syndrome. Lung. 2018;196:609–616. doi: 10.1007/s00408-018-0150-6. [DOI] [PubMed] [Google Scholar]
- 63.Herrero R., Sanchez G., Lorente J.A. New insights into the mechanisms of pulmonary edema in acute lung injury. Ann Transl Med. 2018;6:32. doi: 10.21037/atm.2017.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ikenouchi J., Matsuda M., Furuse M., Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 65.Cao Y., Liu Y., Ping F., Yi L., Zeng Z., Li Y. miR-200b/c attenuates lipopolysaccharide-induced early pulmonary fibrosis by targeting ZEB1/2 via p38 MAPK and TGF-β/smad3 signaling pathways. Lab Invest. 2018;98:339–359. doi: 10.1038/labinvest.2017.123. [DOI] [PubMed] [Google Scholar]
- 66.Gay C.M., Stewart C.A., Park E.M., et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–360.e7. doi: 10.1016/j.ccell.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Byers L.A., Wang J., Nilsson M.B., et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2:798–811. doi: 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L., Gibbons D.L., Goswami S., et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H., Ning S., Ghandi M., et al. The landscape of cancer cell line metabolism. Nat Med. 2019;25:850–860. doi: 10.1038/s41591-019-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fornes O., Castro-Mondragon J.A., Khan A., et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jamieson A.R., Giger M.L., Drukker K., Li H., Yuan Y., Bhooshan N. Exploring nonlinear feature space dimension reduction and data representation in breast Cadx with Laplacian eigenmaps and t-SNE. Med Phys. 2010;37:339–351. doi: 10.1118/1.3267037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Byers L.A., Diao L., Wang J., et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stewart C.A., Gay C.M., Xi Y., et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat Cancer. 2020;1:423–436. doi: 10.1038/s43018-019-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kalu N.N., Mazumdar T., Peng S., et al. Genomic characterization of human papillomavirus-positive and -negative human squamous cell cancer cell lines. Oncotarget. 2017;8:86369–86383. doi: 10.18632/oncotarget.21174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barretina J., Caponigro G., Stransky N., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mak M.P., Tong P., Diao L., et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hmeljak J., Sanchez-Vega F., Hoadley K.A., et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018;8:1548–1565. doi: 10.1158/2159-8290.CD-18-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alqahtani J.S., Oyelade T., Aldhahir A.M., et al. Prevalence, severity and mortality associated with COPD and smoking in patients with COVID-19: a rapid systematic review and meta-analysis. PLoS One. 2020;15 doi: 10.1371/journal.pone.0233147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Di Stadio A., Ricci G., Greco A., de Vincentiis M., Ralli M. Mortality rate and gender differences in COVID-19 patients dying in Italy: a comparison with other countries. Eur Rev Med Pharmacol Sci. 2020;24:4066–4067. doi: 10.26355/eurrev_202004_20980. [DOI] [PubMed] [Google Scholar]
- 80.Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- 81.Yaegashi H., Takahashi T. [Medical changes in preformed host arteries involved in a tumor--a morphometric study of gastric, colonic, and pancreatic carcinomas in man] Gan No Rinsho. 1988;34:1555–1560. [PubMed] [Google Scholar]
- 82.Gay C.M., Balaji K., Byers L.A. Giving AXL the axe: targeting AXL in human malignancy. Br J Cancer. 2017;116:415–423. doi: 10.1038/bjc.2016.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Overgaard C.E., Mitchell L.A., Koval M. Roles for claudins in alveolar epithelial barrier function. Ann N Y Acad Sci. 2012;1257:167–174. doi: 10.1111/j.1749-6632.2012.06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allison Stewart C., Tong P., Cardnell R.J., et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget. 2017;8:28575–28587. doi: 10.18632/oncotarget.15338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peng D.H., Kundu S.T., Fradette J.J., et al. ZEB1 suppression sensitizes KRAS mutant cancers to MEK inhibition by an IL17RD-dependent mechanism. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aaq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peng D.H., Ungewiss C., Tong P., et al. ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene. 2017;36:1925–1938. doi: 10.1038/onc.2016.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le X., Puri S., Negrao M.V., et al. Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC. Clin Cancer Res. 2018;24:6195–6203. doi: 10.1158/1078-0432.CCR-18-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ray P., Wangzhou A., Ghneim N., et al. A pharmacological interactome between COVID-19 patient samples and human sensory neurons reveals potential drivers of neurogenic pulmonary dysfunction. Brain Behav Immun. 2020;89:559–568. doi: 10.1016/j.bbi.2020.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holland S.J., Pan A., Franci C., et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 2010;70:1544–1554. doi: 10.1158/0008-5472.CAN-09-2997. [DOI] [PubMed] [Google Scholar]
- 90.Azulay R.D., Velloso M.B., Suguimoto S.Y., Ishida C.E., Pereira Júnior A.C. Acute disseminated paracoccidioidomycosis. Septic shock. Int J Dermatol. 1988;27:510–511. doi: 10.1111/j.1365-4362.1988.tb00934.x. [DOI] [PubMed] [Google Scholar]
- 91.Cai H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir Med. 2020;8 doi: 10.1016/S2213-2600(20)30117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dave N., Guaita-Esteruelas S., Gutarra S., et al. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J Biol Chem. 2011;286:12024–12032. doi: 10.1074/jbc.M110.168625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zuo W., Zhao X., Chen Y.G. In: Molecular Biology of the SARS-Coronavirus. Lal S., editor. Springer; Berlin, Germany: 2009. SARS coronavirus and lung fibrosis; pp. 247–258. [Google Scholar]
- 94.Jarcho J.A., Ingelfinger J.R., Hamel M.B., D’Agostino R.B., Sr., Harrington D.P. Inhibitors of the renin-angiotensin-aldosterone system and COVID-19. N Engl J Med. 2020;382:2462–2464. doi: 10.1056/NEJMe2012924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Monteil V., Kwon H., Prado P., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bouhaddou M., Memon D., Meyer B., et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712.e619. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dowall S.D., Bewley K., Watson R.J., et al. Antiviral screening of multiple compounds against Ebola virus. Viruses. 2016;8:277. doi: 10.3390/v8110277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.BerGenBio ASA BerGenBio Closes Recruitment Into Trial Assessing Bemcentinib in COVID-19 Patients. Cision PR Newswire. https://www.prnewswire.co.uk/news-releases/bergenbio-closes-recruitment-into-trial-assessing-bemcentinib-in-covid-19-patients-817885187.html Accessed March 5, 2021.
- 99.BerGenBio ASA BerGenBio announces update from investigational phase II trials assessing bemcentinib in hospitalised COVID-19 patients. Cision PR Newswire. https://www.prnewswire.com/news-releases/bergenbio-announces-update-from-investigational-phase-ii-trials-assessing-bemcentinib-in-hospitalised-covid-19-patients-301271176.html Accessed April 20, 2021.
- 100.Dittmar M., Lee J.S., Whig K., et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021;35:108959. doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.